Abstract
Horizontal transmission is recognized as a major infection route for bovine leukemia virus (BLV), and cattle with high viral loads are considered to be a major infectious source in a herd. However, a correlation between viral loads and the risk of infection has been insufficient to use as a foundation for BLV control strategies. In this report, we examined the epidemiology of BLV infection and the infectious source in a local area. In 2013–2014, BLV infection was investigated in 1,823 cattle from 117 farms in two adjacent districts, Miyazaki, Japan. Seropositive samples for BLV were detected with 88 cattle and in 14 farms. Phylogenetic analysis revealed that 94% of the isolates clustered into genotype I and the remaining isolate into genotype III. Among genotype I, genetically distinct strains were spread at each farm, and cattle infected with less than 3 copies/100 cells did not transmit BLV to other cattle for more than thirty months. This is the first report of concrete data of viral load in relation to viral horizontal transmission under the field condition. The data facilitate farmers and veterinarians understanding the status of BLV infected cattle. This research contributes to BLV infection control and the development of effective BLV eradication programs.
Keywords: BLV, EBL, genotype, horizontal transmission, Japanese Black cattle
Bovine leukemia virus (BLV) is an etiological agent of enzootic bovine leukosis (EBL). Although most of infected cattle are asymptomatic carriers of the virus, 1–5% of them develop fatal lymphosarcoma several years after infection [4]. In addition to EBL being a life-threatening illness, cattle with lymphosarcoma are disapproved for human consumption in Japan. Therefore, EBL causes serious economic damage to the Japanese livestock industry. In many western European countries, BLV eradication has been achieved using national control measures and campaigns [1, 22]. Although the seroprevalence of BLV is high and an increase in EBL has been reported in Japan [14, 18, 19], national eradication programs have never been implemented.
BLV is transmitted predominantly by horizontal routes, including via blood-sucking flies, physical contact and blood-contaminated devices [11, 12, 23]. The beef cattle farming system in Japan is characterized by small-scale farm with small number of heads and typically densed. Blood-sucking flies are recognized as an important risk factor for BLV transmission in Japan [10, 24]. However, transmission of the virus from infected to uninfected cattle in a herd or from an affected farm to a neighboring farm remains unclear. Some local veterinarians and farmers suspect that BLV can be transmitted from one infected farm to an adjacent farm.
This study was conducted in the cities of Kawaminami and Tsuno, Miyazaki prefecture, Japan (Fig. 1). This area suffered from foot-and-mouth disease (FMD) in 2010, and all cattle in this area were culled to prevent the spread of FMD virus [20, 21]. After the FMD virus was eradicated from the area, farmers introduced Japanese Black cattle and restarted their stockbreeding in November 2010. In this study, we investigated the epidemiology of BLV infection, the viral load of infected cattle and BLV genotypes. Combining viral load information with BLV genotyping may facilitate the discovery of the probable routes and sources of infection in cattle. The results of this study will aid in the design of strategies for culling infected cattle and in the construction of effective preventive strategies against BLV.
Fig. 1.
A map of Miyazaki prefecture showing Kawaminami and Tsuno.
MATERIALS AND METHODS
Study area, blood sampling and ELISA test: This study was conducted on 117 Japanese Black cattle (Bos taurus) production farms located in the cities of Kawaminami and Tsuno, Miyazaki prefecture, Japan (Fig. 1). In this area, many of cattle were fed by loose housing system. From August 2013 to August 2014, a total of 1,823 blood samples were collected from all the breeding cattle and heifers in each farm using evacuated blood collection tubes containing EDTA. All blood samples were centrifuged (1,500 × g, 5 min), and the plasma was used for BLV enzyme-linked immunosorbent assay (ELISA) tests (JNC Co., Ltd., Tokyo, Japan), according to manufacturer’s instruction with modification. We confirmed that the detection sensitivities of this ELISA tests using plasma and serum as sample are comparable (data not shown).
DNA extraction and viral load quantification by real-time PCR analysis: Out of 1,823 blood samples, genomic DNA was extracted from 88 seropositive samples. Genomic DNA extraction from the blood samples was performed as previously described [16]. Quantitative real-time PCR analyses were performed using an Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, U.S.A.). BLV pol [5] and BoLA-DRA [7] sequence-specific primers were used to quantify proviral loads in infected cattle. The amplifications were performed in 10-µl reaction volumes containing 5 µl of SYBR Premix DimerEraser (TaKaRa Bio Inc., Otsu, Japan), 0.1 µl of ROX Reference Dye, 0.3 µl of primers (3 pmol each), 1 µl of a template DNA sample (50 ng each) and PCR-grade water. The specificity of the PCR products was confirmed by amplicon melting-curve analyses. The relative numbers of proviral and BoLA-DRA copies were calculated using the comparative threshold cycle method [13]. Proviral loads were calculated as the relative number of proviral copies divided by half the relative number of BoLA-DRA copies and represented as the number of proviral copies per 100 cells. To avoid PCR amplification failure due to viral nucleotide variations in the pol primer region, we also amplified the BLV LTR region using the primers LTR256 and LTR453 [9].
Sequencing and phylogenetic analysis of the BLV gp51 env gene: PCR amplification of the full-length BLV gp51 env gene was performed in 10-µl reaction volumes containing 1 µl of 10x Ex Taq buffer, 0.8 µl of dNTP Mix, 0.05 µl of TaKaRa Ex Taq (TaKaRa Bio Inc.), 0.5 µl of primers (5 pmol each), 1 µl of template DNA sample (50 ng each) and PCR-grade water. Primers for BLV gp51 full F (5′-CAA TCG TCG GTG GCT AGG AC-3′) and BLV gp51 full R (5′-GAG GTG AGT CTC TGA TGG CTA AG-3′) were used to amplify the gp51 env gene. The amplified fragments were treated with Illustra ExoProStar (GE Healthcare Life Sciences, Buckinghamshire, U.K.) according to the manufacturer’s instructions. The sequencing reactions were performed using one of the primers listed above together with a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), purified using a BigDye XTerminator Purification Kit (Applied Biosystems) and analyzed with an Applied Biosystems 3730 DNA Analyzer. The nucleotide sequences were merged using the EMBOSS merger program (http://emboss.bioinformatics.nl/cgi-bin/emboss/merger). An alignment of the full-length BLV gp51 env gene (906 bp) and a phylogenetic tree were constructed using MEGA6 software [27]. A phylogenetic tree was constructed using the neighbor-joining method based on Kimura’s two-parameter model.
RESULTS
The seroprevalences of BLV infection among cattle in Kawaminami and Tsuno were 3.0% (31/1013) and 7.0% (57/810), respectively (Table 1). The seroprevalence of BLV-positive farms in these cities was 9.2% (6/65) and 15.3% (8/52), respectively. The seroprevalence of BLV in both cities extremely varied among farms (2.1–100%) (Table 2). Although BLV infection was not found in many small size farms, the average seroprevalence at the BLV infected farms in small size farms was higher than in large and middle size farms (Table 3).
Table 1. The seroprevalence of BLV infection in the cities of Kawaminami and Tsuno.
| City | Number (No.) of infected /examined | |
|---|---|---|
| cattle (%) | farms (%) | |
| Kawaminami | 31/1013 (3.0) | 6/65 (9.2) |
| Tsuno | 57/810 (7.0) | 8/52 (15.3) |
| Total | 88/1823 (4.8) | 14/117 (11.9) |
Table 2. The seroprevalence and genotype of BLV in infected farms.
| Farm ID | BLV seroprevalence (infected /examined) |
BLV genotype | Subgenotype (Accession No.) |
|---|---|---|---|
| T99 | 2.1% (4/195) | I | I-6 (LC007981) |
| I-9 (LC007984) | |||
| I-16 (LC007991) | |||
| K110 | 3.7% (1/27) | I | I-8 (LC007983) |
| K8 | 4.0% (1/25) | I | I-3 (LC007995) |
| T49 | 4.5% (1/22) | - | - |
| T61 | 10.0% (1/10) | I | I-11 (LC007986) |
| T53 | 14.3% (1/7) | I | I-15 (LC007990) |
| T83 | 20.0% (1/5) | I | I-5 (LC007980) |
| K29 | 21.4% (3/14) | III | - (LC007993) |
| T69 | 34.9% (29/83) | I | I-1 (LC007977) |
| I-4 (LC007979) | |||
| I-7 (LC007982) | |||
| T118 | 52.0% (13/25) | I | I-7 (LC007982) |
| I-17 (LC007992) | |||
| K94 | 52.6% (10/19) | I | I-1 (LC007977) |
| I-13 (LC007994) | |||
| K100 | 52.9% (9/17) | I | I-2 (LC007978) |
| T42 | 58.3% (7/12) | I | I-10 (LC007985) |
| I-14 (LC007989) | |||
| K18 | 100% (7/7) | I | I-12 (LC007987) |
Table 3. The relationship between farm size and BLV seroprevalence.
| Farm size (No. of feeding cattle) | Total no. of | Seroprevalence of | Av. seroprevalence at positive farms (%) | ||
|---|---|---|---|---|---|
| farms | cattle | farms (%) | cattle (%) | ||
| <20 | 89 | 632 | 7.8 | 4.5 | 40.2 |
| 20–50 | 23 | 628 | 21.7 | 4.1 | 17.5 |
| >50 | 6 | 563 | 33.3 | 5.8 | 11.8 |
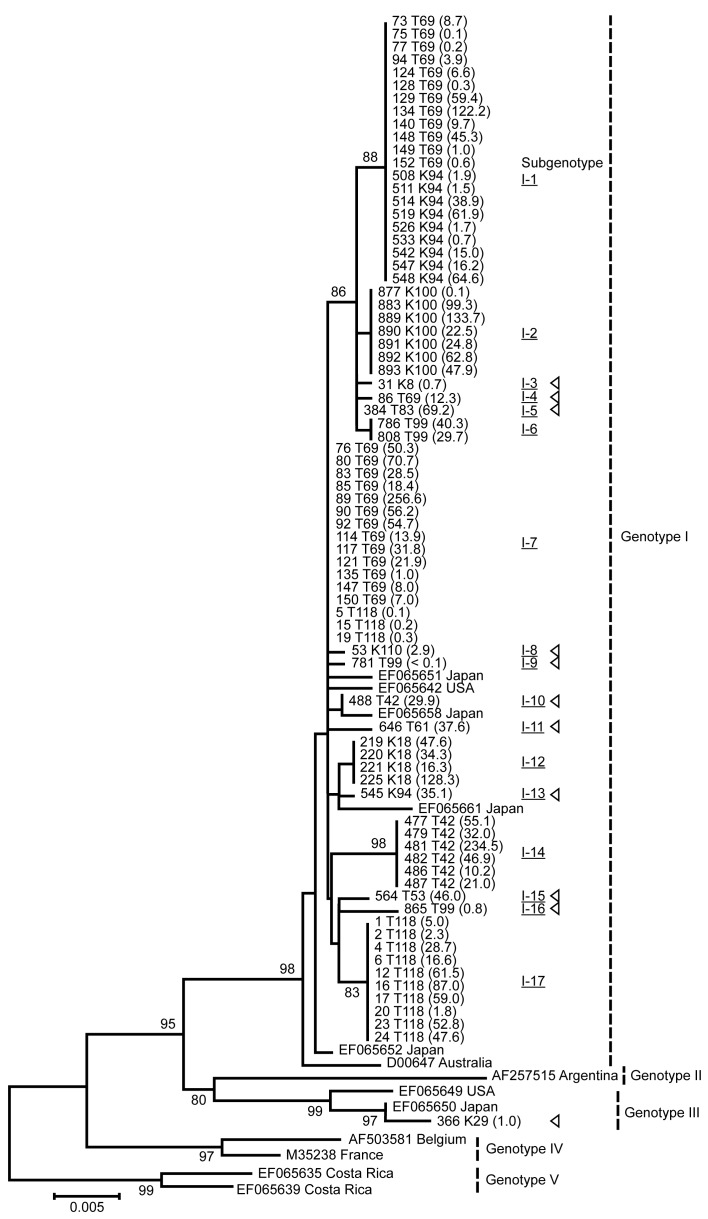
Genotypes I and III were identified in this study, and genotype I accounted for 94% (17/18) of isolates. We classified genotype I into 17 subgenotypes depending on a phylogenetic tree analysis (Fig. 2). Only 1–3 subgenotypes of BLV spread in each farm, and most BLV isolates were genetically different from farm to farm (Table 2). However, the same subgenotypes were isolated from 2 farms (subgenotype I-1 was found on T69 and K94, and I-7 was found on T69 and T118). These farms were geographically separated, and there was no interaction among the personnel of these farms. To understand the horizontal transmission occurring in the field, we quantified the viral loads and combined these data with the phylogenetic tree. Except for less than 20 months of stay or separated cattle, untransmitted strains tended to be found in cattle with less than 3 copies/100 cells (Table 4). This result indicates that cattle with less than 3 copies/100 cells were unlikely to spread the virus for about 30 months in the field. One strain (subgenotype I-14) was not amplified by the pol primer. However, the LTR primer effectively amplified this strain, and nucleotide variations were found in the pol primer region (data not shown).
Fig. 2.
A phylogenetic tree analysis of BLV isolates based on full-length gp51 env gene sequences and the viral loads in infected cattle. BLV isolates are represented by the IDs of the infected cattle and the affected farm. GenBank accession numbers of each isolate are listed in Table 2. The viral loads are given in parentheses. The reference strains are represented by GenBank accession numbers, together with the country of isolation. Subgenotypes are indicated by Roman and Arabic numerals with underline, and genotypes are indicated by Roman numeral [14]. Triangle shapes indicate the subgenotypes observed in only one cattle infected in this study. Out of 88 seropositive cattle, 11 were negative for PCR detection and not included for phylogenic analysis. Horizontal branch lengths are drawn to scale. Bootstrap support values (>80% in 1,000 replications) are shown for key nodes only.
Table 4. Details about the cattle that untransmit BLV to other cattle.
| Viral load (copies/100 cells) |
Cattle ID | BLV subgenotype |
Duration of stay (month) |
Separation |
|---|---|---|---|---|
| 69.2 | 384 T83 | I-5 | 17 | No |
| 46.0 | 564 T53 | I-15 | 10 | No |
| 37.6 | 646 T61 | I-11 | 17 | No |
| 35.1 | 545 K94 | I-13 | 11 | No |
| 29.9 | 488 T42 | I-10 | 33 | Yes |
| 12.3 | 86 T69 | I-4 | 19 | No |
| 2.9 | 53 K110 | I-8 | 36 | No |
| 1.0 | 366 K29 | III | 33 | No |
| 0.8 | 865 T99 | I-16 | 35 | No |
| 0.7 | 31 K8 | I-3 | 30 | No |
| <0.1 | 781 T99 | I-9 | 35 | No |
DISCUSSION
Both cities of Kawaminami and Tsuno suffered from FMD outbreak in late March 2010, and all the cattle and pigs in this area were culled [20, 21]. Stockbreeding was restarted in this area in November 2010. Japanese Black cattle were introduced predominantly from the southern part of Kyusyu (Kumamoto, Kagoshima, Oita and Miyazaki prefectures), and most of the introduced cattle were checked for BLV infection by ELISA. The infected cattle were immediately separated from the non-infected cattle, and only the non-infected cattle were used for breeding. Our study revealed that only 3.0% and 7.0% of cattle and 9.2% and 15.3% of farms were seropositive for BLV in Kawaminami and Tsuno, respectively. The seroprevalence of BLV was higher in Tsuno than in Kawaminami, but this result was influenced by the high seroprevalence of farm T69 in Tsuno. The result of an epidemiological study indicates that BLV screening before their introduction into a herd could not completely prevent the spread of BLV. However, it clearly lowered the prevalence of BLV infection in this area, considering the fact that more than 40% of cattle were infected in the Kyusyu area [18]. An additional testing at later time point is essential for complete prevention of the BLV infection. In small-size farms (less than 20 cattle), the average seroprevalence of cattle in seropositive farms was higher than in large- and middle-size farms. The small size farms tend to feed cattle in higher density environment, and that might increase the opportunity for contact between infected and uninfected cattle.
Previous studies have reported that genotypes I and III have been isolated primarily from Japanese cattle and that genotype I mainly prevails in Japan [2, 3, 15, 17, 24]. In this study, genotype I accounted for 94% (17/18) of isolates. This result may reflect the prevalence of genotype I in Kyusyu area, though our study was conducted in a limited area and further analyses are required. The same subgenotypes (I-1 and I-7) were isolated from different farms. Although the reason is unclear, the result indicates that these subgenotypes might be major strains of genotype I.
In Japan, blood-sucking insects are recognized as an important cause of BLV transmission route [10, 24]. The weather of Kawaminami and Tsuno located in southern part of Kyusyu is warm and humid, and there are a large number of blood-sucking flies. To prevent transmission among herds, 200 m distance has been proposed [26]. However, separating 200 m between two herds is impractical for most of Japanese farms. The result indicates that the viral transmission between two adjacent farms rarely occurs. Although the study did not investigate the number of blood-sucking flies or with or without fly net in farm, these factors could affect efficiency of BLV transmission.
In this study, some cattle were infected with more than 100 copies/100 cells (Fig. 2). Cattle infected with more than 1 copies/cell have been reported in other studies [5, 6]. However, further discussion and research are needed to clear whether BLV is integrated more than one copy per cell. Many studies have suggested that a useful strategy to limit BLV transmission within a herd is to quantify proviral loads and cull cattle with high viral loads [8, 25]. However, concrete data of viral load in relation to viral transmission have not been so far demonstrated. Our results show that more than 3 viral copies/100 cells are a risk factor for horizontal transmission of the virus in the field and it may alter depending on the environment and/or the feeding system. Detection and slaughter of all infected cattle are the fastest way to completely control BLV in a herd. However, this method is economically difficult for many farmers without any financial supports. We suggest that identification of the risk cattle, infected with more than 3 BLV copies/100 cells, and separation of those from uninfected cattle may lead to a successful control of BLV infection in a herd without significant cost. This study contributes to the development of economically feasible BLV eradication programs and control strategies.
Acknowledgments
JSPS KAKENHI Grant Numbers 26850175 and 25450468, the Ito Foundation and the Foundation for Restarting after FMD 2010 in Miyazaki Prefecture supported the research described in this study.
REFERENCES
- 1.Acaite J., Tamosiunas V., Lukauskas K., Milius J., Pieskus J.2007. The eradication experience of enzootic bovine leukosis from Lithuania. Prev. Vet. Med. 82: 83–89. doi: 10.1016/j.prevetmed.2007.05.010 [DOI] [PubMed] [Google Scholar]
- 2.Asfaw Y., Tsuduku S., Konishi M., Murakami K., Tsuboi T., Wu D., Sentsui H.2005. Distribution and superinfection of bovine leukemia virus genotypes in Japan. Arch. Virol. 150: 493–505. doi: 10.1007/s00705-004-0433-5 [DOI] [PubMed] [Google Scholar]
- 3.Balić D., Lojkić I., Periškić M., Bedeković T., Jungić A., Lemo N., Roić B., Cač Z., Barbić L., Madić J.2012. Identification of a new genotype of bovine leukemia virus. Arch. Virol. 157: 1281–1290. doi: 10.1007/s00705-012-1300-4 [DOI] [PubMed] [Google Scholar]
- 4.Burny A., Cleuter Y., Kettmann R., Mammerickx M., Marbaix G., Portetelle D., van den Broeke A., Willems L., Thomas R.1988. Bovine leukaemia: facts and hypotheses derived from the study of an infectious cancer. Vet. Microbiol. 17: 197–218. doi: 10.1016/0378-1135(88)90066-1 [DOI] [PubMed] [Google Scholar]
- 5.Gillet N. A., Gutiérrez G., Rodriguez S. M., de Brogniez A., Renotte N., Alvarez I., Trono K., Willems L.2013. Massive depletion of bovine leukemia virus proviral clones located in genomic transcriptionally active sites during primary infection. PLoS Pathog. 9: e1003687. doi: 10.1371/journal.ppat.1003687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutiérrez G., Alvarez I., Merlini R., Rondelli F., Trono K.2014. Dynamics of perinatal bovine leukemia virus infection. BMC Vet. Res. 10: 82. doi: 10.1186/1746-6148-10-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jimba M., Takeshima S. N., Matoba K., Endoh D., Aida Y.2010. BLV-CoCoMo-qPCR: quantitation of bovine leukemia virus proviral load using the CoCoMo algorithm. Retrovirology 7: 91. doi: 10.1186/1742-4690-7-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juliarena M. A., Gutierrez S. E., Ceriani C.2007. Determination of proviral load in bovine leukemia virus-infected cattle with and without lymphocytosis. Am. J. Vet. Res. 68: 1220–1225. doi: 10.2460/ajvr.68.11.1220 [DOI] [PubMed] [Google Scholar]
- 9.Ikebuchi R., Konnai S., Shirai T., Sunden Y., Murata S., Onuma M., Ohashi K.2011. Increase of cells expressing PD-L1 in bovine leukemia virus infection and enhancement of anti-viral immune responses in vitro via PD-L1 blockade. Vet. Res. 42: 103. doi: 10.1186/1297-9716-42-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi S., Tsutsui T., Yamamoto T., Hayama Y., Kameyama K., Konishi M., Murakami K.2010. Risk factors associated with within-herd transmission of bovine leukemia virus on dairy farms in Japan. BMC Vet. Res. 6: 1. doi: 10.1186/1746-6148-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohara J., Konnai S., Onuma M.2006. Experimental transmission of bovine leukemia virus in cattle via rectal palpation. Jpn. J. Vet. Res. 54: 25–30. [PubMed] [Google Scholar]
- 12.Lassauzet M. L., Thurmond M. C., Johnson W. O., Stevens F., Picanso J. P.1991. Factors associated with transmission of bovine leukemia virus by contact in cows on a California dairy. Am. J. Epidemiol. 133: 164–176. [DOI] [PubMed] [Google Scholar]
- 13.Livak K. J., Schmittgen T. D.2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25: 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 14.MAFF2014. Annual statistics of notifiable animal infectious diseases (1937–2013) (in Japanese).<http://www.maff.go.jp/j/syouan/douei/kansi_densen/pdf/h25_ruinenn_todokede.pdf>Accessed 9 January 2015.
- 15.Matsumura K., Inoue E., Osawa Y., Okazaki K.2011. Molecular epidemiology of bovine leukemia virus associated with enzootic bovine leukosis in Japan. Virus Res. 155: 343–348. doi: 10.1016/j.virusres.2010.11.005 [DOI] [PubMed] [Google Scholar]
- 16.Mekata H., Sekiguchi S., Konnai S., Kirino Y., Honkawa K., Nonaka N., Horii Y., Norimine J.2015. Evaluation of the natural perinatal transmission of bovine leukaemia virus. Vet. Rec. 176: 254. doi: 10.1136/vr.102464 [DOI] [PubMed] [Google Scholar]
- 17.Moratorio G., Obal G., Dubra A., Correa A., Bianchi S., Buschiazzo A., Cristina J., Pritsch O.2010. Phylogenetic analysis of bovine leukemia viruses isolated in South America reveals diversification in seven distinct genotypes. Arch. Virol. 155: 481–489. doi: 10.1007/s00705-010-0606-3 [DOI] [PubMed] [Google Scholar]
- 18.Murakami K., Kobayashi S., Konishi M., Kameyama K., Tsutsui T.2013. Nationwide survey of bovine leukemia virus infection among dairy and beef breeding cattle in Japan from 2009–2011. J. Vet. Med. Sci. 75: 1123–1126. doi: 10.1292/jvms.12-0374 [DOI] [PubMed] [Google Scholar]
- 19.Murakami K., Kobayashi S., Konishi M., Kameyama K., Yamamoto T., Tsutsui T.2011. The recent prevalence of bovine leukemia virus (BLV) infection among Japanese cattle. Vet. Microbiol. 148: 84–88. doi: 10.1016/j.vetmic.2010.08.001 [DOI] [PubMed] [Google Scholar]
- 20.Muroga N., Hayama Y., Yamamoto T., Kurogi A., Tsuda T., Tsutsui T.2012. The 2010 foot-and-mouth disease epidemic in Japan. J. Vet. Med. Sci. 74: 399–404. doi: 10.1292/jvms.11-0271 [DOI] [PubMed] [Google Scholar]
- 21.Nishiura H., Omori R.2010. An epidemiological analysis of the foot-and-mouth disease epidemic in Miyazaki, Japan, 2010. Transbound. Emerg. Dis. 57: 396–403. doi: 10.1111/j.1865-1682.2010.01162.x [DOI] [PubMed] [Google Scholar]
- 22.Nuotio L., Rusanen H., Sihvonen L., Neuvonen E.2003. Eradication of enzootic bovine leukosis from Finland. Prev. Vet. Med. 59: 43–49. doi: 10.1016/S0167-5877(03)00057-6 [DOI] [PubMed] [Google Scholar]
- 23.Ohshima K., Okada K., Numakunai S., Yoneyama Y., Sato S., Takahashi K.1981. Evidence on horizontal transmission of bovine leukemia virus due to blood-sucking tabanid flies. Jpn. J. Vet. Sci. 43: 79–81. doi: 10.1292/jvms1939.43.79 [DOI] [PubMed] [Google Scholar]
- 24.Ooshiro M., Konnai S., Katagiri Y., Afuso M., Arakaki N., Tsuha O., Murata S., Ohashi K.2013. Horizontal transmission of bovine leukemia virus from lymphocytotic cattle, and beneficial effects of insect vector control. Vet. Rec. 173: 527. doi: 10.1136/vr.101833 [DOI] [PubMed] [Google Scholar]
- 25.Rodríguez S. M., Florins A., Gillet N., de Brogniez A., Sánchez-Alcaraz M. T., Boxus M., Boulanger F., Gutiérrez G., Trono K., Alvarez I., Vagnoni L., Willems L.2011. Preventive and therapeutic strategies for bovine leukemia virus: lessons for HTLV. Viruses 3: 1210–1248. doi: 10.3390/v3071210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shettigara P. T., Samagh B. S., Lobinowich E. M.1989. Control of bovine leukemia virus infection in dairy herds by agar gel immunodiffusion test and segregation of reactors. Can. J. Vet. Res. 53: 108–110. [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]