Abstract
Clinically, many chemotherapeutics and ionizing radiation (IR) have been applied for the treatment of various types of human and animal malignancies. These treatments kill tumor cells by causing DNA double-strand breaks (DSBs). Core factors of classical nonhomologous DNA-end joining (C-NHEJ) play a vital role in DSB repair. Thus, it is indispensable to clarify the mechanisms of C-NHEJ in order to develop next-generation chemotherapeutics for cancer. The XRCC4-like factor (XLF; also called Cernunnos or NHEJ1) is the lastly identified core NHEJ factor. The localization of core NHEJ factors might play a critical role in regulating NHEJ activity. The localization and function of XLF have not been elucidated in animal species other than mice and humans. Domestic cattle (Bos taurus) are the most common and vital domestic animals in many countries. Here, we show that the localization of cattle XLF changes dynamically during the cell cycle. Furthermore, EYFP-cattle XLF accumulates quickly at microirradiated sites and colocalizes with the DSB marker γH2AX. Moreover, nuclear localization and accumulation of cattle XLF at DSB sites are dependent on 12 amino acids (288–299) of the C-terminal region of XLF (XLF CTR). Furthermore, basic amino acids on the XLF CTR are highly conserved among domestic animals including cattle, goat and horses, suggesting that the CTR is essential for the function of XLF in domestic animals. These findings might be useful to develop the molecular-targeting therapeutic drug taking XLF as a target molecule for human and domestic animals.
Keywords: cattle, DNA damage, domestic animal, Ku70, XLF
Many chemotherapeutics and ionizing radiation (IR) kill tumor cells by causing DNA double-strand breaks (DSBs). Clinically, cellular resistance to chemotherapy and radiotherapy is a critical component of tumor treatment failure. DNA repair proteins might be key players in those resistances. There are 2 pathways, i.e., homologous recombination (HR) and nonhomologous DNA-end joining (NHEJ), for DSB repair [7, 16]. In human and other mammalian cells, the classical NHEJ (C-NHEJ) process repairs a predominant fraction of DSBs [7, 16]. Thus, to develop next-generation chemotherapeutics for cancer is indispensable to clarify the molecular mechanisms of C-NHEJ.
C-NHEJ repair requires Ku70, Ku80, a DNA-dependent protein kinase catalytic subunit (DNA-PKcs), XRCC4, DNA ligase IV, Artemis and XLF [7, 16]. Studies using laser irradiation to induce DSBs in the nuclei of living cells have shed light on the order of recruitment of core NHEJ factors to DSB sites [16]. Ku70 and Ku80 accumulate at laser-induced DSB sites quickly following irradiation. These are essential for the recruitment of C-NHEJ factors, i.e., XLF, DNA-PKcs and XRCC4, and a HR-related protein (BRCA1) at DSB sites [9, 11, 16, 18, 19]. However, the localization and recruitment of core C-NHEJ factors to DSB sites have not been analyzed in cattle cells.
The localization and accumulation of core C-NHEJ factors at DSB sites might play a crucial role in modulating NHEJ activity [8, 16]. XLF is the lastly identified core C-NHEJ factor and plays critical roles in C-NHEJ [1, 3, 16]. The localization and function of XLF have not been elucidated in animal species other than mice and humans. In general, domestic cattle (Bos taurus) are the most common and important domestic animals in many countries. However, the molecular mechanism underlying DSB repair in cattle is still unknown.
It is important to elucidate the molecular mechanisms underlying the chemosensitivity or radiosensitivity of human and animal cells in order to develop new chemoradiotherapy and next-generation chemotherapeutic drugs for cancer. In this study, we examined the expression and subcellular localization of cattle XLF and its mutants in cattle cells. We also examined whether cattle XLF and its mutants accumulate at DSB sites quickly after irradiation.
MATERIALS AND METHODS
Cell lines, cultures and transfections: A Madin-Darby bovine kidney cell line (MDBK) (HSRRB, Osaka, Japan) was cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum (FCS). A human cervical carcinoma cell line (HeLa) (Riken Cell Bank, Tsukuba, Japan) was cultured as described in previous studies [10, 14]. A cattle XLF gene (NM_001075393.1) with an artificial EcoRI site at the 5′ end and BamHI site at the 3′ end was synthesized. The fragment was confirmed by sequencing and ligated to the EcoRI and BamHI sites of the pEYFP-C1 vector to give the in-frame fusion gene. pEYFP-cattle XLF, pEYFP-cattle XLF (162–299), pEYFP-cattle XLF (162–287) or pEYFP-C1 was transient transfected in cells using FuGene HD (Promega, Madison, WI, U.S.A.) according to the manufacturer’s protocol. The cells were cultured for 2 days and then monitored under an FV300 confocal laser scanning microscope (Olympus, Tokyo, Japan) as previously described [9, 11, 12].
Immunoblotting: The extraction of total lysates and Western blot analysis were conducted based on the previous methods [11, 13]. The blocking step was modified. The membranes were blocked in Blocking One (Nacalai Tesque, Kyoto, Japan) for 30 min. The following antibodies were used: a rabbit anti-XLF polyclonal antibody (A300-730A) (Bethyl Laboratories, Montgomery, TX, U.S.A.), a rabbit anti-GFP polyclonal antibody (FL) (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.), a mouse anti-Ku70 monoclonal antibody (N3H10) (NeoMarkers, Fremont, CA, U.S.A.) or a mouse anti-β-actin monoclonal antibody (Sigma, St. Louis, MO, U.S.A.). Three antibodies (i.e., anti-XLF antibody, anti-Ku70 antibody and anti-GFP antibody) were diluted in Signal Enhancer HIKARI (Nacalai Tesque), respectively. The binding to each protein was visualized using a Select Western blotting detection system (GE Healthcare Bio-Sci. Corp. Piscataway, NJ, U.S.A.), in accordance with the manufacturer’s instructions.
Immunofluorescence staining: Immunofluorescence staining was conducted as previously described [9, 11]. Briefly, the fixed cells were blocked for 10 min using a blocking solution and then incubated for 30 min at room temperature with a mouse anti-γH2AX monoclonal antibody (JBW301) (Upstate Biotechnology Inc., Charlottesville, VA, U.S.A.) or a rabbit anti-XLF polyclonal antibody (X4754) (Sigma). After washing with PBS, detection of each protein was performed using Alexa fluor 568-conjugated secondary antibodies (Molecular Probes, Eugene, OR, U.S.A.).
Local DNA damage induction using laser and cell imaging: Local DNA damage induction using laser and cell imaging was conducted as described previously [9, 11,12,13]. Briefly, a 5–30% power scan (for 1 sec) from a 405 nm laser was used to induce local DSBs. Images of living cells or fixed cells expressing EYFP-tagged cattle proteins or EYFP alone were obtained using an FV300 confocal scanning laser microscopy system (Olympus).
RESULTS
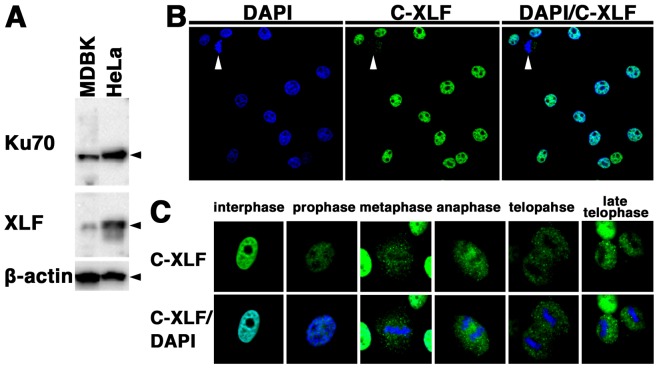
Expression and localization of cattle XLF in cattle cells: We examined the expression and subcellular localization of XLF in cattle cells. First, we examined the expression of XLF and Ku70 in the cattle cell line MDBK and the human cell line HeLa by Western blot analysis using the anti-XLF antibody and anti-Ku70 antibody. As shown in Fig. 1A, a signal of cattle XLF as well as human XLF was detected. In addition, we detected Ku70 in both MDBK and HeLa cells. These results demonstrate that the core NHEJ factors, XLF and Ku70, are expressed in cattle cells.
Fig. 1.
Expression and localization of XLF in cattle cells. (A) Total cell lysates from each cell line (MDBK, 50 µg; HeLa, 10 µg) were analyzed by Western blotting using an anti-XLF antibody, an anti-Ku70 antibody or an anti-β-actin antibody. (B, C) Subcellular localization of XLF in cattle (MDBK) cells during the cell cycle. The cells were fixed and stained with an anti-XLF antibody. Nuclear DNA was counterstained with DAPI. The stained cells were analyzed by confocal laser microscopy. Arrowheads indicate the mitotic phase cells (B). The images shown are a representative example for interphase cells or mitotic phase cells (C).
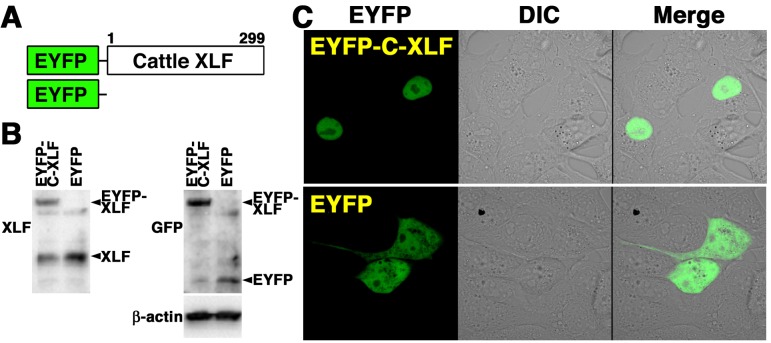
To elucidate the localization of XLF in cattle cells, we studied the distribution of XLF by confocal laser microscopy (Fig. 1B and 1C). Indirect immunofluorescence staining using the anti-XLF antibody showed that fluorescence was detected in the nucleoplasm of MDBK cells during the interphase. On the other hand, the fluorescence was detected throughout the cytoplasm of MDBK cells during the mitotic phase, but not in the condensed chromosomes of the mitotic cells. These observations indicate that the localization of cattle XLF changes dynamically during the cell cycle. To clarify the localization of XLF in living cattle cells during the interphase, we examined the expression and localization of EYFP-cattle XLF in MDBK cells. We generated cells transiently expressing EYFP-cattle XLF in MDBK cells. The expression vector pEYFP-C1 containing cattle XLF (pEYFP-cattle XLF) was transfected into MDBK cells (Fig. 2A). As shown in Fig. 2B, a signal of EYFP-cattle XLF was detected in the transfectants by Western blot analysis using the anti-XLF antibody and anti-GFP antibody. By confocal laser microscopy, we clarified that EYFP-cattle XLF was localized in the nuclei of living interphase cells in EYFP-cattle XLF transfectants (Fig. 2C). Expectedly, in EYFP transfectants, we confirmed that EYFP was distributed throughout the cell excluding the nucleolus (Fig. 2C).
Fig. 2.
Localization of EYFP-cattle XLF in living cattle cells. (A) Schematics of EYFP-cattle XLF chimeric protein and control protein (EYFP). (B) Extracts from cattle (MDBK) cells transiently expressing the EYFP-cattle XLF or EYFP prepared and subjected to Western blotting using the anti-XLF, anti-GFP or anti-β-actin antibody. (C) Imaging of living EYFP-cattle XLF-transfected cells. Living MDBK cells transiently expressing EYFP-cattle XLF or EYFP were analyzed by confocal laser microscopy. EYFP images for the same cells are shown alone (left panel) or merged (right panel) with differential interference contrast images (DIC) (center panel).
EYFP-cattle XLF accumulates quickly at DSBs induced by laser microirradiation: We examined whether EYFP-cattle XLF accumulates quickly at the 405 nm laser-induced DSB sites (Fig. 3A). As shown in Fig. 3B, we observed that EYFP-cattle XLF accumulated at the microirradiated sites in the living cattle cells. Next, we investigated whether cattle XLF accumulated at 405 nm laser-induced DSB sites by immunostaining with an antibody that detects γH2AX. As shown in Fig. 3C, EYFP-cattle XLF colocalized with the DSB marker γH2AX at microirradiated sites in MDBK cells. Next, we carried out time-lapse imaging of EYFP-cattle XLF-transfected MDBK cells. As shown in Fig. 3D, we observed EYFP-cattle XLF accumulation at the microirradiated sites 5 sec after irradiation. In EYFP-cattle XLF-transfected cells, the intensity of the EYFP signal increased quickly at the microirradiated sites. These results reveal that after irradiation, EYFP-cattle XLF quickly accumulates and formes foci at laser-induced DSBs in living cells.
Fig. 3.
EYFP-cattle XLF accumulated quickly at DSBs induced by laser microirradiation. (A) The localization and accumulation of EYFP-cattle XLF at DSBs induced by 405 nm laser irradiation were examined. (B) Imaging of living EYFP-cattle XLF-transfected MDBK cells before (upper panel) and at 1 min after (lower panel) microirradiation. Left panel, EYFP-cattle XLF; right panel, differential interference contrast images (DIC). Arrowheads indicate the microirradiated sites. (C) Immunostaining of microirradiated EYFP-cattle XLF-transfected cells with anti-γH2AX antibody. The cells were fixed and stained with the anti- γH2AX antibody at 5 min postirradiation. Left panel, EYFP-cattle XLF; center panel, γH2AX image; right panel, merged image. (D) Time-dependent EYFP-cattle XLF accumulation in living cells (5-120 sec) after irradiation. Upper panel, EYFP-cattle XLF; lower panel, differential interference contrast images (DIC).
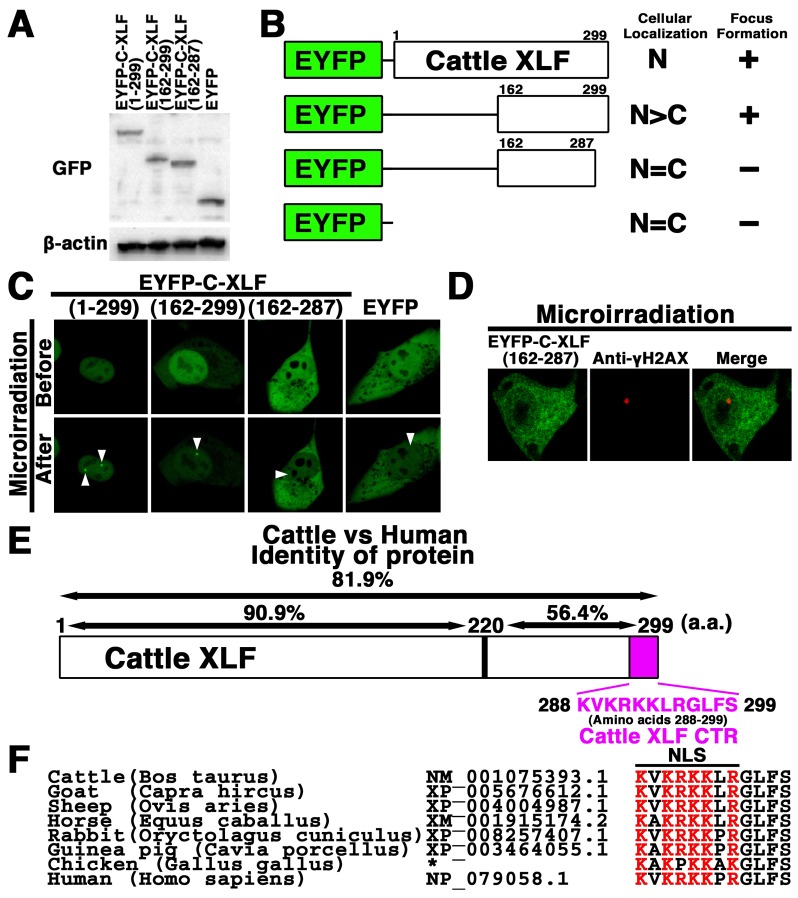
The C-terminal region (CTR) of cattle XLF is essential for the nuclear localization and recruitment of XLF to DSBs in cattle cells: To determine the region essential for nuclear localization of cattle XLF, we investigated the localization of cattle XLF and its mutant. Firstly, the pEYFP-cattle XLF and its mutants were transfected into MDBK cells. As shown in Fig. 4A, a signal of each EYFP-cattle XLF mutant was detected in the extracts of each transfectant by Western blot analysis using the anti-GFP antibody. By confocal laser microscopy, we confirmed that EYFP-cattle XLF was localized in the nuclei of living interphase cells. We observed that N-terminal deletion mutant EYFP-cattle XLF (162–299) localized predominantly in the nuclei, whereas EYFP-cattle XLF (162–287) as well as EYFP was distributed throughout the cell excluding the nucleolus in MDBK cells (Figs. 2C, 4B and 4C). These results indicate that 12 C-terminal amino acids (amino acids 288–299) of cattle XLF are vital for the nuclear localization of XLF in cattle cells (Fig. 4E). To identify which region of cattle XLF is essential for its accumulation at DSBs in vivo, we tested whether the XLF mutant proteins could be recruited to DSBs induced by microirradiation. We observed that a N-terminal deletion mutant EYFP-cattle XLF (162–299) as well as the wild type EYFP-cattle XLF, accumulated at the DSBs sites in the living MDBK cells (Fig. 3C and data not shown). On the other hand, the mutant protein EYFP-cattle XLF (162–287) failed to accumulate at the DSBs sites, which was detected by the DSB marker γH2AX (Fig. 4C and 4D), indicating that deletion of the C-terminal end 12 amino acids abolished the recruitment of cattle XLF to DSBs. The cattle and human XLF proteins are 81.9% identical at the amino acid level, whereas the C-terminal domain (amino acids 220–299) of cattle XLF retains only 56.4% identity to human (Fig. 4E). Interestingly, we confirmed that the basic amino acids in the CTR of XLF are evolutionarily highly conserved among humans and domestic animal species, e.g., cattle, goats, horses and avian, but not in yeast (Fig. 4F and data not shown), which strongly suggests the biological significance of the XLF CTR in domestic animals.
Fig. 4.
The C-terminal region (CTR) is vital for the nuclear localization and recruitment of cattle XLF to DSBs in vivo. (A) Extracts from cattle (MDBK) cells transiently expressing the indicated cattle XLF deletions were prepared and subjected to Western blotting using the anti-GFP or anti-β-actin antibody. (B, C) Identification of essential domain of cattle XLF for nuclear localization and for accumulation at DSBs. EYFP-cattle XLF mutants were expressed in cattle (MDBK) cells. The localization and accumulation of the chimeric proteins at laser-induced DSBs were investigated via live cell imaging. The results are summarized on the right: Cellular localization (N, nucleus; C, cytoplasm) and formation of focus (+, accumulated at microirradiated sites; -, not accumulated at microirradiated sites). Arrowheads indicate the microirradiated sites (C). (D) Immunostaining of microirradiated EYFP-cattle XLF (162–287)-transfected cells with anti-γH2AX antibody. The cells were fixed and stained with the anti-γH2AX antibody at 5 min postirradiation. Left panel, EYFP-cattle XLF (162-287); center panel, γH2AX image; right panel, merged image. (E) Identity between the cattle XLF and human XLF at the amino acid level and the CTR of cattle XLF (amino acids 288–299). (F) The alignment of the primary sequence among homologous XLF proteins. For comparison, the basic (red) or non-basic residues (black) are shown in different colors. The GeneBank accession number for each sequence is mentioned. *, The sequence of CTR of chicken XLF is from Reference [1].
DISCUSSION
To develop next-generation chemotherapeutics for cancer and other disease is important to clarify the molecular mechanisms of C-NHEJ. Human XLF is the most recently identified core NHEJ factor, and it appears to play essential roles in C-NHEJ [1]. Expectedly, XLF-deficient cells derived from human patients and from knockout mice show ionizing radiation sensitivity [3, 15]. In addition, siRNA-mediated downregulation of XLF in human cell lines leads to radiosensitivity and impaired NHEJ. [1]. Homologues of the XLF gene were predicted in several eukaryotic organisms [1]. On the other hand, the expression, function and regulation mechanism of XLF have not been elucidated in animal species other than mice and humans [1, 3, 11, 16, 21]. Domestic cattle are important domestic animals as livestock and draft animals in not only Japan, but also many countries. Recently, cattle have been an ideal animal model for assessing chronic radiation exposure [5, 20]. However, the molecular mechanism of C-NHEJ is still unknown in cattle cells. In this study, we examined the expression and subcellular localization of cattle XLF and its mutants in cattle cell line MDBK. We found that XLF as well as other core NHEJ protein Ku70 is expressed in cattle cells, and the localization of cattle XLF changes dynamically during the cell cycle. In addition, XLF might play a vital role in the repair of DSB immediately after microirradiation of cattle cells. Moreover, our data showed that the CTR of cattle XLF is vital for the nuclear localization of XLF and for the accumulation of XLF at DSBs in vivo. These findings suggest that the mechanisms regulating of the localization and recruitment to DSBs play a key role in the function of cattle XLF.
Cattle XLF (NM_001075393.1) as well as goat XLF (XP_005676612.1) and sheep XLF (XP_004004987.1) is a 299-amino acid protein. The cattle and goat XLF genes are 94.6% identical at the amino acid level. In addition, the cattle and sheep XLF genes are 94.3% identical at the amino acid level. On the other hand, the cattle XLF retains only 81.9% identity to human. Human XLF is a 299-amino acid protein, which contains an N-terminal head domain (amino acids 1–141), a coiled-coil central domain (amino acids 142–230) and a non structured C-terminal domain (amino acids 231–299) [1, 2, 17]. Comparison with other eukaryotic homologues shows a high degree of sequence similarity within the 220 N-terminal amino acids, while the C-terminal domain (amino acids 225–299) is less conserved [1, 2, 17]. On the basis of experimental findings, there are some reports concerning the role of the C-terminal domain of XLF in humans, but not in animals including cattle. Yano et al. reported that a 10-amino-acid deletion at the C-terminal end completely abolishes the Ku-XLF interaction and the accumulation of XLF at DSBs [21]. On the other hand, Malivert et al. have reported that the C-terminal end (amino acids 231–299) of human XLF is dispensable for DNA repair in vivo [17]. In this study, our data showed that a 12-amino-acid deletion at the C-terminal end abolishes the accumulation of cattle XLF at DSBs. Altogether, we conclude that the XLF CTR is important for the accumulation of XLF at DSBs in both human and cattle cells, although the role of the C-terminal region of XLF remains controversial in human cells.
It was demonstrated that there is a general absence of conservation in the 75 C-terminal amino acids among humans and other species, although the extreme C terminus of XLF contains a small conserved basic cluster, which was proposed as a putative NLS (KRKK) [1, 2]. Our data revealed experimentally that the CTR of cattle XLF is critical for the nuclear localization of XLF and recruitment to DSBs, whereas the N-terminal domain is not essential. In addition, basic amino acids in the CTR of cattle XLF are evolutionarily conserved among CTR of domestic animals, which suggests the common biological significance of the XLF CTR in domestic animals. We consider that there is only one NLS (KVKRKKLR) in cattle XLF, and the NLS is a classical monopartite NLS having a single cluster of basic amino acid residues. We speculate that the C-terminus of 75 amino acids is important for a specific function in each species, whereas the XLF CTR is critical for the regulation of common functions in domestic animals. Further studies are needed to clarify this.
In conclusion, we showed that XLF is expressed in cattle cells and the localization of cattle XLF changes dynamically during the cell cycle. In addition, our data showed that the localization and recruitment of cattle XLF to DSB sites at an early stage following irradiation are dependent on the CTR. These basic informations might be useful to develop the molecular-targeting therapeutic drug taking XLF as a target molecule for human and domestic animals. Further studies to elucidate the mechanisms regulating cattle XLF at DSBs will lead to a better understanding of the physiological function of XLF not only in cattle cells, but also in cells of human and other domestic animals. Inherited mutations of core C-NHEJ factors (e.g., DNA-PKcs, DNA ligase IV and XLF), have been discovered in humans [6]. On the other hand, inherited mutations of the DNA-PKcs, which cause SCID, have been identified in not only humans, but also domestic animals, i.e., mice, horses or dogs [4]. Therefore, further comparative studies might provide available information for the development of new clinical medicines and new chemoradiotherapies for humans and domestic animals including cattle.
REFERENCES
- 1.Ahnesorg P., Smith P., Jackson S. P.2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124: 301–313. doi: 10.1016/j.cell.2005.12.031 [DOI] [PubMed] [Google Scholar]
- 2.Andres S. N., Modesti M., Tsai C. J., Chu G., Junop M. S.2007. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol. Cell 28: 1093–1101. doi: 10.1016/j.molcel.2007.10.024 [DOI] [PubMed] [Google Scholar]
- 3.Buck D., Malivert L., de Chasseval R., Barraud A., Fondanèche M. C., Sanal O., Plebani J. L., Stéphan M., Hufnagel F., le Deist A., Fischer A., Durandy J. P., de Villartay A., Revy P.2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124: 287–299. doi: 10.1016/j.cell.2005.12.030 [DOI] [PubMed] [Google Scholar]
- 4.Ding Q., Bramble L., Yuzbasiyan-Gurkan V., Bell T., Meek K.2002. DNA-PKcs mutations in dogs and horses: allele frequency and association with neoplasia. Gene 283: 263–269. doi: 10.1016/S0378-1119(01)00880-0 [DOI] [PubMed] [Google Scholar]
- 5.Fukuda T., Kino Y., Abe Y., Yamashiro H., Kuwahara Y., Nihei H., Sano Y., Irisawa A., Shimura T., Fukumoto M., Shinoda H., Obata Y., Saigusa S., Sekine T., Isogai E., Fukumoto M.2013. Distribution of artificial radionuclides in abandoned cattle in the evacuation zone of the Fukushima Daiichi nuclear power plant. PLoS ONE 8: e54312. doi: 10.1371/journal.pone.0054312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeggo P.2010. The role of the DNA damage response mechanisms after low-dose-radiation exposure and a consideration of potentially sensitive individuals. Radiat. Res. 174: 825–832. doi: 10.1667/RR1844.1 [DOI] [PubMed] [Google Scholar]
- 7.Khanna K. K., Jackson S. P.2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27: 247–254. doi: 10.1038/85798 [DOI] [PubMed] [Google Scholar]
- 8.Koike M.2002. Dimerization, translocation and localization of Ku70 and Ku80 proteins. J. Radiat. Res. (Tokyo) 43: 223–236. doi: 10.1269/jrr.43.223 [DOI] [PubMed] [Google Scholar]
- 9.Koike M., Koike A.2008. Accumulation of Ku80 proteins at DNA double-strand breaks in living cells. Exp. Cell Res. 314: 1061–1070. doi: 10.1016/j.yexcr.2007.11.014 [DOI] [PubMed] [Google Scholar]
- 10.Koike M., Shiomi T., Koike A.2001. Dimerization and nuclear localization of Ku proteins. J. Biol. Chem. 276: 11167–11173. doi: 10.1074/jbc.M010902200 [DOI] [PubMed] [Google Scholar]
- 11.Koike M., Yutoku Y., Koike A.2011. Accumulation of Ku70 at DNA double-strand breaks in living epithelial cells. Exp. Cell Res. 317: 2429–2437. doi: 10.1016/j.yexcr.2011.07.018 [DOI] [PubMed] [Google Scholar]
- 12.Koike M., Yutoku Y., Koike A.2011. Accumulation of p21 proteins at DNA damage sites independent of p53 and core NHEJ factors following irradiation. Biochem. Biophys. Res. Commun. 412: 39–43. doi: 10.1016/j.bbrc.2011.07.032 [DOI] [PubMed] [Google Scholar]
- 13.Koike M., Yutoku Y., Koike A.2013. The C-terminal region of Rad52 is essential for Rad52 nuclear and nucleolar localization, and accumulation at DNA damage sites immediately after irradiation. Biochem. Biophys. Res. Commun. 435: 260–266. doi: 10.1016/j.bbrc.2013.04.067 [DOI] [PubMed] [Google Scholar]
- 14.Koike M., Awaji T., Kataoka M., Tsujimoto G., Kartasova T., Koike A., Shiomi T.1999. Differential subcellular localization of DNA-dependent protein kinase components Ku and DNA-PKcs during mitosis. J. Cell Sci. 112: 4031–4039. [DOI] [PubMed] [Google Scholar]
- 15.Li G., Alt F. W., Cheng H. L., Brush J. W., Goff P. H., Murphy M. M., Franco S., Zhang Y., Zha S.2008. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Mol. Cell 31: 631–640. doi: 10.1016/j.molcel.2008.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahaney B. L., Meek K., Lees-Miller S. P.2009. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 417: 639–650. doi: 10.1042/BJ20080413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malivert L., Callebaut I., Rivera-Munoz P., Fischer A., Mornon J. P., Revy P., de Villartay J. P.2009. The C-terminal domain of Cernunnos/XLF is dispensable for DNA repair in vivo. Mol. Cell. Biol. 29: 1116–1122. doi: 10.1128/MCB.01521-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mari P. O., Florea B. I., Persengiev S. P., Verkaik N. S., Brug-genwirth H. T., Modesti M., Giglia-Mari G., Bezstarosti K., Demmers J. A., Luider T. M., Houtsmuller A. B., Gent D. C.2006. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. U.S.A. 103: 18597–18602. doi: 10.1073/pnas.0609061103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei L., Lan L., Hong Z., Yasui A., Ishioka C., Chiba N.2008. Rapid recruitment of BRCA1 to DNA double-strand breaks is dependent on its association with Ku80. Mol. Cell. Biol. 28: 7380–7393. doi: 10.1128/MCB.01075-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashiro H., Abe Y., Fukuda T., Kino Y., Kawaguchi I., Kuwahara Y., Fukumoto M., Takahashi S., Suzuki M., Kobayashi J., Uematsu E., Tong B., Yamada T., Yoshida S., Sato E., Shinoda H., Sekine T., Isogai E., Fukumoto M.2013. Effects of radioactive caesium on bull testes after the Fukushima nuclear plant accident. Sci. Rep. 3: 2850. doi: 10.1038/srep02850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yano K., Morotomi-Yano K., Lee K. J., Chen D. J.2011. Functional significance of the interaction with Ku in DNA double-strand break recognition of XLF. FEBS Lett. 585: 841–846. doi: 10.1016/j.febslet.2011.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]