Abstract
A third of African Americans with sporadic focal segmental glomerulosclerosis (FSGS) or HIV-associated nephropathy (HIVAN) do not carry APOL1 renal risk genotypes. This raises the possibility that other APOL1 variants may contribute to kidney disease. To address this question, we sequenced all APOL1 exons in 1, 437 Americans of African and European decent, including 464 patients with biopsy-proven FSGS/HIVAN. Testing for association with 33 common and rare variants with FSGS/HIVAN revealed no association independent of strong recessive G1 and G2 effects. Seeking additional variants that might have been under selection by pathogens and could represent candidates for kidney disease risk, we also sequenced an additional 1, 112 individuals representing 53 global populations. Except for G1 and G2, none of the 7 common codon-altering variants showed evidence of selection or could restore lysis against trypanosomes causing human African trypanosomiasis. Thus, only APOL1 G1 and G2 confer renal risk and other common and rare APOL1 missense variants, including the archaic G3 haplotype, do not contribute to sporadic FSGS and HIVAN in the United States population. Hence, in most potential clinical or screening applications, our study suggests that sequencing APOL1 exons is unlikely to bring additional information compared to genotyping only APOL1 G1 and G2 risk alleles.
Keywords: APOL1, FSGS, HIVAN, chronic kidney disease, association, population genetics, selection, trypanolysis, personalized medicine
Introduction
Chronic (CKD) and end stage kidney disease (ESKD) are more prevalent in individuals of African ancestry than in other racial and ethnic groups.1–5 Much of the excess risk is attributed to common APOL1 coding variants, termed G1 and G2, which are restricted to African origin chromosomes and are located in the last exon of the gene. Carrying two APOL1 risk alleles was strongly associated with focal segmental glomerulosclerosis (FSGS, odds-ratio (OR) 17), HIV-associated nephropathy (HIVAN, OR 29), non-diabetic ESKD (OR 7) and accelerated kidney function decline (hazard-ratio 2–3).6–10 As ∼12% of African Americans carry an APOL1 risk genotype (defined by two copies of renal risk alleles: either G1 or G2 homozygosity, or G1/G2 compound heterozygosity), the public health burden in the African American community is substantial.
The prevailing hypothesis is that G1 and to a lesser degree G2 renal risk alleles rose to high frequencies in West Africa due to recent positive selection by Trypanosoma brucei rhodesiense, the causal agent of acute human African trypanosomiasis (HAT or African sleeping sickness).6,11 Carrying two renal risk alleles markedly increases susceptibility to glomerulopathies, while one allele has little or no effect but confers protection against acute HAT, suggesting a process of balancing selection analogous to selection of sickle cell trait by malaria.6,10,12–14
FSGS is a specific histopathological classification of glomerular injury and is caused by a range of genetic, physiologic and environmental factors. HIVAN is a collapsing form of FSGS, and is almost exclusively diagnosed in persons of African ancestry with untreated HIV infection.15 Although statistical evidence strongly supports a causal role for APOL1 G1 and G2 in kidney disease, ∼30% of African Americans with primary sporadic FSGS or HIVAN do not carry a renal risk genotype,7 raising the possibility that other APOL1 variants may contribute to the development of these pathologies, especially in individuals with no or one renal risk allele.16 In this report, we first sought to determine if APOL1 rare and common coding variants were enriched in biopsy-proven sporadic FSGS and HIVAN cases. We sequenced all the APOL1 exons in 1 437 USA individuals, including 464 African (AA) and European (EA) American cases. We also sequenced the last APOL1 exon encoding for the trypanolytic functional domains17 in 1 112 individuals representing 53 distinct human populations to identify variants that might have been under selection by trypanosomes or other pathogens and could therefore, analogously to G1 and G2, represent candidates for kidney disease susceptibility. Finally, we tested plasma containing novel variant APOL1 isoforms for trypanolytic potential against T.b. rhodesiense and T.b. gambiense.
Results
Association of common and rare APOL1 variants with FSGS/HIVAN
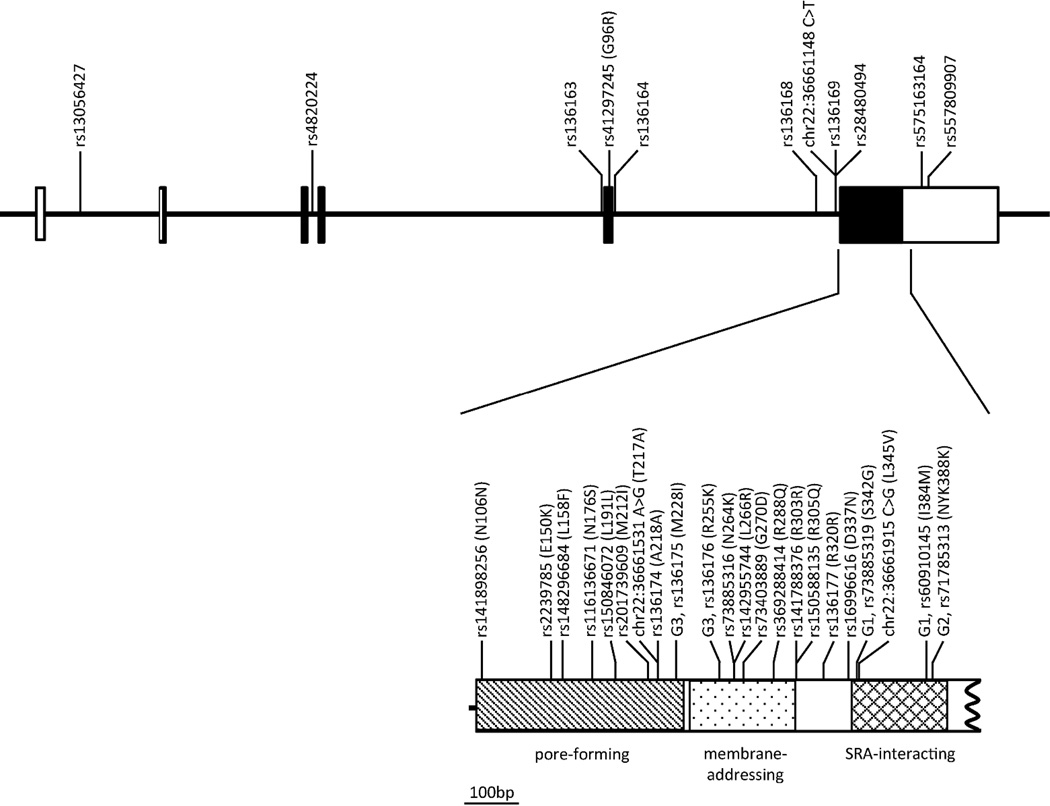
To identify APOL1 variants that might be associated with FSGS and HIVAN, we sequenced all APOL1 exons in 1 437 USA individuals. The study group comprised 241 biopsy-proven sporadic FSGS and 54 biopsy-proven HIVAN AA cases, 169 biopsy-proven sporadic FSGS EA cases, and 651 AA and 322 EA controls. The 33 detected variants comprised 18 missense variants (including the two G1 variants), the G2 in-frame deletion and 3 novel variants (Table 1 and Figure 1). We used three online tools (SIFT, PolyPhen, and MutationAssessor) to predict the functional consequence of the amino acid substitution based on sequence conservation, predicted structure, and annotation of functional domains features (Table 1); four variants are predicted to impact the APOL1 function by at least two algorithms (p.L158F, p.N176S, p.L266R, and p.L345V).
Table 1.
Variant sites identified in African American or European American cases and controls.
| Predicted functional consequence |
African Americans | European Americans | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Position v37 | A1 | A2 | Class | Amino acid change |
S1 | PP2 | MA3 | FSGS/HIVAN (n=295) |
Controls (n=651) |
FSGS (n=169) |
Controls (n=322) |
| rs13056427 | 36,649,802 | T | C | Intron | - | - | - | - | 2.01 | 4.48 | 25.44 | 20.184 |
| rs4820224 | 36,653,275 | A | G | Intron | - | - | - | - | 2.01 | 4.19 | 11.44 | 12.394 |
| rs136163 | 36,657,628 | T | G | Intron | - | - | - | - | 1.94 | 5.18 | 21.86 | 19.574 |
| rs41297245 | 36,657,740 | T | C | NonSyn | G96R | T | B | N | 1.06 | 5.49 | 4.46 | 4.784 |
| rs136164 | 36,657,789 | A | G | Intron | - | - | - | - | 35.28 | 48.47 | 39.22 | 40.00 |
| rs136168 | 36,660,842 | G | A | Intron | - | - | - | - | 63.21 | 48.48 | 23.73 | 21.25 |
| chr22:36661148 C>T | 36,661,148 | T | C | Intron | - | - | - | - | 0.64 | 0.95 | 0.00 | 0.00 |
| rs136169 | 36,661,149 | A | G | Intron | - | - | - | - | 2.58 | 4.83 | 23.51 | 20.03 |
| rs28480494 | 36,661,152 | G | A | Intron | - | - | - | - | 2.58 | 4.74 | 23.51 | 20.03 |
| rs141898256 | 36,661,200 | C | T | Syn | N106N | - | - | - | 0.00 | 0.16 | 0.00 | 0.37 |
| rs2239785 | 36,661,330 | A | G | NonSyn | E150K | T | D | L | 11.30 | 35.71 | 77.27 | 79.15 |
| rs148296684 | 36,661,354 | T | C | NonSyn | L158F | T | D | M | 0.00 | 0.16 | 0.00 | 0.00 |
| rs116136671 | 36,661,409 | G | A | NonSyn | N176S | D | D | M | 1.54 | 5.16 | 0.30 | 0.00 |
| rs1508460725 | 36,661,455 | T | C | Syn | L191L | - | - | - | 0.00 | 0.08 | 0.00 | 0.00 |
| rs2017396096 | 36,661,518 | A | G | NonSyn | M212I | T | B | L | 0.00 | 0.00 | 0.00 | 0.16 |
| chr22:36661531 A>G7 | 36,661,531 | G | A | NonSyn | T217A | T | B | N | 0.18 | 0.00 | 0.00 | 0.00 |
| rs136174 | 36,661,536 | C | A | Syn | A218A | - | - | - | 2.90 | 5.30 | 22.62 | 20.50 |
| rs136175 (G3) | 36,661,566 | G | A | NonSyn | M228I | T | B | N | 2.90 | 5.30 | 22.62 | 20.40 |
| rs136176 (G3) | 36,661,646 | G | A | NonSyn | R255K | T | B | N | 2.47 | 4.72 | 22.16 | 20.66 |
| rs73885316 | 36,661,674 | A | C | NonSyn | N264K | T | PD | L | 0.35 | 2.38 | 0.00 | 0.00 |
| rs142955744 | 36,661,679 | G | T | NonSyn | L266R | D | D | L | 0.17 | 0.15 | 0.00 | 0.00 |
| rs73403889 | 36,661,691 | A | G | NonSyn | G270D | T | B | N | 0.00 | 0.24 | 0.00 | 0.00 |
| rs3692884148 | 36,661,745 | A | G | NonSyn | R288Q | T | B | N | 0.00 | 0.12 | 0.00 | 0.00 |
| rs141788376 | 36,661,791 | T | C | Syn | R303R | - | - | - | 0.27 | 0.12 | 0.00 | 0.00 |
| rs150588135 | 36,661,796 | A | G | NonSyn | R305Q | T | B | N | 0.00 | 0.35 | 0.00 | 0.00 |
| rs136177 | 36,661,842 | G | A | Syn | R320R | - | - | - | 3.18 | 7.16 | 22.92 | 20.56 |
| rs16996616 | 36,661,891 | A | G | NonSyn | D337N | D | B | N | 2.55 | 8.83 | 0.30 | 0.00 |
| rs73885319 (G1) | 36,661,906 | G | A | NonSyn | S342G | T | B | N | 54.10 | 21.81 | 0.00 | 0.00 |
| chr22:36661915 C>G7 | 36,661,915 | G | C | NonSyn | L345V | D | PD | M | 0.00 | 0.12 | 0.00 | 0.00 |
| rs60910145 (G1) | 36,662,034 | G | T | NonSyn | I384M | T | PD | N | 53.06 | 21.27 | 0.00 | 0.00 |
| rs71785313 (G2) | 36,662,046-51 | - | TTATAA | NonSyn | NYK388-389K | - | - | - | 25.42 | 13.13 | 0.00 | 0.00 |
| rs5751631644 | 36,662,425 | A | G | 3’UTR | - | - | - | - | 0.00 | 0.08 | 0.00 | 0.00 |
| rs5578099074 | 36,662,514 | T | C | 3’UTR | - | - | - | - | 0.17 | 0.00 | 0.00 | 0.00 |
The allelic frequencies are reported for the A1 allele in %.
SIFT: (T) Tolerated, (D) Deleterious
PolyPhen: (B) Benign, (PD) Possibly Damaging, (D) Damaging
Mutation Assessor: (N) Neutral, (L) Low, (M) Medium
Due to technical problems, we complemented our EA controls genotypic data with the 1000 Genomes EA data for these four SNPs
Singleton, also reported in the 1000 Genomes Project
Singleton, also reported in the ClinSeq project
Singleton
Singleton, also reported in the Exome Sequencing Project.
Figure 1. Genetic map of the targeted APOL1 regions in the NIH FSGS cohort.
The different functional domains composing the APOL1 protein are coded as follows: hatching for the pore-forming (p.M60-W235), dot for the membrane-addressing (p.A238-P304), and tartan for the SRA-interacting (p.A339-L398) domains. When no rs number is available in dbSNP (build 137), the chromosome position is indicated based on the GRCh37 human genome version. The amino-acid positions refer to isoform a (NP_003652, 398aa). The G1, G2 and G3 variants are labeled. For the sake of clarity, we truncated the last exon and did not represent the full 3’UTR.
Nineteen of the 33 variants had a minor allele frequency (MAF) ≥ 1% in either AA (19) or EA (13) controls allowing for single SNP association analyses. We tested for association with combined sporadic FSGS and HIVAN (FSGS/HIVAN) in AA and sporadic FSGS in EA, adjusting for sex, genetic ancestry, and carriage of APOL1 renal risk genotype (Table 2). In AA, we replicated the strong association of two G1 and/or G2 risk alleles with FSGS/HIVAN (OR=18.31, P=3.3x10−58). After accounting for G1 and G2, a nominally association remained for the intronic rs136163 (OR=1.85, P=2.77x10−2), the coding-changing rs41297245 p.G96R (OR=1.88, P=2.44x10−2), the intronic rs136168 (OR=0.55, P=1.2x10−2) and for the coding-changing rs2239785 p.E150K (OR=0.42, P=3.6x10−2), but none of these associations survived the Bonferroni corrections for multiple testing (P>0.05). The linkage disequilibrium (LD) pattern of the common variants (Figure S1A) shows high linkage between all variants and the G1 (D’ > 0.92) and G2 (D’ > 0.56) polymorphisms. In EA, no common APOL1 variant was significantly associated with FSGS (nominal P>0.05). We also explored additive and dominant genetic models, but no significant associations were identified with sporadic FSGS/HIVAN either in AA or EA (data not shown).
Table 2.
Single SNP recessive associations with sporadic FSGS/HIVAN in African Americans and FSGS in European Americans.
| SNP | African Americans | European Americans | ||||
|---|---|---|---|---|---|---|
| Adj. for sex and ancestry | Adj. for sex, ancestry, and APOL1 G1/G2 |
Adj. for sex and ancestry | ||||
| OR | P | OR | P | OR | P | |
| rs13056427 | 1.74 | 1.52E-02 | 1.67 | 0.07 | 1.83 | 0.53 |
| rs4820224 | 1.36E06 | 0.98 | 1.43E06 | 0.99 | 0.74 | 0.99 |
| rs136163 | 1.96 | 2.88E-03 | 1.85 | 2.77E-02 | 1.25 | 0.78 |
| rs41297245 | 1.96 | 2.98E-03 | 1.88 | 2.44E-02 | 0.00 | 0.99 |
| rs136164 | 2.32 | 1.06E-02 | 1.87 | 0.05 | 2.40 | 0.41 |
| rs136168 | 0.41 | 9.51E-06 | 0.55 | 1.23E-02 | 0.88 | 0.51 |
| rs136169 | 0.54 | 0.59 | 0.63 | 0.72 | 1.77 | 0.20 |
| rs28480494 | 0.73 | 0.79 | 0.76 | 0.83 | 1.77 | 0.20 |
| rs2239785 | 0.14 | 1.20E-06 | 0.42 | 3.56E-02 | 0.95 | 0.80 |
| rs116136671 | 0.00 | 0.98 | 0.00 | 0.98 | . | . |
| rs136174 | 0.69 | 0.75 | 1.89 | 0.59 | 1.67 | 0.24 |
| rs136175 (G3) | 0.69 | 0.75 | 1.89 | 0.59 | 1.67 | 0.25 |
| rs136176 (G3) | 0.67 | 0.73 | 1.73 | 0.64 | 1.66 | 0.25 |
| rs73885316 | 0.00 | 0.97 | 0.00 | 0.98 | . | . |
| rs136177 | 0.00 | 0.97 | 0.00 | 0.98 | 1.68 | 0.24 |
| rs16996616 | 0.00 | 0.97 | 0.00 | 0.98 | . | . |
| rs73885319 (G1) | 9.66 | 9.97E-25 | . | . | . | . |
| rs60910145 (G1) | 9.75 | 9.04E-24 | . | . | . | . |
| rs71785313 (G2) | 5.69 | 3.39E-06 | . | . | . | . |
| 2 APOL1 risk alleles | 18.31 | 3.31E-58 | . | . | . | . |
We further looked for a combined effect of the set of rare and common variants on sporadic FSGS and HIVAN. In AA, a simple burden test showed no increased frequency of rare (MAF<1%) APOL1 variants among FSGS/HIVAN cases compared to controls (P=0.31, Table 3). Seeking a compound heterozygote effect, with a rare variant complementing one APOL1 G1 or G2 risk allele, we looked at the distribution of rare variants among individuals carrying one risk allele: rare variants were non-significantly more frequent among cases (P=0.64). Similarly for EA with sporadic FSGS, there were no associations observed; only two EA controls carried rare variants.
Table 3.
Burden tests for association of rare variants with sporadic FSGS and HIVAN in the US population.
| Ancestry | No. G1/G2 risk alleles |
Subjects carrying rare variants |
Subjects without rare variants |
Odds Ratio (95% CI) |
FET Pvalue |
||
|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | ||||
| AA | 0, 1, or 2 | 6 | 22 | 596 | 1250 | 0.57 [0.19 – 1.46] |
0.31 |
| AA | 0 | 2 | 10 | 62 | 522 | 1.68 [0.18 – 8.16] |
0.38 |
| AA | 1 | 2 | 7 | 106 | 585 | 1.58 [0.16 – 8.43] |
0.64 |
| AA | 2 | 5 | 3 | 425 | 143 | 0.56 [0.11 – 3.66] |
0.42 |
| AA | 0 or 1 | 4 | 21 | 168 | 1103 | 1.25 [0.31 – 3.77] |
0.56 |
| EA | 0 | 0 | 2 | 356 | 646 | 0 [0.0 – 9.69] |
0.54 |
FET, Fisher’s exact test; CI, confidence interval; AA, African American; EA, European American.
Subject numbers are chromosome counts. The analyses were constrained by the number of APOL1 renal risk alleles. The analysis constrained to individuals carrying 1 risk allele was intended to seek for compound heterozygous complementing one G1 or G2 allele.
We then used SKAT to consider more general effects of rare and common variants (Table 4). With default weighting, which emphasizes rare variants, there were no significant associations in AA or EA. Considering that the common assumption that only rare variants can have large effect size is not applicable to APOL1 –as G1/G2 is a clear counterexample– we also ran SKAT with a weighting for predicted functional effect of genetic variants, considering rare and common variants equally; this also revealed no significant association (P=0.25 and P=0.28 in AA and EA respectively).
Table 4.
SKAT tests for association of rare and common variants with sporadic FSGS and HIVAN in the US population.
| Ancestry | No. G1/G2 risk alleles |
Weighting | SKAT Pvalue |
|---|---|---|---|
| AA | 0, 1, or 2 | Default | 0.07 |
| AA | 0, 1, or 2 | Functional prediction | 0.25 |
| AA | 1 | Default | 0.55 |
| AA | 1 | Functional prediction | 0.47 |
| EA | 0 | Default | 0.95 |
| EA | 0 | Functional prediction | 0.28 |
AA, African American; EA, European American.
The analyses were performed in all AA and all EA, as well as restricted to individuals carrying 1 risk allele to seek for compound heterozygous with G1 or G2.
Population genetics of APOL1 variants and haplotypes
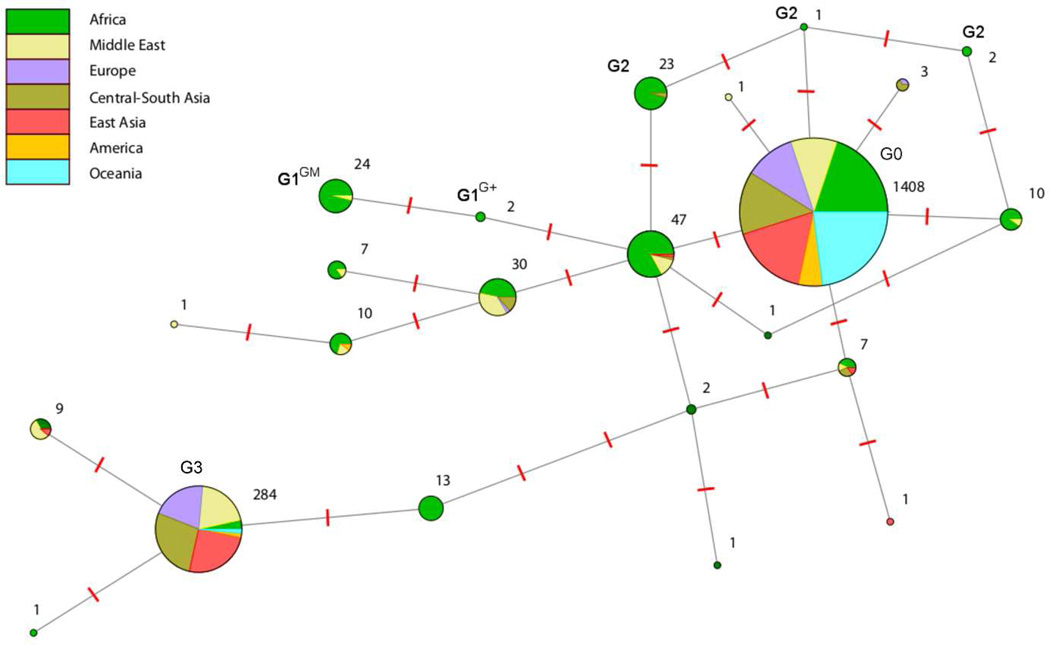
To identify additional APOL1 variants that might associate with renal disease in other populations and which may not be represented in previously reported dataset, we sequenced the polymorphic terminal APOL1 exon in 1 112 individuals from the Human Genome Diversity Project (HGDP)18,19 and International HapMap project,20 representing 53 diverse human populations (Table S1). Because the terminal APOL1 exon encodes the trypanolytic functional domains, evolved rapidly and was under positive selection by pathogens,21 it is thought to be fundamental for APOL1 function. We identified 23 variant sites (Figure S2), including 15 codon-altering variants, and allelic frequencies differed among world populations (Table S2). We then inferred APOL1 haplotypes for all HGDP individuals and defined the G1, G2 and G3 haplotypes as previously reported,7,11,22 and G0 as the most frequent haplotype by exclusion of G1, G2 and G3 (Table S3). We investigated the evolutionary relationship among the APOL1 haplotypes with a minimal network analysis (Figure 2). G0 is carried by 74.6% of HGDP chromosomes and is common in all continental groups. G1 and G2 haplotypes are separated from this basal cosmopolitan haplotype by 1–3 mutations, and represent 2.8% of the HGDP chromosomes. As previously reported,6–8,11,22 G1 and G2 are mostly restricted to Africa, and of the sampled Sub-Saharan populations, only the San Bushmen samples (n=6) carried no G1 and G2 alleles. G3, tagged by the p.M228 and p.R255 variants, is the second most frequent haplotype (15%) and is common throughout Eurasia, but is infrequent in Africa, America, and Oceania (see also Figure S3). This haplotype is separated from G0 by 5 mutations, illustrating the high divergence of this haplotype from the other African haplotypes and suggesting an ancient out-of-Africa separation.
Figure 2. Phylogenetic network for the HGDP APOL1 haplotypes.
Each pie chart represents a haplotype, with the area of the circle a function of the number of haplotypes found in our continental populations (stated next each pie chart). The large separation of G3 from other common haplotypes –indicated by the high number of mutation events (red dashes between two haplotypes)– is consistent with the hypothesis that it was inherited from introgression of archaic populations into modern humans; this is further supported by the fact that it is common in Eurasia but rare in Africa. Correspondingly, G0 plausibly represents the common haplotype in modern humans, common in all continental groups.
Since pathogen-selected variants can also be disease-causing variants (e.g. selection for sickle cell trait by malaria, or for G1 by trypanosomes), we searched for signs of positive selection across the HGDP populations. Fixation index (FST) estimates quantify the population differentiation in allele frequencies among populations; highly differentiated allele frequency may reflect selection on a variant.23 Pairwise FST values suggested population differentiation for G1 (0.12<FST<0.23), and to a lesser extent for G2 (0.10<FST<0.14), when comparing Sub-Saharan Africa with other continental groups (Table S4). No other APOL1 haplotype-tagging variants exhibited population differentiation between continental groups that might suggest a selection event. We next computed pairwise FST between Sub-Saharan populations for the G1 and G2 variants (Table 5). The G2 variant did not show significant frequency differentiation across Sub-Saharan populations. However, the FST estimates for G1 ranked in the significant top 0.16%, 0.25%, and 0.51% of the genome-wide FST distributions when comparing Yorubas with Bantus, Mandenkas, and Biaka Pygmies, respectively, supporting positive selection on the G1 allele in Western Africa.
Table 5.
Pairwise FST estimates between HGDP Sub-Saharan populations.
| G1, rs73885319 | Biaka Pygmy | Mbuti Pygmy | Bantu | San | Yoruba | Mandenka | Bantu South Africa |
|---|---|---|---|---|---|---|---|
| Biaka Pygmy | |||||||
| Mbuti Pygmy | 10–8 | ||||||
| Bantu | 10–8 | 10–8 | |||||
| San | 10–8 | 0 | 10–8 | ||||
| Yoruba | 0.3206 (0.51%) | 0.4015 (1.3%) | 0.3170 (0.16%) | 0.4075 (5.8%) | |||
| Mandenka | 10–8 | 0.0616 | 10–8 | 0.0253 | 0.2036 (0.25%) | ||
| Bantu South Africa | 10–8 | 10–8 | 10–8 | 10–8 | 0.2804 (1.1%) | 10–8 | |
| G2, rs71785313 | Biaka Pygmy | Mbuti Pygmy | Bantu | San | Yoruba | Mandenka | Bantu South Africa |
| Biaka Pygmy | |||||||
| Mbuti Pygmy | 10–8 | ||||||
| Bantu | 10–8 | 10–8 | |||||
| Luhya | 10–8 | 10–8 | 10–8 | ||||
| San | 0.0253 | 10–8 | 10–8 | ||||
| Yoruba | 10–8 | 0.0032 | 10–8 | 0.0325 | |||
| Mandenka | 10–8 | 0.0995 | 0.0829 | 0.1604 | 10–8 | ||
| Bantu South Africa | 10–8 | 0.0692 | 0.0272 | 0.0281 | 10–8 | 10–8 | |
For the top signals, we reported the corresponding top x% of the FST genome-wide distribution in parenthesis.
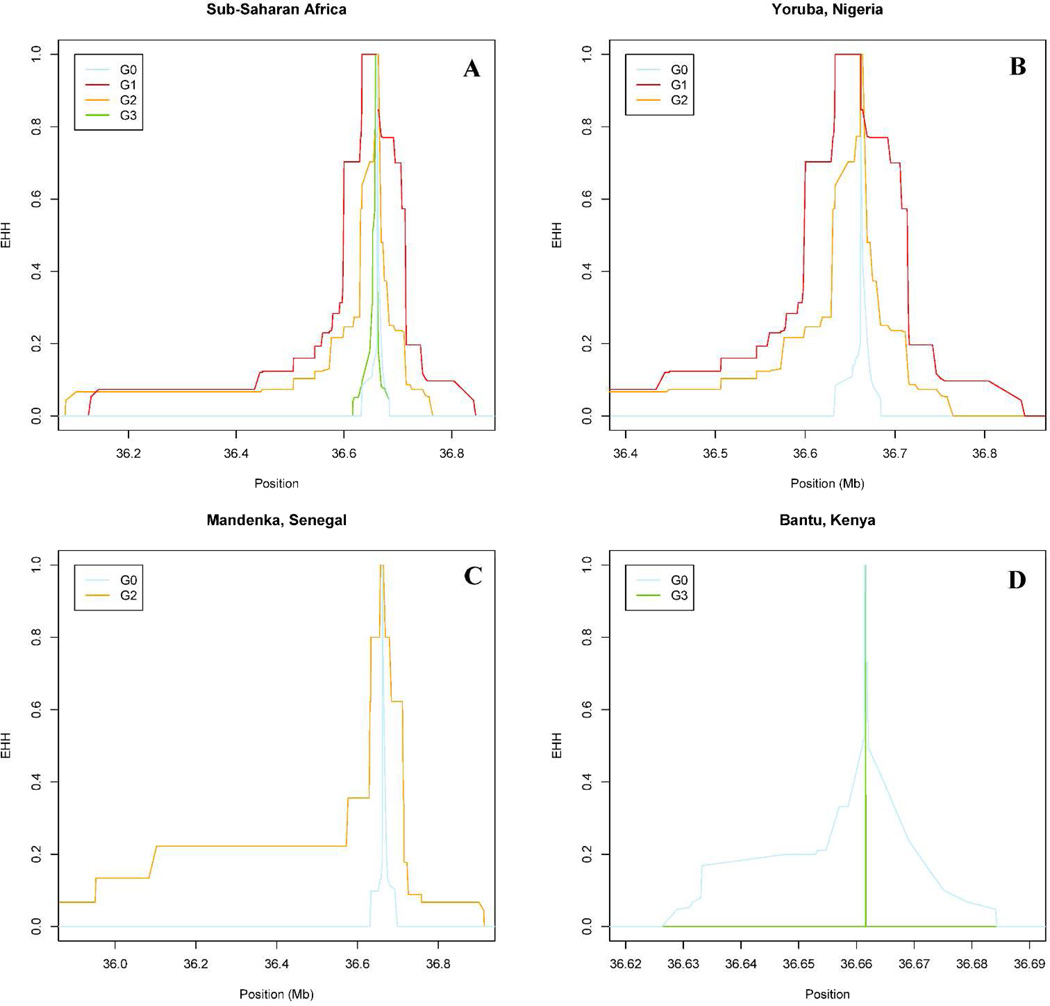
Recent selective sweeps on a variant allele result in longer haplotypes and LD patterns, which can be detected by long haplotype tests using the related iHS (integrated haplotype score) and EHH (extended haplotype homozygosity) metrics.24,25 Significant iHS values were observed for G1 in Sub-Saharan Africa (Table 6, iHS=3.4, top 0.2% of the genome-wide |iHS| distribution), and particularly for Yoruba (top 0.03%), confirming the recent expansion of G1 in West Africa. The extent of LD also tends to be longer for G2 in Sub-Saharan Africa and in Yoruba (Figure 3A and 3B; iHS=1.88, top 5.91%) and a significant sweep was identified in the West African Mandenka population (top 0.83%; see also Figure 3C). A significant iHS was observed for the G3 rs136175 p.M228 tagging variant in Bantus from Kenya (top 0.37%), but the small sample size (n=11), the low MAF (5.6%) and the large frequency confidence interval (95% CI = 0.1–27.3) make this iHS score unreliable, as confirmed by the EHH plot (Figure 3D).
Table 6.
iHS score for HGDP continental (A) and Sub-Saharan (B) populations.
| A) | ||||||||
|---|---|---|---|---|---|---|---|---|
| SNP | North Africa |
Sub-Saharan Africa |
America | East Asia | Central- South Asia |
Middle East | Europe | Oceania |
| rs73885319 (G1) | 3.39 (0.21%) | |||||||
| rs60910145 (G1) | 3.46 (0.18%) | |||||||
| rs71785313 (G2) | 1.88 | |||||||
| rs136175 (G3) | 1.26 | −1.66 | −0.53 | −0.53 | −0.41 | 0.03 | 0.26 | −1.26 |
| rs136176 (G3) | 1.26 | −1.76 | −0.57 | −0.57 | −0.41 | 0.03 | 0.26 | −1.26 |
| B) | |||||||
|---|---|---|---|---|---|---|---|
| SNP | Biaka | Mbuti | Bantu | San | Yoruba | Mandenka | Bantu SA |
| rs73885319 (G1) | 4.34 (0.03%) | n/a | |||||
| rs60910145 (G1) | 4.34 (0.03%) | ||||||
| rs71785313 (G2) | 0.16 | −1.08 | 2.84 (0.83%) | 2.44 | |||
| rs136175 (G3) | 2.19 | −0.28 | −3.37 (0.37%) | n/a | n/a | ||
| rs136176 (G3) | −1.02 | 0.25 | n/a | ||||
iHS score were computed in each population where minor allele frequency of the core variant is >5%. For the top signals, we reported the corresponding top x% of the genome-wide |iHS| distribution (in average, top 1% of the distribution corresponded to |iHS|>2.7). n/a, not available; SA, South Africa.
Figure 3. EHH plots for APOL1 haplotypes in Sub-Saharan Africa (A), Yoruba from Nigeria (B), Mandenka from Senegal (C) and Bantu from Kenya (D).
The genome coordinates refer to the GRCh37 human genome version. APOL1 G1 (red) and to a lesser extent G2 (orange) alleles showed an enlarged LD pattern in Sub-Saharan Africa and in Yoruba populations and G2 also exhibited an extended LD pattern in Mandenkas compared to G0 (light blue), suggesting a recent positive selection in West Africa. On the contrary, APOL1 G3 allele (green) did not show conclusive evidence of long LD pattern in the Bantu population.
Trypanolytic potential of APOL1 isoforms
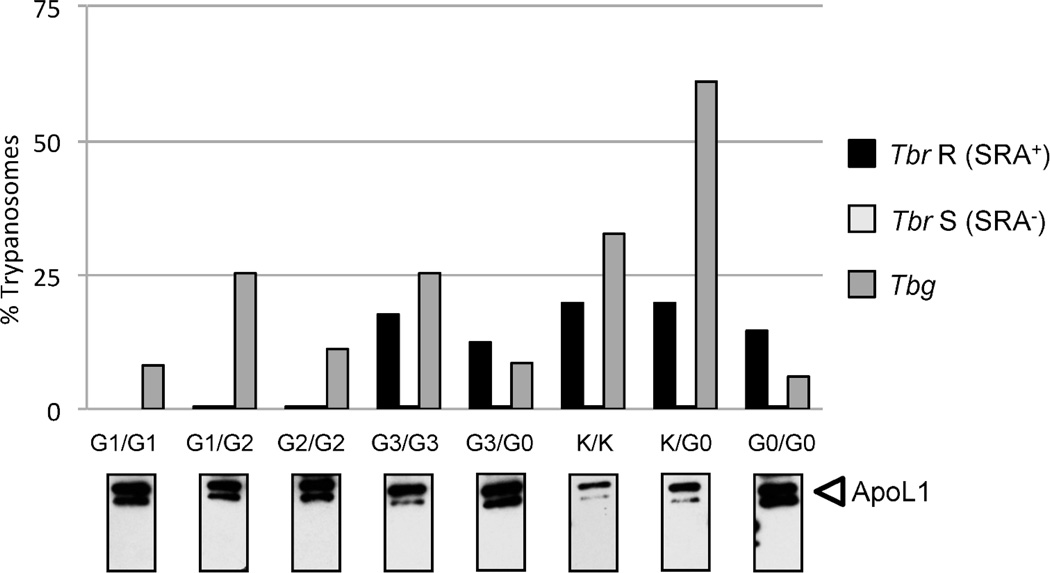
We reasoned that, similarly to APOL1 G1 and G2, trypanolytic variants would be good candidates for renal disease risk variants. We therefore tested plasma from individuals carrying different APOL1 variants for potential to kill T.b rhodesiense or T.b. gambiense, the trypanosomes causing HAT (Figure 4). In order to evaluate the trypanolytic potential of APOL1 isoforms independently of the known trypanocidal activity of G1 and G2 isoforms, we identified individuals carrying only the variants of interest and collected plasma samples for the seven common codon-altering variants that we identified in our extensive sequencing screening. None of the plasma samples killed T.b. gambiense. We used T.b. rhodesiense serum resistance associated (SRA) protein negative (−) sensitive clones as positive controls to confirm that plasma APOL1 could lyse trypanosomes devoid of the SRA protein, which abrogates APOL1-mediated trypanolysis.26 All the plasma samples lysed the T.b. rhodesiense SRA− APOL1-sensitive clone, with the sole exception of p.N337-containing serum although we determine by Western blot that the samples contained a sufficient level of non-degraded APOL1 protein to lyse trypanosomes (data not shown). When testing the T.b. rhodesiense SRA+ resistant clones, only APOL1 G1 and G2 isoforms could lyse the parasite, indicating that G0, G3, p.K150, and p.K264 isoforms could not overcome the SRA-driven APOL1 restriction.
Figure 4. Trypanolytic activity for diverse APOL1 isoforms.
The results are expressed in percent of control growth in fetal calf serum for the following Trypanosoma brucei clones: T.b. rhodesiense SRA+ resistant clone (Tbr R) in black, T.b. rhodesiense SRA sensitive clone (Tbr S) in light gray, and T.b. gambiense (Tbg) in dark grey. Below each individual bar chart, the anti-APOL1 Western blot results confirm the presence of non-degraded APOL1 protein in the plasma sample. We tested the following APOL1 isoforms: G1 in homozygous state (bar chart 1) and heterozygous state with G2 (bar chart 2), G2 (bar chart 3), G3 (bar charts 4 and 5), rs2239785-A; p.150K (K, bar charts 6 and 7), and G0 or WT homozygous (bar chart 8). The only rs73885316-A; p.K264-containing sample was also homozygous for p.150K: the results are therefore similar to bar chart 6. Each bar chart is representative of several experiments. While none of the tested plasma killed T.b. gambiense, all of them killed T.b. rhodesiense devoid of SRA (Tbr S), our positive control. Only plasma from individuals carrying G1 or G2 variants could overcome the SRA-driven inhibition and kill T.b. rhodesiense R.
Discussion
This is the first large study providing sequencing evidence that only APOL1 G1 and G2 variants contribute to APOL1-associated nephropathy. We re-sequenced 1 437 African and European Americans seeking for APOL1 genetic variants that might be enriched in the biopsy-proven sporadic FSGS and HIVAN case groups. We exhaustively analyzed 33 rare and common variants using association analyses, gene-set analyses and compound heterozygous complementation of a single G1 or G2 risk allele analyses, but found no evidence of association with FSGS/HIVAN for other APOL1 variants. In a re-sequencing survey of 2 224 chromosomes from 53 world populations, we identified no APOL1 variants other than G1 or G2 in the trypanolytic domains that showed signature of recent selection: as previously reported in different population samples,6,11 G1 revealed signatures of positive selection by population differentiation (FST) and long haplotype (EHH and iHS) methods; for the first time, we show evidence of selective sweep for the G2 deletion by long haplotype tests in the West African Mandenka population. However, the G2 variant did not exhibit significant population differentiation by the FST estimates, which could suggest a recent selection event that has not yet had time to raise the G2 allele frequency. Finally, none of the tested human plasma containing common codon-altering APOL1 variant other than G1 and G2 restored the ability of APOL1 to lyse T.b. rhodesiense or T.b. gambiense. These results indicate that it is unlikely that additional rare or common APOL1 coding variants contribute to glomerulopathies or that other common APOL1 coding variants have emerged as resistant factors to HAT in Sub-Saharan Africa.
In addition to studying directly the association of APOL1 coding variants with strongly APOL1-related renal outcomes, we intended to characterize additional genetic variants that might have been selected in worldwide populations for protection against pathogens and could therefore, analogously to G1 and G2 with HAT, be new potential candidates for kidney disease pathogenesis. For that, we sequenced worldwide populations, many of which have had historical or recent exposure to trypanosomes, but failed to identify other variants with evidence of selection, including in populations from Central or South America, where Trypanosoma cruzi (the cause of American trypanosomiasis or Chagas disease) is endemic. However, it is important to note that if a pathogen-selected variant can be disease-causing, the presence or absence of selection is not prima faci evidence that a variant is or not disease-causing. Following the G1/G2 analogy, we also tested human plasma samples containing variant APOL1 isoforms for their lytic potential against the two subspecies responsible for HAT. This strategy was engaged to investigate all possible leads that could reveal new potential candidate variant for kidney disease risk, even if there is no evidence to date relating the ability to evade trypanosomes activity with the pathophysiology of renal injury. Finally, if we could not establish a role for common APOL1 coding variants in lysis of T.b. rhodesiense or T.b. gambiense, we cannot exclude that these variants might confer resistance to other pathogen (e.g. Trypanosoma cruzi) as there is mounting evidence for a broad APOL1 role in innate immunity.21,27–30
In parallel to investigating the role of APOL1 coding variants in kidney disease risk, our study also conveys new information on the G3 haplotype. Based on evidence of recent selection by long haplotype test in the Central African Fulani population, Ko et al. had speculated that G3 might afford protection against HAT and contribute to the high prevalence of renal disease reported in African populations.22 First, our analyses strongly suggest that G3 was not selected in a particular African ethnic group, but rather supports the hypothesis of an archaic “out-of-Africa” origin with flow back into Africa from Eurasia:11 (1) our sequencing effort confirmed that G3 is distributed worldwide with high frequency throughout Eurasia; (2) our haplotype network analysis indicates an ancient separation from G0; (3) our selection analysis failed to identify any signs of recent selection. Second, we formally demonstrated that G3 does not confer resistance against trypanosome subspecies causing HAT. Finally, a recent study reported no association for G3 with ESKD in AA.31 Our analysis extends the absence of association for G3 to different kidney pathologies strongly related to APOL1 (i.e. sporadic FSGS and HIVAN) in AA, and for the first time, in EA.
While APOL1 G1 and G2 risk genotypes have been repeatedly associated with non-diabetic CKD and rapid progression to ESKD in African Americans,8 African Americans with low risk APOL1 genotypes remain at greater risk for ESKD than European Americans, suggesting that they may harbor additional, yet unidentified, renal risk variants.9 Collectively, beyond the G1 and G2 risk variants, our extensive re-sequencing effort revealed no additional rare or common APOL1 genetic coding variants that impact the incidence of sporadic FSGS or HIVAN, indicating that if such variants exist, they must be very rare and/or have a weak effect. Future studies will be required to determine whether the variants reported here might influence other renal or cardiac diseases (e.g. Mendelian-inheritance FSGS cases) and/or play a role in a population with a different genetic background (e.g. Asia), and whether regulatory (promoter or intron) variants or non-APOL1 variants (e.g. in other nearby genes) might confer an increased kidney disease risk. Importantly, in a potential translation to personalized medicine, our findings suggests that sequencing APOL1 exons is unlikely to bring additional benefit for kidney transplantation, prevention or clinical management of kidney disease compared to genotyping only APOL1 G1 and G2 risk alleles.
Methods
FSGS/HIVAN study participants
DNA and phenotypic data were available for three case-control groups, as previously described.7,32,33 Cases were enrolled in the NIH FSGS Genetic Study from 22 academic medical centers in the United States and included AA with biopsy-proven sporadic FSGS (n=241) or biopsy-proven HIVAN (n=54), defined histologically as HIV-associated collapsing glomerulopathy, and EA with biopsy-proven sporadic FSGS (n=169). Patients with strong evidence for adaptative FSGS such as a result of reflux nephropathy, reduced renal mass, sickle cell nephropathy, or morbid obesity and FSGS due to medications were excluded. The control groups are composed of AA (n=397) or EA (n=322) controls from the NIH blood bank and the NCI donor pool from Maryland, and of AA hypernormal controls (n=254) from the ALIVE cohort in Baltimore, MD who were HIV-1 infected for 8 or more years with normal kidney function. Institutional review boards at each collaborating medical center approved study protocols and each subject provided written informed consent.
Global population groups
DNA samples representing globally diverse populations were from the Foundation Jean Dausset-CEPH HGDP (Paris, France; n=960 from 52 distinct ethnic groups), and from the International HapMap Project cell lines obtained from the Coriell Repository (Camden, NJ; n=60 unrelated Yorubas, Nigeria and 92 unrelated Luhyas, Kenya). Extensive genotyping data were available from the CEPH-HGDP database (650 000 SNPs)18 and the International HapMap Project (>3 million SNPs in the Phase II).20
Sequencing
For the NIH FSGS cohort, all APOL1 exons and a portion of adjacent introns were sequenced by the Sanger method, while the global population samples were only sequenced for the terminal APOL1 exon (883bp) and a portion of the adjacent intron. Primers and conditions are listed in Table S5. For all samples, APOL1 G1 (rs73885319 and rs60910145), G2 (rs71785313) and G3 (rs136175 and rs136176) SNPs were genotyped by TaqMan (Applied Biosystems, Foster City, CA) to confirm sequencing results. In addition, ancestry informative markers were available for all FSGS/HIVAN cases and controls.7,32
Single SNP association analysis with FSGS/HIVAN
We compared the distribution of common variants (MAF≥1% in controls) among controls and cases for additive, dominant, and recessive models, adjusting for sex, ancestry, and carriage of renal risk genotypes (G1/G1, G2/G2, or G1/G2). Logistic regression was performed using the R glm function.34
Gene-set test for association of rare and common variants with FSGS/HIVAN
We first considered whether rare variants (MAF<1%) were more frequent in cases than controls in a simple burden test. The analyses were performed on all subjects, and also on subsets of subjects determined by the number of APOL1 risk alleles, notably to identify variants complementing one G1 or G2 risk allele as compound heterozygous. For a more general test, allowing for variants to be either deleterious or protective, we used SKAT (R SKAT)35 in two ways: first with default weighting which predominantly considers rare SNPs; secondly weighting for predicted functional consequence of the variants, giving predictions from PolyPhen,36 SIFT,37 and MutationAssessor38 a numerical value for severity, and taking an average, while giving noncoding SNPs a minimal score. Each SKAT analysis was done taking G1/G2 as confounding covariates, as most APOL1 variants have significant LD with these factors in AA.
Evolution and selection analyses of APOL1 haplotypes
APOL1 haplotypes were inferred using the ShapeIT method39 for all the HGDP individuals. The phylogenetic history of the APOL1 haplotypes was examined using a haplotype minimal network determined by the reduced median algorithm implemented in the Network 4.612 program.40 The network was constructed by first connecting haplotypes that differed by single nucleotide changes and next adding increasingly more distant haplotypes. The process was carried on until all haplotypes were included. To detect signature of selection, pairwise FST values were calculated between continental groups and between Sub-Saharan populations as defined in 19 using SmileFinder.41 If selection occurs in a population in a time frame <80 000yrs, the frequency of the selected variant may change, which is reflected by a high FST value.23 For each pairwise comparison, we sampled the HGDP genotypic data across the genome and considered the top 1% (within the 99th percentile) of the FST genome-wide distribution as significant. To further investigate natural selection, we also computed the extended LD or long range haplotype based metrics, EHH and iHS, within each HGDP population using the rehh R package.42 The persistence of a long haplotype with high frequency and a long LD pattern indicates a recent or on-going selective sweep (time frame <10 000yrs). Within each population, we also sampled the HGDP genotypic data of SNPs across the genome with a MAF>5% and considered the top 1% (within the 99th percentile) of the |iHS| genome-wide distribution as significant.
Trypanolytic potential of APOL1 variants
We evaluated the trypanolytic potential of human plasma samples from subjects exhibiting various APOL1 variants on T.b. gambiense LiTat 1.3, T.b. rhodesiense ETat 1.2S and T.b. rhodesiense ETat 1.2R clones as previously described in 43. ETat 1.2S is a SRA− clone sensitive to normal human serum (NHS), and ETat 1.2R is a SRA+ clone resistant to NHS due to the inhibition of APOL1 trypanolytic effect by direct interaction of SRA with APOL1.26 Titration of trypanolytic activity in plasma samples after overnight incubation was expressed as the percentage of survival compared with fetal calf serum control. In parallel, the APOL1 levels in plasma samples were determined by Western Blot with anti-APOL1 antibody (Sigma).
We tested plasma from individuals carrying the G1 and G2 renal risk alleles (3 G1/G1, 3 G1/G2, and 3 G2/G2), the G3 missense variants (4 G3/G3 and 4 G3/WT), the rs2239785-A; p.K150 variant (14 K150/K150 and 5 K150/WT), the rs116136671-G; p.S176 variant (1 S176/WT who carried also p.N337), the rs73885316-A; p.K264 variant (1 K264/WT who was also K150/K150), and the rs16996616-A; p.N337 variant (2 N337/WT). As a control, we also tested a plasma sample from an individual carrying none of these variants (WT/WT or G0/G0).
Supplementary Material
Acknowledgments
Authors would like to thank the reviewers and editorial board for their comments, as we believe they have substantially improved our manuscript. This work has been presented as a poster (FR-PO198) at the American Society of Nephrology annual meeting, Philadelphia PA, 2014. This work has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN26120080001E and by the Intramural Research Programs of Frederick National Lab, Center for Cancer Research, and NIDDK, NIH. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. The work of the Brussels team was financed by WELBIO (Walloon Excellence in Life Science and Biotechnology).
Footnotes
Disclosure. All the authors declared no competing interests.
Supplementary information is available at Kidney International’s website.
REFERENCES
- 1.Coresh J, Selvin E, Stevens LA, et al. Prevalence of chronic kidney disease in the United States. JAMA. 2007;298(17):2038–2047. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- 2.Kopp JB, Winkler C. HIV-associated nephropathy in African Americans. Kidney Int Suppl. 2003;(83):S43–S49. doi: 10.1046/j.1523-1755.63.s83.10.x. [DOI] [PubMed] [Google Scholar]
- 3.System USRD. Annual Data Report: Atlas of of Chronic Kidney Disease and End-Stage Renal Disease in the United States. Bethesda National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; 2011. [Google Scholar]
- 4.Toto RD. Proteinuria and hypertensive nephrosclerosis in African Americans. Kidney Int Suppl. 2004;(92):S102–S104. doi: 10.1111/j.1523-1755.2004.09224.x. [DOI] [PubMed] [Google Scholar]
- 5.Kitiyakara C, Eggers P, Kopp JB. Twenty-one-year trend in ESRD due to focal segmental glomerulosclerosis in the United States. Am J Kidney Dis. 2004;44(5):815–825. [PubMed] [Google Scholar]
- 6.Genovese G, Friedman DJ, Ross MD, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329(5993):841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kopp JB, Nelson GW, Sampath K, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22(11):2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Limou S, Nelson GW, Kopp JB, et al. APOL1 Kidney Risk Alleles: Population Genetics and Disease Associations. Adv Chronic Kidney Dis. 2014;21(5):426–433. doi: 10.1053/j.ackd.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsa A, Kao WH, Xie D, et al. APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med. 2013;369(23):2183–2196. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tzur S, Rosset S, Shemer R, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128(3):345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomson R, Genovese G, Canon C, et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci U S A. 2014;111(20):E2130–E2139. doi: 10.1073/pnas.1400699111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzur S, Rosset S, Skorecki K, et al. APOL1 allelic variants are associated with lower age of dialysis initiation and thereby increased dialysis vintage in African and Hispanic Americans with non-diabetic end-stage kidney disease. Nephrol Dial Transplant. 2012;27(4):1498–1505. doi: 10.1093/ndt/gfr796. [DOI] [PubMed] [Google Scholar]
- 13.Fine DM, Wasser WG, Estrella MM, et al. APOL1 risk variants predict histopathology and progression to ESRD in HIV-related kidney disease. J Am Soc Nephrol. 2012;23(2):343–350. doi: 10.1681/ASN.2011060562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behar DM, Kedem E, Rosset S, et al. Absence of APOL1 risk variants protects against HIV-associated nephropathy in the Ethiopian population. Am J Nephrol. 2011;34(5):452–459. doi: 10.1159/000332378. [DOI] [PubMed] [Google Scholar]
- 15.Rao TK, Filippone EJ, Nicastri AD, et al. Associated focal and segmental glomerulosclerosis in the acquired immunodeficiency syndrome. N Engl J Med. 1984;310(11):669–673. doi: 10.1056/NEJM198403153101101. [DOI] [PubMed] [Google Scholar]
- 16.Atta MG, Estrella MM, Kuperman M, et al. HIV-associated nephropathy patients with and without apolipoprotein L1 gene variants have similar clinical and pathological characteristics. Kidney Int. 2012;82(3):338–343. doi: 10.1038/ki.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Morga D, Vanhollebeke B, Paturiaux-Hanocq F, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. 2005;309(5733):469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- 18.Li JZ, Absher DM, Tang H, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319(5866):1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 19.Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298(5602):2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 20.International HapMap C. Frazer KA, Ballinger DG, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith EE, Malik HS. The apolipoprotein L family of programmed cell death and immunity genes rapidly evolved in primates at discrete sites of host-pathogen interactions. Genome Res. 2009;19(5):850–858. doi: 10.1101/gr.085647.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ko WY, Rajan P, Gomez F, et al. Identifying Darwinian selection acting on different human APOL1 variants among diverse African populations. Am J Hum Genet. 2013;93(1):54–66. doi: 10.1016/j.ajhg.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 24.Sabeti PC, Reich DE, Higgins JM, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419(6909):832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 25.Voight BF, Kudaravalli S, Wen X, et al. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanhamme L, Paturiaux-Hanocq F, Poelvoorde P, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422(6927):83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- 27.Samanovic M, Molina-Portela MP, Chessler AD, et al. Trypanosome lytic factor, an antimicrobial high-density lipoprotein, ameliorates Leishmania infection. PLoS Pathog. 2009;5(1):e1000276. doi: 10.1371/journal.ppat.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sana TR, Janatpour MJ, Sathe M, et al. Microarray analysis of primary endothelial cells challenged with different inflammatory and immune cytokines. Cytokine. 2005;29(6):256–269. doi: 10.1016/j.cyto.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Taylor HE, Khatua AK, Popik W. The innate immune factor apolipoprotein L1 restricts HIV-1 infection. J Virol. 2014;88(1):592–603. doi: 10.1128/JVI.02828-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhaorigetu S, Wan G, Kaini R, et al. ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy. 2008;4(8):1079–1082. doi: 10.4161/auto.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmer ND, Ng MC, Langefeld CD, et al. Lack of Association of the APOL1 G3 Haplotype in African Americans with ESRD. J Am Soc Nephrol. 2014 doi: 10.1681/ASN.2014050444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopp JB, Smith MW, Nelson GW, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40(10):1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orloff MS, Iyengar SK, Winkler CA, et al. Variants in the Wilms’ tumor gene are associated with focal segmental glomerulosclerosis in the African American population. Physiol Genomics. 2005;21(2):212–221. doi: 10.1152/physiolgenomics.00201.2004. [DOI] [PubMed] [Google Scholar]
- 34.Dobson AJ, Barnett A. In: An introduction to generalized linear models. 3rd ed. Chatfield C, Zidek J, editors. London: Chapman & Hall/CRC; 2008. May 12, p. 320. 2008. [Google Scholar]
- 35.Ionita-Laza I, Lee S, Makarov V, et al. Sequence kernel association tests for the combined effect of rare and common variants. Am J Hum Genet. 2013;92(6):841–853. doi: 10.1016/j.ajhg.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reva B, Antipin Y, Sander C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol. 2007;8(11):R232. doi: 10.1186/gb-2007-8-11-r232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10(1):5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 40.Bandelt HJ, Forster P, Sykes BC, et al. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141(2):743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guiblet WM, Zhao K, O’Brien SJ, et al. SmileFinder: a resampling-based approach to evaluate signatures of selection from genome-wide sets of matching allele frequency data in two or more diploid populations. GigaScience. 2015;4(1) doi: 10.1186/2047-217X-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gautier M, Vitalis R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 2012;28(8):1176–1177. doi: 10.1093/bioinformatics/bts115. [DOI] [PubMed] [Google Scholar]
- 43.Lecordier L, Vanhollebeke B, Poelvoorde P, et al. C-terminal mutants of apolipoprotein L-I efficiently kill both Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense. PLoS Pathog. 2009;5(12):e1000685. doi: 10.1371/journal.ppat.1000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.