Abstract
The zinc finger protein ZNF804A rs1344706 variant is a replicated genome-wide significant risk variant for schizophrenia and bipolar disorder. While its association with altered brain structure and cognition in patients and healthy risk allele carriers is well documented, the characteristics and function of the gene in the brain remains poorly understood. Here, we used in situ hybridization to determine mRNA expression levels of the ZNF804A rodent homologue, Zfp804a, across multiple postnatal neurodevelopmental timepoints in the rat brain. We found changes in Zfp804a expression in the rat hippocampus, frontal cortex, and thalamus across postnatal neurodevelopment. Zfp804a mRNA peaked at postnatal day (P) 21 in hippocampal CA1 and DG regions, and was highest in the lower cortical layers of frontal cortex at P1, possibly highlighting a role in developmental migration. Using immunofluorescence, we found that ZFP804A co-localized with neurons and not astrocytes. In primary cultured cortical neurons, we found that Zfp804a expression was significantly increased when neurons were exposed to glutamate [20 μM], but this increase was blocked by the N-methyl-D-aspartate receptor (NMDAR) antagonist MK-801. Expression of Comt, Pde4b, and Drd2, genes previously shown to be regulated by ZNF804A overexpression, were also significantly changed in an NMDA-dependent manner. Our results describe, for the first time, the unique postnatal neurodevelopmental expression of Zfp804a in the rodent brain and demonstrate that glutamate potentially plays an important role in the regulation of this psychiatric susceptibility gene. These are critical steps towards understanding the biological function of ZNF804A in the mammalian brain.
Keywords: ZNF804A, Zfp804a, neurodevelopment, gene expression, in situ hybridization, schizophrenia
1. Introduction
The gene that encodes ZNF804A is a widely studied susceptibility gene for schizophrenia and bipolar disorder. It was the first gene to reach genome-wide significance for psychosis (O'Donovan et al., 2008) and its association with both schizophrenia and bipolar disorder has been subsequently confirmed in multiple genome-wide association studies (GWASs; Riley et al., 2010; Williams et al., 2011; Zhang et al., 2011). Studies of the single nucleotide polymorphism (SNP) rs1344706 in both psychiatric patients and healthy risk allele carriers have demonstrated effects on white and gray matter volume, white matter tract integrity, functional connectivity (Esslinger et al., 2009; Ikuta et al., 2014; Lencz et al., 2010; Voineskos et al., 2011; Wei et al., 2015), and cognition (Chen et al., 2012; Esslinger et al., 2011; Hashimoto et al., 2010; Walter et al., 2011; Walters et al., 2010), indicating potential functional consequences of this gene variant.
While these findings illustrate the potential wide-ranging effects of a ZNF804A polymorphism, little is known about the biological functions of the gene itself. ZNF804A is expressed in the brain and is predicted to encode a protein with a C2H2 zinc finger domain, which suggests a role in DNA or RNA binding (Donohoe et al., 2010). In human post mortem dorsolateral prefrontal cortical tissue, ZNF804A expression is highest fetally, with levels decreasing at a rate of 11% per year on average during infancy and childhood (Tao et al., 2014). These studies provide evidence that ZNF804A has a role in gene regulatory and neurodevelopmental processes, and given the putative gene regulation role of ZNF804A in human neurons (Bernstein et al., 2014), it is critical to know the location and timing of gene expression within the mammalian brain. Moreover, because disorders such as schizophrenia are neurodevelopmental in nature, details about the temporal and spatial changes in ZNF804A transcript levels are especially pertinent. Current knowledge on ZNF804A expression in neurodevelopment is restricted to just a few regions of the brain. Therefore, we performed in situ hybridization on both coronal and sagittal sections from throughout the rat brain at multiple postnatal time points to determine a comprehensive postnatal brain expression profile of the ZNF804A rodent homologue, Zfp804a.
Glutamatergic neurotransmitter activity is an important molecular component of healthy neurodevelopmental processes and there is considerable evidence to suggest that there is glutamatergic neuronal dysfunction in schizophrenia pathophysiology (Javitt, 2012; Olney et al., 1999). In particular, NMDAR hypofunction appears to be an important feature of the disorder, leading to possible secondary effects on dopaminergic systems (Javitt, 2007). We therefore used quantitative polymerase chain reaction (qPCR) to investigate expression levels of Zfp804a in primary cultured cortical neurons treated with glutamate, and also treated with glutamate in the presence of MK-801. Additionally, we quantified the expression levels of three other schizophrenia-relevant genes (Comt, Pde4b, and Drd2) in the same primary cultured cortical neurons.
2. Methods
2.1 Animals
Subjects used for this study were male Long-Evans (Charles River Laboratories, Wilmington, Massachusetts) rats from postnatal 1-86 days of age. Rats had ad libitum access to food and water. All animal procedures were approved by the Feinstein Institute Medical Research Institutional Animal Care and Use Committee and maintained according to National Institutes of Health guidelines.
2.2 In situ hybridization
Rats were sacrificed by decapitation on postnatal day 1 (P1; n=6), P7 (n=6), P21 (n=5), P49 (n=5) or P86 (n=6). The rat pups within each postnatal day group all came from the same litter. Whole brains were rapidly removed and frozen on dry ice. Sections (14 μm) were cut at −20°C on a cryostat (Leica, Nussloch, Germany) and thaw-mounted onto poly-L-lysine-coated glass slides. Each slide had a section from P1, P7, P21, P49 and P86 with equivalent regions of the brain to enable direct comparison. The sections were air-dried for at least 30 min, fixed in 4% paraformaldehyde in 0.1 M PBS for 5 min, rinsed in PBS for 1 min, delipidated in 70% ethanol for 4 min, and stored in 95% ethanol at 4°C.
Hybridizations were performed as previously described (Hall et al., 2001). A cDNA antisense probe (45mer) was synthesized commercially (MWG Operon, Huntsville, AL) complementary to nucleotides 122-166 of Zfp804a mRNA (XM_008775447, 5’-CCTTCAGATCCTCCAGAGCTTTGGCAATAGTGTTCTCCTTCTCAG - 3’). These oligonucleotides were 3'- end-labeled with [β-S35] dATP (1200 Ci/mmol; Perkin Elmer-NEN) in a 30:1 molar ratio of radiolabeled ATP:oligonucleotide using terminal deoxynucleotidyl transferase. Specific activity of the [S35]-labeled probe was between 2.0 ×105 and 3.0 × 105 dpm/μl probe. To define non-specific hybridization, adjacent slide-mounted sections were incubated with radiolabeled oligonucleotide in the presence of an excess (100X) concentration of unlabeled oligonucleotide probe. After hybridization, sections were opposed to Kodak BioMax MR x-ray film for 1–2 weeks to obtain optimal exposure. For silver grains, sections were dipped in K5 photographic emulsion (Ilford, Cheshire, UK) and exposed for 10 weeks at 4°C before being developed and counterstained with 0.01% thionin.
Following development of the autoradiographic film, images were digitized, and gene expression was measured by image densitometry. Optical density was determined using ImageJ imaging software (http://rsbweb.nih.gov/ij/; Schneider et al., 2012). All densitometry measures were corrected by subtracting the corresponding background measures for that specific brain region. For silver grain analysis, images were captured using Leica QWIN imaging software using a 63X objective under oil-immersion. Silver grain density was assessed using a thresholded particle analysis in ImageJ. A specific grain count was then calculated by subtracting total and nonspecific counts. One-way analysis of variance (ANOVA) with Tukey's HSD post hoc tests were used to compare expression differences between groups. Statistical significance was set at a level of P < 0.05.
2.3 Immunofluorescence
16 μM sections from frozen rat brains were fixed using 4% paraformaldehyde and rinsed twice in 1X PBS followed by permeabilization in 1X PBST for 30 mins. Slides were then transferred to a humidified chamber and blocked for 30 mins in 1XPBST/5% heat inactivated donkey serum, followed by incubation in the appropriate primary antibody: goat polyclonal anti-ZNF804A (S-16; Santa Cruz Biotechnology, Santa Cruz, CA) 1:50, rabbit polyclonal anti-GFAP antibody (abcam, Cambridge, MA) 1:1000, rabbit polyclonal anti-NeuN antibody (abcam) 1:500. Slides were incubated overnight at 4°C, washed 3 times in PBST, then blocked in 1XPBST/ 5% heat inactivated donkey serum. Sections were then incubated in secondary antibody and 1XPBST/ 5% heat inactivated donkey serum at 25°C for 2 hours. Secondary antibodies were Alexa Fluor 488 (1:50) and Alexa Fluor 546 (1:500; Molecular probes, Eugene, OR). Sections were then mounted in Vectashield HardSet mounting medium with DAPI (Vector laboratories, Burlingame, CA). Images were acquired on a Leica DMI 4000B inverted wide field fluorescence microscope at 20X. For negative controls, sections were incubated in non-immune serum followed by secondary antibody to check for non-specific binding.
2.4 Neuronal cortical cultures
Cortical neurons were dissected from embryonic day 19 rats and cultured in Neurobasal medium containing 2% B27, 1mM sodium pyruvate, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2mM Glutamax for 10 days before being incubated for 3 hours in either glutamate [20 μM] (Sigma-Aldrich, St Louis, MO), glutamate plus MK-801 [20 μM] (Tocris Biosciences, Bristol, UK) in complete Neurobasal medium.
2.5 RNA isolation and quantitative PCR (qPCR)
mRNA was isolated from cultured cortical neurons using a RNeasy kit (Qiagen, Valencia, California). Reverse transcription was performed using an iScript kit (BioRad, Hercules, California, USA). qPCR was performed using SYBR green (BioRad), as previously described (Zhong et al., 2007). Primers were as follows: Hprt1: Forward 5’-TGTTTGTGTCATCAGCGAAAGTG -3’, Reverse: 5’-ATTCAACTTGCCGCTTTTTA-3’, Zfp804a: Forward 5’-TCAAAGTGCTTCAGCCACAC-3’, Reverse: 5’-ATGGGGTAAGGGTGGAAAAG-3’, Pde4b: Forward: 5’-CAGCTCATGACCCAGATAAGTGG-3’, Reverse: 5’-GTCTGCACAATGTACCATGTTGCG-3’, Comt: Forward: 5’-ATCTTCACGGGGTTTCAGTG-3’ Reverse: 5’-GAGCTGCTGGGGACAGTGAG-3’ and, Drd2: Forward 5’-TGTCCTCCAGGCAACATCAGT-3’, Reverse: 5’-TTGTTTTCCCCCAAATGGTA-3’. All samples were analyzed in triplicate and gene expression normalized to Hprt1. PCR settings were as follows: 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. PCR data was analyzed using the formula 2ΔΔCT.
3. Results
3.1 Postnatal gene expression of Zfp804a throughout the entire rat brain
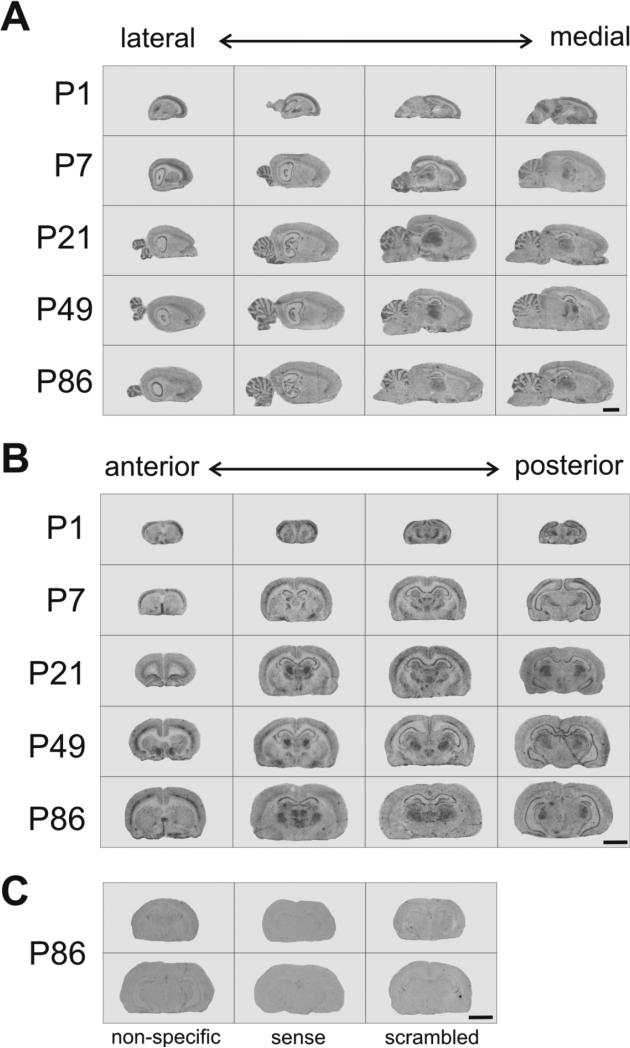
To assess the distribution and quantify levels of Zfp804a in the rodent brain, we performed in situ hybridization at multiple postnatal time points. We found that Zfp804a was widely expressed in the brain, especially in the hippocampus, cortex, thalamus, septum, and cerebellum (Fig. 1). On a macroscopic level, the broad distribution of Zfp804a transcript suggests that the gene potentially has broad importance in the postnatal rodent brain.
Fig. 1.
Zfp804a mRNA is widely expressed throughout the rat brain at multiple postnatal time points. Example in situ hybridization autoradiographic images are shown at five different postnatal days (P) 1, 7, 21, 49, and 86. Panels show representative images of sagittal (A) and coronal (B) and sections from the rat brain. (C) Representative images from P86 brains are shown for non-specific hybridization, sense oligonucleotide, and scrambled oligonucleotide. Scale bars: 4.0 mm.
3.2 Zfp804a expression in rat frontal cortex decreases through postnatal life
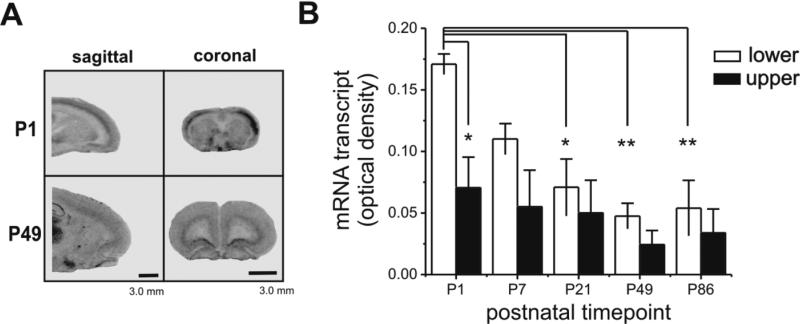
We examined autoradiographically labeled sections from the frontal cortex, a region of the brain important for executive functioning (Fig. 2). There was distinctly higher gene expression in the lower layers (layer V, VI) of the frontal cortex compared to the upper layers (layer I-IV). Compared across the five developmental time points assessed, Zfp804a gene expression was 194% higher overall in the lower layers compared to the upper layers (Fig. 2B, one-way ANOVA, F=5.44, P<0.005). Post hoc Tukey's HSD tests showed that Zfp804a mRNA in P1 lower layers were significantly higher than measured from P21 (P<0.05), P49 (P<0.005), and P86 (P<0.005) lower layers. Furthermore, while there was a significant difference between the P1 lower versus upper layers (Tukey's HSD, P<0.05), this was the only developmental time point that had a statistically significant difference between layers. Therefore, postnatal Zfp804a mRNA in frontal cortex appears to be highest at P1 and then declines steadily throughout postnatal development.
Fig. 2.
Zfp804a mRNA decreases in frontal cortex during postnatal life. (A) Representative autoradiographic images of sagittal and coronal sections through the rat frontal cortex. Note the dramatic difference in transcript levels between the lower and upper layers of frontal cortex. (B) Image densitometry measures of the lower and upper layers of cortex show a pattern of decreasing Zfp804a mRNA levels across postnatal life with P1 being significantly elevated over other time points (Tukey's HSD, P<0.05). Asterisks indicate significant differences: *p<0.05, **, p<0.005. Scale bars: 3.0 mm.
3.3 Sub-regional changes in Zfp804a expression within the hippocampus through postnatal neurodevelopment and neuron-specific ZFP804A expression
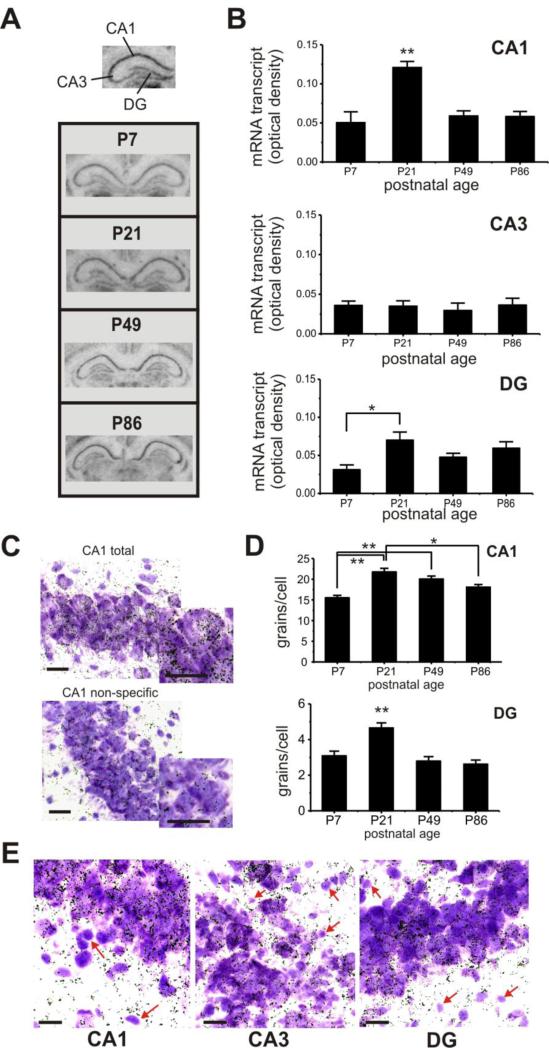
We next examined the hippocampus, which is a critical structure for cognitive processes, by performing image densitometry on three distinct hippocampal subregions (Fig. 3A): area cornu ammonis 1 (CA1), area cornu ammonis 3 (CA3), and the dentate gyrus (DG). Within CA1, there was a peak in postnatal expression of Zfp804a transcript at P21 (Fig. 3B), which was significantly higher than all other time points tested (one-way ANOVA with Tukey's HSD, F=10.9, P<0.005 at least). Levels of Zfp804a mRNA in the CA3 remained stable across postnatal life with no significant differences. Within the DG, Zfp804a transcript was significantly higher at P21 compared to P7 (Tukey's HSD, P< 0.05), but did not differ at other timepoints.
Fig. 3.
Postnatal expression of Zfp804a mRNA varies in hippocampal areas across postnatal life. (A) Representative images of coronal sections showing the hippocampus at different postnatal days. (B) Top, Zfp804a gene expression within area CA1 peaks at P21, which is significantly higher than other time points (Tukey's HSD, P<0.005). Middle, gene expression is fairly low and stable throughout postnatal life in area CA3. Bottom, within the DG, Zfp804a gene expression is significantly lower at P7 compared to P21 (Tukey's HSD, P<0.05), but does not differ from other time points. (C) Representative examples of total and non-specific silver grain labeling within CA1. Dark silver grains show cellular localization of Zfp804a mRNA within thionin-stained hippocampal neurons. Insets show magnified view of labeled neurons. (D) Silver grain density for CA1 and DG confirmed the pattern of elevated mRNA at P21 in both hippocampal regions. (Tukey's HSD, * P<0.001, ** P<0.0005) (E) A subset of smaller, thionin-positive cells that are morphologically consistent with glial cells, show a marked absence of silver grain labeling (red arrows) in all three hipppocampal regions. All scale bars: 10 μm.
To examine cellular localization of Zfp804a transcript, we examined silver grains in thionin-stained sections at high-magnification (Fig. 3C). Consistent with the image densitometry results, we found that grain density was highest at P21 in both CA1 (Fig. 3D; one-way ANOVA with Tukey's HSD, F=13.7, P<0.00001) and DG (one-way ANOVA with Tukey's HSD, F=12.6, P<0.0005). Furthermore, silver grains appeared to consistently cluster on neurons while smaller cellular profiles were unlabeled (Fig. 3E). Based on morphological criteria, these smaller cells may be glial cells, suggesting that Zfp804a expression may be neuron-specific.
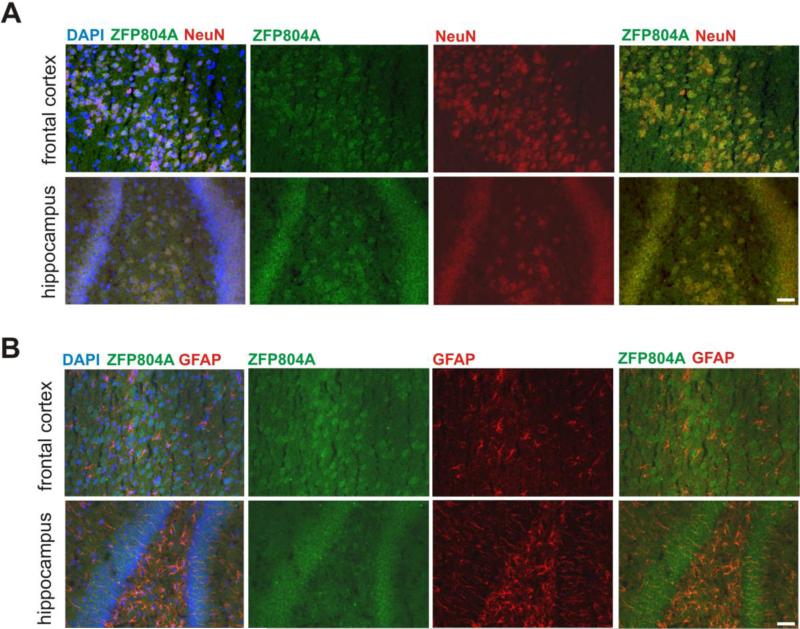
To determine whether Zfp804a is not only transcribed, but also translated in neurons, we performed immunofluorescence staining on frozen brain sections for ZFP804A. We found that ZFP804A expression co-localized with the neuronal marker NeuN (Fig. 4A), but did not overlap with the astrocyte marker GFAP (Fig. 4B). These results suggest that ZFP804A is expressed specifically in neurons.
Fig. 4. Immunofluorescence of ZFP804A in neurons.
Expression of ZFP804A in frontal cortex and hippocampal neurons. (A) Representative immunofluorescence images show ZFP804A expression in neurons labeled with NeuN antibody in the frontal cortex and DG of hippocampus of P21 rats. (B) ZFP804A expression was not observed in GFAP-positive astrocytes. Scale bars: 50 μm.
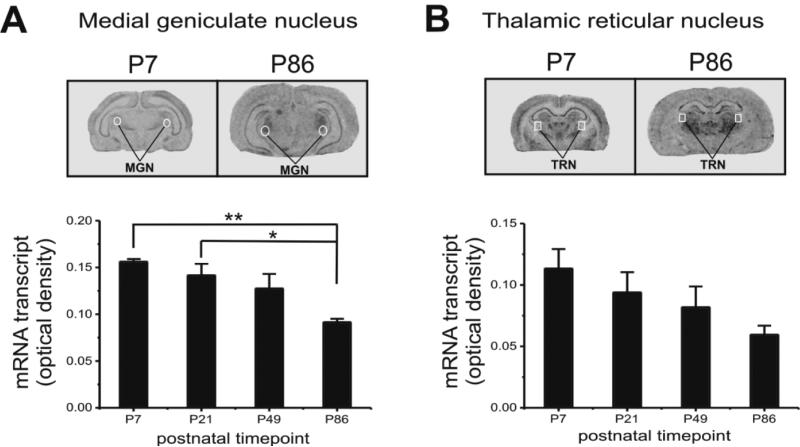
3.4 Zfp804a expression in the thalamic medial geniculate nucleus (MGN) is significantly decreased at P86, but mRNA levels in the thalamic reticular nucleus (TRN) are unchanged
In both coronal and sagittal brain sections of Zfp804a in situ hybridization, the thalamus stood out as a structure with elevated expression levels (Figs. 1, 5), a finding that is concordant with data from the P56 mouse brain (Allen Mouse Brain Atlas, http://mouse.brain-map.org). Therefore, we decided to measure Zfp804a gene expression in two thalamic regions that may have particular relevance to schizophrenia. The MGN, which is involved in the processing of auditory information, had a pattern of expression that was similar to other brain areas measured. Specifically, Zfp804a mRNA levels in MGN were higher in early postnatal tissue compared to adult tissue (Fig. 5A). At P7 and P21, mRNA levels were significantly higher than P86 (Tukey's HSD, P7, P<0.005; P21, P<0.05). Within the TRN, there was a trend towards declining levels of Zfp804a transcript across age, but the changes were not significantly different (Fig. 5B).
Fig. 5.
Zfp804a mRNA in MGN and TRN of thalamus. (A) Top, representative images of sagittal sections through the MGN. Bottom, image densitometry measures within the MGN through postnatal life show that Zfp804a expression in P86 adult rat is significantly lower than earlier P7 and P21 ages (Tukey's HSD, P<0.05). (B) Top, representative images of sagittal sections through the TRN. Bottom, image densitometry measures within the TRN show no significant changes across postnatal life. Scale bars: 4.0 mm.
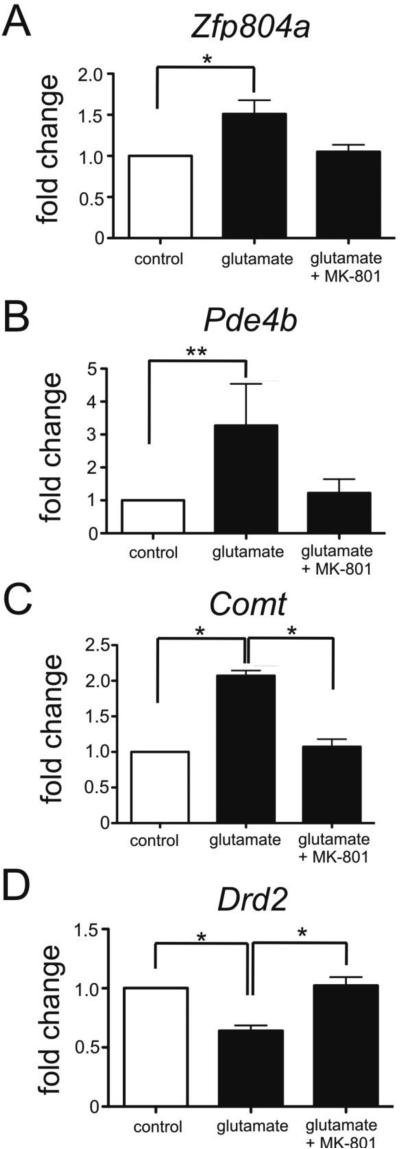
3.5 Zfp804a, Comt, Pde4b, and Drd2 mRNA levels in cortical neurons are modulated by exposure to glutamate [20μM] in an NMDAR-dependent manner
To determine whether Zfp804a levels are linked to glutamate exposure, we assessed Zfp804a expression in cortical neurons after treatment with 20μM glutamate. Untreated cells were used as controls. We found that Zfp804a expression in cortical neurons was increased in glutamate treated cells as compared to controls (Fig. 6A; Mann-Whitney, P < 0.05). We then assessed Comt, Pde4b, Drd2 expression in glutamate treated neurons to determine whether elevated levels of Zfp804a expression correlated with changes in the expression of these genes. Similar to what was previously reported by others (Girgenti et al., 2012), we observed an increase in Comt expression (Mann-Whitney, P < 0.05) and decrease in Drd2 expression (Mann-Whitney, P < 0.05). Pde4b expression was significantly increased by three-fold in the presence of glutamate (Mann-Whitney, P < 0.01). Interestingly, when we blocked NMDA receptors with the antagonist MK-801, we found no differences in expression compared to controls for all four genes tested (Mann-Whitney, P ≥ 0.28). Therefore, glutamate treatment of cortical neurons altered the expression levels of these specific genes that are thought to be involved in cognitive functions in animal models and human studies. Whether Zfp804a itself mediated the expression changes we observed in Comt, Pde4b, and Drd2 remains to be determined.
Fig. 6.
Glutamate exposure upregulates Zfp804a, Pde4b, and Comt expression but decreases levels of Drd2 expression. Plots show quantitative PCR results performed on cultures of rat primary cortical neurons collected at embryonic day 19. All results normalized to the reference gene Hprt1. (A) Cortical neurons exposed to glutamate [20μM] show increased Zfp804a expression. The NMDA receptor antagonist MK-801 blocked this increase. Glutamate exposure led to a four-fold increase in the expression of Pde4b (B) and a two-fold increase in Comt (C) levels in cortical neurons (Student's t-test, . Both increases were blocked by MK-801. (D) Levels of Drd2 were significantly decreased by glutamate exposure, but not in the presence of MK-801. Mean and SEM of three experiments is shown; Mann-Whitney, *P<0.05, **, P<0.01.
4. Discussion
This is the first neurodevelopmental and neurotransmitter-modulated expression data available on Zfp804a in the rat brain. We have shown that expression of Zfp804a is highest postnatally within the frontal cortex at P1, particularly in the lower cortical layers. Levels of Zfp804a expression vary across hippocampal subregions, with peak expression in CA1 and DG at age P21. In cultured primary cortical neurons, Zfp804a expression increased significantly following exposure to glutamate in an NMDA-dependent manner. Finally, cultured cortical neurons had increased mRNA levels of Pde4b and Comt, but decreased Drd2 mRNA in response to glutamate exposure.
While many studies have confirmed the association of ZNF804A polymorphisms to risk for schizophrenia and bipolar disorder in humans, the endogenous temporal and spatial expression of this gene in the mammalian brain has not been sufficiently investigated. We chose to maximize our spatial expression analysis by performing in situ hybridization on both coronal and sagittal brain sections collected throughout the rat brain. We compared expression patterns between postnatal day 1, 7, 21, 49, and 86 as these are rodent ages approximately equivalent to the human third trimester, day of birth, childhood, adolescence, and adulthood, respectively (Rash and Grove, 2006). Our findings provide a potentially important characterization of when and where this gene is expressed in the brain, which is critically important for understanding its specific role in gene regulation and neural development.
Functional imaging studies have demonstrated that individuals with the rs1344706 SNP have altered functional connectivity in the prefrontal cortex and hippocampus (Esslinger et al., 2009; Rasetti et al., 2011), however, the molecular basis for this association remains unknown. Our qPCR results (Fig. 6) suggest that glutamate exposure alone can affect Zfp804a gene expression. This finding, coupled with the neuron-specific expression of ZFP804A in the frontal cortex and hippocampus (Fig. 4), indicates that this gene may be involved in the development of this intermediate phenotype.
Structural abnormalities in the frontal lobe, particularly within the prefrontal cortex, have been a consistent observation in patients with schizophrenia (Benes et al., 1991; Goldman et al., 1997) and may underlie cognitive deficits (Guo et al., 2014; Nakamura et al., 2008). Our in situ hybridization results in the frontal cortex (Fig. 2) show a striking difference in Zfp804a mRNA expression between the lower and upper cortical layers. The increased levels of Zfp804a in the lower layers suggest that Zfp804a could be involved in neurogenesis or neuronal migration (Hill et al., 2012) that is crucial to create the different types of neurons and correctly position the cortical neurons that are necessary for healthy functioning cortical structure (Stansberg et al., 2007). For example, Zfp804a transcripts may be necessary as a protein- or RNA-binding entity (Gamsjaeger et al., 2007) for proper neuronal migration during cortical development in the rat. This is one potential mechanism through which ZNF804A may be involved in schizophrenia pathophysiology.
In healthy adults, ZNF804A is widely expressed in neurons from multiple regions, including the hippocampus (Bernstein et al., 2014), but the expression patterns of this gene in schizophrenia are not well known. The rs1344706 risk allele is associated with allelic expression imbalance of ZNF804A in adult dorsalateral prefrontal cortex samples (Guella et al., 2014) and there are variable cis-effects across different areas of the adult brain, particularly in the hippocampus (Buonocore et al., 2010). It would be interesting to investigate whether rs1344706 carriers have altered ZNF804A expression selectively in the CA1 region of the hippocampus during adolescence, as our results suggest that there is a transient upregulation of Zfp804a at this neurodevelopmental stage. If there was dysregulation of Zfp804a in this region, at this stage, then it could potentially contribute to cognition impairment in individuals with schizophrenia (Walters et al., 2010). rs1344706 risk allele carriers have been shown to have larger hippocampal volumes than non-carriers (Donohoe et al., 2011), suggesting a specific effect on hippocampal structure. We do not know whether Zfp804a brain expression in the rat correlates with any volumetric or behavioral changes.
There are several limitations to this work. The findings are primarily descriptive in nature and we do not show how Zfp804a acts to either regulate developmental or neuronal processes. Furthermore, we did not examine prenatal expression in this study, which could reveal more relevant information on potential neural migration effects of the gene. In humans, ZNF804A expression is highest prenatally, peaking right before birth (Tao et al., 2014). Also, while we have demonstrated an effect of glutamate exposure on Zfp804a expression in cultured neurons, we have not illuminated the molecular mechanism for these changes outside of a potential NMDAR involvement. Lastly, we only studied male rats in this study and there are known differences related to sex in schizophrenia, with males having a consistently earlier onset than females (Seeman, 1982).
Our study provides insight into where one of the most widely confirmed schizophrenia risk genes is expressed at the subregional level throughout the rat brain across postnatal neurodevelopment. While we are not examining tissue from a rat model of schizophrenia in this study, it is important to know the baseline levels of gene expression in these brain regions across postnatal life for future experimental work. Given the neuroanatomical, cognitive, and biological (Hill and Bray, 2012; Hill et al., 2012) influences of ZNF804A rs1344706 in humans, it is important to initially investigate potential molecular mechanisms and functions of this gene in a mammalian animal model. Our findings represent an important step towards comprehending the association between ZNF804A and risk for schizophrenia, bipolar disorder, and psychosis.
Acknowledgements
The authors would like to thank the Feinstein Institute animal facility staff, in particular Roberto Adames and Yanexy Portal, for their expert animal care. We also thank Grace Lee and Sarah Fox for their excellent technical assistance.
Role of funding sources
This work was supported by the Center for Intervention Development and Applied Research (P50MH080173). Funding agencies had no role in the study design, acquisition or interpretation of data, or in writing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributors
AK developed the overall experimental design. EHC, AK, TSC, PB, SH, and VV contributed to the study design, performed the experiments, and analyzed the data. EHC, AK, and AKM wrote the manuscript; TSC, SH, and VV provided revisions to the manuscript. All authors have given final approval of the submitted manuscript.
Conflict of Interest
Dr. Malhotra has been a consultant to Genomind, Forum Pharmaceuticals, and Takeda Pharmaceuticals. The remaining authors do not have financial interests or conflicts of interest to report.
References
- Benes FM, McSparren J, Bird ED, SanGiovanni JP, Vincent SL. Deficits in small interneurons in prefrontal cortex and anterior cingulated cortices of schizophrenic and schizoaffective patients. Arch. Gen. Psychiatry. 1991;48:996–1001. doi: 10.1001/archpsyc.1991.01810350036005. [DOI] [PubMed] [Google Scholar]
- Bernstein HG, Steiner J, Dobrowolny H, Bogerts B. ZNF804A protein is widely expressed in human brain neurons: Possible implications on normal brain structure and pathomorphologic changes in schizophrenia. Schizophr. Bull. 2014;40(3):499–500. doi: 10.1093/schbul/sbt237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonocore F, Hill MJ, Campbell CD, Oladimeji PB, Jeffries AR, Troakes C, Hortobagyi T, Williams BP, Cooper JD, Bray NJ. Effects of cis-regulatory variation differ across regions of the adult human brain. Hum Mol Genet. 2010;19(22):4490–6. doi: 10.1093/hmg/ddq380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Xu Z, Zhai J, Bao X, Zhang Q, Gu H, Shen Q, Cheng L, Chen X, Wang K, Deng X, Ji F, Liu C, Li J, Dong Q, Chen C. Evidence of IQ-modulated association between ZNF804A gene polymorphism and cognitive function in schizophrenia patients. Neuropsychopharmacology. 2012;37(7):1572–1578. doi: 10.1038/npp.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe G, Morris DW, Corvin A. The psychosis susceptibility gene ZNF804A: Associations, functions, and phenotypes. Schizophr. Bull. 2010;36(5):904–909. doi: 10.1093/schbul/sbq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe G, Rose E, Frodl T, Morris D, Spoletini I, Adriano F, Bernardini S, Caltagirone C, Bossù P, Gill M, Corvin AP, Spalletta G. ZNF804A risk allele is associated with relatively intact gray matter volume in patients with schizophrenia. Neuroimage. 2011;54(3):2132–2137. doi: 10.1016/j.neuroimage.2010.09.089. [DOI] [PubMed] [Google Scholar]
- Esslinger C, Kirsch P, Haddad L, Mier D, Sauer C, Erk S, Schnell K, Arnold C, Witt SH, Rietschel M, Cichon S, Walter H, Meyer-Lindenberg A. Cognitive state and connectivity effects of the genome-wide significant psychosis variant in ZNF804A. Neuroimage. 2011;54(3):2514–2523. doi: 10.1016/j.neuroimage.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Esslinger C, Walter H, Kirsch P, Erk S, Schnell K, Arnold C, Haddad L, Mier D, Opitz von Boberfeld C, Raab K, Witt SH, Rietschel M, Cichon S, Meyer-Lindenberg A. Neural mechanisms of a genome-wide supported psychosis variant. Science. 2009;324:605. doi: 10.1126/science.1167768. [DOI] [PubMed] [Google Scholar]
- Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, MacKay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends. Biochem. Sci. 2007;32(2):63–70. doi: 10.1016/j.tibs.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Girgenti MJ, LoTurco JJ, Maher BJ. ZNF804A regulates expression of the schizophrenia-associated genes PRSS16, COMT, PDE4B, and DRD2. PLoS ONE. 2012;7(2):e32404. doi: 10.1371/journal.pone.0032404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RP, Selman LD. Functional and anatomical aspects of prefrontal pathology in schizophrenia. Schizophr. Bull. 1997;23:437–458. doi: 10.1093/schbul/23.3.437. [DOI] [PubMed] [Google Scholar]
- Guella I, Sequeira A, Rollins B, Morgan L, Myers RM, Watson SJ, Akil H, Bunney WE, Delisi LE, Byerley W, Vawter MP. Evidence of allelic imbalance in the schizophrenia susceptibility gene of ZNF804A in human dorsolateral prefrontal cortex. Schizophr. Res. 2014;152(1):111–6. doi: 10.1016/j.schres.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Li J, Wang J, Fan X, Hu M, Shen Y, Chen H, Zhao J. Hippocampal and orbital frontal gray matter volume abnormalities and cognitive deficit in treatment-naïve, first-episode patients with schizophrenia. Schizophr. Res. 2014;152:339–343. doi: 10.1016/j.schres.2013.12.015. [DOI] [PubMed] [Google Scholar]
- Hall J, Thomas KL, Everitt BJ. Cellular imaging of zif268 expression in the hippocampus and amygdala during contextual and cued fear memory retrieval: Selective activation of hippocampal CA1 neurons during the recall of contextual memories. J. Neurosci. 2001;21(6):2186–2193. doi: 10.1523/JNEUROSCI.21-06-02186.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Ohi K, Yasuda Y, Fukumoto M, Iwase M, Iike N, Azechi M, Ikezawa K, Takaya M, Takahashi H, Yamamori H, Okochi T, Tanimukai H, Tagami S, Morihara T, Okochi M, Tanaka T, Kudo T, Kazui H, Iwata N, Takeda M. The impact of a genome-wide supported psychosis variant in the ZNF804A gene on memory function in schizophrenia. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2010;153B(8):1459–1464. doi: 10.1002/ajmg.b.31123. [DOI] [PubMed] [Google Scholar]
- Hill MJ, Bray NJ. Evidence that schizophrenia risk variation in the ZNF804A gene exerts its effects during fetal brain development. Am. J. Psychiatry. 2012;169:1301–1308. doi: 10.1176/appi.ajp.2012.11121845. [DOI] [PubMed] [Google Scholar]
- Hill MJ, Jeffries AR, Dobson RJ, Price J, Bray NJ. Knockdown of the psychosis susceptibility gene ZNF804A alters expression of genes involved in cell adhesion. Hum. Mol. Genet. 2012;21(5):1018–1024. doi: 10.1093/hmg/ddr532. [DOI] [PubMed] [Google Scholar]
- Ikuta T, Peters BD, Guha S, John M, Karlsgodt KH, Lencz T, Szeszko PR, Malhotra AK. A schizophrenia risk gene, ZNF804A, is associated with brain white matter microstructure. Schizophr. Res. 2014;155(1-3):15–20. doi: 10.1016/j.schres.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javitt DC. Glutamate and schizophrenia: phencyclidine, N-methyl-D-aspartate receptors, and dopamine-glutamate interactions. Int. Rev. Neurobiol. 2007;78:69–108. doi: 10.1016/S0074-7742(06)78003-5. [DOI] [PubMed] [Google Scholar]
- Javitt DC. Twenty-five years of glutamate in schizophrenia: are we there yet? Schizophr. Bull. 2012;38(5):911–913. doi: 10.1093/schbul/sbs100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T, Szeszko PR, DeRosse P, Burdick KE, Bromet EJ, Bilder RM, Malhotra AK. A schizophrenia risk gene, ZNF804A, influences neuroanatomical and neurocognitive phenotypes. Neuropsychopharmacology. 2010;35:2284–2291. doi: 10.1038/npp.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Nestor PG, Levitt JJ, Cohen AS, Kawashima T, Shenton ME, McCarley RW. Orbitofrontal volume deficit in schizophrenia and thought disorder. Brain. 2008;131:180–195. doi: 10.1093/brain/awm265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan MC, Craddock N, Norton N, Williams H, Pierce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Hartmann AM, Möller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Williams NM, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Gejman PV, Cichon S, Nöthen MM, Gill M, Corvin A, Rujescu D, Kirov G, Owen MJ, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Cloninger CR, Molecular Genetics of Schizophrenia Collaboration Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat. Genet. 2008;40:1053–5. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Olney JW, Newcomer JW, Farber NB. NMDA receptor hypofunction model of schizophrenia. J. Psychiatr. Res. 1999;33:523–533. doi: 10.1016/s0022-3956(99)00029-1. [DOI] [PubMed] [Google Scholar]
- Rasetti R, Sambataro F, Chen Q, Callicott JH, Mattay VS, Weinberger DR. Altered cortical network dynamics: a potential intermediate phenotype for schizophrenia and association with ZNF804A. Arch. Gen. Psychiatry. 2011;68:1207–1217. doi: 10.1001/archgenpsychiatry.2011.103. [DOI] [PubMed] [Google Scholar]
- Rash BG, Grove EA. Area and layer patterning in the developing cerebral cortex. Curr. Opin. Neurobiol. 2006;16(1):25–34. doi: 10.1016/j.conb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Riley B, Thiselton D, Maher BS, Bigdeli T, Wormley B, McMichael GO, Fanous AH, Vladimirov V, O'Neill FA, Walsh D, Kendler KS. Replication of association between schizophrenia and ZNF804A in the Irish case-control study of schizophrenia sample. Mol. Psychiatry. 2010;15(1):29–37. doi: 10.1038/mp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:621–625. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman MV. Gender differences in schizophrenia. Canadian Journal of Psychiatry. 1982;27:107–112. doi: 10.1177/070674378202700204. [DOI] [PubMed] [Google Scholar]
- Stansberg C, Vik-Mo AP, Holdhus R, Breilid H, Srebro B, Petersen K, Jørgensen HA, Jonassen I, Steen VM. Gene expression profiles in rat brain disclose CNS signature genes and regional patterns of functional specialisation. BMC Genomics. 2007;8:94. doi: 10.1186/1471-2164-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Cousijn H, Jaffe AE, Burnet PW, Edwards F, Eastwood SL, Shin JH, Lane TA, Walker MA, Maher BJ, Weinberger DR, Harrison PJ, Hyde TM, Kleinman JE. Expression of ZNF804A in human brain and alterations in schizophrenia, bipolar disorder, and major depressive disorder: a novel transcript fetally regulated by the psychosis risk variant rs1344706. JAMA Psychiatry. 2014;71(10):1112–1120. doi: 10.1001/jamapsychiatry.2014.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineskos AN, Lerch JP, Felsky D, Tiwari A, Rajji TK, Miranda D, Lobaugh NJ, Pollock BG, Mulsant BH, Kennedy JL. The ZNF804A gene: Characterization of a novel risk mechanism for the major psychoses. Neuropsychopharmacology. 2011;36:1871–1878. doi: 10.1038/npp.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter H, Schnell K, Erk S, Arnold C, Kirsch P, Esslinger C, Mier D, Schmitgen MM, Rietschel M, Witt SH, Nöthen MM, Cichon S, Meyer-Lindenberg A. Effects of a genome-wide supported psychosis risk variant on neural activation during a theory-of-mind task. Mol. Psychiatry. 2011;16(4):462–470. doi: 10.1038/mp.2010.18. [DOI] [PubMed] [Google Scholar]
- Walters JT, Corvin A, Owen MJ, Williams H, Dragovic M, Quinn EM, Judge R, Smith DJ, Norton N, Giegling I, Hartmann AM, Möller HJ, Muglia P, Moskvina V, Dwyer S, O'Donoghue T, Morar B, Cooper M, Chandler D, Jablensky A, Gill M, Kaladjieva L, Morris DW, O'Donovan MC, Rujescu D, Donohoe G. Psychosis susceptibility gene ZNF804A and cognitive performance in schizophrenia. Arch. Gen. Psychiatry. 2010;67(7):692–700. doi: 10.1001/archgenpsychiatry.2010.81. [DOI] [PubMed] [Google Scholar]
- Wei Q, Li M, Kang Z, Li L, Diao F, Zhang R, Wang J, Zheng L, Wen X, Zhang J, Zhao J, Huang R. ZNF804A rs1344706 is associated with cortical thickness, surface area, and cortical volume of the unmedicated first episode schizophrenia and healthy controls. Am J Med Genet B Neuropsychiatr Genet. 2015;168B(4):265–73. doi: 10.1002/ajmg.b.32308. [DOI] [PubMed] [Google Scholar]
- Williams HJ, Norton N, Dwyer S, Moskvina V, Nikolov I, Carroll L, Georgieva L, Williams NM, Morris DW, Quinn EM, Giegling I, Ikeda M, Wood J, Lencz T, Hultman C, Lichtenstein P, Thiselton D, Maher BS, Molecular Genetics of Schizophrenia Collaboration (MGS) International Schizophrenia Consortium (ISC), SGENE-plus, GROUP. Malhotra AK, Riley B, Kendler KS, Gill M, Sullivan P, Sklar P, Purcell S, Nimgaonkar VL, Kirov G, Holmans P, Corvin A, Rujescu D, Craddock N, Owen MJ, O'Donovan MC. Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol. Psychiatry. 2011;16(4):429–441. doi: 10.1038/mp.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lu SM, Qiu C, Liu XG, Gao CG, Guo TW, Valenzuela RK, Deng HW, Ma J. Population-based and family-based association studies of ZNF804A locus and schizophrenia. Mol. Psychiatry. 2011;16(4):360–361. doi: 10.1038/mp.2010.55. [DOI] [PubMed] [Google Scholar]
- Zhong X, Tumang JR, Gao W, Bai C, Rothstein TL. PD-L2 expression extends beyond dendritic cells/macrophages to B1 cells enriched for V(H)11/V(H)12 and phosphatidylcholine binding. European Journal of Immunology. 2007;37:2405–10. doi: 10.1002/eji.200737461. [DOI] [PubMed] [Google Scholar]