Summary
Plant resistance genes (R-genes) harbor tremendous allelic diversity, constituting a robust immune system effective against microbial pathogens. Nevertheless, few functional R-genes have been identified for even the best-studied pathosystems. Does this limited repertoire reflect specificity, with most R-genes having been defeated by former pests, or do plants harbor a rich diversity of functional R-genes whose composite behavior is yet to be characterized? Here, we survey 332 NBS-LRR genes cloned from 5 resistant rice cultivars for their ability to confer recognition of 12 rice blast isolates when transformed into susceptible cultivars. Our survey reveals that 48.5% of the 132 NBS-LRR loci tested contain functional rice blast R-genes, with most R-genes deriving from multi-copy clades containing especially diversified loci. Each R-gene recognized, on average, 2.42 of the 12 isolates screened. The abundant R-genes identified in resistant genomes provide extraordinary redundancy in the ability of host genotypes to recognize particular isolates. If the same is true for other pathogens, many extant NBS-LRR genes retain functionality. Our success at identifying rice blast R-genes also validates a highly efficient cloning and screening strategy.
Keywords: plant resistance genes, Oryza sativa, Magnaporthe oryzae, rice blast, genome-wide survey
Introduction
In the last two decades, great progress has been made in understanding plant resistance to pathogens at the molecular level (Yue et al., 2012; Michelmore et al., 2013; Yang et al., 2015) . We now know that plants harbor a large number of candidate resistance genes, most of which are nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. When the protein encoded by an NBS-LRR recognizes a pathogen effector, either directly or indirectly, a defense response is initiated. The pairwise associations between these R-genes and avirulence (Avr) effectors is termed gene-for-gene resistance (Flor, 1971). Plant NBS-LRR genes are highly diversified within and between individual plants (Yang et al., 2006), and thus the gene-for-gene system has the potential to confer resistance to a great diversity of pathogens .
The specificity implied by the gene-for-gene system suggests that plant-pathogen coevolution should be evolutionarily dynamic. Indeed, resistance bred into agricultural crops is typically overcome in ecological time. Nevertheless, there are some celebrated instances of durable resistance in which major R-genes (or monogenic R-genes) confer stable resistance to specific races of a pathogen (Michelmore et al., 2013). For example, pepper's durable resistance to Xanthomonas vesicatoria is conferred by the major R-gene Bs2 (Tai et al., 1999), presumably because deletion of AvrBs2 in the pathogen imposes a substantial fitness penalty(Wichmann and Bergelson, 2004). In other cases, the pyramiding of R-genes has been shown to enhance the effectiveness of resistance (Houterman et al., 2008). Pyramiding should be especially effective when particular R-genes are able to recognize multiple effectors (Kvitko et al., 2009; Mukhtar et al., 2011) and/or multiple pathogen isolates (Cesari et al., 2013; Mach, 2013). There is a growing perception that a multiple genes-for-genes model is more appropriate for understanding plant-pathogen interaction (Cook et al., 2012; Lozano-Torres et al., 2012) due to the importance of functional redundancy. However, in most instances, it is unclear whether durable resistance results from single R-genes like Bs2 (Wichmann et al., 2005) or multiple, functional R-genes (Spielmeyer et al., 2013). This information is crucial for developing strategies to promote durable resistance in agriculture.
Determining the genetic basis of durable resistance requires analysis of pathogenicity phenotypes in response to a large number of R-genes in the genome. Recently, a high throughput method of R-gene identification (Yang et al., 2013) enables the cloning of many R-genes. This technique contains three essential components: cloning candidate genes, transferring cloned genes into susceptible lines, and confirming the resistance of the transgenic lines through infection with pathogen isolates. The data thus generated allows assessment of the specificity of particular R–genes and the extent of functional redundancy in recognizing a pathogen.
Here, we complete a genome-wide survey of the ability of R-genes in rice to recognize rice blast, the fungus Magnaporthe oryzae. The rice-Magnaporthe interaction is perhaps the best-studied pathosystem to date (Liu et al., 2010; Huang et al., 2014). There are about 480 NBS-LRR genes in a single rice genome and thousands of variants among populations (Yang et al., 2006). Twenty-seven blast R-genes from 11 NBS-LRR loci are known to be effective against isolates of the rice blast disease (Liu et al., 2007; Yang et al., 2013). Historical records also indicate cultivars with effective resistance. In particular, there are two cultivars, Tetep (TTP) and Gumei 2 (GM2), that are well-known to be highly and durably resistant resources to M. oryzae (Ou and Jennings, 1969; Peng et al., 1996). In addition, a moderately resistant indica variety, Tadukan (Kitamura, 1954), a newly bred cultivar Q2436 (Q), and a widely used cultivar called Minghui 63 (MH63) show strong resistance (Lin and Min, 1991). We cloned 332 NBS-LRR genes from five resistant cultivars and confirmed via infection with 12 diverse isolates of M. oryzae that 98 of them are functional rice blast R-genes. These NBS-LRR genes provide redundancy in highly resistant cultivars to recognize each of the isolates. Functional R-genes tend to be located at the loci with high allelic diversity and within multi-copy gene clades. About 15% of the 98 R-genes identified recognize five or more highly diverse isolates of rice blast, and thus provide broad-spectrum resistance.
Results
Cloning candidate NBS-LRR genes
To obtain a representative sample of genes, we categorized all NBS-LRR genes in the rice genome based on their genomic and evolutionary features. We then randomly sampled genes proportionately within each category as candidates for cloning. Copy number variation and allelic diversity are often used to characterize the NBS-LRR genes as conserved or rapidly-evolving (Stahl et al., 1999; Bergelson et al., 2001; Bakker et al., 2006; Yang et al., 2006; Si et al., 2015). A phylogenetic tree of 937 NBS-LRR genes from two cultivars of Oryza sativa, Nipponbare and 9311, was used to provide this information (Yang et al., 2006). In this tree, independent branches were determined by relatively high bootstrap values (>70% in Figure S1) with the branches adjacent to the root denoted as clades (Figure S2 for details). We only considered those clades containing genes from Nipponbare, which is the most complete of the genome sequences. In total, there are 96 single-copy and 115 multi-copy NBS-LRR clades, where we define a multi-copy clade as one containing two or more genes from one genome (Figure S1). For cloning candidate R-genes, we randomly selected half of the clades, 48 that are single-copy and 65 that are multi-copy.
To clone genes, multiple primer pairs were designed for long PCRs in five rice cultivars with varying degrees of resistance to rice blast (Table S1-S2). Due to the difficulty of long PCRs and presence/absence polymorphisms among cultivars, a total of only 332 NBS-LRR candidates (Table S3) were successfully cloned and sequenced (Figure 1a). Our candidates derived from 132 loci (definition in Experimental Procedures) of 56 multi-copy and 39 single-copy clades (Figure S1), in which three clades (Rp1/Pi37, AC134922 and Rp3/Pc clades) were known previously to contain functional rice blast R-genes (Yang et al., 2013). In most cases, we cloned the native promoter and terminator along with the candidate R gene although in 6% of cases we used these elements from Pi9 (details in Table S3).
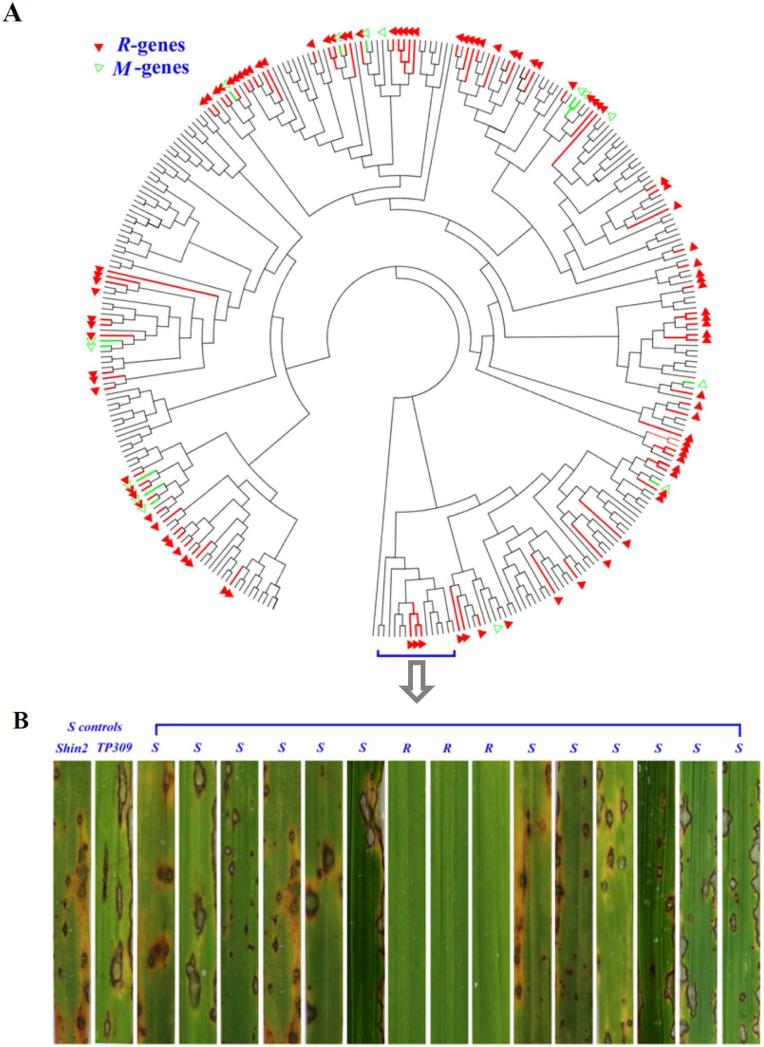
Figure 1. Geneaology of 332 cloned NBS-LRR genes from five resistant cultivars (a) and examples of phenotypes to rice blast disease (b).
Functionally resistant R-genes are indicated in red, and R-genes that confer moderate resistance are indicated in green. A gene that conferred resistance in one susceptible background but moderate resistance in the other is indicated in red. Other NBS-LRR genes do not confer an ability to recognize rice blast; these include 11 pseudogenes. The two leftmost examples in (b) are susceptible cultivars that served as controls. The gene genealogy was constructed using the bootstrap NJ method with p-distance model by MEGA v5.02.
Identification of functional blast R-genes
The evaluation of resistance requires transforming cloned candidate genes into at least one susceptible line for screening the response to infection by pathogen isolates. We selected two susceptible japonica cultivars (S-cultivars), TP309 and Shin2, as transgenic recipients (Table S1). Twelve rice blast isolates from 85 isolates collected in rice fields throughout China were chosen based on their ability to produce large numbers of spores and to span the geographical range of the pathogen (Table S4). Subsequent analysis at 11 loci, including six Avr genes, confirmed that each isolate is distinct (see the phylogenetic tree in Figure S3).
The transformation of these candidates into two S-cultivars resulted in 456 lines with enough T1 or T2 generation seeds to allow screening of the ability to recognize blast isolates. Each recognition assay included two S-cultivars (TP309 and Shin2), two R-cultivars (TTP and GM2), and 28 transgenic lines (>10 plants per line or control), all grown together in a plate until they had four to five true leaves, at which point they were subjected to a standard procedure of infection (Bonman et al., 1986) using each of nine blast isolates (selected from the 12 cultured isolates based on availability of spores on that test date). Seven days later, each transgenic line was scored as either resistant (R) or susceptible (S) based on a visual comparison to the R- and S-controls (see Experimental Procedures for details). Entire plates were replicated three times; for a transgenic line to be designated as R, all three replicates must have shown an R-phenotype. Resistance was further confirmed with a second round of infection using the same isolate, again with three replicates and with negative transgenic controls (transgenic lines containing an empty plasmid). For these confirmations, we additionally included in each plate the S-host genotype transformed with the well-known blast R-genes Pi9 and Pi37 (Qu et al., 2006; Lin et al., 2007). In total, 98 (29.5%) R-genes (Figure 1a-b) from 64 loci were identified as resistant to one or more blast isolates. In our experiment, these R-genes were presented in 122 (26.8%) R-lines (pooling across S backgrounds), including 11 R-lines reported recently (Yang et al., 2013) (Table S5). As expected, 11 pseudogenes, identified by sequencing the 332 cloned candidates, were susceptible to all isolates (Table S5).
We confirmed that the resistance identified from transgenic rice lines was not due to the autoimmunity of NBS-LRR gene transformation. In this scenario, resistant lines would not be responsive against only rice blast. We infected three-week-old seedlings of 11 broad-spectrum resistant lines with two Xanthamonas oryzae pv. Oryzae (Xoo) races (Table S6). Compared with the phenotypes of both resistant (DV85) and susceptible controls (Shin 2 and TP309) (Sun et al., 2004) in the same experiment, all the 11 broad-spectrum resistant lines were susceptible to Xoo, as were the transgenic recipients, TP309 and Shin2, indicating that the identified functional NBS-LRRs are not due to auto-activation.
To test the relevance of our resistance score to resistance under field conditions, panicle blast resistance was tested in a subset of 18 transgenic lines in a paddy field in Nanjing China (Table S7). The field experiments confirmed that the eight out of nine R-lines as determined in our controlled conditions were also resistant to panicle blast disease in the field. All nine S-lines sampled were susceptible to both leaf and panicle blast infection.
Distribution of R-genes in genomes
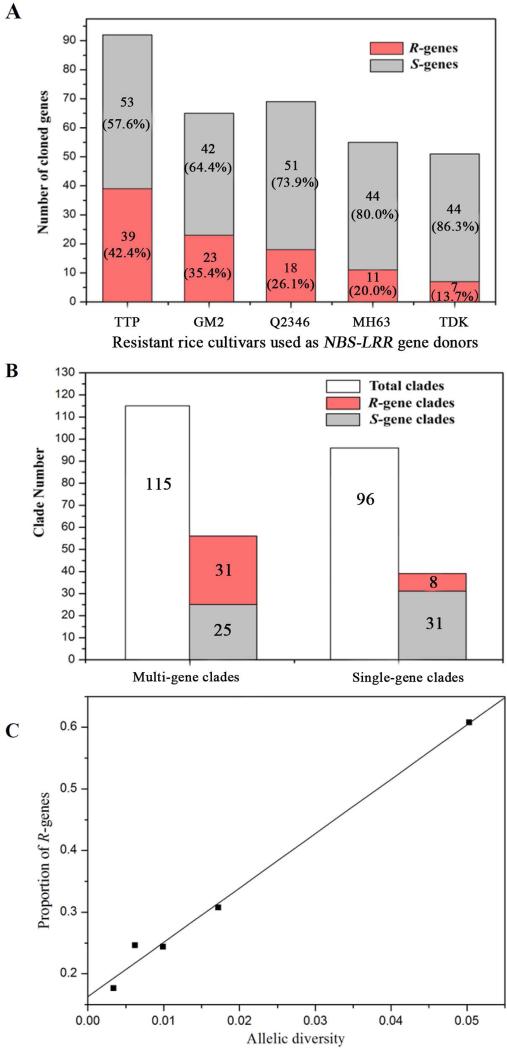
Our survey revealed three patterns. First, a greater proportion of NBS-LRR genes conferred the ability to recognize rice blast when they derived from the most broadly resistant cultivars as based on both published observations (Ou and Jennings, 1969; Peng et al., 1996) and our screen of rice blast isolates (Table S8). In particular, 38.9% of R-genes cloned from TTP and GM2, the well-known durably resistant cultivars (Ou and Jennings, 1969; Peng et al., 1996), encoded an ability to recognize one or more of the 12 isolates whereas only ~20.0% of the other three cultivars did so (χ2 = 7.1, df=1 for a 2 by 2 table, P = 0.008; Figure 2a). Second, functional R-genes are distributed broadly across the genealogy of NBS-LRR genes (Figure 1a) with representatives conferring recognition of rice blast present in 39 out of 95 clades containing cloned genes (Figure S1). The broad distribution of functionally important R-genes suggests that most, if not all, clades have the potential to contribute genes with the ability to recognize pathogen effectors. Third, an analysis of the 25 multi-clades with ≥5 cloned genes (see Table S9 for significance test) revealed a significant clustering of functionally important R-genes. This clustered pattern suggests that closely related NBS-LRR genes have a tendency to recognize the same pathogen.
Figure 2. General patterns of R-gene distributions.
The numbers (percentages) of functionally resistant and susceptible R- genes from (a) five R-cultivars, (b) from multi-copy and single-copy clades and (c) from groups with different levels of allelic diversities. Allelic diversities were calculated for each of 129 cloned loci (excluding three loci that have no 9311 allele). Alleles are the least divergent genes from Nipponbare and 9311 at the same locus. Loci were sorted into five groups (each with 26 loci except the group on the right which is based on 25 loci) based on their levels of allelic diversity.
Abundant functional R-genes occur in multi-gene clades and at diversified loci
We observed redundancy for the ability to recognize each of 12 isolates. Between three and 17 of the 92 NBS-LRR genes cloned from TTP confer resistance to each of the isolates used in this study (Table S5). The R-genes capable of recognizing each isolate are located in at least two independent loci (on different chromosomes), revealing great redundancy in this highly resistant variety. Redundancy was also observed in the two cultivars GM2 and Q2436 with respect to their ability to recognize nine of the 12 isolates, although fewer candidate genes were cloned from these cultivars (Figure 2a).
This redundancy resides largely in multi-copy clades, which are well known to have higher copy number variation, higher polymorphism and higher Ka/Ks ratios relative to single R-genes (Michelmore and Meyers, 1998; Bergelson et al., 2001; Kuang et al., 2004; Yang et al., 2006). These multi-copy clades contribute functional R-genes disproportionately: 55.4% of NBS-LRR genes that we screened from multi-copy clades conferred resistance whereas only 20.5% of NBS-LRR genes from single-copy clades did so (Figure 2b; χ2 =10.14, df=1 for a 2 by 2 table, P =0.001). Furthermore, we observed a positive correlation between allelic nucleotide diversity at a locus and the proportion of functional blast R-alleles at this locus (P<0.001; Figure 2c). The 20% most highly diversified loci in Figure 2c have three-fold more R-genes identified than the 20% with the lowest level of allelic diversity. These features of diversified loci likely enhance the potential to deal with rapidly evolving pathogens such as rice blast (McDonald and Linde, 2002; Valent and Khang, 2010; Chuma et al., 2011; Cook et al., 2012).
Recognition rate of pathogen isolates by R-genes
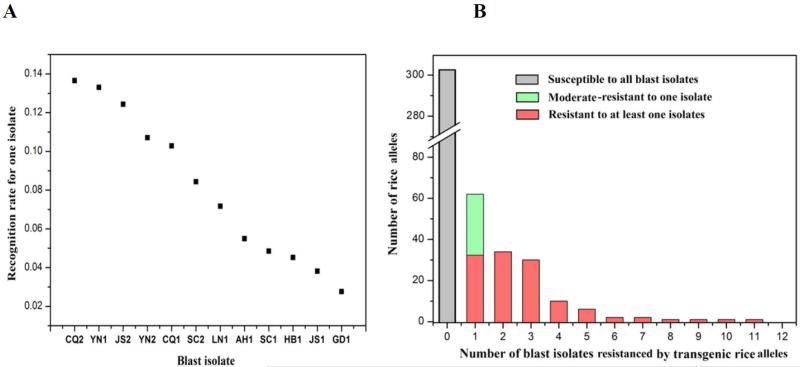
Pathogen isolates varied in the proportion of screenings (2.8% - 13.7%; Figure 3a) that led to recognition (defined here as recognition rate, RR), with an average RR of 8.1%. In other words, roughly 8% of NBS-LRR genes could recognize each pathogen isolate. Consistent with this average, a blind and independent screening of a novel isolate on 110 of our transgenic lines resulted in eight functional R-genes (7.3%), including two new ones (Table S10). We found that 15 R-genes provided broadly effective resistance, leading to the ability to recognize five or more isolates (Figure 3b; Table S5 & 11). One of these R-genes conferred resistance to all of the isolates screened (Os12g36730-TTP). Interestingly, 11 of the 15 broadly effective R-genes derive from the most durably resistant varieties, TTP and GM2, and all 15 are from multi-copy clades (Table S11). Notably, none of the previously identified 27 R-genes (Liu et al., 2010; Yang et al., 2013) fall into clades with broadly effective R-genes, indicating that our systematic cloning could guide discovery of broad-spectrum R-genes in highly resistant cultivars for plant breeding.
Figure 3. The relationship between isolates and cloned genes.
The distribution of recognition rate (RR) for each of 12 isolates by (a) 122 R-lines identified and (b) all 456 transgenic lines screened against the 12 isolates.
The reliability of our identification of R-genes was tested with segregation analysis. The ratio of resistant to susceptible individuals in progeny segregating from the T0 plants should be 3:1 for single insertions and >3:1 for multiple insertions. Infection records of individual plants revealed that segregation ratios did not differ significantly from 3:1 (Chi-square test, P > 0.1; Table S12) in the majority of R-lines, including all the 15 broadly effective R-gene lines.
Expression of cloned genes in transgenic lines
The two japonica S-cultivars used as recipients of transformed R-genes yielded quite consistent results. Of the 332 R-candidate genes that we screened, 124 were successfully transferred into both S-cultivars and screened by the same pathogen strains. Ninety-eight of these 124 NBS-LRR genes gave consistent results in the two backgrounds (24 R and 74 S), suggesting the general reliability of identifying functionally resistant alleles despite variation in genetic backgrounds and insertion sites. In the remaining 21% of comparisons, differences in resistance phenotype could result from either epistatic interactions or differences in the expression level of the transgene.
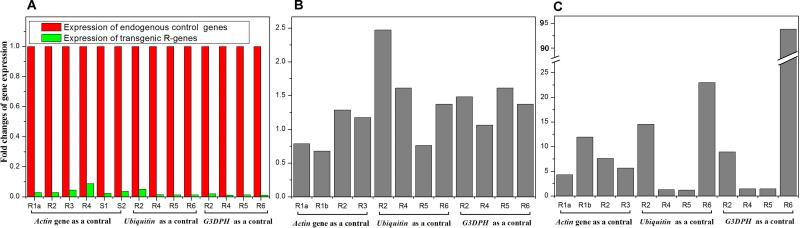
To explore the possibility that differences in the resistance phenotypes were due to differences in expression across the two S- cultivars, we directly measured expression levels of our transgenes. Three sets of comparisons were carried out: between transgenes and an endogenous gene (the Actin, Ubiquitin and G3DPH genes as controls) (Hayashi and Yoshida, 2009), between transgenic lines and donor plants, and between lines before and after pathogen inoculation (Figure 4a-c, respectively). These expression experiments showed that: (i) the expression levels of transgenic NBS-LRR genes are much lower than their endogenous controls, with no significant difference between R- and S-lines (Figure 4a), which is consistent with previous reports that, NBS-LRR genes are typically expressed at low levels relative to endogenous controls (Hulbert et al., 2001); (ii) the expression levels of NBS-LRR genes in transgenic individuals are similar to that in the donor cultivar (Figure 4b); and (iii) the expression levels of NBS-LRR genes in transgenic individuals with pathogen inoculation are still low but higher than in those without infection (Figure 4c), consistent with previously reported results (Levy et al., 2004).
Figure 4. Comparison of gene expression levels in transgenic lines (a), between transgenic lines and donor plants (b), and between the lines after and before pathogen inoculation (c).
R1a, Os06g06390-TTP-SH; R1b, Os06g06390-TTP-TP; R2, Os12g36730-TTP-SH; R3, Os11g35210-TTP-SH; R4, Os08g01580-GM2-TP; R5, Os06g15750-TTP-SH; R6, 08g15880-GM2-SH; S1, 02g27540-TTP-SH; S2, Os12g29170-TTP-SH. Those lines were named as genome position-donor variety-transgenic recipient.
R-genes with moderate resistance
In addition to the functional R-genes discussed above, we discovered a considerable percentage of R-genes with moderate resistance (M-phenotype, as defined in Experimental Procedures). These lines demonstrated weak and less consistent resistance. On average, 30 R-genes showed moderate resistance to a particular isolate; this is almost equivalent to the 32 R-genes that provided consistent resistance to single isolates (Figure 3b). All but one of the moderately resistant R-genes was found in a clade with R-genes providing consistent resistance (marked by green in Figure 1a and Figure S4). Moderate resistance may reflect a beneficial co-evolution between host and pathogen in which host damage is reduced due to limited pathogen growth, but pathogens survive. Indeed, moderate resistance is a practical goal for resistance breeding in crops. The prevalence of moderately resistant R-genes in our experiments suggests that our estimate of the number of R-genes is conservative.
Discussion
Our results demonstrate that a large number of R-loci conferring resistance to rice blast are retained in durable cultivars. On average, 41% of the clades (up to 86 out of 211 clades) or 48.5% of the loci (up to 208 in a total of 430 Nipponbare loci) in a rice genome contribute to rice blast resistance, revealing a highly redundant surveillance system. Indeed, 42.4% of R-gene candidates cloned from TTP were confirmed to be functional, providing insight into the 40 year durability of this well-known resistant germplasm (Ou and Jennings, 1969). In contrast, the two susceptible japonica cultivars that served as transgenic recipients have no resistance to any of the screened isolates in any of our hundreds of experimental trials, although they revealed a range of resistance phenotypes when transformed with indica resistance genes, suggesting that the R-genes identified in our study are highly reliable. Four of the 65 cloned candidate genes from GM2 (Os06g15730, Os06g17880, Os06g17950 and Os06g22460) are located on the rice blast resistance QTL loci identified by (Wu et al., 2005), and all four showed moderate to full resistance. The colocalization of our cloned R-genes with these resistance QTL (Wu et al., 2005) also suggests reliability.
Our rapid identification of 98 R-genes capable of recognizing rice blast demonstrates that systematical cloning is a feasible and efficient method to identify un-reported and broadly effective R-genes. For example, despite their rarity, our study identified 15 broadly effective resistant genes. These R-genes, alone or in combination, may be especially effective in production. To further improve efficiency, the distribution of R-genes can be used to guide the selection of candidate genes by focusing on blast R-genes in multi-gene clades, because these clades contain highly diverse loci with a higher proportion of functional R-genes. Our methods should thus be useful in the search for resistance to a variety of economically and ecologically important diseases, like potato late blight and wheat rust.
Impressively, there is tremendous variation among R-genes in the breadth of rice blast resistance that they encode. While ~15% of R-genes conferred broad resistance to recognize five or more isolates, most of them recognized only 1-3 isolates. The reason for this variation is unknown, but may simply be a function of the prevalence of corresponding Avr genes. Interestingly, the well-known rice blast genes, Pi9 and Pi37, conferred recognition of only two to three of the isolates. These R-genes are thus not broadly resistant (Figure 3b), perhaps explaining their failure to provide durable resistance in agronomic production. It is also interesting that all blast R-genes cloned from maize, sorghum, and brachypodium (Yang et al., 2013) provided a narrow spectrum of resistance (recognizing 1-3 isolates) when transferred into rice, suggesting the rarity of broadly resistant genes in rice relatives. In contrast, our study identified 15 broadly effective resistant genes in a total of 98 R-genes. These R-genes, alone or in combination, could be especially effective in production.
Our genome-wide survey of rice blast R-genes revealed a highly redundant surveillance system for recognizing this rapidly-evolving pathogen. It is tempting to speculate that this pattern reflects a co-evolutionary dynamic particular to rapidly evolving pathogens such as rice blast, and that more conserved R-genes may provide protection against endemic and/or generalist pathogens. Another interesting possibility is that the common proteins or pathways shared by some R-genes contribute to the redundancy we observed (Innes, 1998; Gassmann and Bhattacharjee, 2012). For example, the resistance genes RPM1, RPS2, and RPS5 in Arabidopsis share the NDR1 pathway for conferring resistance. While an interesting possibility, our experiments cannot address this hypothesis. Our results (Figure 2b-c) show that most of NBS-LRR genes in single-gene clades or at the loci with low diversity do not confer resistance to blast disease. This tendency suggests a parallel co-evolution between NBS-LRR genes and their corresponding pathogen effectors: quickly-evolving or diversified loci in hosts may match quickly-evolving or diversified loci in pathogens. Systematic cloning of many R-genes in disparate systems could provide a means of augmenting the set of functional R-genes that guide our understanding of mechanisms and patterns of resistance variation across systems.
Experimental Procedures
Donor cultivars
Five indica cultivars served as donors for cloning of NBS-LRR genes. Two of these cultivars, Tetep and Gumei 2, are well-known to be highly and durably resistant against M. oryzae (Ou and Jennings, 1969; Peng et al., 1996). The other three include the moderately resistant Tadukan, often used as a parent in the 1960s, the newly bred cultivar Q2436, and Minghui 63, the cultivar most widely used in China in the 1980s and early 1990s.
Gene cloning and transformation
Seeds of rice accessions were obtained from various resources (Table S1). Genomic DNA was extracted from fresh leaves using the CTAB method. The genes for cloning were attributed Nipponbare accession numbers because Nipponbare is the only cultivar with complete sequences available. Multiple primer pairs were designed based on the Nipponbare sequence and relatively conserved primer sequences were selected based on a comparison to the 9311 genome (Table S2). The products of long-range PCRs, generally including the native promoter and terminator, were inserted into the binary vector pCAMBIAI3000. Due to the difficulty of PCRs for some clades, a vector, containing a promoter and terminator from the blast R-gene Pi9, was constructed to clone their coding sequences (Table S3). All clones, validated by sequencing, were transferred into blast-susceptible japonica cultivars by Agrobacterium strain EHA105. Transgenic lines (T0) expressing 332 NBS-LRR genes in one or two S-cultivar backgrounds (for a total of 456 plant lines, including 26 lines from 3 multi-copy clades described in (Yang et al., 2013)) were grown to obtain sufficient seeds (T1 or T2). For each transgenic line, 4-12 independently transformed plants were collected for infection assays. For each transgenic T0 plant, the presence of the transgenic DNA fragments was confirmed with PCR. Sequences for all cloned genes have been deposited in the GenBank under accession number of KP401593 to KP401756.
Infection Assays using rice blast strains
Rice blast strains were collected from different areas of China in 2008-2009 (Table S4). T1-T2 plants were used to evaluate blast resistance (Hayashi et al., 2004). For the test of leaf blast resistance, three-week-old seedlings were inoculated by spraying a blast spore suspension on their leaves (5×105 spores/ml) and were then placed in a dew inoculation incubator at 26°C and 100% humidity in the dark for 24h. The plants were then transferred to a greenhouse with a 12/12h light/dark photoperiod at 90% relative humidity for 6-7 days until the disease spots could be seen clearly on the leaves. The disease reaction was scored according to the description of Bonman et al. (Bonman et al., 1986), in which six grades (0-5) were defined. In Bonman's work, scores of 0-3 were considered resistant reactions and scores of 4 - 5 were considered susceptible reactions. To obtain robust results, we applied more stringent criteria: scores of the resistant controls (TTP and GM2) were required to be 0 (no evidence of infection) or 1 (brown specks smaller than 0.5 mm in diameter and no sporulation), whereas the scores of the susceptible controls (the transformation recipient cultivars of SH2 and TP309) were required to be 4 (typical spindle-shaped blast lesions capable of sporulation, 3 mm or longer with necrotic gray centers and water-soaked or reddish brown margins, little or no coalescence of lesions) or 5 (lesions as in 4 but about half of one or two leaf blades killed by coalescence of lesions) in every screening; each transgenic line was screened with three replicates using the same isolate and only these lines with consistent scores of 0 or 1 were defined as R (resistance) lines, 2 or 3 were defined as M-phenotype (moderate resistance), 4 or 5 were defined as S (susceptibility) (Figure S5). The resistant candidate was further confirmed with a second round of infection using the same isolate, again with three replicates. Thus our R-lines have strong resistance and a consistently repeatable R-phenotype whereas our M-lines demonstrated weaker and less consistent resistance.
RNA Extraction and qRT-PCR Analysis
The total RNA was isolated from leaves using the RNAiso Plus (Takara) according to the manufacturer's instructions. First-strand cDNA synthesis was carried out using a PrimeScript 1st Strand cDNA Synthesis Kit (Takara) according to the manufacturer's instructions. Real-time qPCR was carried out using a CFX96 (BioRad) real-time PCR system. Gene-specific primers were designed using Oligo 6 software. All quantitative reverse transcription-PCR was carried out in triplicate. PCR was performed using the following cycling parameters: 95 °C for 2 min, and 40 cycles of 95 °C for 15s and 60 °C for 30s. Gene expression was quantified using the ΔΔCT method in comparison with the endogenous gene controls of Actin, Ubiquitin and G3DPH.
Fold change of gene expression was used to show the relative level of R-gene expression divided by the internal gene (Actin, Ubiquitin and G3DPH gene), the same R-gene in donor plant or the same transgenic plant before inoculation, respectively in Figures 4a-c. Six R-lines and two S-lines were measured in Figure 4a. Six R-genes from TTP or GM2 in transgenic lines were compared with these genes in TTP or GM2 (Figure 4b). The isolate JS2 was used to inoculate the transgenic plants. The leaves after 48 hours inoculation were sampled to compare with these samples before inoculation (Figure 4c).
Statistical Analysis
Chi-square tests were used to detect the difference in R-genes between TTP and GM2 combined versus the other 3 cultivars, or between single- and multi-copy clades (df=1). To detect the significance of an uneven distribution in different clades, the 25 clades with ≥5 cloned genes in Figure S1 were considered. The observed number of R-genes in each clade was tested against the expected as generated by simulating their random distribution among clades, repeated 10,000 times (details in Table S8).
To calculate allelic diversity between Nipponbare and 9311, each Nipponbare gene was denoted as a locus to compare with a 9311 allele. However, when two or more Nipponbare genes co-occurred within a branch at the last layer, the one most similar to an allele from 9311 was used to calculate allelic diversity for this locus (Figure S2).
Supplementary Material
Supporting information
Additional file 1: Figure S1. Phylogenetic tree of 937 NBS-LRR genes from two cultivars. Figure S2. Illustration of a clade or locus in Supplementary Figure S1. Figure S3. Phylogenetic tree of 12 rice blast strains. Figure S4. Phylogenetic trees of cloned NBS-LRR genes from five resistant rice cultivars. Figure S5. Phenotypes of control rice cultivars.
Additional file 2: Table S1. List of rice cultivars used in this study. Table S2. List of the 155 primer pairs of these cloned genes. Table S3. List of the cloned genes and their donors. Table S4. Locations of the collected isolates of M.oryzae used in this study. Table S5. List of 122 R-lines identified and their R-phenotypes specifically to pathogen isolates screened in this study. Table S6. Phenotypic results of transgenic rice lines inoculated by two Xoo races. Table S7. Evaluation of rice panicle blast resistance in an experimental paddy field. Table S8. Phenotypes of five resistant and two highly susceptible rice cultivars screened by 21 isolates. Table S9. Ten clades identified from the cloned 332 candidate genes which contain significantly more blast R-genes relative to th9at expected based from a random distribution among clades. Table S10. Eight R-genes identified from 110 transgenic lines screened independently by another blast isolate. Table S11. Patterns of broadly effective R-genes. Table S12. Co-segregation analysis of the 15 broadly effective R-genes during infection of isolates.
Significance statement.
Resistance genes (R-genes) provide protection against pathogens, but relatively few have been characterized. Here we surveyed the rice genome and identified 98 R-genes capable of recognizing one or more isolates of rice blast, nearly twice the number previously identified against any pathogen. These results provide the most comprehensive picture to date of the architecture of resistance and overturn the perception that most R-genes in genomes are relics that have been overcome by pathogens.
Acknowledgements
This work was supported by National Natural Science Foundation of China (91331205 and 91231102), NSFC of Jiangsu province (BK2011015) and PCSIRT (IRT_14R27) to D.T. or J-Q. C., NIH grant (GM057994) to JB and NSFC (U1131003) to Q. P.
Footnotes
Accession number: KP401593 to KP401756
Conflict of interests
The authors declare that they have no Conflict of interests.
References
- Bakker EG, Toomajian C, Kreitman M, Bergelson J. A Genome-Wide Survey of R Gene Polymorphisms in Arabidopsis. Plant Cell. 2006;18:1803–1818. doi: 10.1105/tpc.106.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Kreitman M, Stahl EA, Tian D. Evolutionary dynamics of plant R-genes. Science. 2001;292:2281–2285. doi: 10.1126/science.1061337. [DOI] [PubMed] [Google Scholar]
- Bonman JM, Dedios TV, Khin MM. Physiologic specialization of Pyricularia oryzae in the Philippines. Plant Dis. 1986;70:767–769. [Google Scholar]
- Cesari S, Thilliez G, Ribot C, et al. The Rice Resistance Protein Pair RGA4/RGA5 Recognizes the Magnaporthe oryzae Effectors AVR-Pia and AVR1-CO39 by Direct Binding. Plant Cell. 2013;25:1463–1481. doi: 10.1105/tpc.112.107201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma I, Isobe C, Hotta Y, et al. Multiple translocation of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 2011;7:e1002147. doi: 10.1371/journal.ppat.1002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Lee TG, Guo X, et al. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science. 2012;338:1206–1209. doi: 10.1126/science.1228746. [DOI] [PubMed] [Google Scholar]
- Flor HH. Current Status of Gene-for-Gene Concept. Annu. Rev. Phytopathol. 1971;9:275–296. [Google Scholar]
- Gassmann W, Bhattacharjee S. Effector-triggered immunity signaling: from gene-for-gene pathways to protein-protein interaction networks. Mol. Plant-Microbe Interact. MPMI. 2012;25:862–868. doi: 10.1094/MPMI-01-12-0024-IA. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Hashimoto N, Daigen M, Ashikawa I. Development of PCR-based SNP markers for rice blast resistance genes at the Piz locus. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2004;108:1212–1220. doi: 10.1007/s00122-003-1553-0. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yoshida H. Refunctionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter. Plant J. 2009;57:413–25. doi: 10.1111/j.1365-313X.2008.03694.x. [DOI] [PubMed] [Google Scholar]
- Houterman PM, Cornelissen BJC, Rep M. Suppression of Plant Resistance Gene-Based Immunity by a Fungal Effector. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Si W, Deng Q, Li P, Yang S. Rapid evolution of avirulence genes in rice blast fungus Magnaporthe oryzae. [July 17, 2015];BMC Genet. 2014 15:45. doi: 10.1186/1471-2156-15-45. Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert SH, Webb CA, Smith SM, Sun Q. Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- Innes RW. Genetic dissection of R gene signal transduction pathways. Curr. Opin. Plant Biol. 1998;1:299–304. doi: 10.1016/1369-5266(88)80050-5. [DOI] [PubMed] [Google Scholar]
- Kitamura SCE. Breeding of blast resistant varieties by hybridization between japonica and indica paddy rices [in Japanese]. J Agric Sci. 1954;9:321–323. [Google Scholar]
- Kuang H, Woo S-S, Meyers BC, Nevo E, Michelmore RW. Multiple Genetic Processes Result in Heterogeneous Rates of Evolution within the Major Cluster Disease Resistance Genes in Lettuce. Plant Cell. 2004;16:2870–2894. doi: 10.1105/tpc.104.025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitko BH, Park DH, Velásquez AC, Wei C-F, Russell AB, Martin GB, Schneider DJ, Collmer A. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 2009;5:e1000388. doi: 10.1371/journal.ppat.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Edelbaum O, Sela I. Tobacco mosaic virus regulates the expression of its own resistance gene N. Plant Physiol. 2004;135:2392–2397. doi: 10.1104/pp.104.044859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Chen S, Que Z, Wang L, Liu X, Pan Q. The blast resistance gene Pi37 encodes a nucleotide binding site leucine-rich repeat protein and is a member of a resistance gene cluster on rice chromosome 1. Genetics. 2007;177:1871–80. doi: 10.1534/genetics.107.080648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Min S. Rice Varieties and Their Genealogy in China. Shanghai Sci. Technol. Press. 1991:414. [Google Scholar]
- Liu J, Liu X, Dai L, Wang G. Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J Genet Genomics. 2007;34:765–76. doi: 10.1016/S1673-8527(07)60087-3. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang X, Mitchell T, Hu Y, Liu X, Dai L, Wang G-L. Recent progress and understanding of the molecular mechanisms of the rice-Magnaporthe oryzae interaction. Mol. Plant Pathol. 2010;11:419–427. doi: 10.1111/j.1364-3703.2009.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Torres JL, Wilbers RHP, Gawronski P, et al. Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. 2012;109:10119–10124. doi: 10.1073/pnas.1202867109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach J. The Direct Approach: Resistance Genes Team up to Recognize Different Fungal Effectors in Rice. Plant Cell. 2013;25:1189–1189. [Google Scholar]
- McDonald BA, Linde C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002;40:349–379. doi: 10.1146/annurev.phyto.40.120501.101443. [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Christopoulou M, Caldwell KS. Impacts of Resistance Gene Genetics, Function, and Evolution on a Durable Future. Annu. Rev. Phytopathol. 2013;51:291–319. doi: 10.1146/annurev-phyto-082712-102334. [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Meyers BC. Clusters of Resistance Genes in Plants Evolve by Divergent Selection and a Birth-and-Death Process. [July 9, 2015];Genome Res. 1998 8:1113–1130. doi: 10.1101/gr.8.11.1113. Available at: [DOI] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis A-R, Dreze M, et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou SH, Jennings PR. Progress in the Development of Disease-Resistant Rice. Annu. Rev. Phytopathol. 1969;7:383–410. [Google Scholar]
- Peng S, FY H, Sun GC, Liu EM, YJ S, RX A, Zhao JX, SZ B, Xiao FH. Studies on Durable Resistance to Blast Disease in Different Latitudes for Rice. Sci Agric Sin. 1996;29:52–28. [Google Scholar]
- Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Dai L, Han B, Wang GL. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics. 2006;172:1901–14. doi: 10.1534/genetics.105.044891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si W, Yuan Y, Huang J, et al. Widely distributed hot and cold spots in meiotic recombination as shown by the sequencing of rice F2 plants. New Phytol. 2015;206:1491–1502. doi: 10.1111/nph.13319. [DOI] [PubMed] [Google Scholar]
- Spielmeyer W, Mago R, Wellings C, Ayliffe M. Lr67 and Lr34 rust resistance genes have much in common – they confer broad spectrum resistance to multiple pathogens in wheat. BMC Plant Biol. 2013;13:96. doi: 10.1186/1471-2229-13-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature. 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 2004;37:517–27. doi: 10.1046/j.1365-313x.2003.01976.x. [DOI] [PubMed] [Google Scholar]
- Tai TH, Dahlbeck D, Clark ET, Gajiwala P, Pasion R, Whalen MC, Stall RE, Staskawicz BJ. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14153–14158. doi: 10.1073/pnas.96.24.14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent B, Khang CH. Recent advances in rice blast effector research. Curr. Opin. Plant Biol. 2010;13:434–441. doi: 10.1016/j.pbi.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Wichmann G, Bergelson J. Effector genes of Xanthomonas axonopodis pv. vesicatoria promote transmission and enhance other fitness traits in the field. Genetics. 2004;166:693–706. doi: 10.1534/genetics.166.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann G, Ritchie D, Kousik CS, Bergelson J. Reduced Genetic Variation Occurs among Genes of the Highly Clonal Plant Pathogen Xanthomonas axonopodis pv. vesicatoria, Including the Effector Gene avrBs2. [July 9, 2015];Appl. Environ. Microbiol. 2005 71:2418–2432. doi: 10.1128/AEM.71.5.2418-2432.2005. Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-L, Fan Y-Y, Li D-B, Zheng K-L, Leung H, Zhuang J-Y. Genetic control of rice blast resistance in the durably resistant cultivar Gumei 2 against multiple isolates. Theor. Appl. Genet. 2005;111:50–56. doi: 10.1007/s00122-005-1971-2. [DOI] [PubMed] [Google Scholar]
- Yang S, Feng Z, Zhang X, Jiang K, Jin X, Hang Y, Chen JQ, Tian D. Genome-wide investigation on the genetic variations of rice disease resistance genes. Plant Mol Biol. 2006;62:181–93. doi: 10.1007/s11103-006-9012-3. [DOI] [PubMed] [Google Scholar]
- Yang S, Li J, Zhang X, Zhang Q, Huang J, Chen J-Q, Hartl DL, Tian D. Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc. Natl. Acad. Sci. 2013;110:18572–18577. doi: 10.1073/pnas.1318211110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang L, Huang J, Zhang X, Yuan Y, Chen J-Q, Hurst LD, Tian D. Parent-progeny sequencing indicates higher mutation rates in heterozygotes. [July 17, 2015];Nature, advance online publication. 2015 doi: 10.1038/nature14649. Available at: http://www.nature.com/nature/journal/vaop/ncurrent/full/nature14649.html. [DOI] [PubMed]
- Yue JX, Meyers BC, Chen JQ, Tian D, Yang S. Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New Phytol. 2012;193:1049–63. doi: 10.1111/j.1469-8137.2011.04006.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Additional file 1: Figure S1. Phylogenetic tree of 937 NBS-LRR genes from two cultivars. Figure S2. Illustration of a clade or locus in Supplementary Figure S1. Figure S3. Phylogenetic tree of 12 rice blast strains. Figure S4. Phylogenetic trees of cloned NBS-LRR genes from five resistant rice cultivars. Figure S5. Phenotypes of control rice cultivars.
Additional file 2: Table S1. List of rice cultivars used in this study. Table S2. List of the 155 primer pairs of these cloned genes. Table S3. List of the cloned genes and their donors. Table S4. Locations of the collected isolates of M.oryzae used in this study. Table S5. List of 122 R-lines identified and their R-phenotypes specifically to pathogen isolates screened in this study. Table S6. Phenotypic results of transgenic rice lines inoculated by two Xoo races. Table S7. Evaluation of rice panicle blast resistance in an experimental paddy field. Table S8. Phenotypes of five resistant and two highly susceptible rice cultivars screened by 21 isolates. Table S9. Ten clades identified from the cloned 332 candidate genes which contain significantly more blast R-genes relative to th9at expected based from a random distribution among clades. Table S10. Eight R-genes identified from 110 transgenic lines screened independently by another blast isolate. Table S11. Patterns of broadly effective R-genes. Table S12. Co-segregation analysis of the 15 broadly effective R-genes during infection of isolates.