Abstract
To elucidate the mechanisms of DNA repair pathway is critical for developing next-generation radiotherapies and chemotherapeutic drugs for cancer. Ionizing radiation and many chemotherapeutic drugs kill tumor cells mainly by inducing DNA double-strand breaks (DSBs). The classical nonhomologous DNA-end joining (NHEJ) (C-NHEJ) pathway repairs a predominant fraction of DSBs in mammalian cells. The C-NHEJ pathway appears to start with the binding of Ku (heterodimer of Ku70 and Ku80) to DNA break ends. Therefore, recruitment of Ku to DSB sites might play a critical role in regulating NHEJ activity. Indeed, human Ku70 and Ku80 localize in the nuclei and accumulate at microirradiated DSB sites. However, the localization and regulation mechanisms of Ku70 and Ku80 homologues in animal models, such as mice and other species, have not been elucidated in detail, particularly in cells immediately after microirradiation. Here, we show that EYFP-tagged mouse Ku70 localizes in the interphase nuclei of mouse fibroblasts and epithelial cells. Furthermore, our findings indicate that EYFP-mouse Ku70 accumulates with its heterodimeric partner Ku80 immediately at laser-microirradiated DSB sites. We also confirmed that the structure of Ku70 nuclear localization signal (NLS) is highly conserved among various rodent species, such as the mouse, rat, degu and ground squirrel, supporting the idea that NLS is important for the regulation of rodent Ku70 function. Collectively, these results suggest that the mechanisms of regulating the localization and accumulation of Ku70 at DSBs might be well conserved between the mouse and human species.
Keywords: DNA damage, Ku70, Ku80, mouse, nuclear localization
Ionizing radiation and many chemotherapeutic drugs, which are used in the treatment of animal and human cancer, kill tumor cells mainly by inducing DNA double-strand breaks (DSBs). It is critical to elucidate the molecular mechanisms of DSB repair pathway in order to develop next-generation radiotherapies and chemotherapeutic drugs for cancer. The classical nonhomologous DNA-end joining (NHEJ) (C-NHEJ) pathway repairs a predominant fraction of DSBs in animal and human cells [2, 18]. The C-NHEJ pathway might start with the binding of Ku (heterodimer of Ku70 and Ku80) to DSB ends.
Human Ku is a heterodimeric nuclear protein (Ku70/Ku80) originally discovered by Mimori et al. as an autoantigen that was recognized by the sera of a Japanese patient with an autoimmune disease [20]. Previously, we showed that the Ku70 gene is localized in mouse chromosome 15 and rat chromosome 7, and the Ku80 gene is localized in mouse chromosome 1 and rat chromosome 9 [16]. Furthermore, both genes were mapped to a region of conserved linkage homology among 5 species, i.e., the Chinese hamster, human, mouse, rat and Syrian hamster [6, 15, 16], suggesting that Ku is important in a function common among these species.
Previously, we reported that Ku70 and Ku80 are mainly localized in the interphase nuclei of normal human diploid lung fibroblasts, e.g., TIG-3 and MRC-5, and human cancer cells, e.g., HeLa and MCF-7 cells [17]. Moreover, the nuclear localization of Ku70 and Ku80 is, at least in part, regulated by their corresponding nuclear localization signals (NLSs) in human cells [13, 14]. On the other hand, it was reported that rodent Ku70 and Ku80 are mainly localized in the cytoplasm [21, 24].
In addition to Ku70 and Ku80, C-NHEJ requires other core factors, i.e., Artemis, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), DNA ligase IV, XLF and XRCC4 [2, 18]. Human Ku70 and Ku80 accumulate at laser-induced DSB sites immediately following irradiation, and these are essential for the accumulation of some core C-NHEJ factors, i.e., DNA-PKcs, XLF and XRCC4, but not Artemis and an HR-related protein (Rad52), at DSB sites [7, 9, 12, 18, 19, 22]. Hence, Ku70 and Ku80 at DSB sites might, at least in part, provide a platform for the selective recruitment of some core C-NHEJ factors. It is also known that Ku is the DSB-sensor protein, which recognizes and binds to the end of DSB, maintaining the stability of the ends of a broken DNA molecule [5].
The localization and function of Ku70 and Ku80 at DSB sites might play a critical role in regulating NHEJ [6, 18]. Thus, it is important to elucidate the localization and accumulation mechanisms of Ku70 and Ku80 at DSB sites. Recent live cell imaging studies using laser irradiation to induce DNA damage and GFP-tagged human proteins have thrown a new light on the order of the recruitment of core C-NHEJ factors to DSB sites [7, 9, 18, 19, 22], whereas there is no report concerning the localization and function of Ku70 homologues at DSB sites in living cells of animal species including mice other than humans. In this work, we examined the expression and subcellular localization of mouse Ku70 in mouse fibroblasts and epithelial cells. We also examined whether mouse Ku70 accumulates at DNA damage sites immediately after laser-microirradiation.
MATERIALS AND METHODS
Cell cultures and plasmid transfections: Ku70-deficient murine lung epithelial (MLE) cell lines established from Ku70 −/− mice and control mice (Ku70 +/−), a human cervical carcinoma cell line (HeLa; Riken Cell Bank, Tsukuba, Japan) and a mouse embryonic fibroblast cell line (NIH3T3; Riken Cell Bank) were cultured as described in previous studies [10, 11, 17]. pEYFP-mouse Ku70 or pEYFP-C1 was transiently transfected in cells using FuGene6 (Roche Diagnostics K.K., Indianapolis, IN, U.S.A.). The transfected mouse cells were cultured for 2 days and then monitored under an FV300 confocal laser scanning microscopy (CLSM) system (Olympus, Tokyo, Japan) as previously described [7].
Immunoblotting: The extraction of total lysates and Western blot analysis were carried out as described previously [7, 9, 12]. The following antibodies were used: a rabbit anti-GFP polyclonal antibody (FL; Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.), a goat anti-Ku70 polyclonal antibody (C-19; Santa Cruz Biotechnology), a goat anti-Ku80 polyclonal antibody (M-20; Santa Cruz Biotechnology) or a mouse anti-β-actin monoclonal antibody (Sigma, St. Louis, MO, U.S.A.). In accordance with the manufacturer’s instructions, the immunocomplexes formed were detected using an Advance Western blotting detection system (GE Healthcare Bio-Sci. Corp., Piscataway, NJ, U.S.A.) and visualized using the ChemiDoc XRS system (Bio-Rad, Hercules, CA, U.S.A.).
Laser microirradiation and cell imaging: Confocal images of living cells or fixed cells expressing EYFP-tagged mouse proteins or EYFP proteins alone were obtained using an FV300 CLSM system. Laser-microirradiation was carried out with an FV300 CLSM system as described previously [7, 9, 10, 12]. A 1–5% power scan (for 1 sec) from a 405 nm laser was used to induce local DSBs.
Immunofluorescence staining: Immunofluorescence staining was carried out as previously described [7, 9]. The fixed cells were first blocked for 10 min using a blocking solution and then incubated for 30 min at room temperature with a mouse anti-γH2AX monoclonal antibody (JBW301; Upstate Biotechnology Inc., Charlottesville, VA, U.S.A.) or a rabbit anti-Ku80 polyclonal antibody (AHP317; Serotec, Oxford, U.K.). After washing with PBS, the cells were incubated with Alexa Fluor 568-conjugated secondary antibodies (Molecular Probes, Eugene, OR, U.S.A.) for 30 min at room temperature. The images were acquired with an FV300 CLSM system.
RESULTS
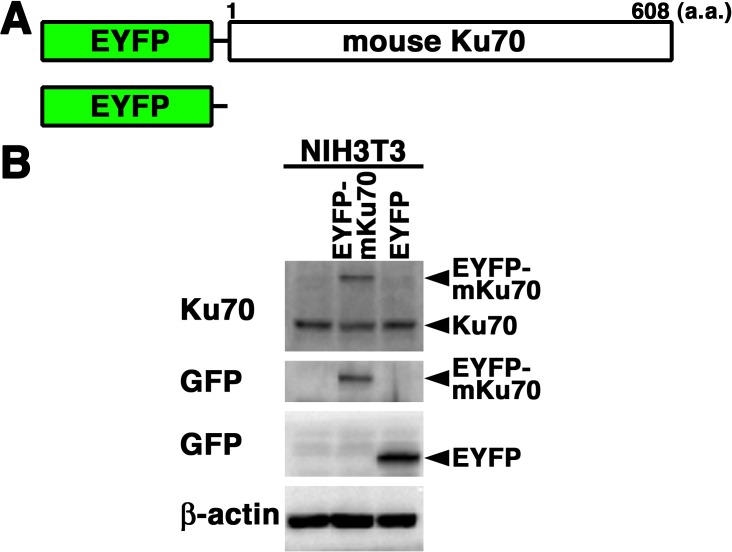
Expression and nuclear localization of EYFP-mouse Ku70 in living mouse fibroblasts: We examined the expression and subcellular localization of EYFP-mouse Ku70 in mouse NIH3T3 fibroblasts. First, we generated cells transiently expressing EYFP-mouse Ku70 in NIH3T3 cells. The expression vector pEYFP-C1 containing full-length mouse Ku70 or pEYFP-C1 alone was transfected into NIH3T3 cells (Fig. 1A). As shown in Fig. 1B, the signal of EYFP-mouse Ku70 was detected in the transfectants by Western blot analysis using the anti-Ku70 antibody and anti-GFP antibody. By confocal laser microscopy, we found that EYFP-mouse Ku70 was localized in the nuclei, but not in the nucleoli, of living interphase cells in EYFP-mouse Ku70 transfectants (Figs. 2 and 3). Predictably, in EYFP transfectants, we observed that EYFP was distributed throughout the cell, but not in the nucleoli (Fig. 2B).
Fig. 1.
Expression of EYFP-mouse Ku70 in mouse cells. (A) Schematics of EYFP-mouse Ku70 chimeric protein (EYFP-mouse Ku70) and control protein (EYFP). (B) Extracts from mouse (NIH3T3) cells transiently expressing the EYFP-mouse Ku70 or EYFP prepared and subjected to Western blotting using the anti-Ku70, anti-GFP or anti-β-actin antibody.
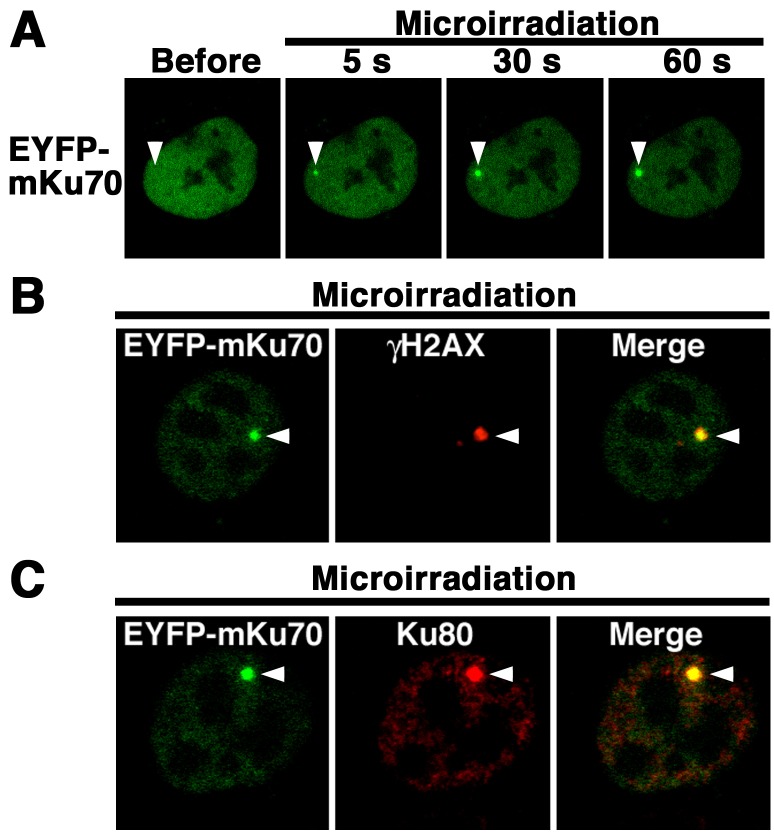
Fig. 2.
EYFP-mouse Ku70 accumulated immediately at DSBs induced by laser microirradiation in the fibroblast cell line NIH3T3. (A) The localization and accumulation of EYFP-mouse Ku70 at DSBs induced by 405 nm laser irradiation were examined. (B) Imaging of living EYFP-mouse Ku70- or EYFP-transfected NIH3T3 cells before and after microirradiation. For the same cells, EYFP images are shown alone (left panel) or merged (right panel) with differential interference contrast (DIC) images. Arrowheads indicate the microirradiated sites.
Fig. 3.
EYFP-mouse Ku70 accumulated with Ku80 immediately at DSBs induced by laser microirradiation. (A) Time-dependent EYFP-Ku70 accumulation in living cells (5–60 sec) after irradiation. (B, C) Immunostaining of microirradiated EYFP-mouse Ku70-transfected cells with the anti-γH2AX antibody (B) or anti-Ku80 antibody (C). At 5 min postirradiation, the cells were fixed and immunostained with each specific antibody. Left panels, EYFP-mouse Ku70 images (B, C); center panels, γH2AX (B) and Ku80 images (C); right panels, merged images (B, C). Arrowheads indicate the microirradiated sites.
EYFP-mouse Ku70 accumulates immediately at DSBs induced by laser microirradiation in NIH3T3 cells: We examined whether EYFP-mouse Ku70 accumulates immediately at the microirradiated sites (Fig. 2A). As shown in Fig. 2B, we found that EYFP-mouse Ku70, but not EYFP alone, accumulated at the 405 nm laser microirradiated sites in living cells. Next, we conducted time-lapse imaging of EYFP-mouse Ku70-transfected NIH3T3 cells. As shown in Fig. 3A, EYFP-mouse Ku70 accumulated at the microirradiated sites 5 sec after irradiation. In EYFP-mouse Ku70-transfected cells, the intensity of the EYFP signal rapidly increased at the microirradiated sites. Then, we tested whether EYFP-mouse Ku70 accumulated at laser-induced DSB sites by immunostaining the cells with an antibody that recognizes γH2AX, which is a golden standard marker of DSBs. As shown in Fig. 3B, EYFP-mouse Ku70 clearly colocalized with γH2AX at the microirradiated sites in NIH3T3 cells, indicating that mouse Ku70 accumulates at 405-nm-laser-induced DSB sites. We also examined whether Ku80, which is a heterodimer partner of Ku70, localizes at the Ku70-accumulation sites. Predictably, we observed that Ku80 clearly localized at the Ku70-accumulation sites (Fig. 3C). Collectivly, these observations demonstrate that after irradiation, EYFP-mouse Ku70 immediately accumulates with Ku80 at laser-induced DSBs in living NIH3T3 cells.
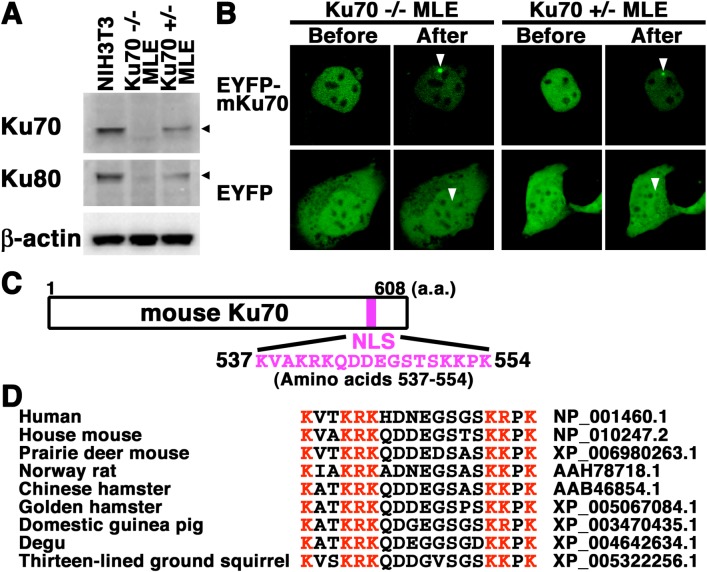
Nuclear localization and accumulation of EYFP-mouse Ku70 immediately at DSBs in mouse epithelial cells with or without endogenous Ku70 expression: We analyzed whether EYFP-mouse Ku70 localizes in the nuclei during the interphase and accumulates rapidly at DSBs induced by laser microirradiation in mouse epithelial cells. First, by Western blot analysis, we investigated the expression of Ku70 and Ku80 in the mouse fibroblast NIH3T3 cells and the 2 mouse epithelial cell lines (Ku70+/− MLE and Ku70−/− MLE) derived from Ku70-knockout mice. As shown in Fig. 4A, we detected Ku70 in NIH3T3 and Ku70+/− MLE cells. In agreement with our previous studies, a signal of mouse Ku70 was not detected in Ku70 −/− MLE cells, which are Ku70-null cells [9, 11]. In addition, we detected Ku80 in all three cell lines examined (Fig. 4A). We also reconfirmed that the Ku80 expression level was clearly associated with the number of functional Ku70 alleles, strongly supporting the idea that the expression of mouse Ku80 protein depends on mouse Ku70 protein [4, 9, 11]. To test whether EYFP-mouse Ku70 localizes in the nuclei of mouse epithelial cells, we transfected expression plasmids into Ku70 +/− MLE and Ku70 −/− MLE cells. By confocal laser microscopy, we found that EYFP-mouse Ku70 was localized in the nuclei, but not in the nucleoli, in the interphase of both living cell lines (Fig. 4B). Predictably, in both EYFP transfectants, we confirmed that EYFP was distributed throughout the cell, but not in the nucleoli (Fig. 4B). We examined whether EYFP-mouse Ku70 accumulates immediately at the microirradiated sites in the 2 epithelial cell lines. As shown in Fig. 4B, we found that EYFP-mouse Ku70, but not EYFP alone (data not shown), accumulated at the microirradiated sites in both the living epithelial cell lines. These findings demonstrated that EYFP-mouse Ku70 localizes in the nuclei and accumulates rapidly at DSBs induced by laser microirradiation in mouse epithelial cells with or without the expression of endogenous Ku70.
Fig. 4.
EYFP-mouse Ku70 accumulated rapidly at DSBs induced by laser microirradiation in mouse lung epithelial cell lines. (A) Expression of Ku70 and Ku80 in total cell lysates from NIH3T3, Ku70 +/– MLE and Ku70 –/– MLE cells. Total cell lysates from the two lung epithelial cell lines and the NIH3T3 cell line were analyzed by Western blotting using the anti-Ku70, anti-Ku80 or anti-β-actin antibody. (B) Imaging of living EYFP-mouse Ku70- or EYFP-transfected mouse lung epithelial cells before and after microirradiation. Arrowheads indicate the microirradiated sites. (C) NLS of mouse Ku70 (amino acids 537–554). (D) Alignment of the primary sequence among human and rodent homologous Ku70 proteins. The basic (red) or nonbasic residues (black) are indicated in different colors for comparison. The GeneBank accession number for each sequence is indicated.
Previously, on the basis of experimental studies, we identified a functional NLS of human Ku70 located at a region composed of 18 amino acid residues (positions 539 to 556) [6, 14]. We also found that the NLS is conserved among the human species and 3 rodent species, i.e., the mouse, rat and Chinese hamster (Fig. 4C and 4D) [6, 14]. To extend these findings, we examined whether the NLS is conserved among other rodent species. As shown in Fig. 4D, the structure and basic amino acids of Ku70 NLS are evolutionarily highly conserved among various rodent species, e.g., mouse, rat, hamster, guinea pig, degu and ground squirrel, strongly suggesting that Ku70 NLS is important for the regulation of rodent Ku70 function.
DISCUSSION
To elucidate the mechanisms of DNA repair pathway of human and animals is critical for developing next-generation chemotherapeutics and radiotherapies for cancer. In mammalian cells, Ku plays a pivotal role in multiple nuclear processes, such as chromosome maintenance, DNA repair, and V(D)J recombination [2, 6, 18, 23]. The mechanism underlying the modulation of all the diverse functions of Ku is still unclear, although the mechanism that controls the localization of Ku70 and Ku80 appears to play a crucial role in regulating the multiple function of Ku [6]. A DSB is the most deleterious type of DNA damage, and binding to the end of the DSB of Ku is required for the DSB repair via the C-NHEJ pathway [18, 22]. In this work, we investigated the expression and localization of mouse Ku70 in mouse cells. We observed that in mouse fibroblasts and epithelial cells, Ku70 localized in the interphase nuclei. In addition, our findings revealed that mouse Ku70 accumulates with Ku80 at DSB sites immediately following microirradiation with a laser in living mouse cells. Moreover, our findings suggest that NLS of Ku70 might be important for the nuclear localization of various rodent Ku70 homologues as well as human Ku70.
Laboratory rodents, such as mice, are a notable animal model for basic and applied research in various fields (e.g., life sciences, pharmacology, basic medical sciences and veterinary medical sciences). However, species differences between humans and mice are known to exist in, at least in part, the requirement for Ku70, although deficiency in Ku70 results in the expected deficits in DSB repair and sensitivities to ionizing radiation in human and mouse cells [3, 4]. For instance, human Ku70-deficient HCT-116 cell lines were not viable [3]. In contrast to the serious phenotype of Ku70 deletion in human cells, Ku70-deficient mice are viable [3, 4]. On the other hand, it is not elucidated in detail whether the mechanism that controls the localization of Ku70 is conserved between human and rodent species, although the mechanism appears to play a critical role in modulating the function of Ku70. It was reported that in rat fibroblasts, Ku70 is observed mainly in the cytoplasm [21], although the localization of Ku70 in rodent cells has not been clarified in detail. Similarly, in mouse fibroblasts, the transfected EGFP-hamster Ku80 mainly localizes in the cytoplasm [24]. On the other hand, recently, we have shown by conventional immunofluorescence staining that mouse Ku70 localizes in the nuclei of MLE cells [11]. In this and previous studies, we also have found that the structure and basic amino acids of Ku70 NLS were conserved among the homologues of various rodent species, e.g., mouse, rat, hamster, guinea pig, degu and ground squirrel [6], supporting the idea that rodent Ku70 localizes in the nuclei. Moreover, we observed by live cell imaging with confocal scanning laser microscopy that the transfected EYFP-mouse Ku70 localized in the interphase nuclei in three living mouse cell lines, i.e., the NIH3T3 cell lines and 2 MLE cell lines. Therefore, we consider that Ku70 mainly localizes in the interphase nuclei of mouse fibroblasts and epithelial cells as well as in human cells under general adherent culture conditions. Furthermore, our findings suggest the importance of NLS in the localization of rodent Ku70
To date, others and we have demonstrated that human Ku70 or Ku80 tagged with GFP or a similar fluorescent protein accumulates at laser-induced DSB sites in hamster and mouse cells [7, 9, 19, 22]. However, it has not been elucidated by live cell imaging whether mouse Ku70 accumulates at the DSB sites immediately following irradiation. Interestingly, some reports have shown that rodent Ku70 and Ku80 are mainly localized in the cytoplasm, and ionizing radiation induces their translocation to the nucleus [21, 24]. For instance, it was reported that EGFP-hamster Ku80 translocates to the nucleus from the cytoplasm 2 hr after γ-irradiation in mouse fibroblasts [24], and rat Ku70 translocates from the cytoplasm to the nucleus 20 min after X-irradiation of rat fibroblasts [21]. In these reports using conventional methods, it appears that the recruitment of Ku70 and Ku80 to DSB sites could not be visualized immediately following irradiation. In this study, using the laser-microirradiation method and real-time imaging technique, we observed that EYFP-mouse Ku70 accumulated in regions of induced DSBs 5 sec after microirradiation and colocalized with mouse Ku80. Altogether, we concluded that mouse Ku70 as well as human Ku70 accumulates with Ku80 at the DSB sites immediately following irradiation.
In conclusion, this and previous findings suggest that the mechanisms regulating the localization and accumulation of Ku70 at DSBs might be well conserved between the mouse and human species and play a key role in the functions of Ku70 common in both species [14]. Ku70 and Ku80 appear to have multiple functions in their monomeric and heterodimeric forms [6, 8]. Most recently, Choi et al. showed that free Ku70 (not bound to Ku80) and free Ku80 (not bound to Ku70) bind to apurinic/apyrimidinic sites, and they suggested that free Ku70 and free Ku80 have a novel role in altering base excision repair [1]. Further studies to elucidate the mechanisms regulating the localization of mouse Ku70 will lead to a better understanding of the physiological function and regulation mechanism of Ku70 not only at a simple DSB site, but also at clustered DNA-damaged sites, where it is predicted to have more complicated responses to DNA damage.
Acknowledgments
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan (M.K).
REFERENCES
- 1.Choi Y. J., Li H., Son M. Y., Wang X. H., Fornsaglio J. L., Sobol R. W., Lee M., Vijg J., Imholz S., Dollé M. E., van Steeg H., Reiling E., Hasty P.2014. Deletion of individual Ku subunits in mice causes an NHEJ-independent phenotype potentially by altering apurinic/apyrimidinic site repair. PLoS ONE 9: e86358. doi: 10.1371/journal.pone.0086358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Downs J. A., Jackson S. P.2004. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 5: 367–378. doi: 10.1038/nrm1367 [DOI] [PubMed] [Google Scholar]
- 3.Fattah F. J., Lichter N. F., Fattah K. R., Oh S., Hendrickson E. A.2008. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proc. Natl. Acad. Sci. U.S.A. 105: 8703–8708. doi: 10.1073/pnas.0712060105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu Y., Seidl K. J., Rathbun G. A., Zhu C., Manis J. P., van der Stoep N., Davidson L., Cheng H. L., Sekiguchi J. M., Frank K., Stanhope-Baker P., Schlissel M. S., Roth D. B., Alt F. W.1997. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 7: 653–665. doi: 10.1016/S1074-7613(00)80386-6 [DOI] [PubMed] [Google Scholar]
- 5.Grundy G. J., Moulding H. A., Caldecott K. W., Rulten S. L.2014. One ring to bring them all–the role of Ku in mammalian non-homologous end joining. DNA Repair (Amst.) 17: 30–38. doi: 10.1016/j.dnarep.2014.02.019 [DOI] [PubMed] [Google Scholar]
- 6.Koike M.2002. Dimerization, translocation and localization of Ku70 and Ku80 proteins. J. Radiat. Res. (Tokyo) 43: 223–236. doi: 10.1269/jrr.43.223 [DOI] [PubMed] [Google Scholar]
- 7.Koike M., Koike A.2008. Accumulation of Ku80 proteins at DNA double-strand breaks in living cells. Exp. Cell Res. 314: 1061–1070. doi: 10.1016/j.yexcr.2007.11.014 [DOI] [PubMed] [Google Scholar]
- 8.Koike M., Shiomi T., Koike A.2001. Dimerization and nuclear localization of Ku proteins. J. Biol. Chem. 276: 11167–11173. doi: 10.1074/jbc.M010902200 [DOI] [PubMed] [Google Scholar]
- 9.Koike M., Yutoku Y., Koike A.2011. Accumulation of Ku70 at DNA double-strand breaks in living epithelial cells. Exp. Cell Res. 317: 2429–2437. doi: 10.1016/j.yexcr.2011.07.018 [DOI] [PubMed] [Google Scholar]
- 10.Koike M., Yutoku Y., Koike A.2011. Accumulation of p21 proteins at DNA damage sites independent of p53 and core NHEJ factors following irradiation. Biochem. Biophys. Res. Commun. 412: 39–43. doi: 10.1016/j.bbrc.2011.07.032 [DOI] [PubMed] [Google Scholar]
- 11.Koike M., Yutoku Y., Koike A.2011. Establishment of Ku70-deficient lung epithelial cell lines and their hypersensitivity to low-dose X-irradiation. J. Vet. Med. Sci. 73: 549–554. doi: 10.1292/jvms.10-0454 [DOI] [PubMed] [Google Scholar]
- 12.Koike M., Yutoku Y., Koike A.2013. The C-terminal region of Rad52 is essential for Rad52 nuclear and nucleolar localization, and accumulation at DNA damage sites immediately after irradiation. Biochem. Biophys. Res. Commun. 435: 260–266. doi: 10.1016/j.bbrc.2013.04.067 [DOI] [PubMed] [Google Scholar]
- 13.Koike M., Ikuta T., Miyasaka T., Shiomi T.1999. Ku80 can translocate to the nucleus independent of the translocation of Ku70 using its own nuclear localization signal. Oncogene 18: 7495–7505. doi: 10.1038/sj.onc.1203247 [DOI] [PubMed] [Google Scholar]
- 14.Koike M., Ikuta T., Miyasaka T., Shiomi T.1999. The nuclear localization signal of the human Ku70 is a variant bipartite type recognized by the two components of nuclear pore-targeting complex. Exp. Cell Res. 250: 401–413. doi: 10.1006/excr.1999.4507 [DOI] [PubMed] [Google Scholar]
- 15.Koike M., Kuroiwa A., Koike A., Shiomi T., Matsuda Y.2001. Expression and chromosome location of hamster Ku70 and Ku80. Cytogenet. Cell Genet. 93: 52–56. doi: 10.1159/000056948 [DOI] [PubMed] [Google Scholar]
- 16.Koike M., Matsuda Y., Mimori T., Harada Y. N., Shiomi N., Shiomi T.1996. Chromosomal localization of the mouse and rat DNA double-strand break repair genes Ku p70 and Ku p80/XRCC5 and their mRNA expression in various mouse tissues. Genomics 38: 38–44. doi: 10.1006/geno.1996.0589 [DOI] [PubMed] [Google Scholar]
- 17.Koike M., Awaji T., Kataoka M., Tsujimoto G., Kartasova T., Koike A., Shiomi T.1999. Differential subcellular localization of DNA-dependent protein kinase components Ku and DNA-PKcs during mitosis. J. Cell Sci. 112: 4031–4039. [DOI] [PubMed] [Google Scholar]
- 18.Mahaney B. L., Meek K., Lees-Miller S. P.2009. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 417: 639–650. doi: 10.1042/BJ20080413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mari P. O., Florea B. I., Persengiev S. P., Verkaik N. S., Brug-genwirth H. T., Modesti M., Giglia-Mari G., Bezstarosti K., Demmers J. A., Luider T. M., Houtsmuller A. B., Gent D. C.2006. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. U.S.A. 103: 18597–18602. doi: 10.1073/pnas.0609061103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mimori T., Akizuki M., Yamagata H., Inada S., Yoshida S., Homma M.1981. Characterization of a high molecular weight acidic nuclear protein recognized by autoantibodies in sera from patients with polymyositis-scleroderma overlap. J. Clin. Invest. 68: 611–620. doi: 10.1172/JCI110295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okui T., Endoh D., Kon Y., Hayashi M.2002. Deficiency in nuclear accumulation of G22p1 and Xrcc5 proteins in hyper-radiosensitive Long-Evans Cinnamon (LEC) rat cells after X irradiation. Radiat. Res. 157: 553–561. doi: 10.1667/0033-7587(2002)157[0553:DINAOG]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 22.Reynolds P., Botchway S. W., Parker A. W., O’Neill P.2013. Spatiotemporal dynamics of DNA repair proteins following laser microbeam induced DNA damage-when is a DSB not a DSB? Mutat. Res. 756: 14–20. doi: 10.1016/j.mrgentox.2013.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuteja R., Tuteja N.2000. Ku autoantigen: a multifunctional DNA-binding protein. Crit. Rev. Biochem. Mol. Biol. 35: 1–33. doi: 10.1080/10409230091169177 [DOI] [PubMed] [Google Scholar]
- 24.Yaneva M., Li H., Marple T., Hasty P.2005. Non-homologous end joining, but not homologous recombination, enables survival for cells exposed to a histone deacetylase inhibitor. Nucleic Acids Res. 33: 5320–5330. doi: 10.1093/nar/gki821 [DOI] [PMC free article] [PubMed] [Google Scholar]