Abstract
Long-chain n-3 polyunsaturated fatty acids (n-3 PUFAs) are beneficial for human health. However, humans and mammals are unable to synthesize n-3 PUFAs because they lack the n-3 desaturase gene fat-1 and must therefore obtain this type of fatty acid through their diet. Through the production of fat-1 transgenic animals, it is possible to obtain animal products that are rich in n-3 PUFAs, such as meat and milk. The aim of this study was to analyze the gene expression profile and the mechanism of lipid metabolism in fat-1 transgenic cattle and to accumulate important basic data that are required to obtain more efficient fat-1 transgenic cattle. Transcriptome profiling of fat-1 transgenic and wild-type cattle identified differentially expressed genes that are involved in 90 biological pathways, eight pathways of which were related to lipid metabolism processes 36 genes of which were related to lipid metabolism. This analysis also identified 11 significantly enriched genes that were involved in the peroxisome proliferator-activated receptor signaling pathway. These findings were verified by quantitative polymerase chain reaction. The information obtained in this study indicated that the introduction of an exogenous fat-1 gene into cattle affects the gene expression profile and the process of lipid metabolism in these animals. These results may provide important insights into how an exogenous fat-1 gene synthesizes n-3 PUFAs in transgenic cattle and other mammals.
Introduction
N-3 polyunsaturated fatty acids (n-3 PUFAs) have been associated with reducing the risk of major diseases, such as cardiovascular diseases, rheumatoid arthritis, diabetes, cancer and so on [1–4]. However, due to the lack of necessary desaturases, n-3 PUFA biosynthetic pathways do not exist in humans and mammals, so n-3 PUFAs must be obtained from the diet [5,6]. The fat-1 gene encoding the n-3 fatty acid desaturase in Caenorhabditis elegans has been cloned and subsequently transfected into mammalian cells, resulting in an increased cellular n-3 PUFA content [7].
Various fat-1 transgenic animals have been generated to analyze the function of this gene. For example, Kang et al. [8] created the first fat-1 transgenic mouse model, which was capable of producing n-3 PUFA from n-6 PUFA through the constitutive expression of the fat-1 gene in vivo, reducing the ratio of n-6/n-3 fatty acids in different tissues and organs. The fat-1 transgenic mouse model also showed that the n-3 PUFAs exert important protective effects in a variety of processes, such as bone development, inflammatory/immune pathology, cancer chemoprevention, and neurological disease [9–12]. It is generally known that the nutritional value of animal meat and milk could be increased by elevating the concentrations of n-3 PUFAs. Therefore, researchers developed other fat-1 transgenic domestic animals. These experiments also showed that it is feasible to yield higher levels of n-3 PUFAs in transgenic animals for the purpose of improving the fatty acid composition of food products [5,13,14].
Although most of the previous studies focused on the role of the fat-1 gene by using various transgenic animals, little is known about the mechanism by which the fat-1 gene promotes the production of n-3 PUFAs in transgenic animals, especially transgenic cattle. Additionally, there has been little reported on the changes in gene expression patterns in fat-1 transgenic cattle. Here, considering that blood is an important metabolic tissue that reflects the health condition of the body, we utilized cDNA microarrays to analyze the blood gene expression profiles in fat-1 transgenic cattle. The objective of this study was to identify the significantly enriched biologically relevant pathways and to gain further insight into these biological pathways in relation to lipid metabolism in fat-1 transgenic cattle. These results will help us accumulate important basic data that are required to obtain more efficient fat-1 transgenic cattle.
Materials and Methods
Ethics statement
All studies adhered to procedures consistent with the National Research Council Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at Inner Mongolia University.
Preparation of the experimental animals
All fat-1 transgenic cattle were obtained by somatic cell nuclear transfer at the key laboratory of mammalian reproductive biology and biotechnology of the ministry of education, Inner Mongolia University, China. A total of 15 cattle (3 transgenic adult cows, 2 transgenic young bulls and 10 wild-type adult cows) were used in this study. Among these 15 cattle, 3 transgenic adult cows were produced by using the same cell line (Holstein cow fetal fibroblasts) and had a similar age (ZK002, 3.5 years; ZK005, 3.0 years; ZK006, 3.0 years) and 2 transgenic young bulls (11 months old) were produced by using another cell line of Holstein bull fetal fibroblasts; the remaining 10 wild-type Holstein cows were between 3.0 and 4.0 years old. All of the cattle were housed in a concrete-sanded cowshed for one month prior to sample collection, fed the same diet (commercial concentrate feed and wet corn silage), and monitored every day to ensure their health. In this study, we first selected 5 transgenic and 5 wild-type cattle for the analysis of blood lipids as a preliminary evaluation of the function of the fat-1 gene, and then, 3 transgenic and 3 wild type cattle were selected for microarray analysis. Lastly, 5 transgenic and 10 wild-type cattle were selected for reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis to validate the microarray results.
Detection of blood lipids
Blood was collected from the jugular vein, and each blood sample was placed into a tube containing 5% ethylene-diaminetetraacetic acid. Plasma was separated by centrifugation at 3500 g/min for 15 min at 4°C. Then, the concentrations of the plasma lipids, including total cholesterol (TC), triglyceride (TG), high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C), were measured using a fully automatic biochemical analyzer Glamour 3000 (Misiones Bernal, Buenos Aires, Argentina).
RNA extraction and purification
RNA was extracted from whole blood using the Trizol extraction protocol and purified using an RNeasy® Mini Kit (QIAGEN, Germany), following the manufacturer’s protocol. For quality control, total RNA was quantified using the NanoDrop ND-2000 spectrophotometer (Thermo Scientific, USA), and the RNA integrity was assessed using an Agilent Bioanalyzer 2100 (Agilent Technologies).
Microarray hybridization
Gene expression profiling was performed using an Agilent Bovine (V2) Gene Expression microarray (4×44K) containing 43,803 probe sets that interrogate approximately 43,711 bovine transcripts from the RefSeq, Unigene, TIGR, and Btau 4.0 databases. Total RNA from each cattle was hybridized to a separate array, as previously described [15]. Briefly, total RNA was reverse-transcribed using a T7 promoter primer, and double-stranded cDNA was generated. Second-strand cDNA was synthesized, in vitro transcribed into cRNA, and labeled with aaUTP. Then, the cRNA was purified using a RNeasy® Mini Kit, labeled with cyanine-3-CTP, purified, and hybridized to the Gene Expression microarray. After hybridization, the microarray slides were washed once with 2×SSC, 0.1% sodium dodecyl sulfate (SDS) at 42°C for 4 min, once with 0.1×SSC, 0.1% SDS at room temperature for 10 min and three times with 0.1×SSC at room temperature for 1 min. The microarray slides were then washed with distilled water and spin-dried. The arrays were then scanned at 5 μm using the Agilent Scanner G2505C (Agilent Technologies).
Microarray data analysis
Feature Extraction software (version10.7.1.1, Agilent Technologies) was used to analyze the array images to obtain raw data, and GeneSpring software was employed to perform the basic analysis. The raw data were normalized using the quantile algorithm. Probes specifying that at least 75% of the samples in any one out of two conditions possessed flags were employed for the selection of the genes to be used in further analyses. The gene expression data were deposited into the NCBI Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo) and can be accessed via the accession number GSE66651. The differentially expressed genes were then identified through fold changes, P values were calculated using the t-test, and the false discovery rate (FDR) was calculated to correct the P values by using the R package. The threshold for up- and down-regulated genes was set as a fold change ≥1.5 and P values and FDR values <0.05. The GO and KEGG analyses were applied to determine the roles of these differentially expressed mRNAs, and the biological functions with a P value <0.05 were considered significant. Hierarchical clustering was performed to display the gene expression patterns among the samples.
RT-qPCR verification
Reverse transcription-qPCR (RT-qPCR) was performed to validate the differentially expressed genes identified by the microarray analysis. Sixteen lipid metabolism-related genes were selected from the gene sets derived from the GO analysis and the PPAR pathway for the RT-qPCR assays. Total RNA from 5 transgenic and 10 wild-type cattle were homogenized to the same concentration. One microgram of homogenized total RNA was reverse-transcribed using random primers and oligonucleotides (dT)18 (GeneCopoeia Inc., USA) for cDNA synthesis, according to the manufacturer’s instructions. Then, qPCR was performed using a SYBRGreen-PCR Master kit (GeneCopoeia Inc.) in a Bio-Rad CFX Connect™ Real-Time PCR Detection System at a final volume of 20 μl. The cycling conditions were 95°C for 10 min, then 40 cycles of the following: 95°C for 10 s, 60°C for 20 s, and 72°C for 15 s. The reactions were carried out in triplicate, and the relative gene expression was expressed as a fold change and calculated by using 2–(ΔΔCq). In this study, two reference genes (GAPDH and β-actin) were selected for the analysis of qPCR, and the calculation of the 2–(ΔΔCq) values was performed according to the previous report of Nuruddin et al. [16]. The concrete methods were: The ΔCq was calculated from the difference in expression between the 16 target genes and the mean expression of the two reference genes (GAPDH and β-actin). The ΔΔCq was calculated by the difference between the ΔCq value of the fat-1 transgenic cattle and the wild-type cattle samples. The mean values of three 2–(ΔΔCq) were calculated as the final relative gene expression fold change. The details of the 18 genes (16 target genes and 2 reference genes), including the gene symbol, accession number, primer sequences, and product size, are listed in S1 Table.
Results and Discussion
Analysis of blood lipids
The blood lipid levels of the fat-1 transgenic and wild-type cattle are shown in Table 1. The results indicated that TG, TC, HDL-C, and the ratio of TC/HDL-C were significantly decreased (p<0.05) in the fat-1 transgenic cattle, but there was no significant influence on LDL-C and the ratio of TG/HDL-C. Several studies have shown that dyslipidemia confers a risk of coronary artery disease (CAD), while higher intakes of n-3 PUFAs are associated with a reduced risk of CAD [17]. In our study, the findings involved into the change of the plasma TG and TC levels are in line with previous studies [18]. However, we observed that the HDL-C levels were lower in fat-1 transgenic cattle than in wild-type cattle; these results seemed to be contradictory with previous studies [19]. Kinosian et al. [20] reported that the TC/HDL-C ratio was a superior measure of risk for coronary heart disease compared with either TC or LDL-C. Therefore, we further observed the ratio of TC/HDL-C, which was significantly decreased in the fat-1 transgenic cattle. This probably explains that the decrease of HDL-C levels is due to the significantly low TC levels in the fat-1 transgenic cattle. In addition, it was also shown that the TG/HDL-C ratio was a significant predictor of cardiovascular disease [21], but we observed the same ratio of TG/HDL-C in the fat-1 transgenic and wild-type cattle. A possible explanation of this finding was that the fat-1 transgenic and wild-type cattle were healthy. However, we obtained preliminary evidence that the fat-1 gene reduced the blood lipid levels of TG, TC, and the ratio of TC/HDL-C in transgenic cattle. These results suggested that the fat-1 transgenic cattle might be healthier than the wild-type cattle.
Table 1. A comparison of the biochemical indexes of the blood lipids between fat-1 transgenic and wild-type cattle.
| Item | TG(umol/L) | TC(umol/L) | HDL-C(umol/L) | LDL-C(umol/L) | TG/HDL-C | TC/HDL-C |
|---|---|---|---|---|---|---|
| Transgenic cattle | 0.14±0.06* | 2.18±0.20* | 1.93±0.16* | 0.21±0.04 | 0.07±0.03 | 1.13±0.08* |
| Wild type cattle | 0.20±0.07 | 3.60±1.22 | 2.94±0.87 | 0.28±0.07 | 0.07±0.03 | 1.21±0.08 |
Serum content of TG, TC, HDL-C, LDL-C in wide type cattle and transgenic cattle (n = 5).
*, P<0.05, transgenic cattle compared with wild-type cattle.
Global expression profile analysis
In this study, we compared the gene expression patterns of the fat-1 transgenic cattle with wild-type cattle. We used 43,711 transcript sequences as probes, and of these, 2042 were identified as being significantly differentially expressed (p<0.05, FDR<0.05), with a greater than 1.5-fold change in expression between the fat-1 transgenic cattle and wild-type cattle. Of the 2042 transcripts, there were 797 transcripts that were significantly up-regulated in the fat-1 transgenic cattle than in the wild-type cattle (Fig 1A, S2 and S3 Tables). Furthermore, we selected the top 1000 expressed genes for hierarchical clustering analysis and found that the transgenic cattle separated from the wild-type cattle, indicating the consistency of the genetic backgrounds for the three transgenic cattle (Fig 1B).
Fig 1. The hierarchical clustering analysis of fat-1 transgenic cattle and wild-type cattle.
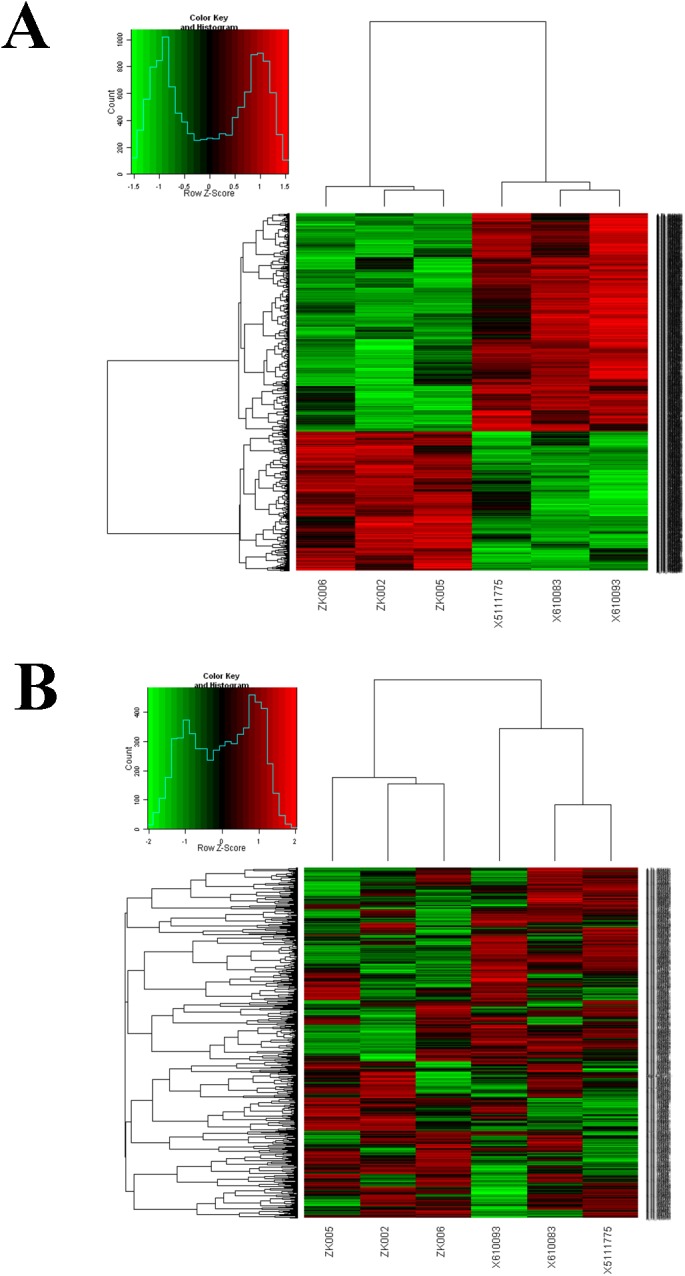
(A) The heat-map of the 2042 differentially expressed genes. (B) The heat-map of the top 1000 expressed genes. The columns and rows in the heat maps represent samples and genes, respectively. Sample names are displayed below the heat maps.
Gene Ontology (GO) functional enrichment analysis of the differentially expressed genes
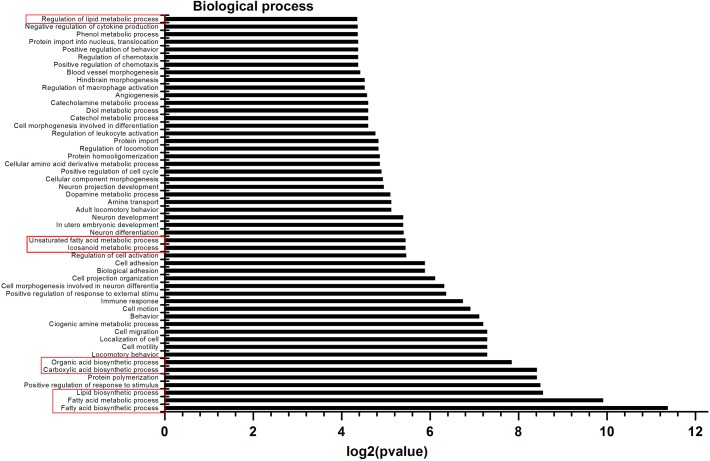
It has been suggested that PUFAs can regulate the expression of genes involved in several metabolic pathways [22]. In the present study, to gain further insight into the metabolic processes that differed between the fat-1 transgenic and wild-type cattle, GO enrichment analysis was performed using 1605 differentially expressed genes from the 2042 significantly differentially expressed transcripts. The three GO categories (biological process, metabolic function, and cell component) were explored using DAVID bioinformatic tools for the overrepresentation of specific GO terms. To extract the most information from our gene expression data, biological functions with a P value <0.05 were considered significant. In total, 1556 differentially expressed genes were annotated in 90 GO functional groups, including 52 groups in biological processes, 15 in cellular components, and 23 in molecular functions (Fig 2, S1 and S2 Figs). In the biological process category, the most important enriched terms were related to lipid metabolism, cell behavior, and immune and nervous systems development (Fig 2). Within the cellular component category, the GO term with the highest level of significance was extracellular, including the extracellular region, extracellular region, extracellular space, and extracellular matrix (S1 Fig). Finally, calcium ion binding, phospholipase D activity, and glycosaminoglycan binding accounted for most of the terms in the molecular function category (S2 Fig). Based on the results of the GO analysis, we focused on eight GO terms for the biological process related to lipid metabolism (Fig 2). Of these, ‘Fatty acid metabolic process’ and ‘Fatty acid biosynthetic process’ are mainly involved in liberating fatty acids from naturally occurring fats and oils by hydrolysis. ‘Lipid biosynthetic process’, ‘Carboxylic acid biosynthetic process’, and ‘Organic acid biosynthetic process’ exert important role in the formation of lipids, carboxylic acids, and organic acids, respectively. ‘Icosanoid metabolic process’ and ‘Unsaturated fatty acid metabolic process’ are the main metabolic processes representing the chemical reactions and pathways involving an unsaturated fatty acid. ‘Regulation of lipid metabolic process’ can modulate the frequency, rate or extent of the chemical reactions and pathways involving lipids. This indicated that these eight lipid metabolic processes may be involved in the function of the fat-1 gene.
Fig 2. Microarray biological process (GO Ontology) classification.
The x-axis indicates the likelihood [−log2(pvalue)] in a category, and the y-axis means the different subcategories of biological process. The GO terms related to lipid metabolism are represented by red boxes.
Genes related to lipid metabolism
To gain further insight into the genes related to lipid metabolism in the fat-1 transgenic cattle, we extracted these genes from the eight GO terms of the lipid metabolism processes obtained by the GO enrichment analysis (S4 Table). We found 36 significantly differentially expressed genes involved in eight lipid metabolism processes; 17 of these genes were up-regulated in the fat-1 transgenic cattle (Table 2) and 19 of these genes were down-regulated (Table 3). Of these 36 differentially expressed genes, eight (CYP51A1, MSMO1, HMGCS1, HMGCR, FDFT1, CYP39A1, CH25H, and APOA1) are involved in cholesterol biosynthetic process. Eight genes (SCD5, LPL, MSMO1, LOC615051, PLP1, AGMO, FASN, and CH25H) are involved in fatty acid biosynthetic process. Four genes (MSMO1, CIDEA, SNCA, and FABP3) are involved in fatty acid metabolic process. Three genes (ACOX1, PEX5, and CPT1B) are involved in fatty acid oxidation. Four genes (CIDEA, AGMO, CH25H, and PTDSS2) are involved in lipid metabolic process. Finally, the remaining genes are involved in the other biological processes related to lipid metabolism. The LOC782922 is involved in the prostaglandin biosynthetic process. The AGPAT41 is involved in the CDP-diacylglycerol biosynthetic process. The STAT5A is involved in lipid storage. This indicated that these significantly differentially expressed genes can influence the same or different biological processes related in lipid metabolism.
Table 2. The significantly up-regulated lipid metabolism-related genes in fat-1 transgenic cattle.
| GeneSymbol | Description | GenbankAccession | p-value | Fold Change |
|---|---|---|---|---|
| CYP51A1 | Bos taurus cytochrome P450,family 51,subfamily A,polypeptide 1 | NM_001025319 | 0.024 | 1.727 |
| *ACOX1 | Bos taurus acyl-CoA oxidase 1,palmitoyl | NM_001035289 | 0.019 | 1.589 |
| *SCD5 | Bos taurus stearoyl-CoA desaturase 5 | NM_001076945 | 0.010 | 1.786 |
| AGPAT4 | Bos taurus cDNA clone IMAGE:8166104 | BC114144 | 0.006 | 2.240 |
| ALOX5AP | Bos taurus arachidonate 5-lipoxygenase-activating protein | NM_001076293 | 0.033 | 1.584 |
| *LPL | Bos taurus lipoprotein lipase | NM_001075120 | 0.028 | 5.407 |
| LOC782922 | Bos taurus prostaglandin F synthetase II-like | NM_001166224 | 0.002 | 5.961 |
| MSMO1 | Bos taurus methylsterol monooxygenase 1 | NM_001098863 | 0.019 | 2.094 |
| HMGCS1 | Bos taurus HMGCS1 protein-like | NM_001206578 | 0.007 | 1.843 |
| PRODH | Bos taurus proline dehydrogenase (oxidase) 1 | NM_001075185 | 0.041 | 1.583 |
| PEX5 | Peroxisomal targeting signal 1 receptor | BT029859 | 0.002 | 8.953 |
| HMGCR | Bos taurus 3-hydroxy-3-methylglutaryl-CoA reductase | NM_001105613 | 0.002 | 1.626 |
| FDFT1 | Bos taurus farnesyl-diphosphate farnesyltransferase 1 | NM_001013004 | 0.001 | 1.692 |
| GGPS1 | Bos taurus geranylgeranyl diphosphate synthase 1 | NM_001079801 | 0.010 | 1.805 |
| CIDEA | Bos taurus cell death-inducing DFFA-like effector | NM_001083449 | 0.0164 | 1.549 |
| IFNG | Bos taurus interferon, gamma | NM_174086 | 0.0234 | 6.972 |
| NR3C1 | Bos taurus nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | NM_001206634 | 0.0352 | 2.106 |
The genes marked with an asterisk are the same as the 11 genes from the PPAR signaling pathways by the KEGG enrichment analysis.
Table 3. The significantly down-regulated lipid metabolism-related genes in fat-1 transgenic cattle.
| GeneSymbol | Description | GenbankAccession | p-value | Fold Change |
|---|---|---|---|---|
| LOC615051 | Uncharacterized protein | XM_002693733 | 0.040 | 1.768 |
| *CPT1B | Bos taurus carnitine palmitoyltransferase 1B (muscle) | NM_001034349 | 0.024 | 1.950 |
| CYP39A1 | Bos taurus cytochrome P450, family 39, subfamily A, polypeptide 1 | NM_001098938 | 0.012 | 1.824 |
| STAT5A | Bos taurus signal transducer and activator of transcription 5A | NM_001012673 | 0.011 | 1.588 |
| MIF | Bos taurus macrophage migration inhibitory factor (glycosylation-inhibiting factor) | NM_001033608 | 0.044 | 1.888 |
| PLP1 | Bos taurus proteolipid protein 1 | NM_174149 | 0.012 | 2.035 |
| ALOX12B | Bos taurus arachidonate 12-lipoxygenase, 12R type | NM_001192038 | 0.005 | 2.651 |
| RNPEP | Bos taurus arginyl aminopeptidase (aminopeptidase B) | NM_001097563 | 0.028 | 1.771 |
| AGMO | Bos taurus alkylglycerol monooxygenase | NM_001192973 | 0.030 | 2.331 |
| PYCR1 | Bos taurus pyrroline-5-carboxylate reductase 1 | NM_001014957 | 0.044 | 1.593 |
| FASN | Bos taurus fatty acid synthase | NM_001012669 | 0.010 | 1.778 |
| EDN1 | Bos taurus endothelin 1 | NM_181010 | 0.036 | 4.768 |
| ISYNA1 | Bos taurus inositol-3-phosphate synthase 1 | NM_001046032 | 0.045 | 1.883 |
| CH25H | Bos taurus cholesterol 25-hydroxylase | NM_001075243 | 0.022 | 2.790 |
| SNCA | Bos taurus synuclein, alpha (non A4 component of amyloid precursor) | NM_001034041 | 0.000 | 2.491 |
| *APOA1 | Bos taurus apolipoprotein A-I | NM_174242 | 0.011 | 2.196 |
| PTDSS2 | PREDICTED: Bos taurus phosphatidylserine synthase 2 | XM_608287 | 0.002 | 4.543 |
| *FABP3 | Bos taurus fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) | NM_174313 | 0.041 | 1.747 |
| PDGFA | Bos taurus platelet-derived growth factor alpha polypeptide | NM_001075231 | 0.036 | 3.215 |
The genes marked with an asterisk are the same as the 11 genes from the PPAR signaling pathways by the KEGG enrichment analysis.
In the present study, we further focused on four key genes (LPL, FASN, SCD5, and ACOX1) related to lipolytic and lipogenic processes. We detected that the expression level of LPL increased by more than five times in fat-1 transgenic cattle relative to wild-type cattle (Table 2). It has been reported that supplementation of n-3 PUFAs in the diet can increase the transcription rate of LPL and accelerate plasma TG clearance [23]. FASN is another key enzyme of lipid metabolism, which plays an important role in de novo lipogenesis in mammals [24]. One study reported that n-3 PUFAs can decrease FASN gene expression and enzyme activity in bovine muscle [25]. Our study shows that there is decreased expression of FASN in fat-1 transgenic cattle compared to wild-type cattle (Table 3). Our study also showed that the other two key genes related to lipid metabolism, SCD5 and ACOX1, were up-regulated in fat-1 transgenic cattle (Table 2), which is consistent with previous reports [26,27]. The regulation of SCD expression by PUFAs has also been observed in brain and immune tissues [28,29]. Furthermore, the over-expression of SCD can change the fatty acid composition, such as increasing the proportion of C18:2 (linoleic acid), C18:3 (linolenic acid), C20:1, C20:3, C20:4 (arachidonic acid), and C20:5 (eicosapentaenoic acid) and reducing the proportion of C14:0 (myristic acid) and total saturated fatty acids in C2C12 cells [26]. Therefore, improving the proportion of PUFAs may require changes in fatty acid metabolism or uptake by the up-regulation of the expression of SCD [26]. ACOX1 is the first enzyme in peroxisomal fatty acid β-oxidation; it is rate-limiting and plays a key role in fatty acid metabolism and fat deposition [30]. In ACOX1 null mice, the expression of ACOX1 was accompanied by increased arachidonic acid (20:4) and DHA (22:6) levels in the serum [31]. Furthermore, older fat-1 mice showed a significant decrease in body weight and epididymal fat mass and an increased expression of ACOX1 [27]. Thus, the function of ACOX1 is potentially related to fat deposition, given its important role in lipid metabolism [31].
Taken together, although we did not analyze and discuss in detail all of the differentially expressed genes related to lipid metabolism, our study indicated that many key genes related to lipid metabolism were influenced in fat-1 transgenic cattle, suggesting that the fat-1 gene could change the expression of lipid metabolism genes.
PPAR pathway affected by the differentially expressed genes
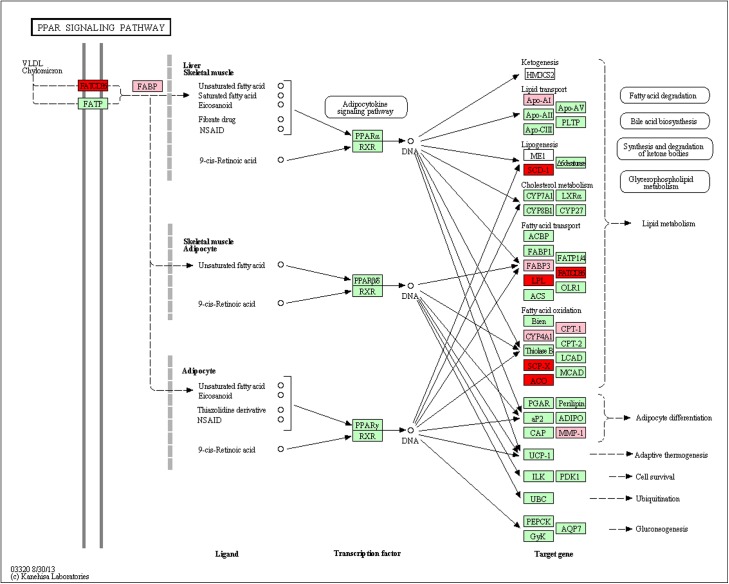
Pathway-based analysis helps to further understand the biological functions of genes. In this study, we performed KEGG analysis and obtained two significantly enriched signal pathways, including the ‘ECM-receptor interaction’ and ‘PPAR signaling pathway’ (p<0.05, FDR<0.2). The ‘PPAR signaling pathway’ is important during adipocyte tissue development and differentiation and the activation of lipogenesis [32]. It is clear that long-chain PUFAs can activate PPARs and subsequently regulate the expression of important genes that are related to lipid metabolism [33]. Therefore, we focused on the ‘PPAR signaling pathway’. We observed that there were 11 significantly differentially expressed genes enriched in the ‘PPAR signaling pathway’ (Fig 3 and S5 Table). When fat-1 transgenic cattle were compared with wild-type cattle, there was a significant (P < 0.05) increase in the expression of six genes related to lipid metabolism (ACOX1, SCP2, FABP2, CD36, SCD5, and LPL) and a significant (P < 0.05) decrease in the expression of five other genes (MMP1, CPT1B, CYP4A22, FABP3, and APOA1) (Fig 3 and S5 Table). At the same time, we found that 6 of the 11 genes (FABP3, APOA1, CPT1B, ACOX1, SCD5, LPL) were same as the 36 genes from the eight GO terms of the lipid metabolism processes obtained by GO enrichment analysis (Table 2, Table 3 and S5 Table).
Fig 3. The significantly enriched genes in the ‘PPAR signaling pathway’.
Red nodes indicate the significantly up-regulated genes in fat-1 transgenic cattle, and the pink nodes indicate the significantly down-regulation genes.
Of the 11 differentially expressed genes enriched in the ‘PPAR signaling pathway’, four (FABP2, FABP3, CD36 and LPL) are involved in fatty acid transport. Four genes (CPT1B, CYP4A22, SCP2 and ACOX1) are involved in fatty acid oxidation. Finally, of the remaining three genes, APOA1, SCD5 and MMP1 are involved in lipid transport, lipogenesis, and adipocyte differentiation, respectively. Our findings showed that fat-1 influences the ‘PPAR signaling pathway’, and the 11 differentially expressed genes point to its important role in the regulation of an extensive network of genes that are involved in lipid metabolism.
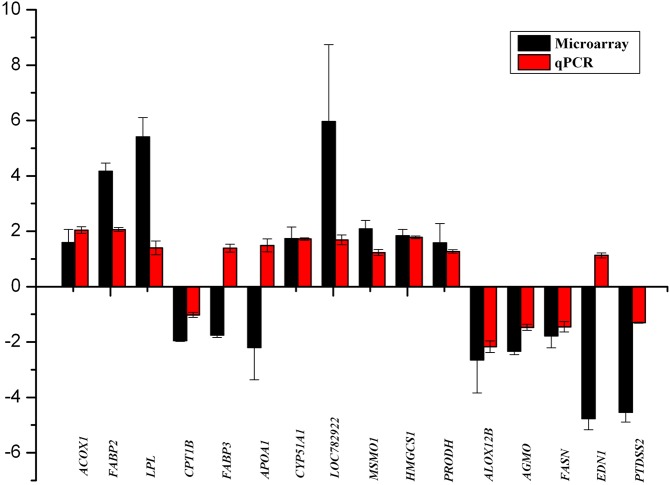
Quantitative PCR validation of the microarray results
To confirm the gene expression patterns obtained by the microarray analysis, 16 genes were examined by RT-qPCR. We first analyzed 6 of the 11 differentially expressed genes in the PPAR pathway (ACOX1, CPT1B, APOA1, FABP3, FABP2, LPL). The results showed that four genes (ACOX1, CPT1B, FABP2, LPL) were in agreement with the microarray data, with the exception of the APOA1 and FABP3 genes (Fig 4). The APOA1 gene encodes apolipoprotein AI (apo AI), which is the major protein component of HDL and plays a key role in lipid metabolism and transport [34]. Despite the discrepancy observed in our study regarding the expression of APOA1, our RT-qPCR results were consistent with the results of previous studies showing that the levels of apolipoprotein increases in the body through the consumption of high n-3 PUFA-containing foods [35]. FABP3 is a small cytoplasmic protein, and it is involved in fatty acid metabolism because it can transport fatty acids into the mitochondria from the cell membrane. This protein is proposed to be involved in early myocardial development and adult myocardial tissue repair and is thought to be responsible for the modulation of cell growth and proliferation [36]. Our RT-qPCR results also demonstrated the high expression of FABP3 in the fat-1 transgenic cattle, suggesting that the fat-1 gene potentially plays a role in the regulation of the expression of the FABP3 genes. We then analyzed 10 of the 32 differentially expressed genes related to lipid metabolism as obtained by GO enrichment analysis (CYP51A1, LOC782922, MSMO1, HMGCS1, PRODH, ALOX12B, AGMO, FASN, EDN1, PTDSS2). Except for EDN1, both the microarray results and the RT-qPCR data showed that CYP51A1, LOC782922, MSMO1, HMGCS1, and PRODH were up-regulated and that ALOX12B, AGMO, FASN, and PTDSS2 were down-regulated in fat-1 transgenic cattle (Fig 4). EDN1 is a potent vasoconstrictor. The beneficial and detrimental physiological roles of EDN1 were reported in different studies. One study reported that high concentrations of EDN1 might lead to the constriction of coronary arteries, thereby impairing the contractility of the heart, resulting in a dilated cardiomyopathy (DCM) phenotype [37]. However, the present study reported that decreasing the expression of EDN1 by as little as 35% caused severely dilated cardiomyopathy, while a threefold increase in the expression of EDN1 only caused slight cardiac hypertrophy, suggesting that cardiac function was sensitive to even modest changes in EDN1 levels [38]. Our RT-qPCR result indicated that the expression of EDN1 was slightly elevated in fat-1 transgenic cattle, which was inconsistent with our microarray results. However, we speculated that the expression increase of EDN1 may be modest and beneficial for the health of the transgenic cattle, especially for maintaining the normal contractile functions of the heart and blood vessels.
Fig 4. Validation of sixteen microarray differentially expressed genes by RT-qPCR.
The fold-change value is expressed as positive when the genes are highly expressed in transgenic cattle and as negative when the genes are highly expressed in wild-type cattle. The gene names are displayed below the histogram.
Taken together, the results showed that 13 of the 16 genes analyzed showed the same expression patterns between the RT-qPCR and the microarray analysis (Fig 4), indicting a high consistency between both analyses, confirming the reliability of the microarray data.
In conclusion, we used microarray technology to analyze the differences in gene expression between fat-1 transgenic cattle and wild-type cattle. We identified 36 differentially expressed genes belonging to eight biological pathways related to lipid metabolism processes. Furthermore, we found that exogenous fat-1 can influence an important lipid metabolism signaling pathway, the PPAR signaling pathway, and 11 genes were significantly enriched in this signaling pathway. Therefore, our findings provide further insight into the function of the fat-1 gene and how to endogenously synthesize n-3 PUFAs in transgenic animals.
Supporting Information
The x-axis indicates the likelihood [−log2(pvalue)] in a category, and the y-axis indicates the different subcategories of cellular components.
(TIF)
The x-axis indicates the likelihood [−log2(pvalue)] in a category, and the y-axis means the different subcategories of molecular function.
(TIF)
(DOC)
(XLS)
(XLS)
(XLS)
(DOC)
Acknowledgments
We thank the Tianjin Ckrg biological technology development Co. Ltd. for providing us with technical assistance in bioinformatics analysis. We also thank Yongwei Nie, Hanlu Fan, and Tao Jin for their assistance in sample collection.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by the National Basic Research Program (Grant No. 2012CB22306) and National Transgenic Animal Program (Grant No. 2013ZX08007-002) of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kris-Etherton PM, Hecker KD, Binkoski AE. Polyunsaturated fatty acids and cardiovascular health. Nutr Rev. 2004; 62:414–426. [DOI] [PubMed] [Google Scholar]
- 2. Lee AL, Park Y. The association between n-3 polyunsaturated fatty acid levels in erythrocytes and the risk of rheumatoid arthritis in Korean women. Ann Nutr Metab. 2013; 63:88–95. 10.1159/000353120 [DOI] [PubMed] [Google Scholar]
- 3. Tárnok A, Marosvölgyi T, Szabó E, Györei E, Decsi T. Low n-3 long-chain polyunsaturated fatty acids in newly diagnosed celiac children with preexisting type-1 diabetes mellitus. J Pediatr Gastroenterol Nutr. 2015; 60:255–258. 10.1097/MPG.0000000000000561 [DOI] [PubMed] [Google Scholar]
- 4. Küllenberg de Gaudry D, Massing U. Importance of long chain omega-3 fatty acids in prostate cancer. Urologe A. 2014; 53:1620–1624. 10.1007/s00120-014-3612-3 [DOI] [PubMed] [Google Scholar]
- 5. Lai L, Kang JX, Li R, Wang J, Witt WT, Yong HY, et al. Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nat Biotechnol. 2006; 24: 435–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu G, Chen H, Wu X, Zhou Y, Lu J, Chen H, et al. A modified n-3 fatty acid desaturase gene from Caenorhabditis briggsae produced high proportion of DHA and DPA in transgenic mice. Transgenic Res. 2008; 17:717–725. 10.1007/s11248-008-9171-x [DOI] [PubMed] [Google Scholar]
- 7. Kang ZB, Ge Y, Chen Z, Cluette-Brown J, Laposata M, Leaf A, et al. Adenoviral gene transfer of Caenorhabditis elegans n-3 fatty acid desaturase optimizes fatty acid composition in mammalian cells. Proc Natl Acad Sci USA. 2001; 98:4050–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kang JX, Wang J, Wu L, Kang ZB. Transgenic mice: fat-1 mice convert n-6 to n-3 fatty acids. Nature. 2004; 427:504 [DOI] [PubMed] [Google Scholar]
- 9. Kang JX. Fat-1 transgenic mice: a new model for omega-3 research. Prostaglandins Leukot Essent Fatty Acids. 2007; 77:263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lau BY, Ward WE, Kang JX, Ma DW. Vertebrae of developing fat-1 mice have greater strength and lower n-6/n-3 fatty acid ratio. Exp Biol Med (Maywood). 2009; 234:632–638. [DOI] [PubMed] [Google Scholar]
- 11. Lebbadi M, Julien C, Phivilay A, Tremblay C, Emond V, Kang JX, et al. Endogenous conversion of omega-6 into omega-3 fatty acids improves neuropathology in an animal model of Alzheimer’s disease. J Alzheimers Dis. 2011; 27:853–869. 10.3233/JAD-2011-111010 [DOI] [PubMed] [Google Scholar]
- 12. MacLennan MB, Clarke SE, Perez K, Wood GA, Muller WJ, Muller WJ, et al. Mammary tumor development is directly inhibited by lifelong n-3 polyunsaturated fatty acids. J Nutr Biochem. 2013; 24:388–395. 10.1016/j.jnutbio.2012.08.002 [DOI] [PubMed] [Google Scholar]
- 13. Zhang P, Liu P, Dou H, Chen L, Chen L, Lin L, et al. Handmade cloned transgenic sheep rich in omega-3 fatty acids. Plos One. 2013; 8:e55941 10.1371/journal.pone.0055941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu X, Ouyang H, Duan B, Pang D, Zhang L, Yuan T, et al. Production of cloned transgenic cow expressing omega-3 fatty acids. Transgenic Res. 2012; 21:537–543. 10.1007/s11248-011-9554-2 [DOI] [PubMed] [Google Scholar]
- 15. Guo T, Liu XF, Ding XB, Yang FF, Nie YW, An YJ, et al. Fat-1 transgenic cattle as a model to study the function of ω-3 fatty acids. Lipids Health Dis. 2011; 10:244 10.1186/1476-511X-10-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nuruddin S, Syverstad GH, Lillehaug S, Leergaard TB, Nilsson LN, Ropstad E, et al. Elevated mRNA-levels of gonadotropin-releasing hormone and its receptor in plaque-bearing Alzheimer’s disease transgenic mice. Plos One. 2014; 9:e103607 10.1371/journal.pone.0103607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tani S, Nagao K, Hirayama A. Association of atherosclerosis-related markers and its relationship to n-3 polyunsaturated fatty acids levels with a prevalence of coronary artery disease in an urban area in Japan. Heart Vessels. 2015; 30:9–19. 10.1007/s00380-013-0442-y [DOI] [PubMed] [Google Scholar]
- 18. Kim EH, Bae JS, Hahm KB, Cha JY. Endogenously synthesized n-3 polyunsaturated fatty acids in fat-1 mice ameliorate high-fat diet-induced nonalcoholic fatty liver disease. Biochem Pharmacol. 2012; 84:1359–1365. 10.1016/j.bcp.2012.08.029 [DOI] [PubMed] [Google Scholar]
- 19. Kinsella JE, Lokesh B, Stone RA. Dietary n-3 polyunsaturated fatty acids and amelioration of cardiovascular disease: possible mechanisms. Am J Clin Nutr. 1990; 52:1–28. [DOI] [PubMed] [Google Scholar]
- 20. Kinosian B, Glick H, Garland G. Cholesterol and coronary heart disease: predicting risks by levels and ratios. Ann Intern Med. 1994; 121: 641–647. [DOI] [PubMed] [Google Scholar]
- 21. Ray S, Talukdar A1, Sonthalia N, Saha M, Kundu S, Khanra D, et al. Serum lipoprotein ratios as markers of insulin resistance: a study among non-diabetic acute coronary syndrome patients with impaired fasting glucose. Indian J Med Res. 2015; 141:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Notarnicola M, Messa C, Refolo MG, Tutino V, Miccolis A, Caruso MG. Polyunsaturated fatty acids reduce fatty acid synthase and hydroxy-methyl-glutaryl CoA-reductase gene expression and promote apoptosis in HepG2 cell line. Lipids Health Dis. 2011; 10:10 10.1186/1476-511X-10-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rudkowska I, Caron-Dorval D, Verreault M, Couture P, Deshaies Y, Barbier O, et al. PPARalpha L162V polymorphism alters the potential of n-3 fatty acids to increase lipoprotein lipase activity. Mol Nutr Food Res. 2010; 54:543–550. 10.1002/mnfr.200900085 [DOI] [PubMed] [Google Scholar]
- 24. Roy R, Taourit S, Zaragoza P, Eggen A, Rodellar C. Genomic structure and alternative transcript of bovine fatty acid synthase gene (FASN): comparative analysis of the FASN gene between monogastric and ruminant species. Cytogenet Genome Res. 2005; 111:65–73. [DOI] [PubMed] [Google Scholar]
- 25. Hiller B, Hocquette JF, Cassar-Malek I, Nuernberg G, Nuernberg K. Dietary n-3 PUFA affect lipid metabolism and tissue function-related genes in bovine muscle. Br J Nutr. 2012; 108:858–863. 10.1017/S0007114511006179 [DOI] [PubMed] [Google Scholar]
- 26. Yu K, Shu G, Yuan F, Zhu X, Gao P, Wang S, et al. Fatty acid and transcriptome profiling of longissimus longissimus dorsi muscles between pig breeds differing in meat quality. Int J Biol Sci. 2013; 9:108–118. 10.7150/ijbs.5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Romanatto T, Fiamoncini J, Wang B, Curi R, Kang JX. Elevated tissue omega-3 fatty acid status prevents age-related glucose intolerance in fat-1 transgenic mice. Biochim Biophys Acta. 2014; 1842:186–191. 10.1016/j.bbadis.2013.10.017 [DOI] [PubMed] [Google Scholar]
- 28. Ntambi JM. Regulation of stearoyl-CoA desaturase by polyunsaturated fatty acids and cholesterol. J Lipid Res. 1999; 40:1549–1558. [PubMed] [Google Scholar]
- 29. Lengi AJ, Corl BA. Identification and characterization of a novel bovine stearoyl-CoA desaturase isoform with homology to human SCD5. Lipids. 2007; 42:499–508. [DOI] [PubMed] [Google Scholar]
- 30. Morais S, Knoll-Gellida A, André M, Barthe C, Babin PJ. Conserved expression of alternative splicing variants of peroxisomal acyl-CoA oxidase 1 in vertebrates and developmental and nutritional regulation in fish. Physiol Genomics. 2007; 28: 239–252. [DOI] [PubMed] [Google Scholar]
- 31. Vluggen A, Andreoletti P, Viswakarma N, Jia Y, Matsumoto K, Kulik W, et al. Functional significance of the two ACOX1 isoforms and their crosstalks with PPARαand RXRα. Lab Invest. 2010; 90:696–708. [DOI] [PubMed] [Google Scholar]
- 32. Cánovas A, Quintanilla R, Amills M, Pena RN. Muscle transcriptomic profiles in pigs with divergent phenotypes for fatness traits. BMC Genomics. 2010; 11:372 10.1186/1471-2164-11-372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nagahuedi S, Popesku JT, Trudeau VL, Weber JM. Mimicking the natural doping of migrant sandpipers in sedentary quails: effects of dietary n-3 fatty acids on muscle membranes and PPAR expression. J Exp Biol. 2009; 212:1106–1114. 10.1242/jeb.027888 [DOI] [PubMed] [Google Scholar]
- 34. Toptas B, Görmüş U, Ergen A, Gürkan H, Keleşoglu F, Darendeliler F, et al. Comparison of lipid profiles with APOA1 MspI polymorphism in obese children with hyperlipidemia. In Vivo. 2011; 25:425–430. [PubMed] [Google Scholar]
- 35. Ahmed AA, Balogun KA, Bykova NV, Cheema SK. Novel regulatory roles of omega-3 fatty acids in metabolic pathways: a proteomics approach. Nutr Metab (Lond). 2014; 11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang S, Zhou Y, Andreyev O, Hoyt RF Jr, Singh A, Hunt T, et al. Overexpression of FABP3 inhibits human bone marrow derived mesenchymal stem cell proliferation but enhances their survival in hypoxia. Exp Cell Res. 2014; 323:56–65. 10.1016/j.yexcr.2014.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matsa LS, Sagurthi SR, Ananthapur V, Nalla, Nallari P. Endothelin 1 gene as a modifier in dilated cardiomyopathy. Gene. 2014; 548:256–262. 10.1016/j.gene.2014.07.043 [DOI] [PubMed] [Google Scholar]
- 38. Hathaway CK, Grant R, Hagaman JR, Hiller S, Li F, Xu L, et al. Endothelin-1 critically influences cardiac function via superoxide-MMP9 cascade. Proc Natl Acad Sci USA. 2015; 112:5141–5146. 10.1073/pnas.1504557112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The x-axis indicates the likelihood [−log2(pvalue)] in a category, and the y-axis indicates the different subcategories of cellular components.
(TIF)
The x-axis indicates the likelihood [−log2(pvalue)] in a category, and the y-axis means the different subcategories of molecular function.
(TIF)
(DOC)
(XLS)
(XLS)
(XLS)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.