Abstract
Full-length de novo sequencing of unknown proteins remains a challenging open problem. Traditional methods that sequence spectra individually are limited by short peptide length, incomplete peptide fragmentation, and ambiguous de novo interpretations. We address these issues by determining consensus sequences for assembled tandem mass (MS/MS) spectra from overlapping peptides (e.g., by using multiple enzymatic digests). We have combined electron-transfer dissociation (ETD) with collision-induced dissociation (CID) and higher-energy collision-induced dissociation (HCD) fragmentation methods to boost interpretation of long, highly charged peptides and take advantage of corroborating b/y/c/z ions in CID/HCD/ETD. Using these strategies, we show that triplet CID/HCD/ETD MS/MS spectra from overlapping peptides yield de novo sequences of average length 70 AA and as long as 200 AA at up to 99% sequencing accuracy.
Keywords: de novo sequencing, tandem mass spectrometry, peptide identification, protein sequencing
INTRODUCTION
In most proteomics studies, proteins are identified by digesting sample proteins into peptides (with an enzyme such as trypsin), generating a tandem mass (MS/MS) spectrum for each peptide precursor, and identifying the peptide sequence of each MS/MS spectrum with a database search tool, such as SEQUEST,1 Mascot,2 MS-GFDB,3 or Spectrum Mill.4 Proteins IDs are then inferred from unique peptide sequence identifications. The utility of protein identification by database search depends upon the existence of a reference database that contains all peptides of interest. But due to mechanisms of sequence variation (such as genetic recombination and somatic hyper-mutation in monoclonal antibodies5) and the existence of unsequenced genomes, many protein sequences remain unknown. Nevertheless, the characterization of monoclonal antibodies and venoms from unsequenced species remains a key step in many therapeutic drug development pipelines.6–9 Historically, only a few low-throughput strategies have been available for de novo protein sequencing. As far back as 1987, Johnson and Biemann manually sequenced a complete protein from rabbit bone marrow using mass spectromtetry.10 Edman degradation is another established approach for sequencing novel proteins but it has experimental bottlenecks that make it unsuitable for sequencing mixtures of proteins, proteins longer than 50 amino acids (AA), or post-translationally modified proteins.11,12 As such, many current applications of de novo sequencing still continue to rely upon manual curation of MS/MS spectra and/or Edman degradation.13–15
Fully automated de novo strategies that interpret MS/MS spectra individually have been less successful compared to database search in part because they are limited by ambiguous interpretations of MS/MS fragmentation.16 Even if both approaches use the same function for scoring peptide-spectrum matches (PSMs), the top scoring peptide in the database for a given MS/MS spectrum may be the second or 7 000 000th highest scoring peptide over all possible de novo peptides, even if it is correct. Thus, de novo peptide sequencing algorithms typically report a ranked list of candidate PSMs for each spectrum where top-scoring PSMs have an accuracy of ~80–90% for low-resolution CID spectra17,18 and ~90–92% for high-resolution CID spectra16 (whereas database search results can typically be validated with 1% false discovery rate, FDR19). To yield these levels of accuracy, de novo tools face a significant trade-off between sequencing accuracy and protein sequence coverage as spectra exhibiting complete peptide fragmentation rarely cover entire proteins, yet are required to reconstruct accurate sequences. De novo peptide sequencing approaches are also limited compared to low-throughput Edman methods in that they can only generate sequences as long as enzymatically digested peptides (8–20 AA) and thus cannot fully sequence protein(s) of interest.
An alternative approach to sequencing individual spectra is to simultaneously interpret multiple MS/MS spectra from overlapping peptides.20 This Shotgun Protein Sequencing (SPS) paradigm has two distinct advantages over per-spectrum strategies. First, the alignment of spectra from overlapping peptides separates true N- and C-terminal ions from noise and leads to more accurate de novo sequences (~95% for high-resolution CID spectra) at almost full sequence coverage (95%).21 Second, the assembly of multiple aligned spectra allows for the extension of longer de novo sequences (up to 40 AA for high-resolution CID spectra).21 Remaining limitations of per-spectrum and SPS-based computational strategies have been addressed by incorporating imperfect databases of known proteins that are homologous to those in the sample. Depending upon the level of similarity between reference and target, an imperfect database can be used to correct de novo sequencing errors and anchor sequences to the reference (as done with Champs22), extend de novo sequences from known to unknown regions (as done with GenoMS23), or reorder de novo sequences to enable nearly full-length sequencing (as done with Comparative SPS, cSPS24).
De novo sequencing techniques have also been improved by utilizing multiple fragmentation modes. Compared to CID, alternative fragmentation strategies such as higher-energy collision dissociation (HCD25) and electron transfer dissociation (ETD26) are known to improve fragmentation and identification of long, highly charged peptides.27 HCD in particular has been shown to improve de novo peptide sequencing accuracy to ~95% and boost interpretations of long peptides, albeit at only 55% sequence coverage of peptides identified by database search.28 When high-resolution CID and HCD spectra were processed with an updated SPS assembly algorithm (called Meta-SPS29), de novo protein sequences were extended to ~100 AA at the maximum and 20 AA on average at 94% sequencing accuracy/65% sequence coverage for a 6-protein sample mixture and 97% sequencing accuracy/89% sequence coverage for a purified monoclonal antibody. ETD has also been shown to improve per-spectrum sequencing length and accuracy,30 but the benefits of ETD for de novo sequencing are perhaps better utilized when it is paired with CID. In this approach, a CID spectrum and an ETD spectrum are acquired for every precursor such that each pair of CID/ETD can be attributed to the same peptide. It is well-known that CID and ETD exhibit complementary fragmentation patterns that, when paired with each other, can yield much richer N/C-terminal ion ladders for a greater variety of peptides.27 Although the decreased scan rate of ETD means fewer MS/MS spectra can be acquired per aliquot of sample material, ETD significantly increases the fraction of identifiable spectra for both database search3 and per-spectrum de novo sequencing,31,32 particularly when used in conjunction with enzymes such as LysC and GluC to acquire spectra from a greater variety of longer peptides (>20 AA).33 However, per-spectrum interpretation of paired fragmentation methods still cannot produce sequences longer than enzymatically digested peptides (13–20 AA depending on the digestion parameters) and has not achieved levels of sequencing accuracy/coverage greater than 95%/65% for high-resolution MS/MS.32 Furthermore, published de novo sequencing tools capable of processing paired CID, HCD, or ETD spectra have not been made publicly available.
Advances in MS/MS instrumentation have enabled fast acquisition of a CID spectrum, HCD spectrum, and ETD spectrum per precursor such that each triplet of CID/HCD/ETD can be attributed to the same peptide. For example, a LTQ Velos Orbitrap instrument can acquire 5 triplets of CID/HCD/ETD MS/MS in a cycle of 1 MS in approximately the same amount of time as a cycle of 1 MS and 5 CID only MS/MS spectra on a prior generation LTQ-Orbitrap instrument. To take advantage of this capability, we describe a fully automated de novo protein sequencing approach that utilizes CID/HCD/ETD triplets from overlapping peptides to yield sequences as long as ~200 AA (~70 AA on average) at 99% sequencing accuracy and 71% sequencing coverage. To this end we updated algorithmic steps of the Meta-SPS29 pipeline to process any combination of high-resolution CID, HCD, and ETD spectra from each peptide. Investigations into separate acquisition of CID, HCD, and ETD have showed promise for database search34–36 but, to the best of our knowledge, this is the first application of triplet CID/HCD/ETD acquisition for de novo protein sequencing. We demonstrate that corroborating evidence of peptide fragmentation observed in CID/ETD pairs and CID/HCD/ETD triplets from overlapping peptides enables near-full length de novo protein sequencing at nearly perfect accuracy.
PROCEDURES
Since Shotgun Protein Sequencing21 interprets spectra from overlapping peptides, sample proteins were digested with multiple enzymes. High-resolution MS/MS CID/HCD/ETD triplets were then acquired on a Thermo LTQ-Orbitrap Velos and run through the updated Meta-SPS pipeline illustrated in Figure 1. To enable support for CID/HCD/ETD spectra we updated our prealignment steps to process and merge any combination of CID/HCD/ETD spectra from each precursor by adding two new stages to the Meta-SPS workflow. First PepNovo+16 was trained to score high resolution CID, HCD, and ETD MS/MS spectra (see section PepNovo+ Training). Since PepNovo+ cannot analyze multiple spectra from the same precursor, a procedure was developed to merge scored CID/HCD/ETD spectra and take advantage of corroborating evidence (see section CID/HCD/ETD Merging).
Figure 1.
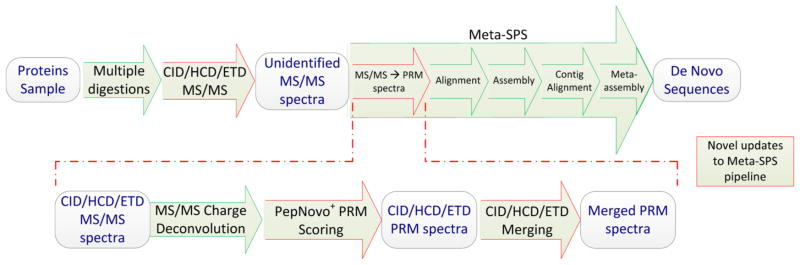
Updated Meta-SPS pipeline. Green arrows denote procedures previously described in refs 29 and 21 while red arrows denote updated procedures described here.
MS/MS ACQUISITION
To benchmark and test this approach, 21901 CID/HCD/ETD triplets (65703 total MS/MS spectra) were separately acquired from aliquots of 7 digests of a mixture of 6 known proteins. An equimolar mixture of 6 commercially purified proteins containing 252 μg of total protein was prepared. Cysteines were reduced with dithiothreitol (DTT) and alkylated with iodoacetamide. Seven 32 ug aliquots were created and used for 7 different digests with Trypsin, Chymotrypsin, Lys-C, Arg-C, Glu-C, Asp-N, or CNBr. The 6 proteins with accompanying molecular weights and Swiss-Prot accession numbers are bovine aprotinin (6.5 kDa, P00974), murine leptin (16 kDa, P41160), horse heart myoglobin (17 kDa, P68082), horseradish peroxidase (39 kDa, P00433), E. coli GroEL (57 kDa, P0A6F5), and human kallikrein-related peptidase (29 kDa, P07288). Details of sample preparation have been described previously.29
Aliquots of each digest (~0.5 μg) were analyzed with an automated nano LC–MS/MS system, consisting of an Agilent 1200 nano-LC system (Agilent Technologies, Wilmington, DE) coupled to an LTQ-Orbitrap Velos Fourier transform mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with generation 2 ion optics (Velos Pro) and a nanoflow ionization source (James A. Hill Instrument Services, Arlington, MA). Peptides were eluted from a 10 cm column (Picofrit 75 um ID, New Objectives) packed in-house with ReproSil-Pur C18-AQ 3 μm reversed phase resin (Dr. Maisch, Ammerbuch Germany) using a 95 min acetonitrile/0.1% formic acid gradient at a flow rate of 200 nL/min to yield ~20 s peak widths. Solvent A was 0.1% formic acid and solvent B was 90% acetonitrile/0.1% formic acid. The elution portion of the LC gradient was 3–6% solvent B in 1 min, 6–31% in 50 min, 31–60% in 13 min, 60–90% in 1 min and held at 90% solvent B for 5 min. Data-dependent LC–MS/MS spectra were acquired in ~3 s cycles; each cycle was of the following form: one full Orbitrap MS scan at 60000 resolution followed by 15 MS/MS scans in the orbitrap at 15000 resolution using an isolation width of 3.0 m/z. The top 5 most abundant precursor ions were each sequentially subjected to CID, HCD, and ETD dissociation. Dynamic exclusion was enabled with a mass width of ±20 ppm, a repeat count of 1, and exclusion duration of 12 s. Charge state screening was enabled along with monoisotopic precursor selection and nonpeptide monoisotopic recognition to prevent triggering of MS/MS on precursor ions with unassigned charge or a charge state of 1. For CID, the normalized collision energy was set to 30 with an activation Q of 0.25 and activation time of 30 ms. For HCD, the normalized collision energy was set to 45. For ETD, fluoranthene was used as the ETD reagent with an anion AGC target of 400 000 ions, supplemental activation was enabled, and the reaction time was dependent on the precursor charge state (precursor charge state – reaction time in msec: +2–100, +3–66.7, +4–50, +5–40, +6–33.3, etc). All MS/MS spectra were collected with an AGC target ion setting of 50000 ions. The instrument control software does not currently allow for separate AGC targets for each dissociation mode. Optimal AGC targets would be closer to 30000 ions for CID, HCD; and 200 000 ions for ETD.36 All mass spectra associated with this paper may be downloaded from ftp://MSV000078436:a@ccms-ftp01.ucsd.edu/.
Spectrum Preprocessing and Notation
Thermo RAW files were converted to mzXML with ProteoWizard37 (version 3.0.3324). To validate de novo sequencing accuracy, all combinations of CID/HCD/ETD pairs/triplets as well as individual CID, HCD, and ETD spectra were searched with MS-GFDB3 against the 6 target proteins and known contaminants with a spectrum-level false discovery rate of 1% (see Supporting Information for parameters used for MS-GFDB). As part of the Meta-SPS pipeline, high-resolution MS/MS peaks were first deconvoluted such that all peaks were converted to charge one.29 The following notation is used below: a peptide MS/MS spectrum S is defined as a collection of peaks where each peak p ∈ S has mass m[p] and intensity i[p]. The parent mass M[S] is the cumulative mass of all amino acids in the peptide sequence and the precursor charge Z[S] is the charge of the peptide precursor ion.
PepNovo+ Training
Rather than processing MS/MS spectra directly, Meta-SPS uses PepNovo+16 to interpret MS/MS fragmentation patterns and convert MS/MS spectra into PRM (prefix residue mass) spectra where peak intensities are replaced with log-likelihood scores and peak masses are replaced by PRMs,38 or Prefix-Residue Masses (cumulative amino acid masses of N-term prefixes of the peptide sequence). Peak scores combine evidence supporting peptide breaks (observed cleavages along the peptide backbone, supported by either N- or C-terminal fragments). N/C-terminal fragments may be observed by b/y ions in CID/HCD and by c/z∘/z∘ ± H39 ions in ETD. Because complementarity between b/y and c/z∘ ions can cause C-terminal MS/MS ions to be misinterpreted as N-terminal ions, PRM spectra also typically contain many SRMs, or Suffix-Residue Masses (cumulative amino acid masses of C-terminal suffixes of the peptide sequence). This approach considers peaks in PRM spectra as both PRMs and SRMs because some spectra may contain predominantly SRMs and on average they make up 30–40% of all true PRMs or SRMs.
In previous work, high-resolution CID and HCD MS/MS spectra were scored with a PepNovo+ scoring model that was not trained to process deconvoluted29 spectra and there was no PepNovo+ scoring model for ETD. In training the new models, we deconvoluted the training spectra because PepNovo+ was optimized to analyze charge 2 and 3 tryptic CID spectra, and thus does not give enough weight to MS/MS peaks of charge 3 or higher in spectra from precursors of charge >3. Here we trained three new scoring models for deconvoluted high-resolution CID, HCD, and ETD MS/MS spectra using multiple data sets. (These new models can only be used to generate PRM spectra, not de novo peptide sequences. Although PepNovo+ PRM models were trained automatically with PSMs from >3,000 unique peptides per precursor charge state, training the rank-boosting47 models needed for peptide sequencing required too many PSMs from unique peptides (>100 000) as well as more extensive modification of PepNovo+ source code.) Due to the limited availability of large sets of annotated CID, HCD, and ETD high-resolution MS/MS spectra from multiple enzymes at the time of this study, only tryptic spectra were used to train the CID model while tryptic and Lys-C spectra were combined to train each of the HCD and ETD models. The first data set consists of high-resolution CID, HCD, and ETD MS/MS spectra from tryptic peptides.36 Another 175 595 tryptic HCD MS/MS spectra were provided by the Zubarev lab at the Karolinska Institute. The third data set consists of high-resolution ETD and HCD MS/MS spectra from Lys-C digestion and SCX fractionation of a yeast lysate collected in conjunction with” the 2011 ABRF-iPRG study (see Supporting Information for description).40 All raw MS/MS spectra then were identified by MS-GFDB at 1% spectrum-level FDR to yield the set of training PSMs. PepNovo+ used these PSMs to automatically learn ion types, intensity ranks, and noise models for each type of spectra and output models which can be used to score unidentified MS/MS spectra of the same type. See Supporting Information for details regarding the MS-GFDB searches and the specific PepNovo+ training procedure.
CID/HCD/ETD Merging
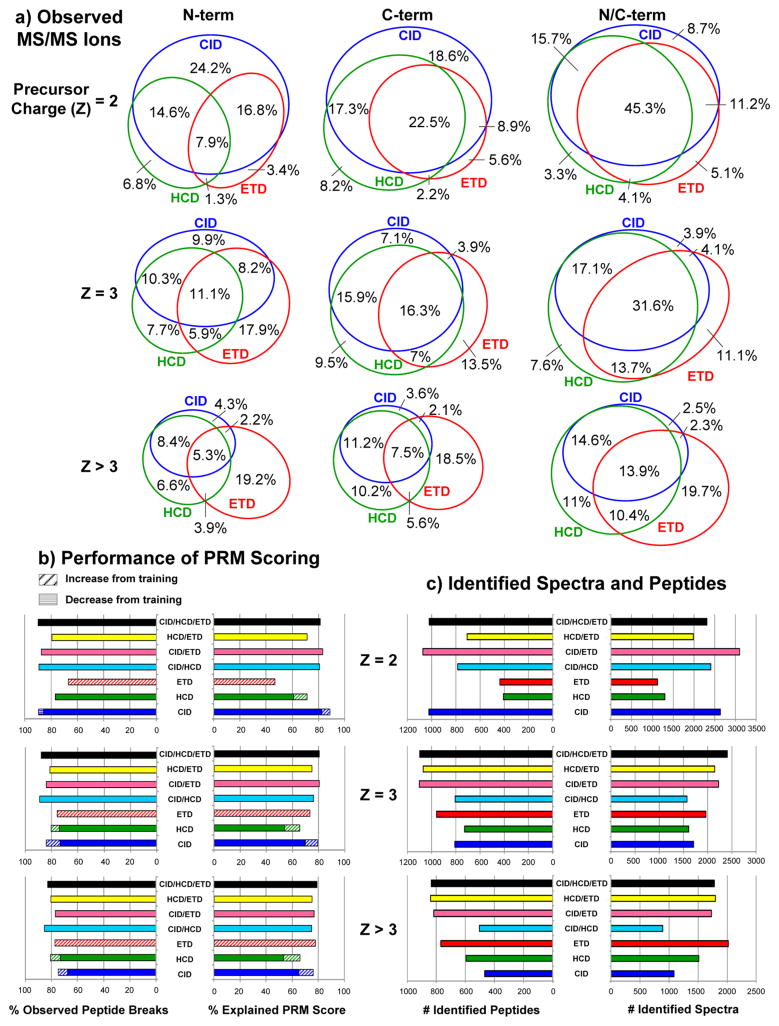
Given a CID (SCID = {c1,…,cn}), HCD (SHCD = {h1,…,hm}), and/or ETD (SETD = {e1,…,eq}) PRM spectrum from the same precursor, the merging procedure generates a single merged PRM spectrum (S = {p1,…,pr}) (with the same parent mass M[S]) for all available spectra. Using the set of training PSMs, the objective is to maximize observed breaks, which is the percentage of all breaks observed as PRMs/SRMs at correct N/C-terminal masses (a measure of sensitivity), while also maximizing explained score, which is the percentage of score in correct PRMs/SRMs relative to the score of all PRMs/SRMs in the same spectrum (a measure of accuracy). PRM spectra typically contain many C-terminal SRM masses along with N-terminal PRM masses. While PRM peaks have no offset from the summed amino acid masses, C-terminal peaks are offset by +18 Da (mass of H2O) from SRMs in CID and HCD spectra.38,41 In ETD spectra, C-terminal peaks are offset by −15 Da (mass of NH) from SRMs.3 Given a PRM or SRM mass m, one can locate the complementary SRM or PRM mass in CID and HCD spectra with the formula twinCID(m,S) = twinHCD(m,S) = M[S] − m + 18, while complementary masses in ETD can be found with twinETD(m,S) = M[S] − m − 15. Using these offsets, one can locate corroborating peaks from CID/ETD and HCD/ETD pairs that support the same peptide break, which are much more likely to explain true peptide breaks than individual PRMs. For example, we found that 92% of the score in peaks from identified ETD PRM spectra with matching peaks at the same (or complementary) mass in CID or HCD spectra was found in true PRMs/SRMs. In contrast, only 70–80% explained score is typically found in individual PRM spectra. Since PepNovo+ does not currently recognize CID/HCD + ETD corroborating evidence when assigning log-likelihood scores, we postprocessed the scores of corroborating PRMs/SRMs into combined scores in the merged PRM spectrum. However, since corroborating PRMs/SRMs only account for 47% of all peptide breaks in identified CID/HCD/ETD triplets, peaks without corroborating evidence must also be added to the merged spectrum.
Since 80% explained score was found to yield high de novo sequencing accuracy (97%) in a previous application of Meta-SPS,29 steps were developed to maximize the percentage of observed breaks at ≥80% explained score for all precursor charge states. First, corroborating PRMs and SRMs from CID/ETD and HCD/ETD pairs were extracted from PRM spectra and the corresponding combined PRMs were inserted into the merged spectrum. This was done in a series of steps to reduce the chances of misinterpreting SRMs as PRMs. But since steps 1–4 only captured PRMs and SRMs explaining 47% of all peptide breaks, the remaining peaks from CID, HCD, and ETD were also added to the merged spectrum in step 5 to bring the percentage of observed breaks to 94%. While this improved sensitivity, it also combined the noise between all three spectra such that the percentage of explained score was only 59% (instead of 91% for PRMs with corroborating evidence). Thus, local rank-based filtering was applied in step 6 to yield 86% observed breaks at 80% explained score over all precursor charge states (Figure 2b). We describe this procedure for merging CID/ETD pairs, but the same method can also be applied to HCD/ETD pairs.
Figure 2.
MS/MS ion statistics and performance of CID/HCD/ETD PRM scoring and merging. (a) Observed MS/MS ions: Percentage of peptide breaks observed by N-terminal ions (b ions in CID/HCD and c ions in ETD) and/or C-terminal ions (y ions in CID/HCD and z∘/z∘+H ions39 in ETD) over all MS/MS CID/HCD/ETD triplets identified by MS-GFDB (considering a 10 ppm peak tolerance). To filter out low-intensity noise peaks, a peak was counted if and only if its intensity was ranked in the top seven over all neighboring peak intensities with in a ± 56 Da radius. Rows separate baseline PSMs by precursor charge of identified triplets. (b) Performance of PRM scoring: Percentage of observed peptide breaks and percentage of explained score (the summed score of all true PRMs over the sum of all scores in the spectrum × 100) was counted over all combinations of merged/unmerged PRM spectra (without clustering) with identified MS/MS spectra. Peaks at N/C-terminal masses indicated peptide breaks in all cases. Each combination of PRM spectra was benchmarked by MS-GFDB IDs of the same combination of MS/MS spectra3 (CID/HCD/ETD PRMs were benchmarked with CID/HCD/ETD IDs, CID/ETD PRMs with CID/ETD IDs, HCD PRMs with HCD IDs, etc). Also indicated is the performance gained by retraining PepNovo+ to individually score high resolution CID, HCD, and ETD spectra. (c) Identified spectra and peptides: The numbers of identified spectra and unique peptides are shown for each combination of MS/MS spectra used to benchmark PRM scores in (b). As expected, incorporation of ETD significantly improves identification rates of spectra from highly charged precursors.
Consider all PRM/PRM matches: Find all pairs of peaks with same mass (ci,ek:m[ci] = m[ek]) and add a peak s to the merged spectrum S with PRM mass m[s] = m[ek]. Whenever a peak is added to the merged spectrum, it only defines a new mass if that mass does not already exist in the merged spectrum within peak tolerance (otherwise the new peak’s score is just added to the existing peak). Also find any complementary SRMs from the set {cx,ez:m[cx] = twinCID(m[s],S)∧m[ez] = twinETD(m[s], S)}. For all of these peaks that were found, assign s the merged score i[s] = 2 × (i[ci] + i[cx] + i[ek] + i[ez]) and remove ci, cx, hj, hy, ek, and ez from SCID and SETD, respectively.
Consider all SRM/SRM matches with at least one PRM: Find all pairs of SRM peaks with mass difference 15 + 18 (cx, ez: m[cx] = m[ez] + 33) and where at least one PRM from the set {ci,ek:m[ek] = twinETD (m[ez],S)∧m[ci] = m[ek]} is found from any spectrum (CID or ETD) for these SRMs. Then add a peak s to the merged spectrum S with the PRM mass m[s]=m[ek], remove all of these peaks from SCID and SETD, and assign s the merged score by the same formula in stage 1.
Consider all PRM/SRM and SRM/PRM pairs: Find all pairs of PRM/SRM peaks (ci ∈ SCID, ez ∈ SETD:m[ci] = twinETD (m[ez], S)) or SRM/PRM peaks (cx ∈ SCID, ek ∈ SETD:m[cx] = twinETD (m[ek], S)). Add a peak s to the merged spectrum with the PRM mass (m[s] = m[ci] for PRM/SRM pairs or m[s] = m[ek] for SRM/PRM pairs), remove all of its supporting peaks from SCID and SETD, and assign s the merged score by the same formula in stage 1.
Consider all SRM/SRM matches without PRMs: Find all pairs of SRM peaks with mass difference 15 + 18 (cx, ez:m[cx] = m[ez] + 33). Then add a peak s to the merged spectrum with the PRM mass m[s] = twinETD (m[ez], S), remove all of its supporting peaks from SCID and SETD, and assign s the merged score by the same formula in stage 1.
Add left over peaks from SCID and SETD to S without changing their scores.
Filter out peaks with low scores in S: a peak is retained if and only if its score is ranked in the top three over all neighboring PRM scores within a ± 56 Da mass range.
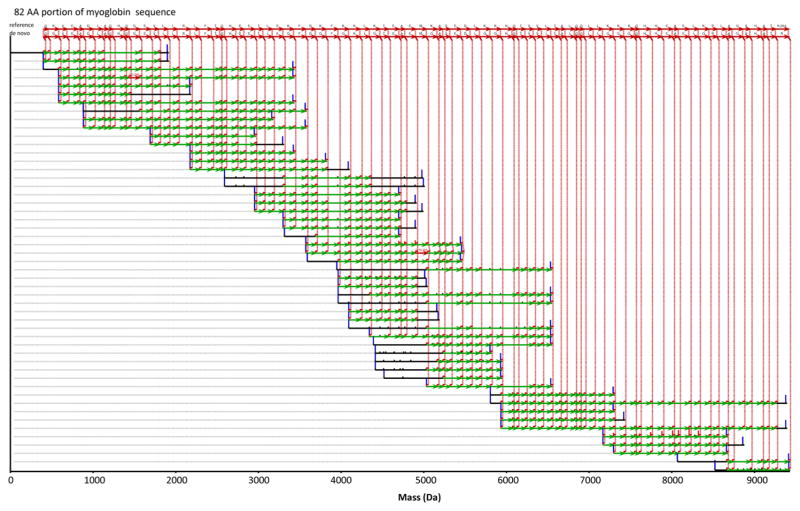
The MS/MS spectra were acquired under conditions yielding mass measurement errors of ±10 ppm. But since PepNovo+ incorporates the parent mass error when assigning PRM masses from C-terminal fragment masses, a fixed 0.04 Da tolerance was used. This corresponds to 400 ppm @ m/z 100, 40 ppm @ m/z 1000, and 10 ppm @ m/z 4000. Merged PRM spectra from the same peptide were then clustered by an approach similar to MSCluster42 (see Supporting Information for description). 21,901 CID/HCD/ETD triplets were combined into 11,325 clusters, each containing one or more triplets. A cluster contains only triplets sharing the same parent mass M[S]. Thus, triplets derived from the same peptide, but in different precursor charge states, were still merged. Replicate triplet spectra exist in the data set for two major reasons. First, given the small number of proteins in the sample and the rapid acquisition rate of the mass spectrometer, the dynamic exclusion time for triggering repeat acquisition of a particular precursor m/z was set to ~1/2 the chromatographic peak width to maximize the chance of collecting MS/MS near each peptide’s chromatographic apex. Second, some of the same peptides can be produced by digestion with two different enzymes. For example some tryptic peptides are also produced by Lys-C or Arg-C digestion. The clustered set of merged PRM spectra was then run through the Meta-SPS pipeline illustrated in Figure 1, which involves two stages of alignment/assembly. PRM spectra were first aligned and assembled into contigs (sets of spectra from overlapping peptides),21 which were further connected to form meta-contigs (sets of overlapping contigs).29 Figure 3 illustrates a resulting de novo protein sequence extracted from the highest-scoring consensus interpretation of a meta-contig. This updated Meta-SPS pipeline along with the newly trained PepNovo+ scoring models are available at http://proteomics.ucsd.edu/Software/MetaSPS.html.
Figure 3.
Assembled meta-contig of CID/HCD/ETD triplets. The topmost sequence is the myoglobin sequence as it is aligned to the de novo sequence below it. Each row denotes a merged PRM spectrum from one or more CID/HCD/ETD triplets where peaks not aligned to other merged PRM spectra from overlapping peptides are removed.21 Red peaks indicate PRMs supporting the de novo sequence and green arrows between red peaks denote 1–2 AA mass differences supporting the consensus de novo sequence. Red vertical dotted lines connect assembled PRMs to each de novo sequence call; black peaks were not assembled into the consensus. Blue bars denote spectrum end points (at mass 0 and parent mass M[S]). The height of each peak corresponds to the merged PRM score from CID, HCD, and ETD. The red labels “[+0.98]” and “[+16.00]” indicate post-translational modification masses that were tolerated during alignment/assembly (without knowing of them in advance). All de novo sequence calls, except the “R” at the end, were verified by database search.
RESULTS
The performance of Meta-SPS on CID/HCD/ETD triplets was assessed in terms of de novo sequencing length, coverage, and accuracy. Coverage and length was determined via modification-tolerant alignment of de novo sequences to the reference protein sequences.24 Sequencing accuracy was also computed as done previously:29 MS-GFDB peptide-spectrum matches were transferred to PRM spectra and then meta-contigs. A sequence call (mass of one or more possibly modified amino acids) was labeled correct if its consecutive flanking peaks are annotated by a MS-GFDB peptide match in the same ion series in the same identified spectrum (i.e., both are annotated as PRMs or SRMs from MS-GFDB’s peptide match). All noncorrect sequence calls from identified spectra are labeled incorrect. Remaining sequence calls whose flanking peaks are not from identified spectra are labeled unannotated. See Supporting Information for details regarding the MS-GFDB searches used to compute performance metrics in Figure 2 and Table 1. Figure 2a shows MS/MS ion statistics over all identified CID/HCD/ETD triplets and Figure 2c shows the numbers of identified spectra and peptides for all combinations of CID/HCD/ETD. Table 1 (top) details the spectrum coverage by MS-GFDB (percent of protein sequence covered by identified peptides) for different combinations of fragmentation methods and Table 1 (bottom) details coverage of all six proteins.
Table 1.
De novo Sequencing Length, Coverage, and Accuracy for Alternative Minimum Meta-contig Size (κ) Cutoffsa
| Sequencing results per combination of fragmentation modes | |||||
|---|---|---|---|---|---|
| fragmentation modes | CID/HCD/ETD | CID/ETD | HCD/ETD | CID/HCD | |
| Spectrum Coverage (%) | 88.3 | 86.9 | 87.9 | 88.3 | |
| Longest Sequence (AA) | 194 | 125 | 131 | 704 | |
| SPS contigs per meta-contig (κ) | |||||
| κ ≥ 5 | Sequencing Coverage (%) | 70.9 | 67.3 | 64.8 | 52.7 |
| # Meta-Contigs | 19 | 22 | 24 | 30 | |
| Average Seq. Length (AA) | 65.9 | 52.1 | 48.5 | 29.8 | |
| Sequencing Accuracy (%) | 98.9 | 99.5 | 96.5 | 91.2 | |
| Unannotated Seq. Calls (%) | 4.1 | 1.3 | 5.2 | 8.2 | |
| κ ≥ 2 | Sequencing Coverage (%) | 79.5 | 75.6 | 76.2 | 73.5 |
| # Meta-Contigs | 33 | 38 | 45 | 50 | |
| Average Seq. Length (AA) | 43.9 | 37.4 | 64.3 | 28.9 | |
| Sequencing Accuracy (%) | 98.9 | 97.8 | 97 | 90.1 | |
| Unannotated Seq. Calls (%) | 4 | 2.2 | 6.6 | 8.7 | |
| κ ≥ 1 | Sequencing Coverage (%) | 83.6 | 80.9 | 82.1 | 81.7 |
| # Meta Contigs | 142 | 139 | 141 | 172 | |
| Average Seq. Length (AA) | 28.3 | 23.8 | 22.7 | 20.3 | |
| Sequencing Accuracy (%) | 97.9 | 97.3 | 96.4 | 88.5 | |
| Unannotated Seq. Calls (%) | 4.2 | 3.4 | 7.5 | 11.4 | |
| Sequencing results per protein (using CID/HCD/ETD fragmentation) | |||||||
|---|---|---|---|---|---|---|---|
| Protein | leptim | kallikrein | groEL | myoglobin | aprotinin | peroxidase | |
| Protein Length (AA) | 167 | 261 | 54.8 | 154 | 100 | 353 | |
| Spectrum Coverage (%) | 94.6 | 90.4 | 99.8 | 99.4 | 61 | 68.8 | |
| Longest Sequence (AA) | 93 | 134 | 194 | 80 | 59 | 58 | |
| SPS contigs per meta-contig (κ) | |||||||
| κ ≥ 5 | Sequencing Coverage (%) | 86.2 | 79.3 | 80.5 | 84.4 | 59 | 39.9 |
| # Meta-Contigs | 3 | 5 | 4 | 2 | 2 | 4 | |
| Average Seq. Length (AA) | 66 | 60.2 | 115.8 | 65 | 39.5 | 35.2 | |
| Sequencing Accuracy (%) | 100 | 98.3 | 99.5 | 99.2 | 89.8 | 100 | |
| Unannotated Seq. Calls (%) | 4.1 | 7.3 | 6.5 | 2.3 | 0 | 9.3 | |
| κ ≥ 2 | Sequencing Coverage (%) | 86.2 | 87.7 | 92.3 | 84.4 | 59 | 53.8 |
| # Meta-Contigs | 6 | 7 | 9 | 2 | 3 | 8 | |
| Average Seq. Length (AA) | 37.5 | 44.5 | 66.9 | 65 | 29.7 | 25.5 | |
| Sequencing Accuracy (%) | 100 | 98.4 | 97.7 | 99.2 | 89.8 | 100 | |
| Unannotated Seq. Calls (%) | 7.3 | 5.9 | 2.8 | 0 | 9.3 | 2.2 | |
| κ ≥ 1 | Sequencing Coverage (%) | 86.2 | 87.7 | 92.5 | 92.2 | 64 | 67.4 |
| # Meta-Contigs | 10 | 12 | 16 | 4 | 5 | 17 | |
| Average Seq. Length (AA) | 26 | 29.5 | 41.9 | 37.5 | 20.4 | 17.1 | |
| Sequencing Accuracy (%) | 100 | 98.5 | 97.7 | 99.3 | 80 | 100 | |
| Unannotated Seq. Calls (%) | 7.3 | 5.6 | 2.8 | 0 | 14.1 | 2.2 | |
Sequencing results per combination of fragmentation modes: Spectrum Coverage is the percent of amino acids in all proteins covered by peptides identified by MS-GFDB at 1% FDR. Sequencing Coverage is the percent of amino acids in all proteins covered by at least one aligned de novo sequence. Average Seq. Length is the average number of amino acids covered by each aligned de novo sequence and Longest Sequence is the maximum number of amino acids covered by a single de novo sequence. Sequencing Accuracy is the percentage of all annotated sequence calls that were labeled correct. Un-annotated Seq. Calls is the percentage of sequence calls that were un-annotated. Each column indicates which combination of MS/MS spectra was used as input to Meta-SPS and database search. Sequencing results per protein (using CID/HCD/ETD fragmentation): The same metrics in the top are shown for each protein in the CID/HCD/ETD data set (cumulative results over all six proteins are shown in the first column of the top).
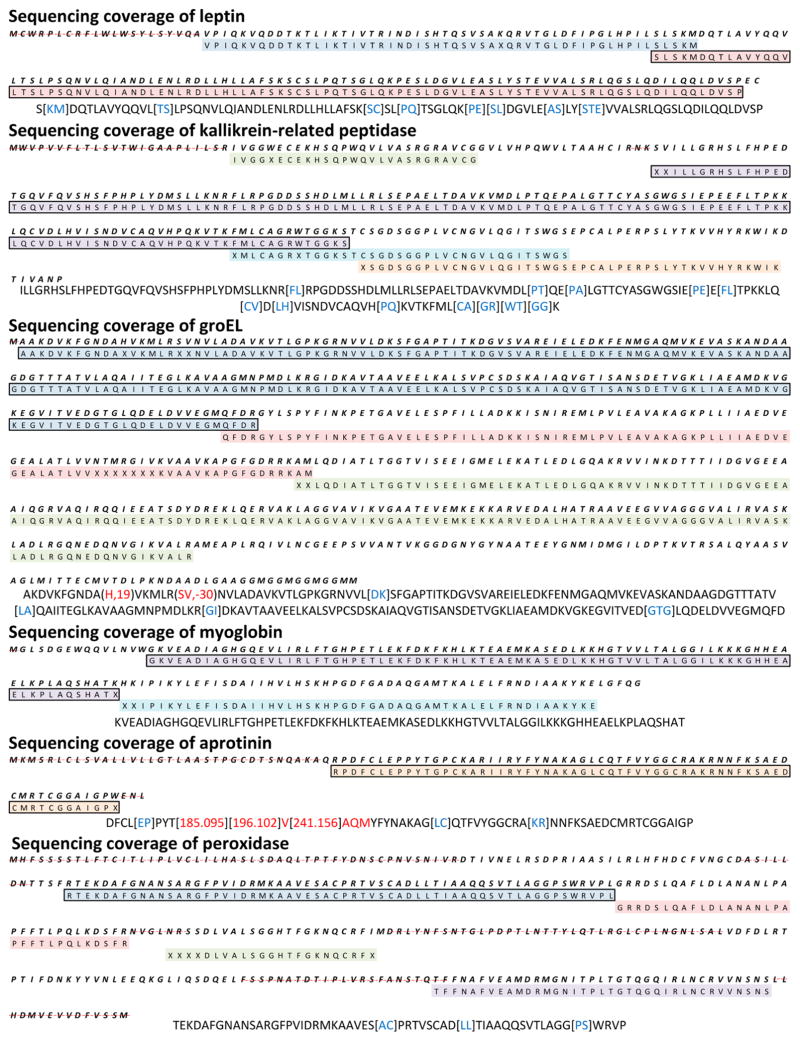
Since Meta-SPS sequencing errors are usually distributed toward the ends of sequences29 we removed the first and last sequence calls from every de novo sequence before computing coverage and accuracy. Resulting meta-contigs were binned by κ, the minimum allowable number of combined SPS contigs per meta-contig, and results are reported for κ ≥ 1, κ ≥ 2, and κ ≥ 5. κ ≥ 5 yields the longest and most accurate subset of meta-contig sequences because each of these must be supported by at least 5 SPS contig sequences, whereas κ ≥ 1 retains unmerged SPS contigs with meta-contigs of all sizes to yield the highest sequencing coverage. At κ ≥ 5, 19 de novo sequences assembling CID/HCD/ETD triplets were returned by Meta-SPS, all of which matched to the reference (with at most two modifications per match) and covered 71% of all six proteins at average length 66 AA (Table 1a). At κ ≥ 1 and κ ≥ 2, minimal losses in sequencing accuracy were sustained (98%) to achieve sequencing coverage (80% and 84%, respectively) closer to the coverage of database search (88%) at 1% FDR. The longest sequence spanned 194 AA and is shown in Figure 4 along with the longest sequences covering each of the six proteins.
Figure 4.
De novo sequencing coverage of six target proteins at κ ≥ 5. Every colored row corresponds to a de novo sequence as separately mapped to the reference protein sequence (information not used by Meta-SPS); each row in the coverage map spans at most 85 AA. Regions of each sequence that were mapped to the reference with unknown modifications have X’s in place of AA letter codes. Below each protein map is the longest de novo sequence covering that protein (also indicated in bold boxes in the coverage maps) following removal of first/last sequence calls. Blue letters correspond to calls that span 2 or more AA in the reference. Red letters indicate incorrect sequence calls as aligned to the reference. Remaining uncolored AA represent sequence calls that match reference amino acid masses. Regions where lack of de novo sequencing coverage was expected (due to lack of coverage by database search) are indicated with a dashed red line. As mentioned in the Results section, these lapses in coverage likely occur because of known cleavage of signal peptides and glycosylation sites.
Although sequences from CID/ETD pairs only (i.e., no HCD) were not as long at the maximum (125 AA), they were still longer than 50 AA on average (at κ ≥ 5) and covered 67–81% of target proteins depending on κ (Table 1, top). HCD/ETD pairs exhibited roughly the same sequence coverage and length as CID/ETD (65–82% coverage, 131 AA maximum length, and 49 AA average length). The highest sequencing accuracy was observed for CID/ETD pairs and CID/HCD/ETD triplets at 99.5% and 98.9%, respectively, while HCD/ETD pairs gave 96.5% accuracy.
ETD provides a significant increase in interpretable MS/MS fragmentation of long, highly charged peptides as well as a gain in PRM scores given to corroborating peaks in CID/ETD and HCD/ETD (Figure 2). Corroborating evidence was a very significant feature of peptide fragmentation as 91.8% of PRM scores was found in true PRMs after stage 1–4 merging. As a result, the combinations of CID/ETD, HCD/ETD, and CID/HCD/ETD gave the highest quality PRM spectra from long peptides, which are especially useful for assembly because they enable the extension of de novo sequences into regions that might not contain overlapping coverage of shorter peptides with precursor charge 2/3 due to either overdigestion or incomplete enzyme digestion. The quality of PRM spectra from long peptides was also improved by training PepNovo+ on high-resolution CID, HCD, and ETD MS/MS spectra (Figure 2b).
Of the 6 proteins analyzed in this work, leptin and GroEL were produced recombinantly in E. coli while kallikrein-related peptidase, aprotinin, myoglobin, and peroxidase were isolated from natural sources. As documented in UniProt, leptin, kallikrein-related peptidase, aprotinin, and peroxidase are each known to contain N-terminal signal peptides that target the proteins for secretion from their cells of origin. Aprotinin and peroxidase further contain propeptide sequences that are cleaved upon activation. While the signal and pro-peptides would be missing from the proteins we analyzed, in Table 1 and Figure 4 we have used the full length gene sequence when calculating coverage by the assembled MS/MS spectra. Leptin contains a signal peptide (amino acids 1–21), that is lacking in the recombinant material obtained from Sigma-Aldrich.43 Kallikrein-related peptidase contains a signal peptide (amino acids 1–17), a propeptide (amino acids 17–24), and known N-linked glycosylation at amino acid 69. Aprotinin contains a signal peptide (amino acids 1–21), and propeptides (amino acids 22–35 and 94–100). Peroxidase contains a signal peptide (amino acids 1–30), a propeptide (amino acids 339–353), and known N-linked glycosylation sites at (amino acids 43, 87, 188, 216, 228, 244, 285, and 298). The sugar microheterogeneity at N-linked glycosylation sites will tend to render any individual proteolytically generated peptide containing that amino acid much less concentrated in the digestion mixture, and if subjected to MS/MS much less likely to yield interpretable fragmentation. These modifications, along with incomplete peptide sampling by the instrument, likely explain why 12% of protein sequences were not covered by database search. Remaining losses of coverage from de novo sequencing can be attributed to lack of spectra from overlapping peptides with sufficient fragmentation.
To determine whether all enzymes were necessary to achieve quality sequencing, seven data sets were generated such that spectra from each of the seven enzymes were separately excluded from the CID/HCD/ETD data. De novo sequencing length, coverage, and accuracy from these runs are shown in Table 2 (top). At κ = 1, each of these data sets exhibited roughly the same sequencing accuracy (97–98%), high maximum sequence length (118–194 AA), yet varying levels of de novo sequencing coverage. All runs yielded 79–83% sequencing coverage except when CNBr spectra were excluded, in which case sequencing coverage dropped to 64%. Table 2 (top) shows that CNBr did not yield spectrum coverage that was missed by other enzymes (MS-GFDB coverage only dropped from 88.3 to 87.4%). However CNBr contributed the most unique peptides from highly charged precursors (Table 2, bottom). Although the most abundant precursor ions in our CNBr data are derived from peptides that span the distance between two methionine residues, much of the data instead consists of peptides bounded by a Met-specific cleavage on one end and a nonspecific hydrolysis cleavage on the other end. This yields sets of overlapping peptides that differ only by short AA truncations on either end. Altogether, these features result in CNBr outperforming Lys-C, Arg-C, Asp-N, and Glu-C digests in terms of generating the most precursors from long overlapping peptides, which are valuable to Meta-SPS for assembling long de novo sequences with high sequencing coverage.
Table 2.
De novo Sequencing and Database Search Results by Enzymea
| Sequencing result per excluded enzyme (using CID/HCD/ETD fragmentation) | ||||||||
|---|---|---|---|---|---|---|---|---|
| excluded enzyme | none | Arg-C | Asp-N | CNBr | Glu-C | Lys-C | trypsin | chymotrypsin |
| Spectrum Coverage (%) | 88.3 | 87.4 | 88.3 | 87.4 | 88.3 | 88.1 | 86.9 | 88 |
| Logest Sequence (AA) | 194 | 194 | 123 | 194 | 118 | 137 | 163 | 143 |
| Sequencing Coverage (%) | 83.6 | 80.1 | 79.7 | 63.9 | 82.5 | 80.9 | 80.9 | 79.5 |
| # Meta-Contigs | 142 | 130 | 148 | 126 | 132 | 141 | 119 | 137 |
| Average Seq. Length (AA) | 28.3 | 28.4 | 24.6 | 25 | 28.4 | 26.2 | 29 | 26.1 |
| Sequencing Accuracy (%) | 97.9 | 97.5 | 97.3 | 97.9 | 98.5 | 96.6 | 98 | 97.5 |
| Unannotated Seq. Calls (%) | 4.2 | 4 | 4.4 | 4.5 | 5.4 | 5.9 | 4 | 5.9 |
| MS-GFDB results per enzyme (using CID/HCD/ETD fragmentation) | |||||||
|---|---|---|---|---|---|---|---|
| enzyme | Arg-C | Asp-N | CNBr | Glu-C | Lys-C | trypsin | chymotrypsin |
| Spectrum Coverage (%) | 64.8 | 67 | 68.7 | 54.6 | 68.3 | 71.2 | 72.3 |
| Sequencing Accuracy (%) | 35 1 | 340 | 442 | 313 | 289 | 390 | 417 |
| Charge >3 Peptides (%) | 42.5 | 38.1 | 47.1 | 37.1 | 41.2 | 24.6 | 27.3 |
Sequencing results per excluded enzyme (using CID/HCD/ETD fragmentation): Each column indicates which spectra (acquired from a specific enzyme digestion) were removed from the full set of triplet spectra. The same metrics as in Table 1 (top) are shown for all contigs (i.e., including meta-contigs and un-merged SPS contigs, κ = 1). Digestion by CNBr was found to contribute the most de novo sequencing coverage to the combined CID/HCD/ETD analysis. MS-GFDB results per enzyme (using CID/HCD/ETD fragmentation): Each column indicates which set of spectra were identified against the six proteins at 1% spectrum-level FDR. Spectrum Coverage is defined in Table 1 (top); Unique Peptides is the number of unique identified peptide sequences (considering PTMs); Charge >3 Peptides is the percentage of unique peptides that were identified by at least one spectrum with precursor charge >3. CNBr was found to contribute the largest set of unique peptide IDs while also having the largest composition of identified peptides from highly charged precursors, which indicates why removing CNBr from the analysis shown in the top yielded the least de novo sequencing coverage.
CONCLUSIONS
Multispectrum acquisition of high resolution CID, HCD, and ETD coupled with the proposed improvements to Meta-SPS enable near full-length automated de novo sequencing of simple protein mixtures at 99% sequencing accuracy. To the best of our knowledge, these are the longest and most accurate de novo sequences ever reported by an automated approach. Although this approach still falls short of fully reconstructing a complete protein, the average sequence length was greater than 60 AA long and approached 200 AA at the maximum, which should potentially enable automated sequencing of small proteins such as venom toxins21,44 and the variable CDR regions of monoclonal antibodies.23,24,45
Related methods for de novo sequencing with complementary fragmentation methods do not consider spectra from overlapping peptides, which limits sequencing length (<10 AA on average), accuracy (<95%), and coverage (<70%).31,32 Still, results could possibly improve from devising more robust probabilistic scoring functions for paired CID/ETD and HCD/ETD MS/MS spectra than described here. Possible ways to do this include the Bayesian networks approach in Spectrum Fusion31 or extensions of the scoring functions used in popular de novo tools like PepNovo+ and PEAKS.
Although our high-resolution MS/MS acquisition enabled ±10 ppm mass tolerance, a fixed 0.04 Da tolerance was used because PepNovo+ and SPS do not yet support ppm tolerance. Allowing for ±0.04 Da mass errors is equivalent to the diminishing mass error tolerance of 400–10 ppm over the increasing mass range of 100–4000 m/z. Implementing ppm tolerance in the Meta-SPS pipeline might allow for reduction alignment thresholds in SPS and Meta-SPS, as the probability of random high scoring matches between spectra from nonoverlapping peptides diminishes with tighter mass tolerance. It would also enable resolving ambiguous interpretations of near isobaric masses (K-Q = 0.03638, K-GA = 0.03638, F-Mox =0.0330, VS-W = 0.02113, and W-DA = 0.1526), which is a common limitation of proteomics mass spectrometry. Other ambiguities, such I/L interpretations, cannot be resolved by mass alone but may be resolved by examination of amino acid-specific fragmentation patterns.46
Here we can report sequencing accuracy because de novo sequencing was done on a set of known proteins. When this method is applied to unknown complex samples, sequencing accuracy may still be approximated with a subset of identified spectra. If the sample is completely unknown, one could anticipate spiking the set of input spectra with a set of spectra acquired under the same experimental conditions from a few known proteins that have no homology to those in the unknown sample. Although this may capture cases where spectra from completely unrelated proteins are assembled into the same meta-contig, it will fail to capture cases where spectra from homologous proteins are combined due to sequence similarity. It remains an open problem to determine whether such sequencing errors and/or false discovery rates can be estimated by de novo assembly of MS/MS spectra.
This approach is mainly limited by instrument peptide sampling bias as a result of hydrophobicity, ionizability, and locations of basic amino acids, which leads to incomplete MS/MS coverage. This can significantly affect the performance of assembly based approaches where full peptide coverage is not usable without sufficient overlap between peptides. As a result, Meta-SPS is currently optimized for data sets where the experimental protocol is expected to yield a high fraction of spectra from overlapping peptides. While this is currently easiest for simple protein mixtures, we would expect that the same methods would apply to more complex samples as long as enough mass spectrometry runs are used to acquire spectra from overlapping peptides. In addition, analysis of more complex mixtures would benefit from faster MS/MS scan rates or analysis of multiple fractions to yield enough coverage with multiple overlapping peptide sequences. The slower scan rate of ETD (~2/3 the rate of HCD) may further limit coverage, but our results suggest that ETD coupled with CID and/or HCD yields much longer and more accurate de novo sequencing than CID or HCD alone (even when considering that more precursors are subjected to MS/MS when fewer dissociation methods are employed), and thus the gains in sequencing outweigh the losses in peptide sampling. We further anticipate improvements in the quality of ETD spectra collected in the CID/HCD/ETD triplet configuration upon revision of the instrument control software to allow for separate AGC targets for each dissociation mode. Currently, we set the ETD AGC target ~4-fold lower than optimal so as not to overly compromise CID and HCD performance.
Acknowledgments
This work was partially supported by the National Institutes of Health Grant 8 P41 GM103485-05 from the National Institute of General Medical Sciences. This work was also supported in part by the Broad Institute of MIT and Harvard, and by grants to Steven A. Carr from the US National Cancer Institute (U24CA160034, part of the Clinical Proteomics Tumor Analysis Consortium initiative) and the National Heart, Lung, and Blood Institute (HHSN268201000033C and R01HL096738). We would like to thank Namrata Udeshi for providing expertise in configuring data acquisition, and the 2011 ABRF-iPRG committee for providing additional data that was generated in conjunction with their 2011 study. We would also like to thank the Zubarev lab at the Karolinska Institute for providing the HCD data set used for training PepNovo+.
Footnotes
Notes
The authors declare no competing financial interest.
Supporting Information
Supplementary data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 2.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Kim S, Mischerikow N, Bandeira N, Navarro JD, Wich L, Mohammed S, Heck AJR, Pevzner PA. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol Cell Proteomics. 2010;9:2840–2852. doi: 10.1074/mcp.M110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agilent Technologies: Santa Clara, CA. http://spectrummill.mit.edu/
- 5.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 6.Maggon K. Monoclonal antibody “gold rush. Curr Med Chem. 2007;14:1978–1987. doi: 10.2174/092986707781368504. [DOI] [PubMed] [Google Scholar]
- 7.Haurum JS. Recombinant polyclonal antibodies: the next generation of antibody therapeutics? Drug Discovery Today. 2006;11:655–660. doi: 10.1016/j.drudis.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Lewis RJ, Garcia ML. Therapeutic potential of venom peptides. Nat Rev Drug Discovery. 2003;2:790–802. doi: 10.1038/nrd1197. [DOI] [PubMed] [Google Scholar]
- 9.Pimenta AM, De Lima ME. Small peptides, big world: biotechnological potential in neglected bioactive peptides from arthropod venoms. J Peptide Sci. 2005;11:670–676. doi: 10.1002/psc.701. [DOI] [PubMed] [Google Scholar]
- 10.Johnson RS, Biemann K. The primary structure of thioredoxin from Chromatium vinosum determined by high-performance tandem mass spectrometry. Biochemistry. 1987;26:1209–1214. doi: 10.1021/bi00379a001. [DOI] [PubMed] [Google Scholar]
- 11.Thoma RS, Smith JS, Sandoval W, Leone JW, Hunziker P, Hampton B, Linse KD, Denslow ND. The ABRF Edman Sequencing Research Group 2008 Study: investigation into homo-polymeric amino acid N-terminal sequence tags and their effects on automated Edman degradation. J Biomol Tech. 2009;20:216–225. [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang B, Walters J, Mawuenyega K, Simpson J, Sandoval W, Smith JS, Hunziker P. Results of the PSRG 2010 Study: Edman and Mass Spectrometric Terminal Sequencing of a Monoclonal Antibody. J Biomol Tech. 2010;21:S18. [Google Scholar]
- 13.Calvete JJ, Ghezellou P, Paiva O, Matainaho T, Ghassempour A, Goudarzi H, Kraus F, Sanz L, Williams DJ. Snake venomics of two poorly known Hydrophiinae: Comparative proteomics of the venoms of terrestrial Toxicocalamus longissimus and marine Hydrophis cyanocinctus. J Proteomics. 2012;75:4091–4101. doi: 10.1016/j.jprot.2012.05.026. [DOI] [PubMed] [Google Scholar]
- 14.Medzihradszky KF, Bohlen CJ. Partial De Novo Sequencing and Unusual CID Fragmentation of a 7 kDa, Disulfide-Bridged Toxin. J Am Soc Mass Spectrom. 2012;23:923–934. doi: 10.1007/s13361-012-0350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huancahuire-Vega S, Ponce-Soto LA, Martins-de-Souza D, Marangoni S. Biochemical and pharmacological characterization of PhTX-I a new myotoxic phospholipase A2 isolated from Porthidium hyoprora snake venom. Comp Biochem Physiol: Toxicol Pharmacol. 2011;154:108–119. doi: 10.1016/j.cbpc.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Frank AM, Savitski MM, Nielsen ML, Zubarev RA, Pevzner PA. De novo peptide sequencing and identification with precision mass spectrometry. J Proteome Res. 2007;6:114–123. doi: 10.1021/pr060271u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank A, Pevzner PA. PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal Chem. 2005;77:964–973. doi: 10.1021/ac048788h. [DOI] [PubMed] [Google Scholar]
- 18.Ma B, Zhang K, Hendrie C, Liang C, Li M, Doherty-Kirby A, Lajoie G. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- 19.Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandeira N, Tang H, Bafna V, Pevzner P. Shotgun protein sequencing by tandem mass spectra assembly. Anal Chem. 2004;76:7221–7233. doi: 10.1021/ac0489162. [DOI] [PubMed] [Google Scholar]
- 21.Bandeira N, Clauser KR, Pevzner PA. Shotgun protein sequencing: assembly of peptide tandem mass spectra from mixtures of modified proteins. Mol Cell Proteomics. 2007;6:1123–1134. doi: 10.1074/mcp.M700001-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Han Y, Yuen D, Ma B. Automated protein (re)sequencing with MS/MS and a homologous database yields almost full coverage and accuracy. Bioinformatics. 2009;25:2174–2180. doi: 10.1093/bioinformatics/btp366. [DOI] [PubMed] [Google Scholar]
- 23.Castellana NE, Pham V, Arnott D, Lill JR, Bafna V. Template proteogenomics: sequencing whole proteins using an imperfect database. Mol Cell Proteomics. 2010;9:1260–1270. doi: 10.1074/mcp.M900504-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bandeira N, Pham V, Pevzner P, Arnott D, Lill JR. Automated de novo protein sequencing of monoclonal antibodies. Nat Biotechnol. 2008;26:1336–1338. doi: 10.1038/nbt1208-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 26.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci US A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guthals A, Bandeira N. Peptide identification by tandem mass spectrometry with alternate fragmentation modes. Mol Cell Proteomics. 2012;11:550–557. doi: 10.1074/mcp.R112.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chi H, Sun RX, Yang B, Song CQ, Wang LH, Liu C, Fu Y, Yuan ZF, Wang HP, He SM, Dong MQ. pNovo: de novo peptide sequencing and identification using HCD spectra. J Proteome Res. 2010;9:2713–2724. doi: 10.1021/pr100182k. [DOI] [PubMed] [Google Scholar]
- 29.Guthals A, Clauser KR, Bandeira N. Shotgun protein sequencing with meta-contig assembly. Mol Cell Proteomics. 2012;10:1084–1096. doi: 10.1074/mcp.M111.015768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Shan B, Xin L, Ma B. Better score function for peptide identification with ETD MS/MS spectra. BMC Bioinform. 2010;11:S4. doi: 10.1186/1471-2105-11-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Datta R, Bern M. Spectrum Fusion: Using Multiple Mass Spectra for De Novo Peptide Sequencing. J Comput Biol. 2009;16:1169–1182. doi: 10.1089/cmb.2009.0122. [DOI] [PubMed] [Google Scholar]
- 32.Savitski MM, Nielsen ML, Kjeldsen F, Zubarev RA. Proteomics-grade de novo sequencing approach. J Proteome Res. 2005;4:2348–2354. doi: 10.1021/pr050288x. [DOI] [PubMed] [Google Scholar]
- 33.Swaney DL, Wenger CD, Coon JJ. Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J Proteome Res. 2010;9:1323–1329. doi: 10.1021/pr900863u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Y, Tolić N, Xie F, Zhao R, Purvine SO, Schepmoes AA, Moore RJ, Anderson GA, Smith RD. Effectiveness of CID, HCD, and ETD with FT MS/MS for degradomic-peptidomic analysis: comparison of peptide identification methods. J Proteome Res. 2011;10:3929–3943. doi: 10.1021/pr200052c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen Y, Tolić N, Purvine SO, Smith RD. Improving collision induced dissociation (CID), high energy collision dissociation (HCD), and electron transfer dissociation (ETD) fourier transform MS/MS degradome–peptidome identifications using high accuracy mass information descriptions of Dat. Proteome. 2012;11:668–677. doi: 10.1021/pr200597j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frese CK, Altelaar AFM, Hennrich ML, Nolting D, Zeller M, Griep-Raming J, Heck AJR, Mohammed S. Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J Proteome Res. 2011;10:2377–2388. doi: 10.1021/pr1011729. [DOI] [PubMed] [Google Scholar]
- 37.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dancík V, Addona TA, Clauser KR, Vath JE, Pevzner PA. De novo peptide sequencing via tandem mass spectrometry. J Comput Biol. 1999;6:327–342. doi: 10.1089/106652799318300. [DOI] [PubMed] [Google Scholar]
- 39.Savitski MM, Kjeldsen F, Nielsen ML, Zubarev RA. Hydrogen rearrangement to and from radical z fragments in electron capture dissociation of peptides. J Am Soc Mass Spectrom. 2007;18:113–120. doi: 10.1016/j.jasms.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 40.Clauser KR, Askenazi M, Bandeira N, Chalkley RJ, Deutsch E, Lam H, McDonald WH, Neubert T, Rudnick P, Martens L. iPRG 2011: A study on the identification of electron transfer dissociation (ETD) mass spectra. Association of Biomolecular Resource Facilities; Bethesda, MD: 2011. Proteome Informatics Research Group 2011 study. Available from: http://www.abrf.org/index.cfm/group.show/ProteomicsInformaticsResearchGroup.53.html. [Google Scholar]
- 41.Taylor JA, Johnson RS. Implementation and uses of automated de novo peptide sequencing by tandem mass spectrometry. Anal Chem. 2001;73:2594–2604. doi: 10.1021/ac001196o. [DOI] [PubMed] [Google Scholar]
- 42.Frank AM, Bandeira N, Shen Z, Tanner S, Briggs SP, Smith RD, Pevzner PA. Clustering millions of tandem mass spectra. J Proteome Res. 2008;7:113–122. doi: 10.1021/pr070361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sigma-Aldrich. 2013 http://www.sigmaaldrich.com/
- 44.Bhatia S, Kil YJ, Ueberheide B, Chait BT, Tayo L, Cruz L, Lu B, Yates JR, Bern M. Constrained de novo sequencing of conotoxins. J Proteome Res. 2012;11:4191–4200. doi: 10.1021/pr300312h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castellana NE, McCutcheon K, Pham VC, Harden K, Nguyen A, Young J, Adams C, Schroeder K, Arnott D, Bafna V, Grogan JL, Lill JR. Resurrection of a clinical antibody: template proteogenomic de novo proteomic sequencing and reverse engineering of an anti-lymphotoxin-α antibody. Proteomics. 2011;11:395–405. doi: 10.1002/pmic.201000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta K, Kumar M, Chandrashekara K, Krishnan KS, Balaram P. Combined Electron Transfer Dissociation-Collision-Induced Dissociation Fragmentation in the Mass Spectrometric Distinction of Leucine, Isoleucine, and Hydroxyproline Residues in Peptide Natural Products. J Proteome Res. 2011;11:515–522. doi: 10.1021/pr200091v. [DOI] [PubMed] [Google Scholar]
- 47.Frank AM. A ranking-based scoring function for peptide-spectrum matches. J Proteome Res. 2009;8:2241–2252. doi: 10.1021/pr800678b. [DOI] [PMC free article] [PubMed] [Google Scholar]