Abstract
Although alcohol dependence (AD) is approximately 50% heritable, little is known about how specific genetic loci affect AD risk. In a genome-wide association study (GWAS), we identified highly significant associations between two population-specific functional variants in the alcohol dehydrogenase 1B gene (ADH1B) and AD in African-Americans (AAs; rs2066702) and European-Americans (EAs; rs1229984). In the current study, we determined which specific diagnostic criteria contributed to the observed associations of ADH1B SNPs with AD. Our analysis included both the DSM-IV and DSM-5 diagnostic systems. We also investigated the relationship of ADH1B variants to the maximum number of drinks consumed in a 24-hour period (MaxDrinks), a presumed intermediate phenotype of AD. We found that, although all criteria made strong individual contributions to the associations, the largest contributions came from those reflecting neuroadaptation: tolerance (rs2066702) and withdrawal (rs1229984). Overall, evidence for association with DSM-5 criteria was slightly stronger than for DSM-IV criteria. For rs2066702, results were similar for DSM-IV and DSM-5 criteria. However, the most significant DSM-5 criterion associated with rs1229984 was alcohol-related social/interpersonal problems. Both ADH1B variants were associated with MaxDrinks, a measure of innate tolerance, and MaxDrinks mediated the associations between ADH1B and alcohol outcomes. We replicated the findings for rs2066702 and tolerance in an independent sample of AAs. Taken together, these results suggest that variation in ADH1B affects the adaptation to heavy drinking, highlighting population-specific differences in genetic risk for AUD. They also suggest that the revisions reflected in DSM-5 AUD may enhance the utility of that diagnosis for gene finding.
Keywords: Alcohol dehydrogenase 1B, DSM, tolerance
Introduction
Alcohol dependence (AD) is a complex psychiatric disorder with an estimated heritability of about 50% (Goldman et al, 2005). Understanding the genetic influences on AD could inform diagnosis, treatment, and prevention efforts. However, relatively little is known regarding the actions of specific genetic loci on AD risk. A major focus of the AD candidate gene literature has been on genes that encode alcohol-metabolizing enzymes, such as alcohol dehydrogenase 1B (ADH1B), which catalyzes the oxidation of alcohol to acetaldehyde. Several ADH1B variants alter the enzyme’s kinetics, including the missense single nucleotide polymorphisms (SNPs) rs2066702 (C→T; Arg370Cys) and rs1229984 (G→A; Arg48His). Both of these substitutions result in enhanced ADH1B enzymatic activity that, following alcohol consumption, increases the production of acetaldehyde, producing such aversive effects as facial flushing, tachycardia, and nausea (Crabb et al, 2004; Edenberg, 2007). The rs2066702*T and rs1229984*A alleles are relatively uncommon in European-American (EA) and African-American (AA) populations. The rs1229984*A allele is prevalent in Asian populations, where it was first shown to decrease risk for AD (Chen et al, 1999; Thomasson et al, 1991; Li et al, 2011; Thomasson et al, 1991). Subsequent studies demonstrated that these alleles are also protective from AD in AA and EA populations (Bierut et al, 2012; Li et al, 2011; Luo et al, 2006; Meyers et al, 2013).
We recently published a genome-wide association study (GWAS) of AD in groups of AA and EA subjects (Gelernter et al, 2014). Two approaches were employed: a case-control model that used a dichotomous AD diagnosis from the fourth edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV; American Psychiatric Association, 1994) as the phenotype, and an ordinal trait model where the phenotype was the number of DSM-IV criteria endorsed, adjusted for the number of criteria endorsed for other major substance dependence diagnoses (cocaine, opioid, and nicotine). Rs2066702 and rs1229984 were among the top genome-wide significant findings for both models, and consistent with the literature, we found that the rs2066702*T (in AAs) and rs1229984*A (in EAs) alleles protected against AD. Thus, although collectively, results from candidate gene and genome-wide studies demonstrate that ADH1B variants reduce AD risk, the question remains as to whether these variants influence specific aspects of AD and if so, which ones. The primary goal of the present study was to assess the contributions of specific diagnostic criteria to the observed associations of ADH1B SNPs with AD.
A second phenotype related to alcohol metabolic activity is the maximum number of drinks consumed in a 24-hour period (“MaxDrinks”; Bierut et al, 2012; Kapoor et al, 2013; Saccone et al, 2000). Because as an intermediate phenotype, MaxDrinks may be more closely related to the specific genetic mechanisms and biological pathways underlying AD risk, its use may enhance gene identification (Goldman and Ducci, 2007). Although other phenotypes have been used to examine the genetic influences on drinking behavior, such as the frequency of heavy drinking (Heath et al, 2011), MaxDrinks has been the most widely studied and has the advantage of being relatively easy to measure.
As expected of an intermediate phenotype, MaxDrinks is correlated with AD (Dick and Bierut, 2006; Dick et al, 2011; Grant et al, 2009; Kendler et al, 2010) and also has a heritability of about 50% (Saccone et al, 2000). In a study of adolescents, MaxDrinks was significantly correlated with the score on the Self-Rating of the Effects of Alcohol (SRE) First 5, a measure of initial sensitivity to alcohol, described as reflecting “innate tolerance” (Schuckit et al, 2005). In addition to its association with AD, ADH1B has been associated with MaxDrinks (rs1229984; Bierut et al, 2012; Macgregor et al, 2009; Meyers et al, 2013), as was a null mutation in another well-studied gene encoding a metabolic enzyme, ALDH2, in a Chinese population (Quillen et al, 2014). Thus, investigation of the relationship between ADH1B variants and MaxDrinks could help to elucidate the mechanisms underlying the observed association with AD.
Changes in alcohol-related diagnoses could also influence associations with variation in alcohol metabolizing genes. The DSM-IV (American Psychiatric Association, 1994) differentiated alcohol use disorders (AUDs) into AD (based on the endorsement of three of seven criteria) and alcohol abuse (based on the endorsement of any one of four criteria). In the recent revision of the DSM (DSM-5; American Psychiatric Association, 2013), alcohol abuse and dependence criteria were combined to yield a single diagnosis of alcohol use disorder (AUD). This decision was based on findings from item response theory analyses that consistently demonstrated that 10 of the 11 combined criteria map onto a unidimensional continuum of severity (Borges et al, 2010; Proudfoot et al, 2006; Saha et al, 2006). The utility of the DSM-5 diagnosis for gene identification has not yet been tested.
Here, we used our GWAS sample of EA and AA subjects that was carefully assessed for substance use disorder diagnoses to gain insight into the mechanism by which ADH1B variants influence AD risk. We hypothesized that specific criteria contributed to the observed associations with AD, which we sought to identify. We also investigated the impact of the change from DSM-IV AD to DSM-5 AUD on the strength of association of ADH1B variants. A secondary aim of the study was to explore the relationship between ADH1B variants and MaxDrinks, a proposed intermediate phenotype for AD.
Materials and Methods
Subjects and phenotyping procedures
Subjects were recruited from five sites for studies of the genetics of drug and alcohol dependence: Yale University School of Medicine (APT Foundation; New Haven, CT), the University of Connecticut Health Center (Farmington, CT), McLean Hospital (Harvard Medical School; Belmont, MA), the Medical University of South Carolina (Charleston, SC), and the University of Pennsylvania (Philadelphia, PA). Subjects gave written informed consent as approved by the institutional review board at each site, and certificates of confidentiality were obtained from NIDA and NIAAA. The current study was restricted to AA and EA subjects (with Hispanic subjects assigned genetically to one of those groups). Based on the findings of the published GWAS, analyses of rs2066702 were conducted in the AA part of the sample (P=2.18×10−9 in the GWAS, not significant in EAs (probably because of minimal information)) and analyses of rs1229984 were conducted in the EA part of the sample (P=6.75×10−14 in the GWAS, not significant in AAs (probably because of minimal information)). Although the sample consisted largely of unrelated individuals, there was a subset of subjects from small nuclear families in both populations. Subjects were interviewed with the computer-assisted Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA; Pierucci-Lagha et al, 2005; 2007), a psychiatric interview that assesses physical, psychological, social, and psychiatric manifestations of substance use disorders. This part of the sample constituted most of the “discovery” sample for our published AD GWAS.
The following DSM diagnostic criteria were evaluated with the SSADDA: giving up or reducing important social, occupational, or recreational activities because of alcohol use (“activities given up”); a great deal of time spent in activities necessary to obtain, use, or recover from alcohol use (“much time spent using”); continuing alcohol use despite persistent or recurrent physical or psychological problems (“physical/psychological problems”); persistent desire or unsuccessful efforts to cut down or control alcohol use (“repeated attempts to quit”); a need for markedly increased amounts of alcohol to achieve intoxication or desired effect or markedly diminished effect with continued use of the same amount of alcohol (“tolerance”); using alcohol in larger amounts or over a longer period than was intended (“used larger amounts/longer”); the characteristic withdrawal syndrome for alcohol or alcohol is used to relieve or avoid withdrawal symptoms (“withdrawal”); a strong desire or urge to use alcohol (“craving”); recurrent alcohol use in situations in which it is physically hazardous (“hazardous use”); recurrent alcohol use resulting in a failure to fulfill major role obligations at work, school, or home (“neglected major roles”); and continued alcohol use despite having persistent or recurrent social or interpersonal problems caused or exacerbated by the effects of the alcohol (“social/interpersonal problems”). During the interview, subjects were also asked “In your lifetime, what is the largest number of drinks you have ever had in a 24-hour period?” (“MaxDrinks”).
Genotyping and quality control
Samples were genotyped on the Illumina HumanOmni1-Quad v1.0 microarray at the Center for Inherited Disease Research and the Yale Center for Genome Analysis. Following quality control, 5,697 individuals and 889,659 SNPs were available for imputation. Detailed genotyping and quality control methods are given in Gelernter et al (2014). Both rs2066702 and rs1229984 were well imputed and genotype dosages were rounded to the nearest integer for this analysis (i.e., 0, 1, 2).
We verified subjects’ self-reported ancestry with principal component analysis (PCA) implemented by the SmartPCA component of EIGENSOFT (Patterson et al, 2006; Price et al, 2006). The first principal component separated AA subjects from EA subjects (with Hispanic subjects clustering with one of these groups). We then conducted PCA analyses in each sample separately and used the first three principal components as covariates in the statistical analyses to correct for residual population structure.
Statistical analyses
Chi-squared tests were used to evaluate the differences in DSM diagnosis and criterion prevalence in each population group. Tetrachoric and biserial correlations among DSM criteria and among DSM criteria and MaxDrinks, respectively, were computed with the psych package in R (Revelle, 2014). To account for within-family correlations in responses (MaxDrinks, rs2066702, and rs1229984), all of our analyses used robust sandwich standard errors, implemented through the geepack package (Halekoh et al, 2006) in R (R Development Core Team, 2014), with an exchangeable correlation structure specified. For each model, age, gender, and the first three ancestry PCs were included as covariates. In both the AA and EA samples, the MaxDrinks phenotype was Winsorized to address overestimation of self-reported values using a cutoff of 50 drinks. For both SNPs, due to the small number of minor-allele homozygotes, we combined the heterozygote and homozygote minor-allele groups.
Using a logistic regression model, we tested for association between rs2066702 in AAs and rs1229984 in EAs and the number of DSM-IV AD criteria and DSM-5 AUD criteria. We did not test for association with DSM-IV AD or DSM-5 AUD diagnoses, but used the criterion count regardless of clustering. SNP genotype group was the dichotomous response variable and the 7 (DSM-IV) or 11 (DSM-5) criteria were the explanatory variables. We used the fmsb package in R (Nakazawa, 2012) to calculate the percentage of variance explained by these SNPs (Nagelkerke’s R2). We note that we could not account for family structure in the calculations of Nagelkerke’s R2. We tested for association between rs2066702 and rs12229984 and MaxDrinks using a linear regression model, where MaxDrinks was the response variable and SNP genotype group was the dichotomous explanatory variable.
We also ran sequential logistic regression analyses, with the dichotomous rs2066702 and rs1229984 genotypes as responses, to determine the relative contribution of each DSM criterion (for either the set of 7 DSM-IV dependence criteria or 11 DSM-5 AUD criteria). In Step 1, each criterion was entered into the model separately without adjustment for other criteria. In Step 2, models were adjusted for the criterion that yielded the most significant Wald z-statistic in Step 1. This process was repeated until no other criteria contributed significantly to the model (defined as P<0.05). Model fit was assessed with the Quasi-likelihood under Independence Model Criterion (QIC) goodness of fit statistic (Pan, 2001) calculated with the MESS package in R. To ensure that our results were not confounded by the severity of the disorder, we performed a secondary analysis comparing the fit of the model including all criteria versus the fit of the models where one criterion was dropped out (i.e., 7 models for DSM-IV and 11 models for DSM-5). In all analyses, the major allele group (T/T for rs2066702 and G/G for rs1229984) served as the reference; the major allele for both SNPs was also the risk allele. Thus, in Tables 2 through 5, an odds ratio greater than one for a criterion indicates that a subject endorsing that criterion was more likely to have the risk allele than a subject not endorsing that criterion (all else being equal).
Table 2.
Results from the sequential logistic regression analysis of rs2066702 genotype on DSM-IV AD criteria in African-Americans.
| Step 1 (unadjusted) | Step 2 (adjusted for tolerance) | Step 3 (adjusted for tolerance and much time spent using) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Criterion | OR (95% CI) |
Wald test (z-ratio) |
P | QIC | OR (95% CI) |
Wald test (z-ratio) |
P | QIC | OR (95% CI) |
Wald test (z- ratio) |
P | QIC |
| Tolerance | 1.48 (1.28-1.70) |
28.58 | 9.0×10−8 | 4207.36 | - | - | - | - | - | - | - | - |
| Much time spent using |
1.48 (1.27-1.72) |
26.12 | 3.2×10−7 | 4205.41 | 1.30 (1.10-1.53) |
9.17 | 0.002 | 4198.70 | - | - | - | - |
| Activities given up to use |
1.43 (1.23-1.65) |
23.10 | 1.5×10−6 | 4210.42 | 1.25 (1.07-1.47) |
7.66 | 0.006 | 4201.57 | 1.15 (0.95-1.38) |
2.12 | 0.145 | 4198.91 |
| Withdrawal | 1.47 (1.25-1.73) |
21.75 | 3.1×10−6 | 4211.31 | 1.28 (1.07-1.52) |
7.28 | 0.007 | 4201.90 | 1.17 (0.96-1.42) |
2.57 | 0.109 | 4198.45 |
| Used larger amounts/ longer |
1.38 (1.19-1.60) |
18.23 | 2.0×10−5 | 4213.45 | 1.17 (0.98-1.39) |
3.14 | 0.076 | 4205.12 | 1.08 (0.89-1.30) |
0.68 | 0.409 | 4199.70 |
| Physical/ psychological problems |
1.37 (1.17-1.59) |
16.19 | 5.7×10−5 | 4217.09 | 1.18 (0.99-1.39) |
3.55 | 0.059 | 4205.81 | 1.07 (0.89-1.29) |
0.54 | 0.463 | 4200.37 |
| Repeated attempts to quit |
1.26 (1.10-1.45) |
10.44 | 0.001 | 4221.03 | 1.07 (0.92-1.26) |
0.78 | 0.377 | 4207.81 | 0.99 (0.84-1.17) |
0.01 | 0.909 | 4200.65 |
Abbreviations: AD, alcohol dependence; OR, odds ratio; CI, confidence interval; P, p-value; QIC, quasi-likelihood under the independence model criterion
Table 5.
Results from the sequential logistic regression analysis of rs1229984 genotype on DSM-5 AUD criteria in European-Americans.
| Step 1 (unadjusted) | Step 2 (adjusted for social/interpersonal problems) | Step 3 (adjusted for social/interpersonal problems and withdrawal) | Step 3 (adjusted for social/interpersonal problems, withdrawal, and tolerance) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Criterion | OR (95% CI) |
Wald test (z- ratio) |
P | QIC | OR (95% CI) |
Wald test (z- ratio) |
P | QIC | OR (95% CI) |
Wald test (z- ratio) |
P | QIC | OR (95% CI) |
Wald test (z- ratio) |
P | QIC |
| Social/ interpersonal problems |
2.13 (1.67- 2.71) |
37.66 | 8.4× 10−10 |
1833.83 | - | - | - | - | - | - | - | - | - | - | - | - |
| Withdrawal | 2.35 (1.78- 3.10) |
35.98 | 2.0× 10−9 |
1835.52 | 1.28 (1.07- 1.52) |
14.70 | 1.3 × 10−4 |
1821.52 | - | - | - | - | - | - | - | - |
| Used larger amounts/ longer |
2.07 (1.62- 2.65) |
33.58 | 6.8× 10−9 |
1843.72 | 1.52 (1.12- 2.04) |
7.20 | 0.007 | 1827.77 | 1.35 (0.99- 1.84) |
3.63 | 0.057 | 1819.54 | 1.27 (0.92- 1.75) |
2.04 | 0.153 | 1818.83 |
| Tolerance | 1.96 (1.55- 2.47) |
31.17 | 2.4× 10−8 |
1845.21 | 1.51 (1.15- 1.98) |
8.89 | 0.003 | 1826.84 | 1.70 (1.24- 2.33) |
4.50 | 0.034 | 1818.96 | - | - | - | - |
| Much time spent using |
2.08 (1.60- 2.69) |
30.57 | 3.2× 10−8 |
1844.16 | 1.57 (1.17- 2.11) |
9.00 | 0.003 | 1827.51 | 1.30 (0.94- 1.79) |
2.58 | 0.108 | 1821.12 | 1.22 (0.88- 1.70) |
1.44 | 0.230 | 1819.60 |
| Repeated attempts to quit |
1.95 (1.53- 2.50) |
28.19 | 1.1× 10−7 |
1842.80 | 1.47 (1.10- 1.95) |
6.95 | 0.008 | 1827.10 | 1.25 (0.94- 1.68) |
2.29 | 0.130 | 1820.07 | 1.18 (0.87- 1.60) |
1.12 | 0.291 | 1819.10 |
| Physical/ psychological problems |
1.99 (1.53- 2.57) |
27.13 | 1.9× 10−7 |
1844.96 | 1.51 (1.12- 2.04) |
7.39 | 0.007 | 1827.47 | 1.19 (0.85- 1.68) |
1.05 | 0.306 | 1822.21 | 1.14 (0.81- 1.62) |
0.57 | 0.450 | 1820.30 |
| Craving | 2.14 (1.61- 2.86) |
27.07 | 2.0×10−7 | 1848.69 | 1.28 (1.07- 1.52) |
9.82 | 0.002 | 1826.99 | 1.00 (0.99- 1.01) |
2.75 | 0.097 | 1821.48 | 1.30 (0.90- 1.88) |
2.01 | 0.157 | 1819.67 |
| Neglected major roles |
1.88 (1.47- 2.40) |
25.04 | 5.6× 10−7 |
1849.32 | 1.32 (0.96- 1.80) |
2.89 | 0.089 | 1832.94 | 0.99 (0.98- 1.00) |
0.24 | 0.627 | 1823.31 | 1.02 (0.73- 1.43) |
0.02 | 0.897 | 1820.99 |
| Activities given up to use |
1.78 (1.39- 2.28) |
21.05 | 4.5× 10−6 |
1851.79 | 1.27 (0.93- 1.74) |
2.33 | 0.127 | 1832.39 | 1.04 (0.74- 1.46) |
0.05 | 0.823 | 1823.61 | 0.98 (0.69- 1.39) |
0.02 | 0.902 | 1821.45 |
| Hazardous use |
1.74 (1.36- 2.22) |
19.49 | 1.0× 10−5 |
1856.26 | 1.21 (0.90- 1.61) |
1.59 | 0.208 | 1833.60 | 0.99 (0.98- 1.01) |
0.68 | 0.408 | 1822.38 | 1.06 (0.77- 1.43) |
0.09 | 0.759 | 1820.74 |
Abbreviations: AD, alcohol dependence; OR, odds ratio; CI, confidence interval; P, p-value; QIC, quasi-likelihood under the independence model criterion
Using path analyses implemented in Mplus version 7.3 (Muthén and Muthén, 1998-2012), we tested for indirect effects of ADH1B on alcohol outcomes with MaxDrinks as a mediator variable. Path analysis allows for the parsing of indirect and direct effects of a predictor on an outcome. Individuals with missing values for MaxDrinks were excluded from the analysis (n=101 AAs, 50 EAs). A maximum likelihood estimator was used for continuous outcomes (DSM-IV and DSM-5 criterion counts) and a weighted least squares means and variance adjusted parameter estimator was used for binary outcomes (individual criteria). Age, sex, and the first three ancestry principal components were used as covariates in the analyses. The significance of the indirect effect of ADH1B on alcohol phenotypes via MaxDrinks was estimated by multiplication of the regression coefficients of ADH1B on MaxDrinks and of MaxDrinks on the alcohol phenotype. To address non-normality of indirect effects, bias-corrected bootstrapped 95% confidence intervals were estimated from 1,000 draws.
Replication analyses
We sought to replicate our findings in publicly available data from the Study of Addiction: Genetics and Environment (SAGE). Data were obtained from the database of Genotypes and Phenotypes (dbGaP, accession number phs000092.v1.p1). The sample consists of 1,103 unrelated AA subjects and 2,733 unrelated EA subjects. We used statistical methods largely identical to those described above, with some exceptions. Because not all required criteria for DSM-5 were available, we tested for replication using only DSM-IV AD criteria. Additionally, we did not include ancestry PCs as covariates for this analysis, as they were not available. We did not embed models in GEE because, in contrast to the primary study sample, there were no related subjects in the SAGE sample. In the replication mediation analyses, we excluded subjects with missing data (n=24 AAs, 21 EAs).
Results
Sample characteristics
A summary of sample demographics and characteristics is presented in Table 1. The GWAS sample consisted of 3,318 African-American (AA) and 2,379 European-American (EA) subjects. Due to missing data, 17 AAs and 11 EAs were excluded from the analyses presented here. Both samples were, on average, approximately 40 years old, largely male, never married, with a high school education, and mostly unemployed at the time of the study. The EA sample had a higher prevalence of DSM-IV AD, DSM-IV alcohol abuse, and DSM-5 severe AUD (P<0.001, Table S1). Of the 11 DSM-5 AUD criteria, the most commonly endorsed criterion in both populations was “used larger amounts/longer” (64% in AAs, 70.5% in EAs, Table S1). With the exception of the “repeated attempts to quit” criterion, all criteria showed significantly higher endorsement in EAs (P<0.001, Table S1).
Table 1.
Sample demographics and characteristics.
| AA (n=3,301) | EA(n=2,368) | |
|---|---|---|
| Age (yrs) Mean (SD) |
41.5 (9.1) | 38.1 (10.5) |
| Sex (% male) |
52.0 | 58.8 |
| Marital Status (% married) (% divorced/separated) (% widowed) (% never married) |
13.9 23.1 2.3 60.7 |
13.6 28.8 2.0 55.6 |
| Maximum education level (yrs) Mean (SD) |
12.0 (1.99) |
11.8 (2.32) |
| Employment (% currently employed (% full time)) |
36.5 (59.7) |
32.5 (61.4) |
| Maximum drinks consumed in a 24-hr period Mean (SD) |
18.2 (14.0) |
23.6 (14.0) |
| DSM-IV/DSM-5 criterion count Mean (SD) |
3.1 (2.5)/4.8(4.0) |
3.6 (2.6)/5.8 (3.9) |
| rs2066702 genotype counts n=C/C, n=C/T+T/T |
2162, 1139 | - |
| rs1229984 genotype counts n=G/G, n=A/G+A/A |
- | 2044, 324 |
Abbreviations: AA, African-American; EA, European-American; AD, alcohol dependence; AUD, alcohol use disorder
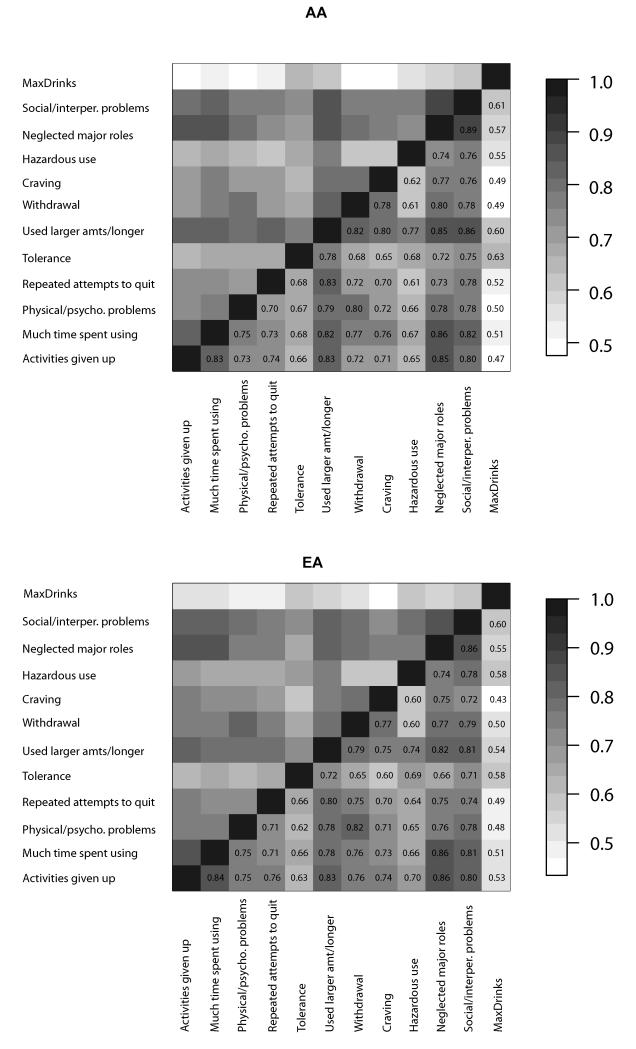
In the phenotype data, we observed substantial collinearity among the 11 DSM-5 criteria (which include all 7 of the DSM-IV dependence criteria), with tetrachoric correlations ranging from 0.61-0.89 in AAs and 0.60-0.86 in EAs (all correlations P<2×10−16, Figure 1). MaxDrinks was moderately correlated with each criterion (biserial, 0.47-0.63 in AAs, 0.43-0.60 in EAs; Figure 1) and with the overall number of criteria endorsed (DSM-IV: r=0.55, P<2×10−16 and DSM-5: r=0.57, P<2×10−16).
Figure 1.
Collinearity among DSM-IV alcohol dependence and DSM-5 alcohol use disorder criteria and MaxDrinks in African-Americans (AA) and European-Americans (EA). Tetrachoric correlations are given for the dichotomous criteria; biserial correlations are reported between MaxDrinks and DSM criteria. High correlations were observed among DSM-5 AUD criteria in both populations; moderate correlations were observed between MaxDrinks and DSM criteria.
Association of ADH1B variants with MaxDrinks and number of DSM criteria
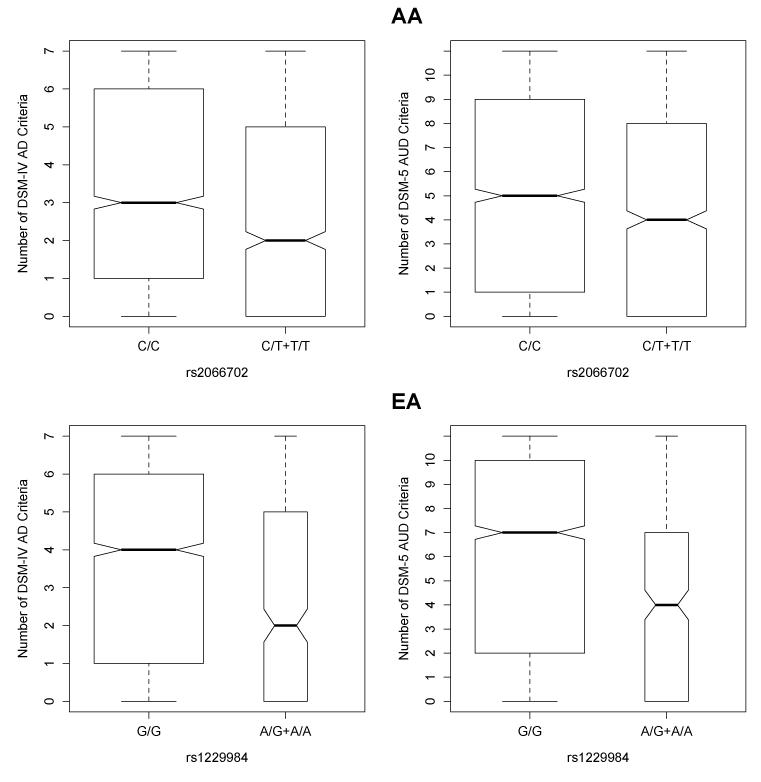
Consistent with our prior findings (Gelernter et al, 2014), we observed statistically significant associations between the number of DSM-IV criteria and rs2066702 in AAs (ß=0.09, SE=0.014, Wald test P=1.9×10−9, R2 = 0.028, Figure 2) and rs1229984 in EAs (ß=0.18, SE=0.024, Wald P= 1.4×10−13, R2 = 0.029, Figure 2). Subjects with the rs2066702*C/C genotype or the rs1229984*G/G genotype (i.e., the major allele homozygote groups for both SNPs) had higher criterion counts than the combined heterozygote and minor allele homozygote group. We also observed significant associations of both ADH1B variants and the number of DSM-5 criteria endorsed (rs2066702 ß=0.06, SE=0.009, Wald test P=1.4×10−9, R2=0.067, Figure 2; rs1229984 ß=0.12, SE=0.009, Wald test P = 5.3×10−14, R2=0.067, Figure 2). Interestingly, we also observed that EAs endorsed more criteria than AAs (DSM-IV P=8.7×10−11; DSM-5 P<2×10−16). Neither of these analyses was reported by Gelernter et al (2014).
Figure 2.
Association of ADH1B single nucleotide polymorphisms (SNPs) with the number of DSM-IV alcohol dependence (AD) and DSM-5 alcohol use disorder (AUD) criteria endorsed in African-Americans (AA) and European-Americans (EA). Top panel: AA individuals homozygous for the major allele of rs2066702 (C/C) showed higher number of DSM-IV (left) and DSM-5 (right) criteria endorsed (P=1.9×10−9 and P=1.4×10−9). Bottom panel: EA individuals homozygous for the major allele of rs12289984 (G/G) showed higher number of DSM-IV (left) and DSM-5 (right) criteria endorsed (P=1.4×10−13 and P = 5.3×10−14).
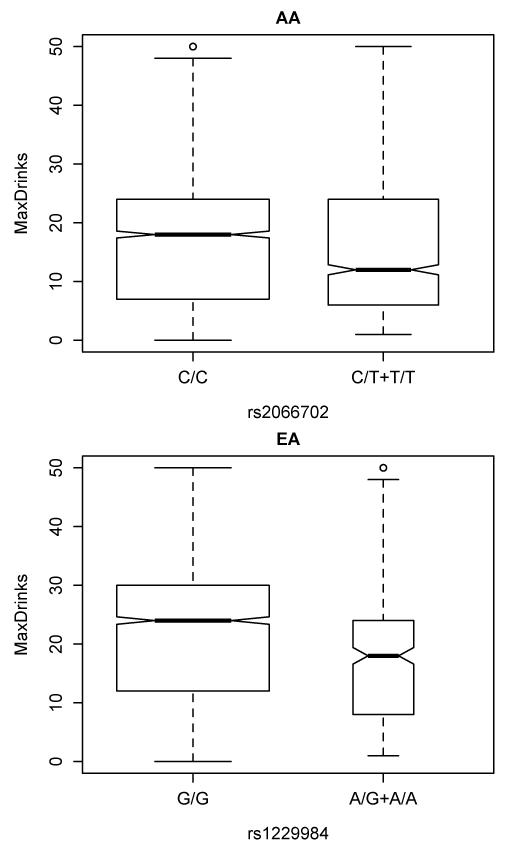
Our hypothesis that these SNPs would be associated with the maximum number of drinks consumed in a single 24-hour period (MaxDrinks) was also supported. Individuals in the major allele homozygote groups (rs2066702 T/T and rs1229984 G/G) for both ADH1B SNPs reported greater MaxDrinks (rs2066702 ß=2.67, SE=0.49, Wald P=6.4×10−8, Figure 3; rs1229984 ß=5.17, SE=0.75, Wald P=5.2×10−12, Figure 3). Similar to what we found for the number of DSM-IV and DSM-5 criteria, EAs had higher MaxDrinks than AAs (P<2.0×10−16).
Figure 3.
Association of ADH1B variants with the maximum number of drinks consumed in a 24-hour period (MaxDrinks) in African-Americans (AA) and European-Americans (EA). Top panel: AA individuals homozygous for the major allele of rs2066702 (C/C) reported greater MaxDrinks (P=6.4×10−8). Bottom panel: EA individuals homozygous for the major allele of rs12289984 (G/G) reported greater MaxDrinks (P=5.2×10−12).
Sequential logistic regression analyses: rs2066702 in AAs
We hypothesized that, despite collinearity among the DSM criteria, a limited number of them were primarily responsible for the overall association between the ADH1B SNPs and the number of DSM-IV or DSM-5 criteria endorsed. To test this hypothesis, we performed sequential logistic regression analyses, evaluating the contributions of each criterion to the model, adjusted for age, sex, and the first three ancestry PCs (base model, QIC=4230.84). In Table 2, we present the results for DSM-IV criteria and rs2066702 in AAs. In Step 1, when the criteria were entered into the base model separately (without adjustment for other criteria), all were significantly associated with rs2066702 genotype, with tolerance being the strongest predictor (Wald z-ratio=28.58, P=9.0×10−8). In Step 2, when we repeated the same analysis after adjusting for tolerance, much time spent using was the strongest predictor (Wald z-ratio=9.17, P=0.002 after adjustment for tolerance). In Step 3, after adjusting for both tolerance and much time spent using, no other criterion significantly predicted genotype. Thus, differences in tolerance and much time spent using largely accounted for the association between rs2066702 and AD. This was underscored by the finding that the model fit was better after adjustment for tolerance and much time spent using than after adjustment for all DSM-IV criteria (QIC=4198.70 vs. 4204.80).
We obtained very similar results for AAs using the DSM-5 criteria (Table 3). Again, tolerance was the strongest predictor in Step 1 (Wald z-ratio=28.58, P=9.0×10−8) and much time spent using was the strongest predictor in Step 2 (Wald z-ratio=9.17, P=0.002), with no significant contributors in Step 3. In addition, the fit for the model adjusted for tolerance and much time spent using was better than the fit for all DSM-5 criteria (QIC=4198.70 vs. 4211.60).
Table 3.
Results from the sequential logistic regression analysis of rs2066702 genotype on DSM-5 AUD criteria in African-Americans.
| Step 1 (unadjusted) | Step 2 (adjusted for tolerance) | Step 3 (adjusted for tolerance and much time spent using) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Criterion | OR (95% CI) |
Wald test (z-ratio) |
P | QIC | OR (95% CI) |
Wald test (z-ratio) |
P | QIC | OR (95% CI) |
Wald test (z-ratio) |
P | QIC |
| Tolerance | 1.47 (1.28-1.70) |
28.58 | 9.0×10−8 | 4207.36 | - | - | - | - | - | - | - | - |
| Much time spent using |
1.48 (1.27-1.72) |
26.12 | 3.2×10−7 | 4205.41 | 1.29 (1.09-1.53) |
9.17 | 0.002 | 4198.70 | - | - | - | - |
| Activities given up to use |
1.43 (1.23-1.65) |
23.10 | 1.5×10−6 | 4210.42 | 1.25 (1.07-1.47) |
7.66 | 0.006 | 4201.57 | 1.15 (0.95-1.38) |
2.12 | 0.145 | 4198.91 |
| Withdrawal | 1.47 (1.25-1.73) |
21.75 | 3.1×10−6 | 4211.31 | 1.28 (1.07-1.52) |
7.28 | 0.007 | 4201.90 | 1.17 (0.96-1.42) |
2.57 | 0.109 | 4198.45 |
| Used larger amounts/ longer |
1.38 (1.19-1.60) |
18.23 | 2.0×10−5 | 4213.45 | 1.17 (0.98-1.39) |
3.14 | 0.076 | 4205.12 | 1.08 (0.90-1.30) |
0.68 | 0.409 | 4199.70 |
| Hazardous use |
1.37 (1.18-1.59) |
17.68 | 2.6×10−5 | 4212.99 | 1.19 (1.01-1.40) |
4.34 | 0.037 | 4203.78 | 1.00 (0.99-1.01) |
1.67 | 0.196 | 4198.54 |
| Neglected major roles |
1.46 (1.25-1.69) |
17.14 | 3.5×10−5 | 4207.50 | 1.27 (1.07-1.51) |
7.77 | 0.005 | 4200.91 | 1.00 (0.99-1.01) |
1.93 | 0.165 | 4198.95 |
| Social/ interpersonal problems |
1.36 (1.17-1.57) |
17.06 | 3.6×10−5 | 4212.69 | 1.16 (0.98-1.37) |
2.90 | 0.088 | 4204.78 | 1.00 (0.99-1.01) |
0.24 | 0.621 | 4200.10 |
| Craving | 1.47 (1.25-1.73) |
16.60 | 4.6×10−5 | 4215.33 | 1.28 (1.07-1.52) |
4.58 | 0.032 | 4204.02 | 1.00 (0.99-1.01) |
1.06 | 0.303 | 4199.60 |
| Physical/ psychological problems |
1.36 (1.17-1.59) |
16.19 | 5.7×10−5 | 4217.09 | 1.18 (0.99-1.39) |
3.55 | 0.059 | 4205.81 | 1.07 (0.89-1.29) |
0.54 | 0.463 | 4200.37 |
| Repeated attempts to quit |
1.26 (1.09-1.45) |
10.89 | 9.7×10−4 | 4221.03 | 1.07 (0.92-1.25) |
0.78 | 0.377 | 4207.81 | 0.99 (0.84-1.17) |
0.01 | 0.909 | 4200.65 |
Abbreviations: AD, alcohol dependence; OR, odds ratio; CI, confidence interval; P, p-value; QIC, quasi-likelihood under the independence model criterion
Consistent with our findings in AAs that tolerance contributed most to the association in the sequential logistic regression analyses, when we included all criteria in the model (either DSM-IV or DSM-5), tolerance was the only criterion that was significant (DSM-IV Wald z-ratio=6.22, P=0.013; DSM-5 Wald z-ratio=5.01, P=0.025; Table S2).
Sequential logistic regression analyses: rs1229984 in EAs
We present the results for DSM-IV criteria and rs1229984 for EAs in Table 4. In Step 1, withdrawal was the strongest predictor of rs1229984 genotype (Wald z-ratio=35.98, P=2.0×10−8). In Step 2, after adjusting for withdrawal, used larger/amounts longer was the next best predictor (Wald z-ratio=10.95, P=9.3×10−4). In Step 3, only tolerance contributed significantly to the model that was adjusted for withdrawal and used larger amounts/longer (Wald z-ratio=4.52, P=0.033). In Step 4, after adjustment for withdrawal, used larger amounts/longer, and tolerance, no other criteria contributed significantly. Confirming that withdrawal, used larger amounts/longer, and tolerance largely explained the association with DSM-IV AD in EAs, the fit for the model that adjusted for these predictors was better than that for the model incorporating all DSM-IV criteria (QIC=1821.38 vs. 1826.00).
Table 4.
Results from the sequential logistic regression analysis of rs1229984 genotype on DSM-IV AD criteria in European-Americans.
| Step 1 (unadjusted) | Step 2 (adjusted for withdrawal) | Step 3 (adjusted for withdrawal and used larger amounts/longer) |
Step 3 (adjusted for withdrawal, used larger amounts/longer, and tolerance) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Criterion | OR (95% CI) |
Wald test (z- ratio) |
P | QIC | OR (95% CI) |
Wald test (z- ratio) |
P | QIC | OR (95% CI) |
Wald test (z- ratio) |
P | QIC | OR (95% CI) |
Wald test (z- ratio) |
P | QIC |
| Withdrawal | 2.35 (1.78- 3.10) |
35.98 | 2.0×10−9 | 1835.52 | - | - | - | - | - | - | - | - | - | - | - | - |
| Used larger amounts/ longer |
2.07 (1.62- 2.65) |
33.58 | 6.8× 10−9 |
1843.72 | 1.58 (1.20- 2.08) |
10.95 | 9.3× 10−4 |
1824.30 | - | - | - | - | - | - | - | - |
| Tolerance | 1.95 (1.54- 2.47) |
31.17 | 2.4× 10−8 |
1845.21 | 1.52 (1.18- 1.95) |
10.75 | 0.001 | 1825.79 | 1.76 (1.28- 2.41) |
4.52 | 0.033 | 1821.38 | - | - | - | - |
| Much time spent using |
2.07 (1.60- 2.69) |
30.57 | 3.2× 10−8 |
1844.16 | 1.50 (1.12- 2.01) |
7.58 | 0.006 | 1829.17 | 1.34 (0.98- 1.82) |
3.39 | 0.065 | 1822.71 | 1.26 (0.91- 1.73) |
1.97 | 0.160 | 1821.24 |
| Repeated attempts to quit |
1.95 (1.53- 2.50) |
28.19 | 1.1× 10−7 |
1842.80 | 1.45 (1.10- 1.91) |
7.19 | 0.007 | 1826.98 | 1.24 (0.92- 1.68) |
2.06 | 0.151 | 1823.00 | 1.17 (0.86- 1.60) |
1.03 | 0.310 | 1821.54 |
| Physical/ psychological problems |
1.99 (1.53- 2.57) |
27.13 | 1.9×10−7 | 1844.96 | 1.37 (1.00- 1.87) |
3.93 | 0.047 | 1831.96 | 1.20 (0.86- 1.68) |
1.19 | 0.275 | 1824.51 | 1.16 (0.82- 1.63) |
0.70 | 0.404 | 1822.34 |
| Activities given up to use |
1.78 (1.39- 2.28) |
21.05 | 4.5× 10−6 |
1851.79 | 1.26 (0.94- 1.69) |
2.41 | 0.121 | 1833.14 | 1.05 (0.76- 1.46) |
0.09 | 0.765 | 1826.09 | 0.99 (0.70- 1.40) |
0.00 | 0.971 | 1823.69 |
Abbreviations: AD, alcohol dependence; OR, odds ratio; CI, confidence interval; P, p-value; QIC, quasi-likelihood under the independence model criterion
Table 5 displays the results for DSM-5 criteria in EAs. In contrast to what we observed for the DSM-IV criteria, the social/interpersonal problems criterion was the strongest predictor of rs1229984 genotype (Wald z-ratio=37.66, P=8.4×10−10). In Step 2, after adjusting for social/interpersonal problems, withdrawal was the next best predictor (Wald z-ratio=14.70, P=1.3×10−4). In Step 3, tolerance was the best predictor, similar to what we observed for the DSM-IV model (Wald z-ratio=4.50, P=0.034). After adjustment for social/interpersonal problems, withdrawal, and tolerance, no other criteria contributed significantly. The fit for these three criteria was better than that for the model incorporating all DSM-5 criteria (QIC=1818.96 vs. 1830.30)
When we included all DSM-IV criteria in the model, withdrawal was the only criterion that remained significant (Wald z-ratio=4.91, P=0.027; Table S3). However, in the full DSM-5 model, no individual criterion was significant.
To ensure that our results were not confounded by the severity of AD/AUD, we performed an alternative analysis in which we compared the fit of the model that included all criteria to the fit of the models in which one criterion was dropped out (i.e., 7 models for DSM-IV and 11 models for DSM-5). For both SNPs and both diagnostic systems, we found that the major criteria identified by adding in criteria to the model were the same that impacted the model fit most when they were dropped from the full model (tolerance, much time spent using for rs2066702; withdrawal, used larger amounts/longer for rs1229984; Table S4).
Mediation by MaxDrinks of the associations between ADH1B SNPs and alcohol phenotypes
Using path analysis, we tested the hypothesis that MaxDrinks mediated the relationship between ADH1B and the DSM criterion count and individual criteria. The results for the mediation analyses are shown in Table S7 (rs2066702 in AAs) and Table S8 (rs1229984 in EAs). In AAs, we found significant indirect effects of rs2066702 via MaxDrinks on the DSM-IV criterion count, the DSM-5 criterion count, and individual criteria (Table S7), suggesting that MaxDrinks partially mediates the associations between rs2066702 and alcohol phenotypes. For some criteria, the direct effect of rs2066702 was no longer significant when MaxDrinks was included in the model (used larger amounts/longer, neglected major roles, physical/psychological problems, and repeated attempts to quit), suggesting that in these cases mediation by MaxDrinks is substantial. Similarly, in EAs the indirect effect of ADH1B via MaxDrinks was significant for all alcohol phenotypes (Table S8). However, in this population, the inclusion of MaxDrinks did not render any direct associations non-significant, suggesting that the mediation by MaxDrinks may be weaker in EAs.
Replication results
Consistent with the findings in our sample, we observed statistically significant associations between both ADH1B SNPs and both the number of DSM-IV criteria endorsed (rs2066702: ß=0.08, SE=0.026, P=0.002; rs1229984: ß=0.006, SE=0.002, P=0.003; Figure S1) and MaxDrinks (rs2066702: ß=2.21, SE=0.89, P=0.013; rs1229984: ß=2.88, SE=0.90, P=0.001; Figure S1).
We present the results for the sequential logistic regression replication analyses in Tables S5 and S6. Consistent with the findings in our sample, we observed that tolerance was able to explain the association between rs2066702 and number of DSM-IV criteria in the SAGE AA replication sample. After adjusting for tolerance in Step 2, no other criteria were significant (Table S5). In contrast, we did not replicate our findings for rs1229984, i.e., withdrawal did not contribute to the association (Table S6).
Results for the mediation analyses in the SAGE sample are found in Tables S9 and S10. Similar to the findings in our sample, there was a significant indirect effect of ADH1B SNPs on the DSM-IV criterion count and individual criteria via MaxDrinks in both populations. For rs2066702 in AAs, the direct effect of ADH1B was no longer significant when MaxDrinks was present in the model for nearly all phenotypes (excluding tolerance). For rs1229984 in EAs, we also observed that the direct effects of ADH1B were no longer significant for nearly all phenotypes (excluding repeated attempts to quit).
Discussion
The results of this study demonstrate that a limited number of diagnostic criteria contributed to the associations observed between ADH1B variants and AD in our recent GWAS (Gelernter et al, 2014). In each model, once these criteria were accounted for, the addition of other criteria had little impact. Specifically, we found that tolerance and much time spent using largely explained the association between rs2066702 and the number of DSM-IV AD criteria endorsed by AAs. In contrast, withdrawal, used larger amounts/longer, and tolerance explained the association between rs1229984 and the number of DSM-IV AD criteria endorsed by EAs. Extension of these results to the DSM-5 criteria for AUD yielded findings that for rs2066702 were very similar to those for DSM-IV AD, but for rs1229984 social/interpersonal problems was the most significant DSM-5 criterion. We also found that both of the ADH1B SNPs (rs2066702 in AAs and rs1229984 in EAs) were associated with the maximum number of drinks consumed in a 24-hour period (also reported in Sartor et al, 2014 and observed by K. Xu, personal communication, June 15, 2014). We found that MaxDrinks was moderately correlated with DSM criterion counts and that it partially mediated the associations between ADH1B SNPs and the DSM-IV criterion count, the DSM-5 criterion count, and individual DSM criteria, consistent with it being an intermediate phenotype of the DSM diagnosis of AD/AUD.
We sought to replicate our findings in the SAGE sample, which is the only publicly available GWAS dataset for AD other than our own. For both SNPs, we replicated the associations that we observed in our sample with the number of AD criteria endorsed and MaxDrinks (Figure S1) and found that MaxDrinks partially mediated the associations between ADH1B SNPs and the DSM-IV criterion count and individual criteria (Table S7 and S8). We also replicated our finding that tolerance explained the association between rs2066702 and DSM-IV AD in AAs (Table S5). However, we did not replicate the finding of an association between withdrawal and rs1229984 in EAs (Table S6). One possible explanation for the lack of association of rs1229984 with alcohol withdrawal was that the SAGE EA sample was much less severely affected than our EA sample. This was evidenced by the difference in mean number of AD criteria endorsed in the risk genotype group (4 criteria in our sample, which is moderately severe vs. 2 criteria in SAGE, which is subthreshold for DSM-IV AD). The replication effort was also limited by the fact that we were unable to test the DSM-5 criteria, as these were not available in the SAGE dataset. Thus, this finding must be validated in other samples.
A recently published article examined the relationship between rs1229984 and individual DSM-IV AUD criteria in an Israeli population-based sample (Kilcoyne et al, 2014). The authors found that tolerance, repeated attempts to quit, used larger amounts longer, physical/psychological problems, hazardous use, and social problems were significantly associated with ADH1B. Although the methods differed somewhat from those used in our study, it is notable that this study did not identify an association between withdrawal and rs1229984. However, similar to the SAGE sample, the Israeli sample was less severely affected than our sample and the overall prevalence of withdrawal was much higher in our sample (40.8% in our sample vs. 14.7% in the Israeli sample), suggesting that the contrasting findings may be a result of differences in AUD severity.
Results from the sequential logistic regression analyses, the analyses of the effect of genotype on MaxDrinks, and the MaxDrinks mediation analyses converge on a role for ADH1B in influencing heavy alcohol consumption. Tolerance and withdrawal were the top criteria contributing to the associations with rs2066702 and rs1229984, respectively (though the finding for withdrawal did not replicate in the SAGE sample). These criteria are thought to be manifestations of adaptation (metabolic and/or neuronal) to sustained alcohol intake, but they appear to have different underlying mechanisms (Littleton, 1998). These criteria showed moderate correlation in both samples (AA: r=0.68, EA: r=0.65, Figure 1). Tolerance was the only criterion that emerged as a significant predictor of genotype in all models tested, and for rs2066702, it was the only criterion significant in the full model for both DSM-IV and DSM-5 criteria (Table S2). The significant contribution of tolerance to the models presented here is also noteworthy in light of the association with MaxDrinks, which is shown in Figure 3. MaxDrinks is correlated with innate tolerance, or the initial lack of sensitivity to alcohol’s stimulant and sedative effects (Chung and Martin, 2009; Schuckit et al, 2005). While one model postulates that increased tolerance to alcohol’s effects predicts future risk for AD (Schuckit, 1980), recent longitudinal studies have shown that greater sensitivity to the subjective effects of alcohol may predict AD risk (King et al, 2014). Irrespective of the direction of the effect, it is clear that tolerance is an important aspect of AUD and based on our findings, further investigation of the effects of ADH1B on tolerance is warranted.
The influence of ADH1B on alcohol consumption and subsequent alcohol outcomes is further underscored by the results of our mediation analysis. We found that for all outcomes (the DSM-IV and DSM-5 criterion counts, individual criteria), MaxDrinks significantly mediated the association with ADH1B polymorphisms, suggesting that ADH1B may exert its effect on risk for AUD through alcohol consumption. In their recent study Kilcoyne et al also examined the role of MaxDrinks in mediating the effects of rs1229984 on alcohol outcomes in their Israeli sample, They found that, for criteria significantly associated with ADH1B, the effect of MaxDrinks explained 23-74% of the associations (Kilcoyne et al, 2014). Although the methods from our study differed from Kilcoyne et al (path analysis vs. multiple regression), both studies support the notion that alcohol consumption is an important mediator of the associations between ADH1B and AUD.
Our analysis of the association of rs1229984 and DSM-5 criteria in EAs showed that the social/interpersonal problems criterion was the most important predictor of genotype, suggesting that ADH1B genotype may predict social consequences of alcohol use. A study in Asian-Americans found that ALDH2 genotype predicted social consequences of drinking, however this was not observed for ADH1B in that sample (Hendershot et al, 2009). The grouping of alcohol dependence and abuse criteria together has been criticized for the addition of three additional psychosocial criteria to the AD diagnosis (Meyer, 2011), which are thought to be less informative for diagnosis than the biological criteria that form the basis for the dependence syndrome. However, the reliability of the social/interpersonal problems is among the highest of all criteria for AUD (test-retest κ=0.77, inter-rater κ=0.60; C. Denis, personal communication, May 2, 2014). Additionally, in a staging analysis, heavy drinking with associated social problems was found to be one of the earliest symptoms to occur in the development of AUDs (Martin et al, 1995). Our results suggest that psychosocial consequences are an important aspect of AUDs that are potentially influenced by variation in ADH1B.
Because both of the variants examined in this study were in ADH1B, the differential association of the variant with diagnostic criteria provides evidence for either population-specific or variant-specific effects. Because each variant is sufficiently informative for analysis in only one population, the specificity of effect attributable to the variants themselves cannot be evaluated. This extends the findings from the original GWAS, in which rs2066702 was not significantly associated with DSM-IV AD in EAs and rs1229984 was not significantly associated with DSM-IV AD in AAs. However, it is important to note that the minor allele frequencies of rs2066702 and rs1229984 were low in EAs and AAs, respectively (1% and 2%; Gelernter et al, 2014). Although tolerance was a recurring theme for both variants, it showed a stronger influence on the association of rs2066702 in AAs than for rs1229984 in EAs. The findings reported here suggest that the structure of the syndromes (AD in DSM-IV and AUD in DSM-5) differs substantially by population. Furthermore, we found that AA subjects had lower DSM-IV and DSM-5 criterion counts and a lower number of MaxDrinks than EAs. These results corroborate findings from the 2001-2002 National Epidemiologic Survey on Alcohol and Related Conditions (NESARC), which reported a lower incidence of alcohol dependence among AAs than EAs (3.29% vs 5.10%; Grant et al, 2004). But unlike the NESARC study, which was representative of the U.S. population, we cannot exclude ascertainment bias as an influence on this observation in our sample.
The associations of ADH1B with DSM-5 criteria tended to be slightly stronger than those with DSM-IV criteria (Figure 2). These results are interesting because they argue against the contention that excess sensitivity and inadequate specificity in DSM-5 could lead to over diagnosis and less reliable, noisier phenotypes (Meyer, 2011). Although in one study (Mewton et al, 2011), there was a 61.7% increase in the prevalence of AUD from DSM-IV to DSM-5, others have found only modest increases (11.3%: Agrawal et al, 2011; 5.1%: Edwards et al, 2013; 0.8%: Peer et al, 2013). In a large sample of twin pairs, Edwards et al. (2013) noted similar heritability estimates and genetic correlations for DSM-IV and DSM-5 diagnoses. Furthermore, a recent study found higher test-retest and inter-rater reliabilities for DSM-5 substance use disorder diagnoses than DSM-IV diagnoses in a sample interviewed using the SSADDA, the diagnostic instrument used in the current study (C. Denis, personal communication, May 2, 2014). Taken together, the findings suggest that the revision of the diagnostic categories and criteria in DSM-5 may increase the utility of the AUD diagnosis for gene finding.
There are limitations to our study that should be considered. First, as demonstrated in the correlation analyses presented in Figure 1, there was substantial collinearity among the 11 DSM-5 diagnostic criteria. Although this is consistent with the results from the IRT analyses showing that the criteria lie along a single continuum (Borges et al, 2010; Proudfoot et al, 2006; Saha et al, 2006), it may have limited our ability to discern which criteria contribute to the observed associations. In addition, MaxDrinks was based entirely on self-report, which may not reflect the true value in all cases. Furthermore, estimates of MaxDrinks on one occasion may be less reliable than estimates of alcohol-related criteria, which have minimum frequency and/or duration thresholds. The operationalization of certain criteria in the SSADDA may have affected our results. Tolerance is operationalized as a dichotomous variable based on a cutoff (“after drinking for some years, needed 50% more alcohol to get an effect”), which may be subject to interpretation. Similarly, while equivalent to the DSM-5 criterion, the operationalization of the craving criterion in the SSADDA (“a strong desire or urge to use alcohol”) as a single item may not be optimal for use in diagnoses and genetic analyses (Agrawal et al., 2014). Nevertheless, the reliabilities of these items in the SSADDA were fair to excellent (tolerance: test-retest κ=0.88, inter-rater κ=0.54; craving: test-retest κ=0.69, inter-rater κ=0.69; C. Denis, personal communication, May 2, 2014). Lastly, the affected sample that we analyzed had a variety of co-occurring substance dependence diagnoses (i.e., those involving cocaine, nicotine, marijuana, and opioids). In our analyses, to avoid over-fitting of the models, we included the individual alcohol criteria as predictors without controlling for the other major substance use disorder criteria that were endorsed by subjects.
Our study had several strengths. We analyzed a large dataset that allowed us to detect significant effects in a GWAS (Gelernter et al, 2014). Furthermore, the phenotype data analyzed here were obtained using a diagnostic instrument with good diagnostic and criterion-level reliability (Pierucci-Lagha et al, 2005; 2007). As a result of our previous GWAS, we had very strong prior associations to consider here, providing a firm foundation for an examination of the specific criteria contributing to the observed associations. Additionally, our study was strengthened by the analysis of specific variants in two different populations, which allowed us to examine population-specific effects. Lastly, the comparison of DSM-IV and DSM-5 results was a novel aspect of the study that addresses the utility of the changes in the diagnostic system for research on AUD.
This study represents an effort to further elucidate the effects of ADH1B on AUD, with the aim of better understanding the specific effects of polymorphisms associated with AUD, and the ultimate goal of generating biomarkers for risk. It should be noted that the observed effects of ADH1B variants are small, which is consistent with findings from the majority of GWAS for complex traits. Further studies will be necessary to evaluate the clinical utility of these findings. Presumably other variants show specific association with other DSM criteria, and it would be of great interest to identify them. Additionally, further research that examines the effects of ADH1B variation on adaptations resulting from heavy alcohol consumption is warranted.
Supplementary Material
Acknowledgments
We appreciate the work in recruitment and assessment provided at Yale University School of Medicine and the APT Foundation by James Poling, PhD; at McLean Hospital by Roger Weiss, M.D., at the Medical University of South Carolina by Kathleen Brady, M.D., Ph.D., and at the University of Pennsylvania by David Oslin, M.D. Genotyping services for a part of our GWAS study were provided by the Center for Inherited Disease Research (CIDR) and Yale University (Center for Genome Analysis). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University (contract number N01-HG-65403). We are grateful to Ann Marie Lacobelle, Catherine Aldi, and Christa Robinson for their excellent technical assistance, to the SSADDA interviewers, led by Yari Nuñez and Michelle Slivinsky, who devoted substantial time and effort to phenotype the study sample, to John Farrell and Alexan Mardigan for database management assistance, and to Richard Feinn, PhD for assistance with the mediation analysis. This study was supported by National Institutes of Health grants RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, R01 AA017535, T32 DA028874, and the VA Connecticut and Philadelphia VA MIRECCs. Although unrelated to the current study, Dr. Kranzler has been a consultant or advisory board member for Alkermes, Lilly, Lundbeck, Pfizer, and Roche. He is also a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which is supported by Abbvie, Ethypharm, Lilly, Lundbeck, and Pfizer.
Footnotes
Author contributions: ABH, KGL, and HRK were responsible for the study concept and design. ABH performed the data analysis. HRK assisted with interpretation of findings. ABH and HRK drafted the manuscript. KGL, LF, and JG provided critical revision of the manuscript for important intellectual content. All authors critically reviewed content and approved the final version for publication.
No other authors declare possible conflicts.
References
- Agrawal A, Heath AC, Lynskey MT. DSM-IV to DSM-5: the impact of proposed revisions on diagnosis of alcohol use disorders. Addiction. 2011;106:1935–1943. doi: 10.1111/j.1360-0443.2011.03517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Wetherill L, Bucholz KK, Kramer J, Kuperman S, Lynskey MT, Nurnberger JI, Jr, Schuckit M, Tischfield JA, Edenberg HJ, Foroud T, Bierut LJ. Genetic influences on craving for alcohol. Addict Behav. 2013;38:1501–1508. doi: 10.1016/j.addbeh.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic and statistical manual of mental disorders: DSM-IV. American Psychiatric Association; Washington, DC: 1994. [Google Scholar]
- American Psychiatric Association . Diagnostic and statistical manual of mental disorders: DSM-5. American Psychiatric Association; Washington, DC: 2013. [Google Scholar]
- Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, Agrawal A, Bucholz KK, Grucza R, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J, Porjesz B, Saccone NL, Schuckit M, Tischfield J, Wang JC, Foroud T, Rice JP, Edenberg HJ. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry. 2012;17:445–450. doi: 10.1038/mp.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges G, Ye Y, Bond J, Cherpitel CJ, Cremonte M, Moskalewicz J, Swiatkiewicz G, Rubio-Stipec M. The dimensionality of alcohol use disorders and alcohol consumption in a cross-national perspective. Addiction. 2010;105:240–254. doi: 10.1111/j.1360-0443.2009.02778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-C, Lu R-B, Chen Y-C, Wang M-F, Chang Y-C, Li T-K, Yin S-J. Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. Am J Hum Genet. 1999;65:795–807. doi: 10.1086/302540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T, Martin CS. Subjective stimulant and sedative effects of alcohol during early drinking experiences predict alcohol involvement in treated adolescents. Journal of Studies on Alcohol and Drugs. 2009;70:660–667. doi: 10.15288/jsad.2009.70.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb DW, Matsumoto M, Chang D, You M. Overview of the role of alcohol dehydrogenase and aldehyde dehydrogenase and their variants in the genesis of alcohol-related pathology. Proc Nutr Soc. 2004;63:49–63. doi: 10.1079/pns2003327. [DOI] [PubMed] [Google Scholar]
- Dick DM, Bierut LJ. The genetics of alcohol dependence. Curr Psychiatry Rep. 2006;8:151–157. doi: 10.1007/s11920-006-0015-1. [DOI] [PubMed] [Google Scholar]
- Dick DM, Meyers JL, Rose RJ, Kaprio J, Kendler KS. Measures of current alcohol consumption and problems: Two independent twin studies suggest a complex genetic architecture. Alcoholism: Clinical and Experimental Research. 2011;35:2152–2161. doi: 10.1111/j.1530-0277.2011.01564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Research & Health. 2007;30:5. [PMC free article] [PubMed] [Google Scholar]
- Edwards AC, Gillespie NA, Aggen SH, Kendler KS. Assessment of a modified DSM-5 diagnosis of alcohol use disorder in a genetically informative population. Alcoholism: Clinical and Experimental Research. 2013;37:443–451. doi: 10.1111/j.1530-0277.2012.01954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D. Genome-wide association study of alcohol dependence: significant findings in African-and European-Americans including novel risk loci. Mol Psychiatry. 2014 doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D, Ducci F. Deconstruction of vulnerability to complex diseases: enhanced effect sizes and power of intermediate phenotypes. The Scientific World JOURNAL. 2007;7:124–130. doi: 10.1100/tsw.2007.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6:521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Grant BF, Stinson FS, Dawson DA, Chou SP, Dufour MC, Compton W, Pickering RP, Kaplan K. Prevalence and co-occurrence of substance use disorders and independent mood and anxiety Disorders: results from the national epidemiologic survey on alcohol and related conditions. Arch Gen Psychiatry. 2004;61:807–816. doi: 10.1001/archpsyc.61.8.807. [DOI] [PubMed] [Google Scholar]
- Grant JD, Agrawal A, Bucholz KK, Madden PAF, Pergadia ML, Nelson EC, Lynskey MT, Todd RD, Todorov AA, Hansell NK, Whitfield JB, Martin NG, Heath AC. Alcohol consumption indices of genetic risk for alcohol dependence. Biological Psychiatry. 2009;66:795–800. doi: 10.1016/j.biopsych.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halekoh U, Højsgaard S, Yan J. The R package geepack for generalized estimating equations. Journal of Statistical Software. 2006;15:1–11. [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PA, Montgomery GW. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biological Psychiatry. 2011;70:513–518. doi: 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot CS, Collins SE, George WH, Wall TL, McCarthy DM, Liang T, Larimer ME. Associations of ALDH2 and ADH1B genotypes with alcohol-related phenotypes in Asian young adults. Alcoholism: Clinical and Experimental Research. 2009;33:839–847. doi: 10.1111/j.1530-0277.2009.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Wang J-C, Wetherill L, Le N, Bertelsen S, Hinrichs AL, Agrawal A, Bucholz K, Dick D, Harari O, Hesselbrock V, Kramer J, Nurnberger JI, Rice J, Saccone N, Schuckit M, Tischfield J, Porjesz B, Edenberg HJ, Bierut L, Foroud T, Goate A. A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum Genet. 2013;132:1141–1151. doi: 10.1007/s00439-013-1318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Myers J, Dick D, Prescott CA. The relationship between genetic influences on alcohol dependence and on patterns of alcohol consumption. Alcoholism: Clinical and Experimental Research. 2010;34:1058–1065. doi: 10.1111/j.1530-0277.2010.01181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilcoyne B, Shmulewitz D, Meyer JL, Aharonovich E, Greenstein E, Frisch A, Weizman A, Spivak B, Edenberg HJ, Gelernter J, Hasin DS. Alcohol consumption mediates the relationship between ADH1B and DSM-IV alcohol use disorder and criteria. Journal of Studies on Alcohol and Drugs. 2014;75:635–642. doi: 10.15288/jsad.2014.75.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AC, McNamara PJ, Hasin DS, Cao D. Alcohol challenge responses predict future alcohol use disorder symptoms: a 6-year prospective study. Biological Psychiatry. 2014;75:798–806. doi: 10.1016/j.biopsych.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao H, Gelernter J. Strong association of the alcohol dehydrogenase 1B gene (ADH1B) with alcohol dependence and alcohol-Induced medical diseases. Biological Psychiatry. 2011;70:504–512. doi: 10.1016/j.biopsych.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littleton J. Neurochemical mechanisms underlying alcohol withdrawal. Alcohol Health Res World. 1998;22:13–24. [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Schork NJ, Gelernter J. Diplotype trend regression analysis of the ADH gene cluster and the ALDH2 gene: multiple significant associations with alcohol dependence. Am J Hum Genet. 2006;78:973–987. doi: 10.1086/504113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PAF, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum Mol Genet. 2009;18:580–593. doi: 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CS, Langenbucher JW, Kaczynski NA, Chung T. Staging in the onset of DSM-IV alcohol symptoms in adolescents: survival/hazard analyses. Journal of Studies on Alcohol and Drugs. 1995;57:549–558. doi: 10.15288/jsa.1996.57.549. [DOI] [PubMed] [Google Scholar]
- Mewton L, Slade T, McBride O, Grove R, Teesson M. An evaluation of the proposed DSM-5 alcohol use disorder criteria using Australian national data. Addiction. 2011;106:941–950. doi: 10.1111/j.1360-0443.2010.03340.x. [DOI] [PubMed] [Google Scholar]
- Meyer RE. A commentary on “addiction and dependence in dsm-v”. Addiction. 2011;106:873–874. doi: 10.1111/j.1360-0443.2010.03238.x. [DOI] [PubMed] [Google Scholar]
- Meyers JL, Shmulewitz D, Aharonovich E, Waxman R, Frisch A, Weizman A, Spivak B, Edenberg HJ, Gelernter J, Hasin DS. Alcohol-metabolizing genes and alcohol phenotypes in an Israeli household sample. Alcoholism: Clinical and Experimental Research. 2013;37:1872–1881. doi: 10.1111/acer.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M. fmsb: Functions for Medical Statistics Book with Some Demographic Data. R Package Version 0.3.4. 2012 http://cran.r-project.org/web/packages/fmsb/index.html
- Pan W. Akaike’s information criterion in generalized estimating equations. Biometrics. 2001;57:120–125. doi: 10.1111/j.0006-341x.2001.00120.x. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS genetics. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer K, Rennert L, Lynch KG, Farrer L, Gelernter J, Kranzler HR. Prevalence of DSM-IV and DSM-5 alcohol, cocaine, opioid, and cannabis use disorders in a largely substance dependent sample. Drug Alcohol Depend. 2013;127:215–219. doi: 10.1016/j.drugalcdep.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Chan G, Arias A, Cubells JF, Farrer L, Kranzler HR. Reliability of DSM-IV diagnostic criteria using the semi-structured assessment for drug dependence and alcoholism (SSADDA) Drug Alcohol Depend. 2007;91:85–90. doi: 10.1016/j.drugalcdep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, Farrer L, Kranzler HR. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Proudfoot H, Baillie AJ, Teesson M. The structure of alcohol dependence in the community. Drug Alcohol Depend. 2006;81:21–26. doi: 10.1016/j.drugalcdep.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Quillen EE, Chen X-D, Almasy L, Yang F, He H, Li X, Wang XY, Liu TQ, Hao W, Deng HW, Kranzler HR, Gelernter J. ALDH2 is associated to alcohol dependence and is the major genetic determinant of “daily maximum drinks” in a GWAS study of an isolated rural chinese sample. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:103–110. doi: 10.1002/ajmg.b.32213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Development Team . A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. [Google Scholar]
- Revelle W. psych: Procedures for Personality and Psychological Research, R Package Version 1.4.5. 2014 http://CRAN.R-project.org/package=psych
- Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, Li TK, Begleiter H, Reich T, Rice JP. A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet. 2000;96:632–637. doi: 10.1002/1096-8628(20001009)96:5<632::aid-ajmg8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Saha TD, Chou SP, Grant BF. Toward an alcohol use disorder continuum using item response theory: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Psychol Med. 2006;36:931–941. doi: 10.1017/S003329170600746X. [DOI] [PubMed] [Google Scholar]
- Sartor C, Wang Z, Xu K, Kranzler H, Gelernter J. The joint effects of ADH1B variants and childhood adversity on alcohol related phenotypes in African-American and European-American women and men. Alcohol Clin Exp Res. 2014;38:2907–2914. doi: 10.1111/acer.12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA. Self-rating of alcohoI intoxication by young men with and without family histories of alcoholism. Journal of Studies on Alcohol and Drugs. 1980;41:242–249. doi: 10.15288/jsa.1980.41.242. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Beltran I, Waylen A, Horwood J, Davis JM. Performance of a self-report measure of the level of response to alcohol in 12- to 13-year-old adolescents. Journal of Studies on Alcohol and Drugs. 2005;66:452–458. doi: 10.15288/jsa.2005.66.452. [DOI] [PubMed] [Google Scholar]
- Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48:677–681. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.