Abstract
We developed Copy Number Segmentation by Regression Tree in Next Generation Sequencing (CONSERTING), a novel algorithm for detecting somatic copy number alteration (CNA) using whole-genome sequencing (WGS) data. CONSERTING performs iterative analysis of segmentation by read depth change and localized structural variation detection, achieving high accuracy and sensitivity. Analysis of 43 pediatric and adult cancer genomes revealed novel oncogenic CNAs, complex re-arrangements and subclonal CNAs missed by alternative approaches.
Somatically acquired gains or losses of DNA segments, known as copy number alterations (CNAs), are an important class of genetic lesions that contribute to cancer initiation, progression and relapse1. Whole genome sequencing (WGS) of tumor samples2 should greatly improve the ability to detect somatic (tumor-acquired) CNAs relative to what is possible with methods such as array comparative genome hybridization and SNP array, because it avoids signal saturation in high-level amplification, has greater capability for detecting focal events that may span <1 kilobases, and can define CNA boundaries at base-pair resolution. However, despite the availability of many analysis algorithms (e.g. SegSeq3, CNV-Seq4, FREEC5, CNVnator6 and BIC-seq7), accurate identification of CNAs remains problematic. Although large CNAs can be reliably identified, bona fide focal changes are often missed outright or embedded among hundreds or thousands of false CNAs, many of which arise due to coverage bias, WGS mapping ambiguity in repetitive regions, or library construction artifacts.
As part of the St. Jude/Washington University Pediatric Cancer Genome Project (PCGP)8, we developed CONSERTING (Copy Number Segmentation by Regression Tree in Next Generation Sequencing), a novel algorithm for improving somatic CNA analysis using high-coverage WGS data (Supplementary Software). The core component of the CONSERTING pipeline (Fig. 1 and Supplementary Fig. 1) was designed to integrate read-depth change with structural variation (SV) identification through an iterative process of segmentation by read depth, segment merging, and localized SV detection. CONSERTING employs recursive partitioning techniques to find the transition point for read depth changes. The computing efficiency of regression tree analysis enables CONSERTING to run read depth segmentation using both log ratio signal and normalized read depth difference of the paired tumor-normal WGS data with a 100-bp window size in a reasonable time (50 minutes per iteration of read depth analysis). This implementation ensures true integration of read depth segmentation and SV breakpoint analysis so that CNAs with subtle read-depth changes can be detected without incurring a high error rate. CONSERTING can be freely downloaded from http://www.stjuderesearch.org/site/lab/zhang with a user manual and test data. Alternatively, a pre-configured cloud version of CONSERTING can be launched from Amazon Web Services (AWS) with parallel implementation of SV analysis (Online Methods).
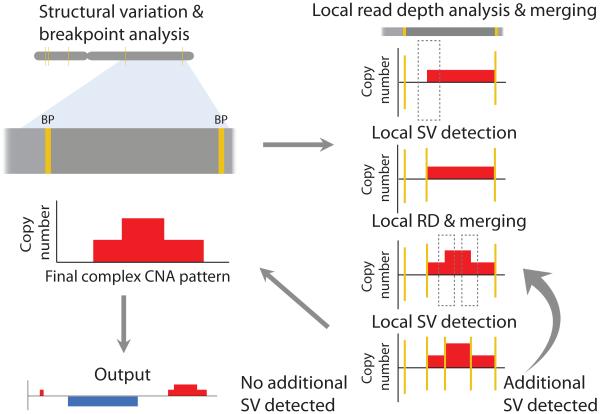
Figure 1.
Strategy for CNA detection used by CONSERTING. CNAs are identified through iterative analysis of (i) local segmentation by read depth (RD) within boundaries identified by structural variation (SV) breakpoints followed by (ii) segment merging and local SV analysis. Yellow vertical bars mark SV breakpoints. Dotted boxes indicate the candidate breakpoint regions for local SV analysis, which display RD changes but are not reported in global SV analysis.
In this study, we employed CONSERTING along with four existing somatic CNA analysis methods (CNV-Seq, SegSeq, FREEC and BIC-seq) to analyze somatic CNAs in 43 paired tumor-normal WGS data sets. These included pediatric T-cell precursor acute lymphoblastic leukaemia (T-ALL)9, B-progenitor acute lymphoblastic leukemia (B-ALL)10, retinoblastoma11, low-grade glioma12, adult glioblastoma13 and one adult melanoma cancer cell line (COLO-829) which was diluted with its matching normal (COLO-829BL) for evaluation of subclonal CNA analysis (Supplementary Table 1). CNAs derived from non-sequencing methods were used to compare the performance of CONSERTING with the existing CNA analysis methods (Fig. 2 and Supplementary Figs 2,3).
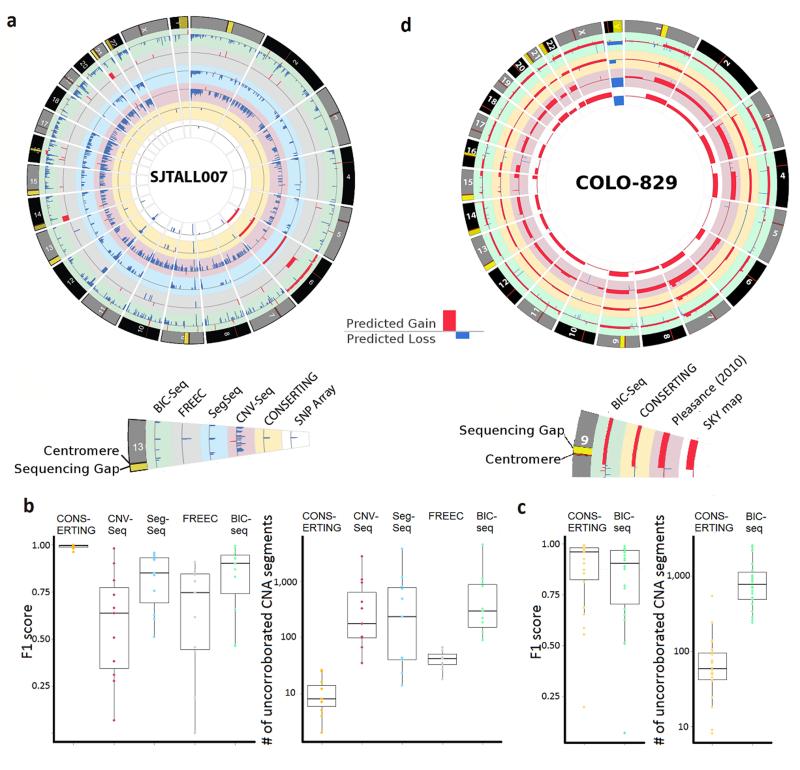
Figure 2.
Comparison of WGS CNAs detected by CONSERTING and four other methods. (a) A Circos plot which displays CNAs found by all six methods in one of the 12 ETP-ALL samples, SJTALL007. (b) Box plots showing F1 scores of WGS CNAs (compared against CNAs curated from SNP arrays) and the number of CNA segments uncorroborated by SNP arrays in the 12 ETP-ALL samples. The box represents the interquartile range (IQR) while whiskers extend to the most extreme data point which is no more than 1.5-fold of IQR away from the box. (c) Box plot of F1 score of WGS CNAs and SNP-array CNAs and number of CNA segments uncorroborated by SNP array analysis in the 22 TCGA-GBM samples. (d) A Circos plot which displays CNAs found by CONSERTING and BIC-seq in the diluted COLO-829 (scale adjusted for dilution effects), published COLO-829 CNA from un-diluted COLO-82914 and SKY map data. The 1-copy gain of chromosome X was found only in the diluted sample by both CONSERTING and BIC-seq, which is consistent with the SKY data as there are 2 chromosome X in this cell line derived from a male patient.
For pediatric cancer, we used manually-curated somatic CNAs derived from paired SNP array analysis of 12 T-ALL tumors (Supplementary Table 2) for benchmarking analysis. These CNAs were selected because they were obtained via an independent assay, are expected to be highly accurate based on prior studies1, and in many cases were validated using orthogonal technology. To summarize the accuracy for each CNA analysis method, we calculated the F1 score (Online Methods) between WGS and SNP array and the number of WGS-CNA segments that are uncorroborated by SNP array. The results (Fig. 2b and Supplementary Table 3) demonstrate that among all paired CNA methods, CONSERTING has the highest consistency with SNP array with a median F1 score of 0.99 and a median of 8 (range 2-26) uncorroborated CNA segments per genome. BIC-seq ranks second with a median of F1 score of 0.90, but with a much higher number of uncorroborated CNA segments per tumor (median 294, range 89-4521). Receiver operating curveanalysis for the T-ALL samples (Supplementary Fig. 4) also suggests that CONSERTING achieved near optimal CNA calling performance.
In the analysis of adult TCGA-GBM WGS data, CONSERTING also shows higher consistency with SNP array compared to BIC-seq: median F1 scores are0.96 and 0.90 for CONSERTING and BIC-seq, respectively (Fig. 2c, Supplementary Fig. 3, Supplementary Table 4 and Supplementary Data 1). The median uncorroborated CNA segments per genome is 58 by CONSERTING, which is significantly fewer (p = 3.0 * 10−6 by Wilcoxon signed rank test) than the 778 by BIC-seq. The melanoma cancer cell line COLO-829 analyzed by our dilution experiment has approximately 40% tumor purity. Although the global CNA profile generated from both CONSERTING and BIC-seq matches well with the published SKY mapping result of the undiluted COLO-82914 (Fig. 2d), there is a 10-fold difference in the number of CNA segments predicted by CONSERTING and BIC-seq. While 76% of the 104 CNA segment boundaries predicted by CONSERTING match validated SVs reported in literature14, 15 (Supplementary Table 5), only 4.4% of the 1,004 CNAs generated by BIC-seq match the validated SVs (Online Methods).
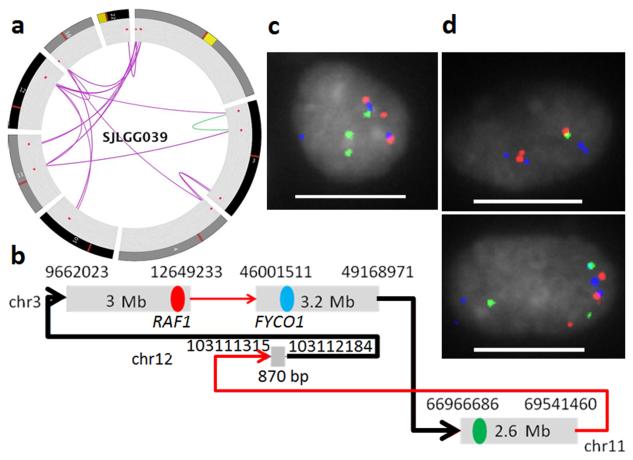
Chromothripsis has recently been recognized as a mechanism that can generate multiple CNAs through a massive, single-step genomic rearrangement16. Accurate identification of chromothripsis requires evidence from both CNAs and SVs. Local SV analysis implemented in CONSERTING runs the SV analysis algorithm CREST15 at putative CNA segment boundaries using sensitive parameters so that SVs in repetitive regions or SVs with weak signatures (due to e.g. tumor heterogeneity, tumor purity or WGS coverage bias) can be identified without incurring a high genomewide false positive rate. In the pediatric cancer genomes analyzed in this study, CONSERTING identified 20 CNAs estimated to have 0.25-0.3-fold amplification in SJLGG039, a low grade glioma tumor with an estimated purity of 30%12. Among them, only 2 CNAs were identified by SNP array (Supplementary Fig. 2f). While these CNAs were dispersed, their boundaries were inter-connected through inter-chromosomal translocations involving seven chromosomes (Fig. 3a). Twenty percent of the SVs detected by CONSERTING were missed by genome-wide SV analysis as a result of low tumor purity. We designed a 3-color fluorescence in situ hybridization assay for one inter-connected amplicon involving 3 CNA segments (Fig. 3b). The three targeted segments were expected to be physically adjacent in red-blue-green order based on the SV graph, and indeed 36% of the 100 nuclei exhibited the red-blue-green fusion signal (Fig. 3c). Importantly, the SV verified by the red-blue fusion signal produces an in-frame fusion of FYCO1-RAF1, and RAF1 fusions are known driver lesions in low grade glioma12. Interestingly, various 2-color co-localizations such as red-blue and red-green were also noted in a subset of the cells (Fig. 3d), suggesting that some of the tumor cells may have subsequently undergone additional re-arrangement. Subclonal CNAs and SVs resulting from complex re-arrangements were also found in retinoblastoma tumor SJRB003. Only a subset of these events were found by standalone CNA or SV analysis due to intra-tumor heterogeneity (Supplementary Fig. 5 and Supplementary Data 2).
Figure 3.
A complex re-arrangement in a pediatric low grade glioma sample identified by CONSERTING. (a) Circos plot of 8 chromosomes (1, 3, 4, 10, 11, 12, 16 and 22) with inter-chromosomal SVs (purple lines) and intrachromosomal SVs (green lines) connecting the amplification CNAs (red dots). (b) SV graph constructed from CNAs and SVs identified on chromosomes 3, 11 and 12. The black lines indicate SVs detected only by CONSERTING. The red, blue and green dots mark the three BAC clones selected for FISH assay. The SV represented by red-blue fusion represents an in-frame FYCO1-RAF1 fusion. (c) Red, green, blue fusion signal found in 36% of the nuclei. d) Various 2-color fusion signals including red-blue fusion and red-green fusion (scale bar: 10 μm).
In the analysis of adult GBM data, CNA and SV profiles computed by CONSERTING have shown that double minute chromosomes generated by complex re-arrangments resulted in high-level amplifications of EGFR, MDM2, MDM4, PDGFRA and CDK4 (Supplementary Fig. 6). This included three previously reported cases (06-0648-01A, 06-0145-01A and 06-0152-01A)17 as well as 10 additional cases identified in this study. Nine tumors had a chromothripsis-like CNA/SV profile (Supplementary Fig. 6), and two of these (06-0211-01A and 06-0211-02A) had multiple SVs in EGFR which may result in multiple EGFR isoforms.
The complete genomic landscapes of three ALL samples, SJTALL015 (T-ALL), PALETF and PALJDL (B-ALL) have not previously been reported. Therefore, to verify novel CNAs identified by CONSERTING but not by SNP array in gene coding regions for these three tumors, we used custom capture or Sanger sequencing of PCR amplicons encompassing the CNA breakpoints. Ten such CNA segments (range10-80kb) were found and 9 were validated (Supplementary Table 6 and Supplementary Data 3), most notably a 10 kb deletion spanning exons 14 – 27 of NOTCH1 in SJTALL015 (Supplementary Fig.7). CONSERTING predicted an in-frame intragenic deletion of NOTCH1, which was confirmed by Sanger sequencing of both genomic DNA and cDNA. The resulting mutant protein is predicted to lose amino acids 774-1687 of NOTCH1 which encode several calcium-binding EGF-like repeats, the Lin-12/Notch repeat domain, and the heterodimerization domain. Expression of stabilized intracellular NOTCH1 was confirmed by western blotting of this tumor.
Using high-coverage WGS datafrom 43 paired tumor/normal samples, we demonstrated that CONSERTING has much higher sensitivity and accuracy compared with existing CNA analysis methods and also enhances SV detection by identifying breakpoints with weak SV signatures caused by tumor heterogeneity or low tumor purity. The high concordance of CNAs and SVs detected in the diluted and undiluted adult cancer cell line COLO-829 demonstrates that CONSERTING’s increased sensitivity does not come at the cost of an elevated false discovery rate. Methods designed for characterization of genome deletion polymorphisms in large populations such as Genome STRiP also integrate read depth with rearrangement analysis to impove specificity and sensitivity18. However, identification of somatic CNAs is considered a distinct analysis for several reasons: somatic CNAs are occasionally exceedingly complex (including chromothripsis events), they may exist in a tumor subclone, and recurrence at base-pair resolution across multiple individuals is exceedingly rare.
While CONSERTING represents a substantial improvement over existing CNA detection methods, it still has difficulty distinguishing bona-fide CNAs from mapping artifacts in regions with high repeat content such as telomeres and centromeres. In addition, the SV analysis implemented in CONSERTING requires high coverage (>20x) and long WGS reads of ≥75bp which may limit its use with earlier WGS data sets with shorter read length or genomic regions with poor sequence coverage. A subset of WGS data in our study have excessive read-depth changes reminiscent of “fractured” genome without matching SVs in CONSERTING output, and further analysis attributed this phenomenon to library construction artifacts (Supplentary Fig. 8 and Supplementary Data 4). Nevertheless, the comprehensive analysis of the 43 cancer genomes presented in this study shows that our unique approach of iterative analysis of RD segmentation coupled with SV detection has resulted in a significant improvement in CNA detection in WGS. In addition to the 43 cases presented here, CONSERTING has been used to carry out CNA analyses for 700 paired tumor-normal WGS data sets across 21 subtypes of pediatric cancer generated by PCGP. Recent major findings enabled by CONSERTING included chromothripsis-driven recurrent C11orf95-RELA fusion in supratentorial ependymomas19 and multiple kinase fusions in pediatric high-grade glioma20. These examples demonstrate that high accuracy and sensitivity coupled with base-pair precision enables CONSERTING to make effective use of high-coverage WGS data, which in turn enhances our understanding of the genetic landscape of cancer genomes.
ONLINE METHODS
Code availability
CONSERTING software, user manual and test data can be downloaded from http://www.stjuderesearch.org/site/lab/zhang. Alternatively, a pre-configured cloud version of CONSERTING can be launched from Amazon Web Services (AWS) with parallel implementation of SV analysis. Instructions on running CONSERTING on the AWS cloud is available at http://www.stjuderesearch.org/site/docs/conserting/conserting-ami-steps.pdf.
Input data for CONSERTING analysis
The input for CONSERTING analysis is BAM files, the compressed binary version of the Sequence Alignment/Map (SAM) format21, which store the alignment of WGS reads to the reference human genome. Read depth is summarized from aligned bases with quality score ≥ 15 for each base-pair position of the reference genome using the Coverage module of the program Bambino22. A user-defined fixed-size window is used to obtain the mean coverage for each window. The default window size is 100bp, which was used for all analyses presented in this study. The mean read-depth per window was then normalized to a set of reference diploid chromosomal regions selected by the following criteria: no loss of heterozygosity (LOH) signal within a 1 Mb region and the coverage of the 1 Mb regions is within 1.25x median of all 1Mb non-LOH regions. Alternatively, reference diploid genomic regions may be provided by the user. The read-depth difference and the log2 ratio of the tumor and its matching normal were further normalized for GC content by linear regression.
Regression Tree Segmentation
Regression tree models are popular alternatives to global regression models because they can recursively partition sample spaces into smaller regions using one of the predictor variables until a constant estimate can be fit for each small region23. The predictive model of a regression tree T with m leaves (Li, i = 1, 2, …, m) for a dataset with n observations with predictor variables X and a response variable Y could be described as:
where
and I(.) is the indictor function. The deviance of T is:
and the Bayesian information criterion (BIC)24 of the model is:
where k is the number of estimated parameters in the tree, which equals to the number of leaf nodes (constant for each leaf) plus one (the constant error variance).
CONSERTING utilizes an open-source regression tree implementation (The “tree” R package, version 1.0-28 and above).
Local SV detection
Local SV detection runs CREST in the 20 kb flanking regions of each segmentation breakpoint with the following sensitive settings: -max_rep_cover 2000 -min_hit_len 15 -min_percent_hq 40 -m 1 -min_one_side_reads 2.
Segment merging
Segment merging is performed by (1) pruning the initial tree using the prune.tree function in R, which produces a nested sequences of subtrees with various size, selecting the optimal subtree based on Bayesian information criterion (BIC) criteria (smallest BIC); (2) recursively merging adjacent CNA segments with the most significant t-test P value within each chromosomal region defined by SV breakpoints until all breakpoints reach a genome-wide family-wise error rate (FWER) of less than 0.05 (the P value is adjusted by the Bonferroni correction with the number of tests estimated before the segmentation and remains constant throughout the run); (3) recursively merging adjacent segments with signal differences less than a pre-specified threshold across the whole chromosome (default threshold for difference signal: 0.125, log-ratio signal: 0.170); and (4) calculating a heuristic quality score for each breakpoint using the gap ratio (based on the mapability track in the UCSC genome browser25), segment length, ratio between observed and expected number of heterozygous germline SNPs, coverage of neighboring segments in the normal sample, SV support, difference between the read-count and difference between the log-ratio of read count at the breakpoint. Breakpoints with quality score less than a user-specified score are recursively merged.
Default parameter selections
The default parameters work optimally for NGS whole-genome sequencing of tumor specimens at 30X coverage, a standard adopted by the community since the whole-genome sequencing of the first cancer genome2. We set the default threshold for signal difference to 0.125 as this allows us to identify coverage change across a breakpoint with 0.25 copy gain or loss relative to a normal diploid region, which is 4-read difference in 30X coverage. A lower threshold could be considered for samples with higher coverage.
Running time analysis
Total running time of CONSERTING comprises 3 portions: 1) the presprocessing of input data, i.e, converting BAMs to BW files using scripts from the UCSC genome browser project and converting BW files to input file for CONSERTING using custom java code (O(l) where l is the genome length); 2) the RD segmentation (O(iter*nlog(n)) where iter is number of RD/SV iterations used and n is the number of windows, ~50 minutes on Amazon Web Services cloud per iteration using 100 bp windows); and 3) the SV detection (O(n2)) where n2 is the number of local SV runs. Running time analysis using the TCGA-GBM dataset without SV parallelization (median running time: 22 CPU hours on an Intel Xeon E5-2670 processor @2.60 GHz with 128 GB RAM, excluding data preprocessing) showed that the number of local SV detection runs is the significant predictor for running time (p = 1.80 * 10−9, R2 = 0.82). The median memory usage for CONSERTING is 19,352 MB in the TCGA GBM dataset. UCSC’s wigToBigWig program uses close to 40 GB of RAM during preprocessing.
Definition of corroboration
A genomic position is considered to have the corroborated CNA call if its computed CNA type (amplification/deletion) matches the curated CNA from SNP Array. A CNA segment computed from one platform is corroborated in the other platform if ≥ 90% positions of this segment are corroborated in the other platform. F1 score, or the harmonic mean of precision and recall between WGS and SNP array is used to summarize the accuracy for each CNA analysis method.
Rationale for not using the reciprocal overlap rule
The reciprocal 50% overlap is a commonly used criterion in comparing CNA calls from different algorithms. However, this criterion may not be appropriate when two CNA calls are derived from platforms with dramatically different power in detecting focal CNAs. In this study, with a significantly larger average distance between adjacent probes (kbs in SNP Array vs. 100 bp in CONSERTING), SNP array derived CNA calls have an inherently lower resolution than the WGS based CNA calls, which was proven by the detection of focal events with CONSERTING that were missed by SNP Array (Supplementary Table 3). When a focal CNA occurs on top of a large CNA fragment (such as the homozygous SH2B3 deletion in PALJDL), it breaks the region into multiple segments. While all WGS CNA fragments in the region corroborate with the SNP array calls, at most one of these fragments will satisfy the reciprocal 50% overlap rule. Consequently, we did not apply the reciprocal 50% overlap rule in this study.
Preparation and sequencing of diluted COLO-829 DNA
COLO 829 (ATCC® CRL-1974™) and COLO 829BL (ATCC® CRL-1980™), were obtained from ATCC (American Type Culture Collection) and separately expanded in culture per ATCC instructions (RPMI-1640 media with 10% FBS at 37°C, 5% CO2). Cells were not tested for mycoplasma contamination.
Genomic DNA was extracted from both tumor (COLO 829) and normal (COLO 829BL) cells using phenol-chloroform and treated with RNase A to remove residual RNA. DNA integrity and concentration were assessed by E-Gel® agarose gel electrophoresis (Life Technologies) and Qubit® dsDNA BR Assay (Life Technologies), respectively. A 50% dilution of the tumor was then obtained by mixing the tumor (COLO-829) and normal (COLO-829BL) genomic DNA in equal concentrations.
Whole Genome Sequencing (WGS) libraries of the 50% COLO-829 tumor dilution and the matched normal DNA were constructed using the TruSeq DNA PCR-Free sample preparation kit (Illumina, Inc) following the manufacturer’s instructions for 1 μg genomic DNA input and 350bp insert size. Briefly, 1 μg of genomic DNA was sheared by acoustic fragmentation using a Covaris E210 (Covaris). The fragments were end-repaired, adenylated by adding “A” bases to the 3’ end of the DNA fragments and an indexing-specific paired-end adapter was ligated to the fragments. The adapter-ligated library was then purified using the sample purification beads provided in the kit.
The resulting WGS libraries were assessed for quality using the Agilent 2200 TapeStation (Agilent Technologies). Library concentrations (nM) were determined using the Kapa NGS library quantification kit with Illumina library-specific primers and external standards (Kapa Biosystems) and analyzed on the Eco Real-Time PCR System (Illumina, Inc). Libraries were diluted to 2nM, denatured with sodium hydroxide and clustered on the cBot (Illumina, Inc) using the HiSeq PE Cluster Kit v4-cBot Kit (Illumina, Inc) according to the manufacturer's instructions. Sequencing was performed on HiSeq 2500 instruments with paired-end (2 × 126 bp) sequencing using Illumina’s HiSeq SBS v4 chemistry (Illumina, Inc). The 50% COLO-829 dilution was sequenced across 3 lanes and the matched normal across 4 lanes. DNA extraction, library preparation and next generation sequencing were performed according to standard operating procedures in our CAP/CLIA laboratory.
SKY mapping for COLO 829 was retrieved from http://www.path.cam.ac.uk/~pawefish/OtherCellLineDescriptions/COLO829.html.
SV support of CNA breakpoints in COLO-829
Validated SVs were downloaded from ref14, 15 and lifted-over to hg19. A predicted CNA breakpoint is considered to have SV support if: 1) one of the validated SV breakpoints falls within a specified distance (500 bp for CONSERTING, 5 kb for BIC-seq) to the CNA breakpoint, 2) there are no other CNA breakpoints between the SV breakpoint and the CNA breakpoint and 3) the orientation of the SV breakpoint supports the predicted coverage change across the CNA breakpoint.
Paired tumor/normal whole-genome sequencing data
WGS data for the 12 ETP ALL cases (SJTALL001-009 and 011-013), four retinoblastoma cases (SJRB001-004), B-ALL cases (PALETF and PALJDL) and TCGA GBM data were obtained from dbGaP (http://www.ncbi.nlm.nih.gov/gap) under the accession numbers phs000340.v1.p1, phs000352.v1.p1, phs000218.v1.p1 and phs000178.v8.p7, respectively. The low grade glioma sample (SJLGG039) is available at EBI under accession EGAS00001000255. TCGA GBM data downloaded from dbGaP included a total of 46 samples. A subset of the samples (24/46) showed patterns of fractured genome (Supplentary Fig. 8 and Supplementary Data 4) and were excluded from WGS-SNP array comparison. WGS data of diluted COLO-829 cell line along with its matching normal has been submitted to EBI under accession EGAS00001001050. WGS data for SJTALL015 have been deposited in EGA under accession EGAS00001001202.
FISH
Multi-color interphase FISH was performed from formalin-fixed paraffin-embedded tissue. Probes were derived from BAC clones (BACPAC Resources, Oakland, CA) and labeled with AlexaFluor-488, Rhodamine or SpectrumAqua fluorochromes. Probes were co-denatured with target cells on a hotplate at 90°C for 12 minutes. The slides were incubated overnight at 37°C and then washed in 4M urea/2xSSC at 25°C for 1 minute. Nuclei were counterstained with DAPI (200ng/ml; Vector Labs).
The following BACs were used to evaluate copy number abnormalities and any gene fusion: They are RP11-455K5 located chr3:12.4Mb (rhodamine, red), RP11-91P19 & RP11-979G9 located at chr3:46.6Mb (aqua, blue), and RP11-482O1 located at chr11:67.1Mb (AlexaFluor-488, green).
Experimental validation
Structural variations identified by CONSERTING were validated by genomic PCR and Sanger sequencing of whole genome amplified leukemic cell DNA. Oligonucleotide primers were designed within the 1000bp flanking sequences of each boundary using Primer 326 (Supplementary Table 6). PCR reactions were set up with 1 μl DNA template, 0.25 μM forward and reverse primer, 5X Phusion HF buffer, 0.2 mM dNTPs and 0.4 units of Phusion DNA polymerase (New England Biolabs). Reactions were performed on an Eppendorf thermocycler with cycling conditions consisting of a denaturation step at 98°C for 1 min, followed by 33 cycles of [98°C for 10 sec, 66°C for 15 sec, 72°C for 1 min] and a final extension step at 72°C for 10 min. PCR products were visualized with GelRed (Biotium, Inc.) on a 1.5% agarose gel run at 110V for 1 hour, purified using the Wizard PCR purification Kit (Promega) and fusion transcripts were confirmed by direct Sanger sequencing.
Validation of intragenic NOTCH1 deletion by RT-PCR
To amplify the region spanning the intragenic NOTCH1 deletion, 0.5μg of total leukemic cell RNA was reverse-transcribed using SuperScript III First-Strand kit (Life Technologies). The resulting cDNA was PCR-amplified using AccuPrime polymerase (Life Technologies) and primers were designed within the 500 bp flanking sequences of the boundaries using Primer 326 (Supplementary Table 6). The purified PCR product was subjected to bi-directional Sanger sequencing.
Western blotting of cleaved intracellular NOTCH1
Western blotting for ICN was performed for the T-ALL cell lines MOLT3, HPBALL, DND41, PF382, TALL-1 and LOUCY (Supplementary Fig 7). The murine fibroblast cell line GPE-86 was included as a negative control. Cells were lyzed in RIPA buffer containing protease inhibitors, followed by a protein concentration measurement using the BCA Protein Assay Kit (Pierce). Forty micrograms of protein was separated by electrophoresis in NuPAGE 4-12% Bis-Tris gels (Life Technologies) and transferred to nitrocellulose membranes (Whatman). Membranes were probed with an antibodiesy specific for cleaved intracellular NOTCH1 (ICN; #2421, Cell Signaling) or α-tubulin (DM1A; #3873, Cell Signaling), followed by staining with HRP-conjugated donkey-anti-rabbit (ICN) or donkey-anti-mouse (α-tubulin) secondary antibodies (Thermo Fisher). After washing ICN and tubulin proteins were visualized using the Femto Chemiluminescent Kit (Thermo Fisher
BIC-seq analysis
We used the recommended lambda value and used a bin size of 100 for all analyses presented in this study including the paired tumor/normal WGS data. To determine the threshold for amplification and deletion, we first removed the segments that do not reach P≤0.05 after Bonferroni correction. We then tested thresholds between 0.17 and 0.3 for each sample and selected the 0.22 for final report as it gives the highest F1 score across all ETP-ALL samples.
SegSeq
To prepare input files for SegSeq, we extracted the chromosomal coordinates (defined as the middle point between start and end of a read) and orientation of each uniquely mapped read with mapping quality ≥ 35 from the BAM files. SegSeq was run using the default parameters except for the local window size set to 300 as this window size shows the highest sensitivity. Only CNAs with copy ratio >0.2 were retained.
CNV-seq
We used uniquely mapped reads with mapping quality ≥ 35 as the input to the CNV-seq and first calculated the theoretical minimum window size according to a preset threshold of P<0.001 and log2 copy number ratio of 0.5 for each pair of tumor and normal samples. For each window, the number of reads was replaced with the mean coverage of the sample if it was less than that number before global normalization and calculations of the log2 ratio of tumor vs. normal and the P value. We then used circular binary segmentationto segment the log2 ratio values per chromosome and identify candidate gain and loss regions using the following cutoffs: abs(seg.mean)≥0.5; ≥8 markers per segment; and median CNV-seq P values for a segment ≤0.001. Finally, we merged the above filtered segments where the inter-segment distance is less than 500kb and copy number difference < 0.25.
CNVnator
A single sample CNV method (CNVnator) was applied independently on the tumor genomes and matching normal genomes of SJTALL002 and SJTALL007. Somatic CNAs were determined by subtracting the matching normal genome copy number from the tumor genome copy number. We followed the manual in the CNVnator and used a bin size of 100 bp. The optimal threshold for calling CNAs was determined as in BIC-seq analysis except that we tested a range of (0, 0.5].
FREEC
We constructed the SAM pileup files from the BAM files and followed the instruction in the FREEC package for paired BAM analyses. We set the ploidy as 2 for all samples analyzed and a window size of 300 bp, due to the long run time (>5 days) for the window size of 100.
Supplementary Material
Acknowledgement
This study was supported by St. Jude Children's Research Hospital–Washington University Pediatric Cancer Genome Project, Cancer Center support grant P30 CA021765 from the US National Cancer Institute and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital. C.G.M. is supported as a Pew Scholar in the Biomedical Sciences and is a St. Baldrick’s Scholar.
Footnotes
ACCESSION CODES
Diluted COLO-829 cell line along with its matching normal have been deposited at EBI (https://www.ebi.ac.uk/ega/) under accession EGAS00001001050.
Contribution
X.C. and J.Z. conceived and designed the CONSERTING algorithm. X.C., P.G.and J.W. implemented the algorithm. J. Z., S.J.B., M.A.D., J.R.D., D.W.E. and C.G.M. designed the experiment. X.C., J.W., J.D.D., M.P., J.M., M.R., W.G., A.P., S.E., S.Pounds. and J.Z. analyzed the data. K.R., J.D.D., S.Patel., L.H., D.P. and J.E performed validation and functional assay. J.N. and S.S. generated COLO-829 whole-genome sequencing data. X.C. and J.Z. wrote the manuscript.
Competing financial Interests
The authors declare no competing financial interests.
REFERENCES
- 1.Mullighan CG, et al. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley TJ, et al. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiang DY, et al. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie C, Tammi MT. BMC Bioinformatics. 2009;10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeva V, et al. Bioinformatics. 2011;27:268–269. doi: 10.1093/bioinformatics/btq635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abyzov A, Urban AE, Snyder M, Gerstein M. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xi R, et al. Proc Natl Acad Sci U S A. 2011;108:E1128–1136. doi: 10.1073/pnas.1110574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Downing JR, et al. Nat Genet. 2012;44:619–622. doi: 10.1038/ng.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, et al. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts KG, et al. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, et al. Nature. 2012;481:329–334. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, et al. Nat Genet. 2013;45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brennan CW, et al. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pleasance ED, et al. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, et al. Nat Methods. 2011;8:652–654. doi: 10.1038/nmeth.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephens PJ, et al. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanborn JZ, et al. Cancer Res. 2013;73:6036–6045. doi: 10.1158/0008-5472.CAN-13-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Handsaker RE, Korn JM, Nemesh J, McCarroll SA. Nat Genet. 2011;43:269–276. doi: 10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker M, et al. Nature. 2014;506:451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu G, et al. Nat Genet. 2014;46:444–450. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, et al. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edmonson MN, et al. Bioinformatics. 2011;27:865–866. doi: 10.1093/bioinformatics/btr032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breiman L, Friedman JM, Olshen R, Stone C. Classification and Regression Trees, Edn. 1. (Chapman and Hall/CRC, 1984) [Google Scholar]
- 24.Schwarz G. The Annals of Statistics. 1978;6:461–464. [Google Scholar]
- 25.Kent WJ, et al. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rozen S, Skaletsky H. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.