Summary
Cooperative systems are susceptible to invasion by selfish individuals that profit from receiving the social benefits but fail to contribute. These so-called “cheaters” can have a fitness advantage in the laboratory, but it is unclear whether cheating provides an important selective advantage in nature. We used a population genomic approach to examine the history of genes involved in cheating behaviors in the social amoeba Dictyostelium discoideum, testing whether these genes experience rapid evolutionary change as a result of conflict over spore-stalk fate. Candidate genes and surrounding regions showed elevated polymorphism, unusual patterns of linkage disequilibrium, and lower levels of population differentiation, but they did not show greater between-species divergence. The signatures were most consistent with frequency-dependent selection acting to maintain multiple alleles, suggesting that conflict may lead to stalemate rather than an escalating arms race. Our results reveal the evolutionary dynamics of cooperation and cheating and underscore how sequence-based approaches can be used to elucidate the history of conflicts that are difficult to observe directly.
Results
The social amoeba Dictyostelium discoideum is a model system for cooperation and conflict [1, 2]. Upon starvation, up to hundreds of thousands of amoebae converge, forming a multicellular slug and eventually a fruiting body. Some cells form the stalk of the fruiting body and die, while others rise to the top, form resistant spores, and disperse. Stalk formation is altruistic, because death of the stalk cells enhances the survival and dispersal of the spores. However, because fruiting bodies can contain multiple, genetically different clones, selection can favor cheaters – individuals that avoid forming the stalk themselves, yet benefit from its production by others [2].
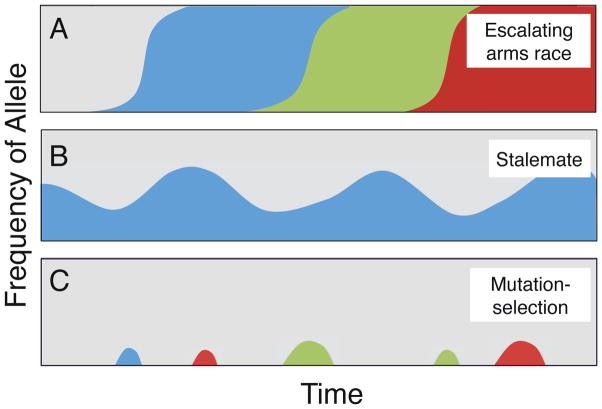
Consistent with the prediction of cheating, cheaters can be readily found in natural populations of D. discoideum [1–3]. However, whether individuals that cheat are evolutionarily successful is unclear, and several hypotheses have been proposed. One hypothesis is that cheating selects for resistance, and resistance in turn selects for greater cheating. Social conflict could thus drive an escalating arms race of adaptations and counter-adaptations, reminiscent of the arms races between hosts and pathogens or predators and prey [4–7]. An alternative possibility is that cheaters have a selective advantage only when rare. For example, as cheaters increase in frequency in a population, they potentially displace the very victims on which they depend, or face other trade-offs [8]. This negative frequency-dependence predicts that cheaters and cooperators can be maintained as a balanced polymorphism, effectively leading to a stalemate (Fig. 1B) [9, 10]. Finally, some have suggested that there is no selective advantage to cheating [11]. Cheating might be selected against if relatedness among the strains in a fruiting body is high, such that cheaters primarily cheat their own relatives [12]. In this case, cheating might persist in populations as a ‘cheating load’, analogous to a genetic load for deleterious mutations (Fig. 1C). Alternatively, cheating might also not be favored if the multicellular stage occurs only rarely in nature [13], such that there is little selection for or against these phenotypes.
Figure 1. Example scenarios for the evolutionary dynamics of cheating behaviors.
Shaded areas are proportional to the frequencies of different alleles (colors) in a population. (A) Escalating arms races, where epidemics of cheating and/or resistance sweep through populations, (B) Stalemates, where cheaters invade but neither fix nor become lost from the population, resulting in endemic cheating, (C) Mutation-selection balance, where new mutations that produce cheating behaviors are continually introduced into a population, but selection removes them.
Crucially, these different hypotheses about the long-term success of social cheating make unique, testable predictions about variation in the genes that mediate these conflicts ([14]; Table 1). To distinguish among these different possibilities, we took advantage of a previous screen that identified over 150 loci in D. discoideum that impact cheating behaviors [5]. We used whole genome sequencing and molecular evolution to ask whether genes that mediate cheating behaviors show distinctive signatures of molecular evolution that differ from the rest of the genome, distinguishing among the different hypotheses described in Table 1.
Table 1.
Predicted sequence patterns for cooperation and cheating genes under different evolutionary scenarios.
| Scenario | Description | Within-species polymorphism | Between-species divergence | Additional Signatures |
|---|---|---|---|---|
| Escalating Arms Race (Directional Selection) | Repeated selective sweeps of cheating alleles through populations remove variation within populations and drive rapid divergence between species | Decreased | Increased | Elevated population structure (higher FST), excess of high frequency derived alleles (negative Fay and Wu’s H) |
| Stalemate (Balancing Selection) | Negative frequency-dependence maintains both cheaters and cooperators within populations | Increased | Decreased | Reduced population structure (lower FST), excess of intermediate frequency alleles (positive Tajima’s D), elevated linkage disequilibrium (lower Ĉ), haplotype structure (higher Wall’s B and Wall’s Q) |
| Relaxed Selection | Cheating behaviors do not experience strong selection, possibly because the multicellular (social) stage is rare in nature | Increased | Increased | Allele frequency skew closer to zero (Tajima’s D=0) |
| Purifying Selection | Cheaters are selected against, for example if relatedness is high | Decreased | Decreased | Excess of low frequency alleles (negative Tajima’s D) |
Candidate Genes Show Elevated Polymorphism
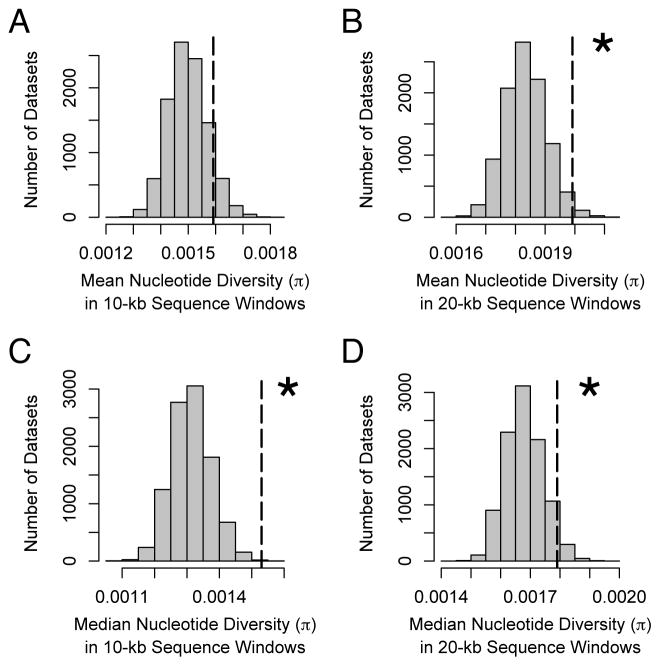
The different evolutionary scenarios for cheating alleles make unique predictions about the levels of polymorphism versus divergence (Table 1). For example, an escalating arms race driven by repeated selective sweeps of cheating alleles should reduce variation within species while elevating the sequence divergence between species, whereas the stalemate model makes the opposite prediction. To test these possibilities, we first examined levels of polymorphism in regions surrounding candidate genes, comparing these values to a null hypothesis based on other regions of the genome. We observed higher polymorphism in candidate genes as a group compared to randomly chosen regions, which was significant for both mean and median levels at 20 kb (Fig. 2). Compared to other genes, sequence variation was also disproportionately non-synonymous (higher pN/pS; Table S1). Higher levels of polymorphism might occur if genes important for cheating behaviors show limited expression (e.g., if they are expressed in an infrequent portion of the life cycle [13]), but analysis of the published transcriptome of the lab strain [15] indicated no difference in their timing or levels of expression compared to other genes in the genome (Table S2).
Figure 2. Comparison of polymorphism in regions surrounding candidate loci compared to the rest of the genome.
Dotted lines show the mean or median nucleotide diversity in sequence windows of 10- or 20-kb compared to the null distribution based on 10,000 data sets of the same size where sequence windows were chosen randomly. Asterisks indicate statistically significant results. (A) Mean nucleotide diversity in 10-kb sequence windows, (B) mean nucleotide diversity in 20-kb sequence windows, (C) median nucleotide diversity in 10-kb sequence windows, and (D) median nucleotide diversity in 20-kb sequence windows.
No Evidence of Elevated Sequence Divergence between Species
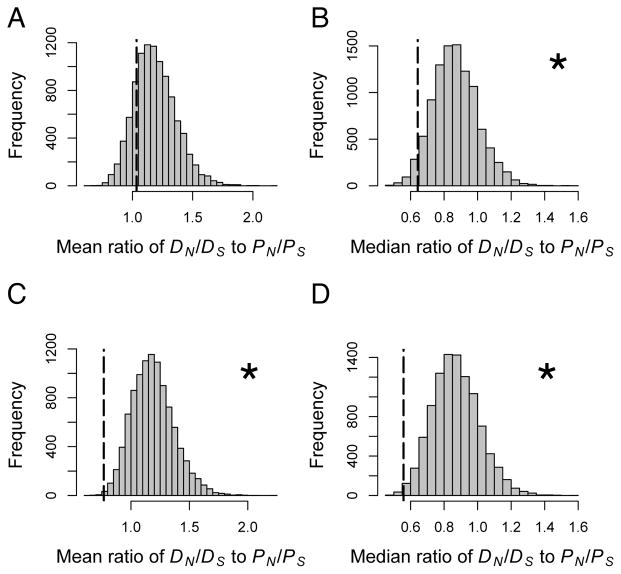
The arms race hypothesis also predicts elevated rates of divergence between species, so we compared D. discoideum to its sister species D. citrinum at all identified orthologs. These analyses revealed lower rates of non-synonymous to synonymous substitution (dN/dS) for candidate genes compared to other genes in the genome (Table S1), which was inconsistent with the predictions of an arms race (Table 1). We reached a similar conclusion using McDonald-Kreitman tests [16], which compare PN/PS to DN/DS for each gene individually. Given very low levels of sequence polymorphism within D. discoideum, there was little resolution to detect significant deviations in this ratio for each locus individually. Nevertheless, two candidate genes (DDB_G0285541 and chtC) had strongly significant McDonald-Kreitman tests that were also in the extreme tail of the genome-wide distribution. These genes showed elevations in DN/DS relative to PN/PS, indicative of directional selection driving sequence divergence. In the remainder of candidate genes (that is, removing these two genes), the ratio was strongly significant and opposite in direction, indicating an excess of non-synonymous polymorphism to non-synonymous divergence compared to other genes (Fig. 3). Taken together, genes mediating social cheating did not show the elevated rates of amino-acid substitution predicted under an escalating arms race or relaxed selection scenario.
Figure 3. McDonald-Kreitman tests.
Histograms show the mean or median ratio of DN/DS to PN/PS for 10,000 randomly generated gene sets, and the dotted line shows the observed value of this ratio for candidate genes. Asterisks indicate statistically significant results. (A) Mean ratio, (B) median ratio, (C and D) mean and median ratios after removing two candidate genes with extreme values. See text for details.
Additional Signatures of Selection Support Stalemates
At first glance, elevated non-synonymous polymorphism, combined with low amino acid divergence, is potentially consistent with the stalemate model of frequency-dependent selection, where novel alleles can invade and establish in populations, but ultimately fail to take over. Several additional tests support this interpretation. First, scaled to gene length, we observed significantly fewer haplotypes than expected and high levels of intragenic linkage disequilibrium (lower Ĉ; Fig. S1), indicating divergent alleles, a signature of balancing selection. In addition, two metrics of balancing selection, Wall’s B and Wall’s Q [17], were significantly elevated in sequence windows surrounding candidate loci (Table S3).
A common test for balancing selection is to examine the distribution of allele frequencies – whereas positive or purifying selection produce a strongly skewed distribution, balancing selection can maintain multiple alleles at intermediate frequencies. Surprisingly, given our results above supporting balancing selection, candidate loci showed greater skew, indicated by more negative values for two metrics of the site frequency spectrum (Table S4). Candidate genes as a group also showed a significant excess of high frequency derived alleles (Fay and Wu’s H: −0.002; P=0.03), which can indicate incomplete selective sweeps. The significant, negative Fay and Wu’s H test for candidate genes suggests that variants rise to high frequency quickly and that these genes experience stronger-than-expected selection for sequence changes.
Finally, we calculated the index of population structure (FST) at each segregating site in the genome, comparing SNPs in close proximity to candidate genes against the rest of the genome. Unusually high FST can indicate that different alleles predominate in different geographic locations (in this case, Texas and Virginia) and can be caused by geographically restricted selective sweeps. On the other hand, unusually low FST values indicate alleles attain similar frequencies across geographically distinct populations, with each subpopulation maintaining multiple divergent alleles—a signature of negative frequency-dependent selection [18]. These results revealed lower-than-expected FST at candidate compared to non-candidate loci (Fig. S2). The elevation in polymorphism in these genes, combined with significant reductions in population structure, argues against divergent alleles caused by local sweeps and suggests a role for negative frequency-dependent selection, with selection maintaining the same variants across subpopulations.
Evidence for Multiple Modes of Selection
By examining the molecular evolution patterns of candidate genes as a group, we could determine whether genes that mediate social conflict have general, recognizable patterns indicating unique forms of selection. Analyzing these genes as a group also provided greater sensitivity, which was helpful given the low levels of polymorphism in this species. However, these results reflect only average differences between candidate and non-candidate genes – and in fact, a diversity of dynamics is possible. Moreover, for many metrics, candidate genes might be extreme in opposite directions, such that we may fail to observe a strong signal of selection because these effects average out. We addressed this possibility in two ways. First, for each evolutionary metric, we asked whether the variance was higher for candidate genes compared to groups where genes are chosen randomly, indicating extremes in opposite directions. However, we did not observe elevated variance for any metric (Table S5). We also asked whether there was overrepresentation of candidate genes in both tails of the genome-wide distribution for each metric, but we observed no such cases (Table S6). While there was no overrepresentation in either tail of the genome-wide distribution for the McDonald-Kreitman test, two genes (DDB_G0285541 and chtC) showed extreme signatures of positive (directional) selection. The remaining genes showed the opposite pattern, an average excess of within-species non-synonymous polymorphism compared to non-synonymous divergence. Thus, while the average signature in conflict-related genes was elevated polymorphism and other patterns suggesting the selective maintenance of multiple alleles, other signatures were evident as well, including signatures of strong directional selection that could reflect escalating arms races at these particular loci.
Discussion
In D. discoideum, a model system for the study of social conflict, it has long been noted that cheating behaviors are present among natural isolates [2, 3], but there is little understanding of why selfish behaviors arise, whether they persist, and whether the prevalence of cheaters in natural populations signifies long-term evolutionary success of this social strategy. We have indications of three main signatures of balancing selection on these genes as a class compared to other genes. First, they show higher levels of polymorphism, as expected when at least one SNP is under balancing selection and increases diversity at linked neutral loci. However, they did not show the concomitant elevation in amino acid substitution expected if polymorphism simply reflected relaxed selection. Second, they showed lower FST values, which can occur if selection is maintaining the same balanced polymorphisms across geographically different subpopulations. Third, another indicator of the deeper coalescence times characteristic of balancing selection is the significantly fewer haplotypes and higher linkage disequilibrium values, including significantly elevated Wall’s B and Wall’s Q.
The observation of elevated levels of polymorphism surrounding genes implicated in social cheating, combined with other signatures of recent selection, argue that D. discoideum experiences ongoing selection at these loci and is consistent with frequency-dependent selection allowing multiple types (or alleles) to coexist. Notably, we failed to observe the molecular signatures of a simple arms race – these dynamics are expected to reduce genetic diversity and drive long-term sequence divergence between populations and/or species. Rather, our results are reminiscent of ‘trench warfare’, an alternative arms race scenario where alternative alleles do not rapidly displace one another, resulting in a prolonged stalemate [19]. Negative-frequency dependence is also a hallmark of Red Queen dynamics, a form of coevolutionary arms race where alleles continually cycle but rarely fix [20].
The finding of balancing selection is also consistent with evolutionary theory about the role of frequency-dependence in social interactions [21]. Many social behaviors are inherently frequency-dependent, where the fitness of a given strategy (e.g., cheat or cooperate) is dependent on whether an individual’s social partners employ the same strategy or not. Experimental studies of bacterial mutants that exhibit cheating behaviors suggest frequency-dependence might be common [22–26]. In Dictyostelium, frequency-dependent fitness was shown for the fbxA- strain, a mutant that allocates fewer cells to the stalk but produces disproportionately many spores when co-developed with another strain, which it cheats [12].
Like social conflict, conflicts between the sexes over optimal levels of mating and between parents and offspring over optimal provisioning are also hypothesized to result in antagonistic coevolution [20, 27–29]. While some studies have shown directional selection on genes underlying these other forms of intraspecific conflict, many others have found signatures of balancing selection, diversifying selection, or a combination of both [30–33]. Thus taken together, our results not only indicate stalemate as a possible outcome of social conflict, but add to a growing body of evidence that stalemates may be a common outcome in conflict-driven systems more generally. While identification of these polymorphisms should open the door to investigation into the functional consequences of this variation for cheating and resistance behaviors, the population genomic approach used here provides insight into the long-term consequences of social conflict and highlights the possibility of an ongoing, dynamic interaction at these loci.
Experimental Procedures
Strains
We re-sequenced the genomes of 20 natural isolates of D. discoideum, primarily from two locations: Houston, TX (six strains) and Mountain Lake, VA (nine strains), as well as 5 additional strains from different geographic locations (two sites in Texas, and one site in each of Massachusetts, Kentucky, and Illinois) using 454 or Illumina. Sequencing reads were aligned to the Ax4 reference genome (Assembly/GFF3 file generated June 9 2010, available at dictybase.org) using MAQ for Illumina sequencing reads and ATLAS-SNP for 454 data. Detailed mapping and SNP calling procedures are available in the Supplemental Information.
Molecular Evolution Analyses
Nucleotide diversity, Tajima’s D, Fu and Li’s D*, Hudson’s C (recombination, or rho), haplotype diversity, Fay and Wu’s H, and haplotype number were determined for all genes in the genome and in sequence windows using “compute” (available at molpopgen.org). Levels of non-synonymous (pN) and synonymous (pS) diversity were calculated using the program “gestimator”, and the McDonald-Kreitman tests were obtained using “MKtest” (both available at molpopgen.org). FST was calculated for all segregating sites using scripts written in Ruby and Python. Resampling analyses were performed using R. More details are available in the Supplemental Information.
Supplementary Material
Highlights.
Molecular evolution analyses reveal the history of social conflict
Genes that mediate social conflict show signatures of frequency-dependent selection
Balanced polymorphisms suggest that cheating may be stable and endemic
Acknowledgments
We thank Rasmus Nielsen for discussions of the data and comments on an earlier version of this manuscript.
Footnotes
Author Contributions
Cell culture and DNA extraction: EAO, CD, MKK, DB; Conceived and designed the experiments: EAO, JES, DCQ, SR, KCW, RAG, RS; Library preparation and sequencing: FL-G, SLL, CK, HD, VK, LJ, SP, YH, LC, DMM, SR, RAG, KW; Analyzed the data: EAO, YS, XT, RS, SR; Assembly and annotation of D. citrinum: JQ, HJ, KW; Analysis of D. citrinum and D. discoideum: EAO, XT; Wrote the paper: EAO, JES, AK, DCQ.
References
- 1.Strassmann JE, Queller DC. Evolution of cooperation and control of cheating in a social microbe. Proc Natl Acad Sci USA. 2011;108(Suppl 2):10855–10862. doi: 10.1073/pnas.1102451108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strassmann JE, Zhu Y, Queller DC. Altruism and social cheating in the social amoeba Dictyostelium discoideum. Nature. 2000;408:965–967. doi: 10.1038/35050087. [DOI] [PubMed] [Google Scholar]
- 3.Buttery NJ, Rozen DE, Wolf JB, Thompson CRL. Quantification of social behavior in D. discoideum reveals complex fixed and facultative strategies. Curr Biol. 2009;19:1373–1377. doi: 10.1016/j.cub.2009.06.058. [DOI] [PubMed] [Google Scholar]
- 4.Khare A, Santorelli LA, Strassmann JE, Queller DC, Kuspa A, Shaulsky G. Cheater-resistance is not futile. Nature. 2010;461:980–982. doi: 10.1038/nature08472. [DOI] [PubMed] [Google Scholar]
- 5.Santorelli LA, Thompson CRL, Villegas E, Svetz J, Dinh C, Parikh A, Sucgang R, Kuspa A, Strassmann JE, Queller DC, et al. Facultative cheater mutants reveal the genetic complexity of cooperation in social amoebae. Nature. 2008;451:1107–1110. doi: 10.1038/nature06558. [DOI] [PubMed] [Google Scholar]
- 6.Hollis B. Rapid antagonistic coevolution between strains of the social amoeba Dictyostelium discoideum. Proc Biol Sci. 2012;279:3565–3571. doi: 10.1098/rspb.2012.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang QG, Buckling A, Ellis RJ, Godfray HCJ. Coevolution between cooperators and cheats in a microbial system. Evolution. 2009;63:2248–2256. doi: 10.1111/j.1558-5646.2009.00708.x. [DOI] [PubMed] [Google Scholar]
- 8.Sathe S, Khetan N, Nanjundiah V. Interspecies and intraspecies interactions in social amoebae. J Evol Biol. 2014;27:349–362. doi: 10.1111/jeb.12298. [DOI] [PubMed] [Google Scholar]
- 9.Maynard Smith J. Evolution and the Theory of Games. Cambridge: Cambridge University Press; 1982. [Google Scholar]
- 10.Doebeli M, Hauert C, Killingback T. The evolutionary origin of cooperators and defectors. Science. 2004;306:859–862. doi: 10.1126/science.1101456. [DOI] [PubMed] [Google Scholar]
- 11.Van Dyken JD, Linksvayer TA, Wade MJ. Kin selection–mutation balance: A model for the origin, maintenance, and consequences of social cheating. Am Nat. 2011;177:288–300. doi: 10.1086/658365. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert OM, Foster KR, Mehdiabadi NJ, Strassmann JE, Queller DC. High relatedness maintains multicellular cooperation in a social amoeba by controlling cheater mutants. Proc Natl Acad Sci USA. 2007;104:8913–8917. doi: 10.1073/pnas.0702723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linksvayer TA, Wade MJ. Genes with social effects are expected to harbor more sequence variation within and between species. Evolution. 2009;63:1685–1696. doi: 10.1111/j.1558-5646.2009.00670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Dyken JD, Wade MJ. Detecting the molecular signature of social conflict: theory and a test with bacterial quorum sensing genes. Am Nat. 2012;179:436–450. doi: 10.1086/664609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parikh A, Miranda ER, Katoh-Kurasawa M, Fuller D, Rot G, Zagar L, Curk T, Sucgang R, Chen R, Zupan B, et al. Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol. 2010;11:R35. doi: 10.1186/gb-2010-11-3-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 17.Wall JD. Recombination and the power of statistical tests of neutrality. 1999;74:65–79. [Google Scholar]
- 18.Glémin S. Balancing selection in the wild: testing population genetics theory of self-incompatibility in the rare species Brassica insularis. Genetics. 2005;171:279–289. doi: 10.1534/genetics.104.035915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature. 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- 20.Brockhurst MA, Chapman T, King KC, Mank JE, Paterson S, Hurst GDD. Running with the Red Queen: the role of biotic conflicts in evolution. Proc Biol Sci. 2014;281:20141382. doi: 10.1098/rspb.2014.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Queller DC. Expanded social fitness and Hamilton’s rule for kin, kith, and kind. Proc Natl Acad Sci USA. 2011;108:10792–10799. doi: 10.1073/pnas.1100298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 23.Ross-Gillespie A, Gardner A, West SA, Griffin AS. Frequency dependence and cooperation: theory and a test with bacteria. Am Nat. 2007;170:331–342. doi: 10.1086/519860. [DOI] [PubMed] [Google Scholar]
- 24.Dugatkin LA, Perlin M, Lucas JS, Atlas R. Group-beneficial traits, frequency-dependent selection and genotypic diversity: an antibiotic resistance paradigm. Proc Biol Sci. 2005;272:79–83. doi: 10.1098/rspb.2004.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis RJ, Lilley AK, Lacey SJ, Murrell D, Godfray HCJ. Frequency-dependent advantages of plasmid carriage by Pseudomonas in homogeneous and spatially structured environments. ISME J. 2007;1:92–95. doi: 10.1038/ismej.2007.11. [DOI] [PubMed] [Google Scholar]
- 26.MacLean RC, Gudelj I. Resource competition and social conflict in experimental populations of yeast. Nature. 2006;441:498–501. doi: 10.1038/nature04624. [DOI] [PubMed] [Google Scholar]
- 27.Arnqvist G, Rowe L. Sexual Conflict. Princeton: Princeton University Press; 2005. [Google Scholar]
- 28.Rice WR, Holland B. The enemies within: intergenomic conflict, interlocus contest evolution (ICE), and the intraspecific Red Queen. Behav Ecol Sociobiol. 1997;41:1–10. [Google Scholar]
- 29.Trivers RL. Parent-offspring conflict. Am Zool. 1974;14:249–264. [Google Scholar]
- 30.Hamm D, Mautz BS, Wolfner MF, Aquadro CF, Swanson WJ. Evidence of amino acid diversity–enhancing selection within humans and among primates at the candidate sperm-receptor gene PKDREJ. Am J Hum Gen. 2007;81:44–52. doi: 10.1086/518695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelleher ES, Clark NL, Markow TA. Diversity-enhancing selection acts on a female reproductive protease family in four subspecies of Drosophila mojavensis. Genetics. 2011;187:865–876. doi: 10.1534/genetics.110.124743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawabe A, Fujimoto R, Charlesworth D. High diversity due to balancing selection in the promoter region of the Medea Gene in Arabidopsis lyrata. Curr Biol. 2007;17:1885–1889. doi: 10.1016/j.cub.2007.09.051. [DOI] [PubMed] [Google Scholar]
- 33.Miyake T, Takebayashi N, Wolf DE. Possible diversifying selection in the imprinted gene, MEDEA, in Arabidopsis. Mol Biol Evol. 2009;26:843–857. doi: 10.1093/molbev/msp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.