Abstract
An avian paramyxovirus (APMV) isolated from goose feces (APMV/Shimane67) was biologically, serologically and genetically characterized. APMV/Shimane67 showed typical paramyxovirus morphology on electron microscopy. On hemagglutination inhibition test, antiserum against APMV/Shimane67 revealed low reactivity with other APMV serotypes and vice versa. The fusion (F) protein gene of APMV/Shimane67 contained 1,638 nucleotides in a single open reading frame encoding a protein of 545 amino acids. The cleavage site of F protein contained a pair of single basic amino acid (VRENR/L). The nucleotide and deduced amino acid sequences of the F gene of APMV/Shimane67 had relatively low identities (42.9–62.7% and 28.9–67.3%, respectively) with those of other APMVs. Phylogenetic analysis showed that APMV/Shimane67 was related to NDV, APMV-9 and APMV-12, but was distinct from those APMV serotypes. These results suggest that APMV/Shimane67 is a new APMV serotype, APMV-13.
Keywords: APMV, avian paramyxovirus, F gene, phylogenetic analysis, serotype
Avian paramyxovirus (APMV) belongs to the genus Avulavirus in the Paramyxovirinae subfamily of the family Paramyxoviridae. Members of family Paramyxoviridae are characterized by pleomorphic enveloped particles that contain a single-stranded, negative sense RNA genome [10]. APMV is classified into nine distinct serotypes based on serological tests, such as hemagglutination inhibition (HI) and neuraminidase inhibition (NI) tests [1]. APMV-1, also known as Newcastle disease virus (NDV), causes severe disease in chickens and huge economic losses for poultry industries. Thus, APMV-1 is the most extensively characterized APMV serotype. In addition to APMV-1, APMV-2, 3, 6 and 7 are reportedly associated with disease in poultry. APMV-2 causes respiratory disease in chickens and turkeys, while APMV-3, 6 and 7 cause respiratory disease or egg production disorder in turkeys. On the other hand, APMV-4, 5, 8 and 9 have not been reported to infect poultry. APMV-4, 8 and 9 were mainly isolated from waterfowl, such as ducks and geese, while APMV-5 was isolated from budgerigar and is associated with diarrhea and high mortality [1]. Three APMVs isolated from the rockhopper penguin in the Falkland Islands, the common snipe in France and the Eurasian wigeon in Italy have recently been proposed to be new serotypes of APMV, APMV-10, 11 and 12, respectively [4, 12, 19]. The intracerebral pathogenicity index test using one-day-old chicks suggested that APMV-10 and 12 showed little or no virulence in chickens, resembling the lentogenic NDV.
Since 1979, we have surveyed for influenza A viruses and APMVs in the San-in district of western Japan. During this surveillance, numerous influenza A viruses and APMVs have been isolated from the feces of migratory waterfowl flying from Siberia or northern China [5, 16, 20]. APMV/Shimane/67/2000 (APMV/Shimane67) examined in this study was isolated from the feces of geese collected in 2000 in Shimane prefecture. Here, we report the biological, serological and genetic analyses of APMV/Shimane67. The results suggested that APMV/Shimane67 belongs to a new serotype APMV-13.
MATERIALS AND METHODS
Viruses and cells: APMV/Shimane67 was isolated from goose fecal samples collected in Shimane prefecture, the wintering location for wild waterfowl, such as ducks, geese and swans. Virus isolation was performed as described previously [16].
Electron microscopy: Purification of APMV/Shimane67 was conducted by differential centrifugation and two successive sedimentations through a 10 to 50% sucrose gradient as described previously [6]. The purified virus particles stained with 2% phosphotungstic acid (pH 6.8) were observed under the JEM-100CX electron microscope (JEOL, Tokyo, Japan) at 80 kV.
Hemagglutination (HA) and hemagglutination inhibition (HI) test: HA and HI tests were performed using established procedures as described previously [5]. For HI test, chicken hyperimmune sera against APMV/Shimane67, NDV/goose/Alaska/415/91 (NDV/AK/415), APMV-2/chicken/California/Yucaipa/56 (APMV-2/Yucaipa), APMV-3/turkey/Wisconsin/68 (APMV-3/WI), APMV-4/duck/Mississippi/320/75 (APMV-4/MS), APMV-6/duck/Hong Kong/D199/77 (APMV-6/HK/D199) and APMV-7/dove/Tennessee/4/75 (APMV-7/TN) were prepared as described previously [20].
Propagation of APMV/Shimane67 in cultured cells: Madin-Darby canine kidney (MDCK), Madin-Darby bovine kidney (MDBK), African green monkey kidney (Vero) and baby hamster kidney (BHK21) cells were grown in Eagle’s minimal essential medium (E-MEM) containing 10% fetal bovine serum (FBS). Human embryonic kidney (293) and chicken embryo fibroblast (DF-1) cells were grown in Dulbecco’s modified Eagle’s medium (D-MEM) containing 10% FBS. Cell monolayers prepared in 6-well plates were infected with 104 50% egg infectious dose (EID50) of APMV/Shimane67. After adsorption for 1 hr at 37°C, cells were washed three times with phosphate buffered saline (PBS, pH7.2) and were incubated at 37°C in E-MEM or D-MEM containing 0.3% bovine serum albumin and 0.5 µg of TPCK-treated trypsin (Sigma-Aldrich, St. Louis, U.S.A.). The virus titer of each cell culture supernatant was determined as follows. Ten-fold diluted supernatant (100 µl) was inoculated into the allantoic cavities of five 10-day-old embryonated chicken eggs and were incubated at 37°C for 3 days unless death of the embryo was detected. After the inoculated eggs were chilled at 4°C, their allantoic fluids were tested for hemagglutination activity. The virus titer was calculated using the method of Reed and Muench [13].
Pathogenicity test: The mean death time (MDT) at the minimum lethal dose for chicken embryos and the intracerebral pathogenicity index (ICPI) in 1-day-old chickens were measured in order to assess the virulence of this virus, as described previously [2].
Nucleotide sequencing and phylogenetic analysis: Viral RNAs were extracted from purified APMV/Shimane67 using the QIAamp Viral RNA Mini Kit (QIAGEN, Tokyo, Japan), and were transcribed to cDNA using F10 primer and RAV-2 reverse transcriptase (TaKaRa Bio, Otsu, Japan) in accordance with the manufacturer’s instructions. F10 primer was designed to bind with the gene-stop signal region of the matrix gene and gene-start signal of the F gene of NDV/AK/415 [21]. PCR was carried out using Pwo DNA polymerase (Roche, Mannheim, Germany) and a cycling sequence of 94°C for 2 min, followed by 35 cycles of 94°C for 30 sec, 55°C for 30 sec and 72°C for 2 min, which was then followed by a final extension at 72°C for 5 min. PCR products were purified using a QIAquick Gel Extraction Kit (QIAGEN) and were sequenced using the Thermo Sequenase Cy5.5 Dye Terminator Cycle Sequencing Kit (Amersham Pharmacia Biotech, Piscataway, NJ, U.S.A.) and a Gene Rapid DNA sequencer (Amersham Pharmacia Biotech). Nucleotide sequences of PCR and sequencing primers are shown in Table 1. The nucleotide and deduced amino acid sequences of APMV/Shimane67 F gene were compared to those of published APMVs by Clustal X [11]. Phylogenetic trees were generated using the Neighbor-Joining (NJ) method with 1,000 bootstraps.
Table 1. Oligonucleotide primers used in this study.
| Primer | Nucleotide sequence (5′ to 3′) | |
|---|---|---|
| For PCR | ||
| F10 | ATTAGAAAAAACACGGGTAGAA | |
| FR1215 | CAACGACAGATGACCTGTTG | |
| F270 | GAAACATTATCCCGCATTCTAAC | |
| HNR39 | GAATGAGCAATTTGATAACCCCAG | |
| For Sequencing | ||
| F10 | ATTAGAAAAAACACGGGTAGAA | |
| FR258 | GCAATGATTGTTTGGCACAATC | |
| F270 | GAAACATTATCCCGCATTCTAAC | |
| F588 | TAACCCTTGAATCCAGGCTAGG | |
| F905 | GTTCAACCTTACTAGAAACATTAGC | |
| F1090 | CATGTCTCAATGGAAATCTAAGTGA | |
| FR1215 | CAACGACAGATGACCTGTTG | |
| FR1545 | GTGATTGCAATTATTAAGCATACCACTG | |
| F1689 | ACTCCCCCATCCAGCCACAT | |
| FR1746 | ACCTACATGTTGGAGTGCCA | |
| HNR39 | GAATGAGCAATTTGATAACCCCAG | |
RESULTS
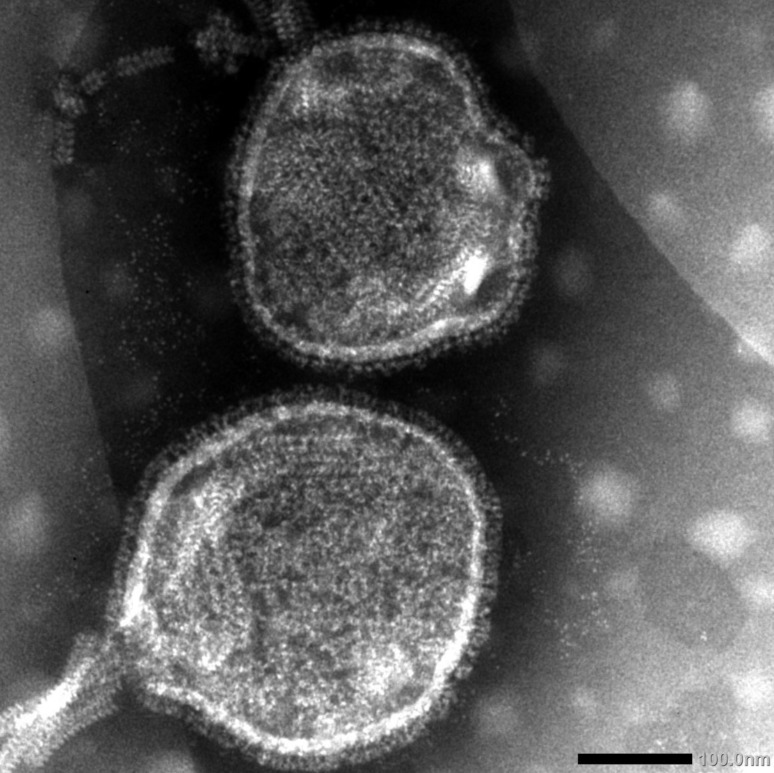
Electron microscopy: Electron microscopy of APMV/Shimane67 showed typical characteristics of a paramyxovirus. Virions were spherical or pleomorphic, enveloped particles (250–300 nm in diameter), and virion surfaces were covered with spiked projections of about 8–10 nm in length (Fig. 1).
Fig. 1.
Negative contrast electron micrograph of APMV/Shimane67 virus showing a partially disrupted particle with nucleocapsid emerging. × 200,000.
HI test: In order to investigate the antigenic relationships between APMV/Shimane67 and other serotypes of APMV, HI tests were carried out (Table 2). As APMV-5 has no HA activity, and APMV-8 to 12 were not available in our laboratory, HI tests against these 6 APMVs were not performed. Antiserum against APMV/Shimane67 showed the highest HI titer (1:1,280) with the homologous antigen, but little or no HI activity against other APMV serotypes. Reactivity of antisera against NDV/AK/415, APMV-2/Yucaipa, APMV-3/Wisconsin, APMV-4/Mississippi, APMV-6/Hong Kong and APMV-7/Tennessee on HI test was highest with each homologous antigen, while these antisera showed lower activity against APMV/Shimane67 (1:<40 − 1:1,280).
Table 2. Antigenic analysis of APMV/Shimane67 by HI tests with antisera against representative APMV strains.
| Antigen | Antiserum to | ||||||
|---|---|---|---|---|---|---|---|
| APMV/Shimane67 | NDV/AK/415 | APMV-2/Yucaipa | APMV-3/WI | APMV-4/MI | APMV-6/HK | APMV-7/TN | |
| APMV/Shimane67 | 1280a) | 1,280 | <40 | <40 | 640 | <40 | 320 |
| NDV/AK/415 | <40 | 5,120 | |||||
| APMV-2/Yucaipa | <40 | 160 | |||||
| APMV-3/WI | <40 | 160 | |||||
| APMV-4/MI | <40 | 5,120 | |||||
| APMV-6/HK | <40 | 640 | |||||
| APMV-7/TN | <40 | 2,560 | |||||
a) HI titer represents the reciprocal of the serum dilution inhibiting the activity of 4 hemagglutinating units of virus antigen. Blank: not tested.
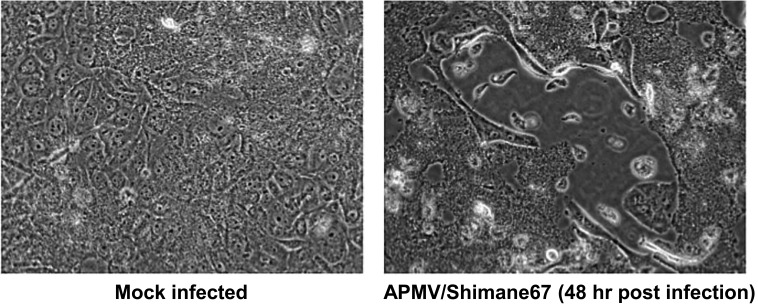
Growth in cell culture and pathogenicity of APMV/Shimane67: Growth of APMV/Shimane67 in avian and mammalian origin cell lines (DF-1, MDCK, MDBK, BHK, Vero and 293 cells) was measured. In the presence of trypsin, regardless of cell lines, APMV/Shimane67 efficiently grew and reached approximately 105.0 EID50/0.1 ml at 72 hr post-infection. Syncytium formation, a typical characteristic of paramyxovirus cytopathic effects, was clearly observed, particularly in MDBK cells (Fig. 2). In DF-1 cells without trypsin, titers of APMV/Shimane67 reached about 103.5 EID50/0.1 ml at 72 hr post-infection. These results indicate that external proteases are necessary for efficient cleavage of the F protein.
Fig. 2.
MDBK cells infected with APMV/Shimane67.
The MDT and ICPI of APMV/Shimane67 were 120 hr < and 0.0, respectively.
Nucleotide sequencing and phylogenetic analysis: A total of 1,878 nucleotides of the genome sequence of APMV/Shimane67 were determined. The putative initiation codon, which better matched the Kozak consensus sequence ((G/A) NNATGG) [8], was the third ATG from 5′-end of the F10 primer binding site (data not shown). It contained a long open reading frame (ORF) composed of 1,638 nucleotides and encoded 545 amino acids. The nucleotide sequence of this ORF showed similarities with the F genes of NDVs and APMV-12/wigeon/Italy/3920_1/2005 (APMV-12/ITA) by discontiguous MegaBLAST search [24], indicating that this ORF was the F gene of APMV/Shimane67. The nucleotide and deduced amino acid identities between the F gene of APMV/Shimane67 and other APMV serotypes ranged from 42.9% with APMV-3/NLD to 62.7% with APMV-12/ITA, and from 28.9% with APMV-3/NLD to 67.3% with APMV-12/ITA, respectively (Table 3). The deduced amino acid sequence of the putative cleavage site of the APMV/Shimane F gene was QVRENR/LVG (Fig. 3). This resembled the motif, a pair of single basic residues, of the lentogenic NDV. The phylogenetic tree of the F gene constructed with APMVs and members of subfamily Paramyxovirinae revealed that APMV/Shimane67 is a member of genus Avulavirus and is grouped with NDV, APMV-9 and APMV-12, while distinct from these APMV serotypes (Fig. 4).
Table 3. Nucleotide (upper right) and deduced amino acid (lower left) identities (%) among the fusion protein gene of avian paramyxoviruses.
| Virus | Shimane 67 | NDV | APMV-2 | APMV-3 | APMV-4 | APMV-5 | APMV-6 | APMV-7 | APMV-8 | APMV-9 | APMV-10 | APMV-11 | APMV-12 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AK/ 415 | LaSota | SF02 | Yucaipa | Bangor | NLD | WI | HK/D3 | KR/YJ | Kunitachi | TWN/Y1 | ITA/ 4524-2 | TN | DE/1053 | Wakuya | NY/22 | ITA/5709 | FLK/324 | FRA/ 100212 | ITA/ 3920_1 | ||
| APMV/Shimane67 | - | 53.6 | 56.1 | 54.3 | 45.6 | 44.6 | 42.9 | 43.0 | 43.7 | 43.3 | 46.2 | 44.6 | 45.3 | 43.5 | 46.6 | 46.8 | 53.3 | 53.4 | 46.3 | 43.8 | 62.7 |
| NDV/AK/415 | 52.8 | - | 71.8 | 71.5 | 46.4 | 46.8 | 43.3 | 41.0 | 42.8 | 42.0 | 54.4 | 44.6 | 45.0 | 44.7 | 47.0 | 47.4 | 58.5 | 58.4 | 45.6 | 43.1 | 55.1 |
| NDV/LaSota | 53.7 | 87.3 | - | 84.4 | 47.5 | 46.6 | 42.3 | 41.9 | 41.6 | 41.4 | 46.8 | 44.4 | 45.0 | 43.5 | 46.7 | 46.9 | 58.2 | 60.3 | 46.1 | 42.1 | 55.5 |
| NDV/SF02 | 52.6 | 84.8 | 88.6 | - | 47.5 | 48.0 | 42.5 | 42.2 | 42.7 | 41.7 | 45.3 | 44.9 | 44.4 | 44.6 | 47.3 | 47.3 | 58.4 | 58.7 | 45.5 | 43.2 | 54.8 |
| APMV-2/Yucaipa | 37.3 | 40.4 | 40.6 | 40.4 | - | 69.8 | 40.7 | 41.1 | 42.8 | 41.8 | 50.2 | 50.9 | 51.8 | 47.2 | 60.5 | 60.9 | 46.3 | 47.2 | 59.2 | 48.5 | 46.7 |
| APMV-2/Bangor | 36.5 | 40.7 | 41.3 | 41.3 | 79.1 | - | 41.0 | 41.5 | 42.4 | 41.8 | 50.4 | 51.1 | 52.3 | 48.3 | 60.5 | 60.0 | 46.1 | 46.5 | 59.8 | 50.0 | 46.1 |
| APMV-3/NLD | 28.9 | 31.2 | 31.2 | 30.7 | 30.8 | 31.5 | - | 66.8 | 47.8 | 47.4 | 42.3 | 42.7 | 42.4 | 41.3 | 43.4 | 43.2 | 40.7 | 40.1 | 40.8 | 42.0 | 43.2 |
| APMV-3/WI | 29.4 | 30.0 | 30.2 | 30.0 | 30.0 | 30.3 | 70.1 | - | 46.8 | 47.1 | 43.2 | 42.3 | 42.6 | 42.5 | 41.6 | 41.7 | 41.4 | 41.2 | 41.5 | 44.3 | 41.6 |
| APMV-4/HK/D3 | 32.3 | 31.0 | 31.3 | 31.0 | 33.6 | 34.0 | 32.0 | 33.5 | - | 92.3 | 43.5 | 42.9 | 43.5 | 40.8 | 44.2 | 44.1 | 41.9 | 42.4 | 43.3 | 41.9 | 40.9 |
| APMV-4/KR/YJ | 32.1 | 30.8 | 31.3 | 30.6 | 33.6 | 34.2 | 32.2 | 33.1 | 97.7 | - | 42.5 | 43.1 | 44.0 | 40.6 | 44.1 | 43.7 | 41.1 | 42.4 | 42.4 | 41.0 | 41.3 |
| APMV-5/Kunitachi | 39.3 | 40.1 | 41.0 | 41.4 | 46.5 | 45.7 | 31.0 | 30.9 | 33.9 | 33.5 | - | 56.5 | 57.0 | 47.9 | 50.5 | 50.7 | 44.9 | 45.0 | 52.1 | 46.9 | 44.6 |
| APMV-6/TWN/Y1 | 36.9 | 36.8 | 37.8 | 37.8 | 48.8 | 49.4 | 31.6 | 29.8 | 33.6 | 33.0 | 53.7 | - | 72.6 | 47.3 | 49.3 | 49.4 | 45.1 | 44.9 | 52.3 | 48.9 | 45.6 |
| APMV-6/ITA/4524-2 | 37.3 | 37.1 | 37.4 | 37.6 | 49.2 | 48.6 | 31.2 | 29.5 | 33.8 | 33.6 | 54.1 | 85.7 | - | 47.0 | 50.4 | 50.0 | 44.8 | 45 | 52.2 | 49.2 | 45.1 |
| APMV-7/TN | 35.7 | 38.1 | 39 | 38.8 | 38.1 | 38.8 | 28.5 | 30.1 | 30.1 | 29.3 | 38.0 | 37.0 | 36.9 | - | 49.2 | 49.0 | 42.8 | 43.7 | 48.8 | 53.0 | 44.4 |
| APMV-8/DE/1053 | 36.4 | 40.1 | 41.1 | 41.1 | 63.7 | 63.7 | 31.8 | 30.6 | 35.9 | 35.9 | 45.5 | 48.2 | 47.3 | 38.3 | - | 97.1 | 47.3 | 47.6 | 59.5 | 48.2 | 46.8 |
| APMV-8/Wakuya | 36.2 | 40.1 | 41.1 | 41.1 | 63.7 | 63.7 | 31.8 | 30.6 | 35.9 | 35.7 | 45.5 | 48.2 | 47.3 | 38.1 | 99.4 | - | 47.5 | 48.0 | 59.5 | 48.4 | 46.4 |
| APMV-9/NY/22 | 48.4 | 56.4 | 55.9 | 56.4 | 37.8 | 39.1 | 28.2 | 28.3 | 28.4 | 28.6 | 36.5 | 37.4 | 36.6 | 35.6 | 37.9 | 37.9 | - | 89.2 | 45.5 | 42.8 | 54.3 |
| APMV-9/ITA/5709 | 50.1 | 58.1 | 58.6 | 58.4 | 37.8 | 39.6 | 29.3 | 29.2 | 28.6 | 28.8 | 37.8 | 37.9 | 37.5 | 36.6 | 39.4 | 39.4 | 92.7 | - | 46.5 | 42.6 | 55.2 |
| APMV-10/FLK/324 | 37.8 | 39.5 | 39.9 | 39.5 | 61.7 | 60.3 | 29.0 | 28.1 | 32.4 | 32.2 | 46.3 | 48.1 | 48.0 | 38.2 | 61.4 | 61.4 | 38.9 | 39.1 | - | 48.0 | 46.5 |
| APMV-11/FRA/100212 | 31.6 | 32.6 | 33.9 | 33.9 | 42.4 | 41.8 | 31.0 | 33.0 | 32.5 | 32.2 | 39.9 | 39.7 | 41.7 | 36.0 | 40.4 | 40.4 | 32.7 | 33.3 | 39.4 | - | 43.9 |
| APMV-12/ITA/3920_1 | 67.3 | 55.1 | 54.6 | 53.8 | 38.1 | 37.3 | 29.5 | 28.5 | 32.1 | 31.9 | 37.4 | 37.7 | 37.9 | 36.5 | 36.1 | 36.1 | 52.3 | 53.2 | 37.8 | 32.7 | - |
Fig. 3.
Amino acid sequences of F protein cleavage site from APMVs. NDV (lentogenic); strain LaSota, NDV (velogenic); strain Herts 33, APMV-4/HK/D3; APMV-4/duck/Hong Kong/D3/1975, APMV-5/Kunitachi; APMV-5/budgerigar/ Japan/Kunitachi/1974, APMV-6/TWN/Y1; APMV-6/duck/Taiwan/Y1/1998, APMV-8/DE; APMV-8/goose/Delaware/1053/1976, APMV-9/NY; duck/New York/22/1978, APMV-10/FLK/324; APMV10/penguin/Falkland Islands/324/2007, APMV-11/FRA/100212; APMV-11/common_snipe/France/100212/2010. Abbreviations of other APMV are given in the text.
Fig. 4.
Phylogenetic tree of F gene sequences from subfamily Paramyxovirinae. The phylogenetic tree was generated using the neighbor-joining algorithm with 1,000 bootstrap replicates in Clustal X.
DISCUSSION
The typing of APMVs has mainly been conducted by HI test [1]. In addition, because the nucleotide sequence data from various APMVs have been collected in DNA databases, genetic analysis has also been attempted to classify new APMV isolates, and the results obtained have demonstrated the effectiveness of this approach [4, 12, 19]. On HI test, the anti-APMV/Shimane67 serum reacted strongly (HI titer of 1:1,280) with homologous virus and did not react with the other APMVs (HI titer of 1: <40). In addition, sera against other APMVs (NDV, APMV-2, 3, 4, 6 and 7) showed weaker reactivity with APMV/Shimane67 than with homologous viruses (Table 2). These results indicate that APMV/Shimane67 is serologically distinct from NDV, APMV-2, 3, 4, 6 and 7. Anti-NDV, APMV-4 and 7 sera showed some cross reactivity (HI titer of 1:320–1:1,280) with APMV/Shimane67. It is possible that this cross reactivity is caused by high titers of hyperimmune sera (HI titer of more than 1:2,560 against homologous viruses). The sera used in this study were prepared by multiple immunization of APMV antigen. Miller et al. [12] also reported cross reactivity on HI test of APMVs using hyperimmune serum.
Analysis of the F gene of APMV/Shimane67 also revealed the relationship of this virus with other APMVs. The ORF of the APMV/Shimane67 F gene was 1,638 nucleotides long and encoded 545 amino acids. Although these nucleotide and amino acid lengths for the F gene were the same as those of APMV-6/duck/Italy/4524-2/07 [23], the identities of nucleotide and amino acid sequences between APMV/Shimane67 and APMV-6/duck/Italy/4524-2/07 were low (45.3% and 37.3%, respectively) (Table 3). The amino acid sequence at the F protein cleavage site of APMV/Shimane67 was deduced as being QVRENR/LVG, which resembles the motif of lentogenic NDV (Fig. 3). This motif was observed in the F proteins of APMV/Shimane and NDV, APMV-9 and 12, and interestingly, these four APMVs containing conserved amino acid sequence motifs at the F protein cleavage site also formed one phylogenetic cluster (Fig. 4). The phylogenetic tree showed that APMV/Shimane67 is a member of genus Avulavirus, is distinct from other APMVs and showed close relationship with APMV/wigeon/Italy/3920-1/2005 (APMV-12/ITA/3920-1). APMV-2, −3 and −6 viruses have antigenic differences constituting the antigenic subgroup within each serotype [3, 9, 18]. Similar situations in phylogenetic analysis were also observed in the nucleotide and amino acid identities of the F gene (Table 3). APMV/Shimane67 exhibited the highest nucleotide and amino acid identities (62.7% and 67.3%, respectively) with APMV-12. These identities were lower than those observed between the subgroups, APMV-2/Yucaipa and APMV-2/Bangor (69.8% in nucleotide and 79.1% in amino acid sequence), and APMV-3/NLD and APMV-3/WI (66.8% in nucleotide and 70.1% in amino acid sequence) (Table 3). These observations confirm a distant relationship corresponding to APMV serotype between APMV/Shimane67 and APMV-12/ITA/3920-1.
The pathogenicity of APMV/Shimane67 to chickens was evaluated by MDT and ICPI tests. MDT (120 hr <) and ICPI (0.0) of APMV/Shimane67 were applicable to the lentogenic NDV and revealed that the virus had no virulence in chickens. The amino acid sequence at the F protein cleavage site is the major determinant of NDV pathogenicity to chickens. As described above, the putative cleavage site of the APMV/Shimane F protein is QVRENR/LVG, which resembles lentogenic NDV. Non-virulence of APMV/Shimane67 may be affected. The introduction of an amino acid sequence of velogenic NDV at the F protein cleavage site by reverse genetics did not increase the virulence of APMV-2, 4 and 7 in chickens [7, 17, 22]. However, it has been reported that lentogenic NDV isolated from wild geese changed to velogenic virus by amino acid substitutions in the F protein cleavage site during serial passage in chickens [15]. Whether the virulent variant of APMV/Shimane67 will emerge with multiple passaging of APMV/Shimane67 in chickens is of interest.
Recently, whole genome sequences from various APMV serotypes have been reported, and the classification of APMVs using those whole genome sequence data has been proposed [4, 12, 14, 19]. In this study, only the F gene of APMV/Shimane67 was identified; however, data obtained have suggested that APMV/Shimane67 is a novel APMV serotype, APMV-13. To emphasize this idea, the full-length genome sequence of APMV/Shimane67 needs to be determined.
REFERENCES
- 1.Alexander D. J.2003. Newcastle disease, other avian paramyxoviruses, and pneumovirus infections. pp. 63–99. In: Diseases of Poultry, 11th ed. (Saif, Y. M., Barnes, H. J., Glisson, J. R., Fadly, A. M., McDougald, L. R. and Swayne, D. E. eds.), Iowa State Press, Ames. [Google Scholar]
- 2.Allan W. H., Lancaster J. E., Toth B.1978. pp. 74–79. In: Newcastle Disease Vaccines. Food and Agriculture Organisation of the United Nations, Rome. [Google Scholar]
- 3.Anderson C., Kearsley R., Alexander D. J., Russel P. H.1987. Antigenic variation in avian paramyxovirus type 3 detected by mouse monoclonal antibodies. Avian Pathol. 16: 691–698. doi: 10.1080/03079458708436416 [DOI] [PubMed] [Google Scholar]
- 4.Briand F. X., Henry A., Massin P., Jestin V.2012. Complete genome sequence of a novel avian paramyxovirus. J. Virol. 86: 7710. doi: 10.1128/JVI.00946-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujimoto Y., Ito H., Shivakoti S., Nakamori J., Tsunekuni R., Otsuki K., Ito T.2010. Avian influenza virus and paramyxovirus isolation from migratory waterfowl and shorebirds in San-in district of western Japan from 2001 to 2008. J. Vet. Med. Sci. 72: 963–967. doi: 10.1292/jvms.10-0012 [DOI] [PubMed] [Google Scholar]
- 6.Kida H., Yanagawa R.1981. Classification of avian paramyxovirus by immunodiffusion on the basis of antigenic specificity of their M protein antigens. J. Gen. Virol. 52: 103–111. doi: 10.1099/0022-1317-52-1-103 [DOI] [PubMed] [Google Scholar]
- 7.Kim S. H., Xiao S., Shive H., Collins P. L., Samal S. K.2013. Mutations in the fusion protein cleavage site of avian paramyxovirus serotype 4 confer increased replication and syncytium formation in vitro but not increased replication and pathogenicity in chickens and ducks. PLoS ONE 8: e50598. doi: 10.1371/journal.pone.0050598 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Kozak M.1987. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 196: 947–950. doi: 10.1016/0022-2836(87)90418-9 [DOI] [PubMed] [Google Scholar]
- 9.Kumar S., Nayak B., Samuel A. S., Xiao S., Collins P. L., Samal S. K.2010. Complete genome sequence of avian paramyxovirus-3 strain Wisconsin: Evidence for the existence of subgroups within the serotype. Virus Res. 149: 78–85. doi: 10.1016/j.virusres.2009.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamb R. A., Parks G.2007. Paramyxoviridae: the viruses and their replication. pp. 1449–1496. In: Fields Virology, 5th ed. (Knipe, D. M., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B. and Straus, S. E. eds.), Lippincott Williams &Wilkins, Philadelphia. [Google Scholar]
- 11.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G.2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. doi: 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 12.Miller P. J., Afonso C. L., Spackman E., Scott M. A., Pedersen J. C., Senne D. A., Brown J. D., Fuller C. M., Uhart M. M., Karesh W. B., Brown I. H., Alexander D. J., Swayne D. E.2010. Evidence for a new avian paramyxovirus serotype 10 detected in rockhopper penguins from the Falkland Islands. J. Virol. 84: 11496–11504. doi: 10.1128/JVI.00822-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reed L. J., Muench H.1938. A simple method for estimating fifty percent endpoints. Am. J. Hyg. 27: 493–497. [Google Scholar]
- 14.Samuel A. S., Palduri A., Kumar S., Collins P. L., Samal S. K.2010. Complete genome sequence of avian paramyxovirus (APMV) serotype 5 completes the analysis of nine PMV serotypes and reveals the longest APMV genome. PLoS ONE 5: e9269. doi: 10.1371/journal.pone.0009269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shengqing Y., Kishida N., Ito H., Kida H., Otsuki K., Kawaoka Y., Ito T.2002. Generation of velogenic Newcastle disease viruses from a nonpathogenic waterfowl isolate by passaging in chickens. Virology 301: 206–211. doi: 10.1006/viro.2002.1539 [DOI] [PubMed] [Google Scholar]
- 16.Shengqing Y., Shinya K., Otsuki K., Ito H., Ito T.2002. Isolation of myxoviruses from migratory waterfowls in San-in district, western Japan in winters of 1997–2000. J. Vet. Med. Sci. 64: 1049–1052. doi: 10.1292/jvms.64.1049 [DOI] [PubMed] [Google Scholar]
- 17.Subbiah M., Khattar S. K., Collins P. L., Samal S. K.2011. Mutations in the fusion protein cleavage site of avian paramyxovirus serotype 2 increase cleavability and syncytium formation but do not increase viral virulence in chickens. J. Virol. 85: 5394–5405. doi: 10.1128/JVI.02696-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subbiah M., Nayak S., Collins P. L., Samal S. K.2010. Complete genome sequences of avian paramyxovirus serotype 2 (APMV-2) strains Bangor, England and Kenya: Evidence for the existence of subgroups within serotype 2. Virus Res. 152: 85–95. doi: 10.1016/j.virusres.2010.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terregino C., Aldous E. W., Heidari A., Fuller C. M., De Nardi R., Manvell R. J., Beato M. S., Shell W. M., Monne I., Brown I. H., Alexander D. J., Capua I.2013. Antigenic and genetic analyses of isolate APMV/wigeon/Italy/3920–1/2005 indicate that it represents a new avian paramyxovirus (APMV-12). Arch. Virol. 158: 2233–2243. doi: 10.1007/s00705-013-1735-2 [DOI] [PubMed] [Google Scholar]
- 20.Tsubokura M., Otsuki K., Kawaoka Y., Yanagawa R.1981. Isolation of influenza A viruses from migratory waterfowls in San-in District, Western Japan in 1979–1980. Zentralbl. Bakteriol. Mikrobiol. Hyg. [B] 173: 494–500. [PubMed] [Google Scholar]
- 21.Tsunekuni R., Ito H., Otsuki K., Kida H., Ito T.2010. Genetic comparisons between lentogenic Newcastle disease virus isolated from waterfowl and velogenic variants. Virus Genes 40: 252–255. doi: 10.1007/s11262-009-0427-1 [DOI] [PubMed] [Google Scholar]
- 22.Xiao S., Khattar S. K., Subbiah M., Collins P. L., Samal S. K.2012. Mutation of the f-protein cleavage site of avian paramyxovirus type 7 results in furin cleavage, fusion promotion, and increased replication in vitro but not increased replication, tissue tropism, or virulence in chickens. J. Virol. 86: 3828–3838. doi: 10.1128/JVI.06765-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao S., Subbiah M., Kumar S., Nardi R. D., Terregino C., Collins P. L., Samal S. K.2010. Complete genome sequences of avian paramyxovirus serotype 6 prototype strain Hong Kong and a recent novel strain from Italy: Evidence for the existence of subgroups within the serotype. Virus Res. 150: 61–72. doi: 10.1016/j.virusres.2010.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z., Schwartz S., Wagner L., Miller W.2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7: 203–214. doi: 10.1089/10665270050081478 [DOI] [PubMed] [Google Scholar]