Abstract
Background and Aims
A new gene expression profile test may distinguish eosinophilic esophagitis (EoE) and gastroesophageal reflux disease (GERD), but the optimal tissue preparation and biopsy location are unknown. We aimed to determine if formalin-fixed paraffin-embedded (FFPE) and RNA-later (RNAL) preserved specimens from newly diagnosed EoE patients have equivalent gene expression scores and whether scores vary by esophageal biopsy location.
Methods
We analyzed prospectively collected and banked esophageal biopsies from EoE patients and GERD controls. Paired FFPE and RNAL samples from the distal, mid, and proximal esophagus were used. RNA was extracted, and gene expression for a previously constructed 96 gene panel was quantified with a summary expression score. Scores were compared between EoE and GERD patients, between FFPE and RNAL samples, and between the different esophageal locations.
Results
A total of 72 samples, representing paired FFPE and RNAL specimens from 9 EoE cases and 3 GERD controls, were analyzed. Overall median gene expression scores were similar between FFPE and RNAL (238 vs 227; p=0.64), correlation was excellent between FFPE and RNAL (Spearman’s rho=0.90; p<0.001), and there were no differences by biopsy level. Median gene scores distinguished EoE from controls (134 vs 402; p=0.02), and overall agreement between preservation methods and EoE case status was perfect (kappa=1.0; p<0.001).
Conclusions
Gene expression scores were equivalent in FFPE and RNAL, and were also similar across three esophageal locations. This implies that a single biopsy in either FFPE or RNAL from anywhere in the esophagus may have the potential for genetic diagnosis of EoE.
Keywords: Eosinophilic esophagitis, gene expression, RNA, paraffin, biopsy, transcriptome
Introduction
Eosinophilic esophagitis (EoE) is a chronic immune-mediated clinicopathologic condition [1]. In order to diagnose EoE, current criteria (EoE) require symptoms of esophageal dysfunction and persistent esophageal eosinophilia (at least 15 eosinophils per high power field [eos/hpf]) after a high-dose proton pump inhibitor (PPI) trial, and with other potential causes of eosinophilia excluded [2, 3]. While these appear to be straightforward, in clinical practice the differentiation between EoE and GERD is difficult. There are no pathognomonic features of EoE, symptoms such as dysphagia, heartburn, and chest pain can both be present in both GERD and EoE, and even high levels of esophageal eosinophilia are not specific [1, 4–12]. Moreover, there is a complicated relationship between EoE and GERD, and both conditions can coexist in some patients [13].
Because of this difficulty, there has been extensive research interest in distinguishing the two conditions. To date, there has been examination of symptom scores [9, 14–16], tissue biomarkers [10, 17–24], and non-invasive biomarkers [25–28], but few have been clinically validated and none are in routine practice. Recently, a molecular diagnostic approach has been reported [29]. Based on the previously described EoE transcriptome [30], this new test selected 96 of the most differentially expressed genes in EoE and created a summary score that is highly accurate for separating EoE from GERD, even with a single biopsy [29]. However, the optimal strategy for tissue preparation and the location for obtaining the biopsy in the esophagus are unknown.
The aim of this study was to determine if formalin-fixed paraffin-embedded (FFPE) and RNA-later preserved (RNAL) specimens from newly diagnosed EoE patients would have equivalent results on the gene expression profile panel, and whether gene expression scores would vary by biopsy location in the proximal, mid, or distal esophagus. We hypothesized that there would be no differences between the two tissue preservation methods, but that differences might be detected by biopsy location.
Methods
Study subjects and specimen collection
This was a case-control study analyzing biospecimens that were prospectively obtained and stored in the University of North Carolina (UNC) EoE Patient Registry and Biobank. This resource was created and maintained during prospective investigations of EoE from 2009–2014 [20, 31–33], where subjects were enrolled if they had symptoms of dysphagia, gastroesophageal reflux disease (GERD), or suspected EoE. These studies were approved by the UNC IRB, and all study subjects provided informed consent for participating prior to undergoing endoscopy, and this included consent for future use of stored specimens. The present studying analyzing the banked specimens was also approved by the UNC IRB.
Patients with EoE were diagnosed as per consensus guidelines [2, 3]. Specifically, they had to have symptoms of esophageal dysfunction (dysphagia, food impaction, heartburn, chest pain), esophageal biopsies with 15 eos/hpf that persisted after a high-dose PPI trial (20–40 mg twice daily of any of the available PPIs, prescribed at the discretion of their clinician), and exclusion of other potential causes of esophageal eosinophilia. GERD controls were patients who did not meet criteria for EoE diagnosis, but who had heartburn- or reflux-predominant symptoms.
At the time of the endoscopy, research protocol biopsies from all subjects were obtained in order to bank tissue for future use. We obtained specimens from the distal (3 cm above the gastroesophageal junction [GEJ]), mid (10 cm above the GEJ), and proximal (15 cm above the GEJ) esophagus using standard large capacity forceps (RJ4; Boston Scientific, Marlborough, MA). At each level, one biopsy fragment was placed in formalin and subsequently embedded in paraffin, and one was placed in RNA Later (RNAL; Life Technologies/Thermo-Fisher Scientific, Grand Island, NY), frozen, and stored at −80°C. For this study, paired FFPE and RNAL samples from each esophageal level were selected for each patient (6 samples per patient).
In addition to tissue, patient demographics, symptoms, endoscopic findings, and final diagnoses were also recorded prospectively. Further, the esophageal eosinophil counts were determined based on our previously validated methodology [34]. The maximum eosinophil density (eos/mm2) was quantified in five high power fields and then converted to an eosinophil count (eos/hpf) based on a microscopy field size of 0.24mm2, the most commonly reported size in the literature [35].
RNA extraction and gene expression
For the FFPE tissue, 5 sections 10 microns thick were cut for RNA extraction. For the RNAL samples, the entire biopsy (approximately 8mm3) was used. Each sample was processed with standard techniques to extract RNA. In brief, for RNAL samples, the miRNeasy mini RNA extraction kit (Qiagen, Valencia, CA) was used. The biopsies were transferred to a microtube and macerated in a small volume of QIAzol Lysis Reagent with a micropestle (Fisher Sci, Pittsburgh, PA). After addition of chloroform, mixing, and centrifugation, the aqueous phases were mixed with ethanol and then transferred to a QIAcube (Qiagen) preloaded with RNeasy mini columns for purification. For FFPE samples, samples were de-paraffined with xylene and and digested with Proteinase K and lysis buffer (miRNeasy FFPE kit, Qiagen). The digested samples were then passed thru gDNA eliminator columns and then transferred to the QIAcube preloaded with RNeasy MinElute columns for purification. RNA extraction from FFPE samples was 100% successful provided that there was adequate tissue, defined as the equivalent of 1 standard-sized biopsy specimen. RNA concentration purity was measured by NanoDrop spectrometry (ThermoFisher) and then reverse transcribed into cDNA using iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA) as per the manufacturer’s instructions.
Next, gene expression for a previously constructed 96 gene panel [29] was quantified (Eosinophilic Diagnostic Panel [EDP], Diagnovus, Nashville, TN). Specifically, a set of TaqMan probes for the 96 genes (including two housekeeping genes, GAPDH and 18S rRNA), were pre-spotted on 384 well fluidic cards (TaqMan Low Density Array Cards; TLDA, Life Technologies, Foster City, CA). The qPCR was performed on a ViiA7 cycler (Life Technologies) to determine gene expression levels measured as Ct.
Finally, using this expression data, a summary score was calculated by subtracting the housekeeping gene from the Ct value of each gene of interest to acquire the ΔCT and then summing their absolute values of the upregulated and downregulated genes separately, as previously described [29]. A difference of the two sums was used to calculate the EDP score with a score <333 diagnostic for EoE.
Statistical analysis
Descriptive statistics were used to summarize demographic, endoscopic, and histologic characteristics. The median of the maximum esophageal eosinophil counts were calculated both overall and for each esophageal level. Median gene expression scores were also calculated overall and for each esophageal level, for both FFPE and RNAL preservation methods. The overall score in a given individual was defined as the mean of the scores from all levels. Using non-parametric methods (Wilcoxon rank-sum; Wilcoxon sign-rank) the median gene scores were compared between the EoE and GERD groups, between FFPE and RNAL samples, and between the different esophageal locations. We also assessed for differences in individual gene expression by biopsy location and preservation method, requiring any differentially expressed genes to pass false discovery rate [36]. Spearman’s correlation was performed between the FFPE and RNAL gene scores, as well as between the gene scores and esophageal eosinophil counts, both overall and by esophageal level. Finally, agreement between EoE case status (as defined by the consensus diagnostic guidelines [2, 3]) and gene score was determined using the kappa coefficient, both overall and by esophageal level. All analyses were performed with Stata 9.2 (StataCorp, College Station, TX).
Results
Patient characteristics and samples
A total of 72 samples, representing paired FFPE and RNAL specimens from the proximal, mid, and distal esophagus from each of 9 EoE cases and 3 GERD controls, were analyzed for this study. Those with EoE were younger than GERD controls (median 34 vs 62 years; p = 0.03), all had dysphagia, and typical endoscopic findings such as rings, furrows, plaques, and edema were common (Table 1). The median of the maximum eosinophil count was 80 eos/hpf in the EoE group and 0 eos/hpf in the controls (p < 0.001). For the controls, two subjects had normal biopsies with no esophageal eosinophilia, and one had a biopsy showing 6 eosh/hpf. For the EoE cases, eosinophil counts were high at all esophageal levels (50, 60, and 49 eos/hpf for the proximal, mid, and distal esophagus, respectively).
Table 1.
Patient characteristics and overall gene scores
| GERD (n = 3) | EoE (n =9) | p* | |
|---|---|---|---|
| Age (median, IQR) | 62 (39–74) | 34 (23–49) | 0.03 |
| Male (n, %) | 0 (0) | 4 (44) | 0.49 |
| White (n, %) | 3 (100) | 8 (89) | 1.0 |
| Symptoms (n, %) | |||
| Dysphagia | 2 (67) | 9 (100) | 0.25 |
| Heartburn | 3 (100) | 3 (33) | 0.18 |
| Endoscopic findings (n, %) | |||
| Rings | 0 (0) | 9 (100) | 0.005 |
| Furrows | 0 (0) | 9 (100) | 0.005 |
| Plaques | 0 (0) | 4 (44) | 0.49 |
| Edema | 0 (0) | 4 (44) | 0.49 |
| Stricture | 2 (67) | 6 (67) | 1.0 |
| Maximum overall eosinophil count (median eos/hpf, IQR) | 0 (0–6) | 80 (75–100) | 0.01 |
| Maximum eosinophil counts by esophageal level (median eos/hpf, IQR) | |||
| Proximal | 0 (0–0) | 50 (35–80) | 0.01 |
| Mid | 0 (0–4) | 65 (21–75) | 0.01 |
| Distal | 0 (0–6) | 49 (38–100) | 0.01 |
| Gene panel scores (medians, IQR) | |||
| RNALater | 402 (341–513)** | 134 (115–264)** | 0.02 |
| FFPE | 362 (358–374) | 223 (194–247) | 0.01 |
Medians were compared between groups with Wilcoxon rank-sum, and proportions were compared with Fisher’s exact test.
No differences detected for comparisons between medial RNALater and FFPE scores for GERD controls (p =0.29) or EoE cases (p = 0.21) using Wilcoxon signed-rank
Gene scores by preservation methods and biopsy location
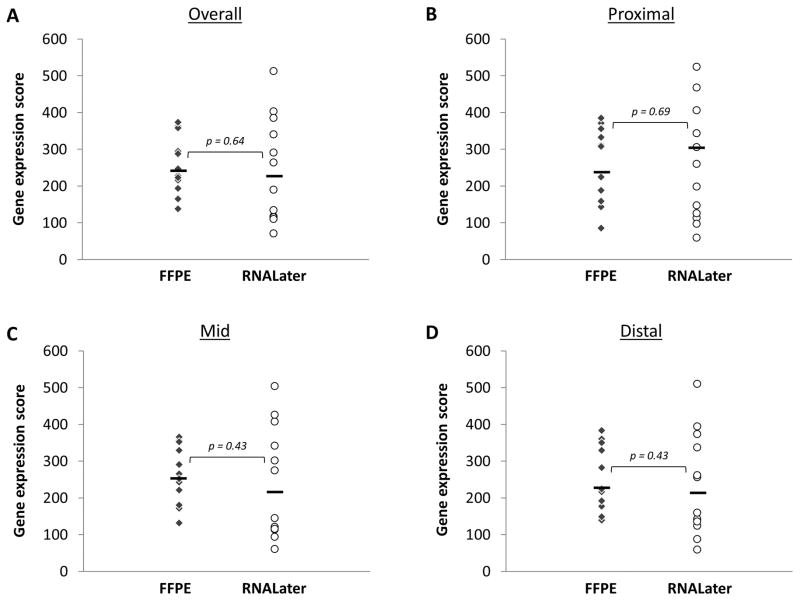
When RNAL samples were compared to FFPE samples for the entire study population, there were no significant differences in the median gene scores either overall or by biopsy location (Figure 1). For example, overall median gene scores were 227 for RNAL and 238 for FFPE (p = 0.64), and scores in the mid esophagus were 242 and 264, respectively (p = 0.43).
Figure 1.
Comparison of gene expression scores for samples persevered in FFPE (diamonds) vs RNALater (circles). (A) Overall score. (B) Score for the proximal esophageal biopsy. (C) Score for the mid esophageal biopsy. (D) Score for the distal esophageal biopsy. For all graphs, the solid black line represents the median value.
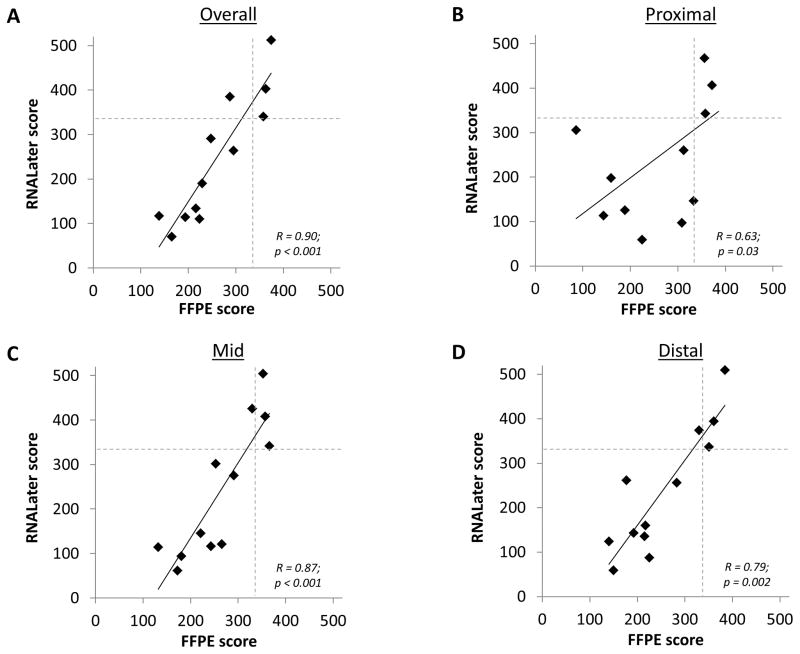
In addition, there were statistically significant correlations between the RNAL and FFPE scores, both overall and by biopsy location (Figure 2). For example, Spearman’s rho was 0.90 for the overall correlation (p < 0.001), and 0.87 in the mid esophagus (p < 0.001). There were no differences in the expression of individual genes (none passed false detection rate) either by biopsy location or by tissue preservation type (gene array data are shown in the Supplemental Figure).
Figure 2.
Correlation of gene expression scores measured in FFPE and RNALater. (A) Overall score. (B) Score for the proximal esophageal biopsy. (C) Score for the mid esophageal biopsy. (D) Score for the distal esophageal biopsy. For all graphs, the dotted lines show the score cut point (333) below which a score is consistent with a diagnosis of EoE.
EoE diagnosis by preservation method and biopsy location
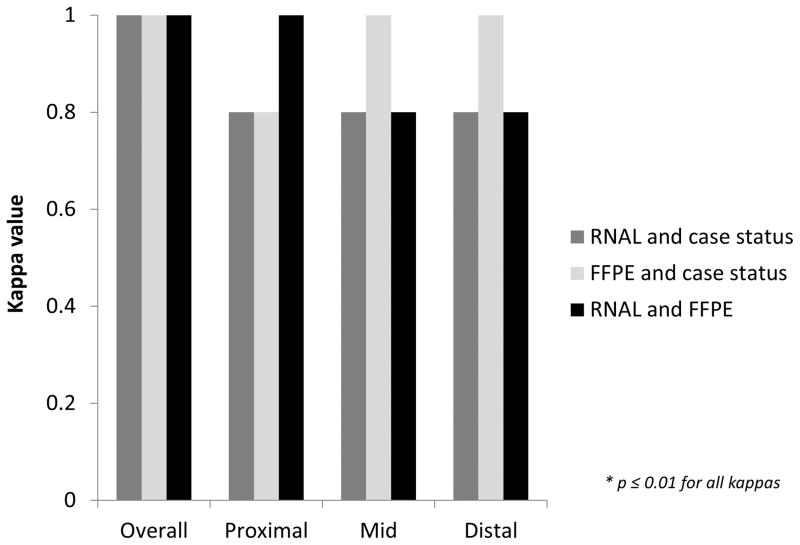
Median gene scores were significantly different between the EoE cases and the controls (Table 1). For RNAL, EoE cases had a median score of 134 compared to 402 in the controls (p = 0.02), and for FFPE, the median scores were 223 and 362, respectively (p = 0.01). Additionally, there were no differences in these scores by preservation method among cases alone (p = 0.21) or among controls alone (p = 0.29). Using a score <333 as the threshold for diagnosis of EoE, overall agreement between preservation methods and EoE case status was perfect, with a kappa of 1.0 (p < 0.001; Figure 3). Agreement was also excellent to perfect at each esophageal level, with kappas ranging from 0.8 to 1.0 (p ≤ 0.01 for all comparisons).
Figure 3.
Agreement between case status and tissue preservation method, as measured by kappa, for the overall gene score, as well as gene score by esophageal level. Dark gray bars show agreement between case status and RNALater, light gray bars show agreement between case status and FFPE, and black bars show agreement between RNALater and FFPE.
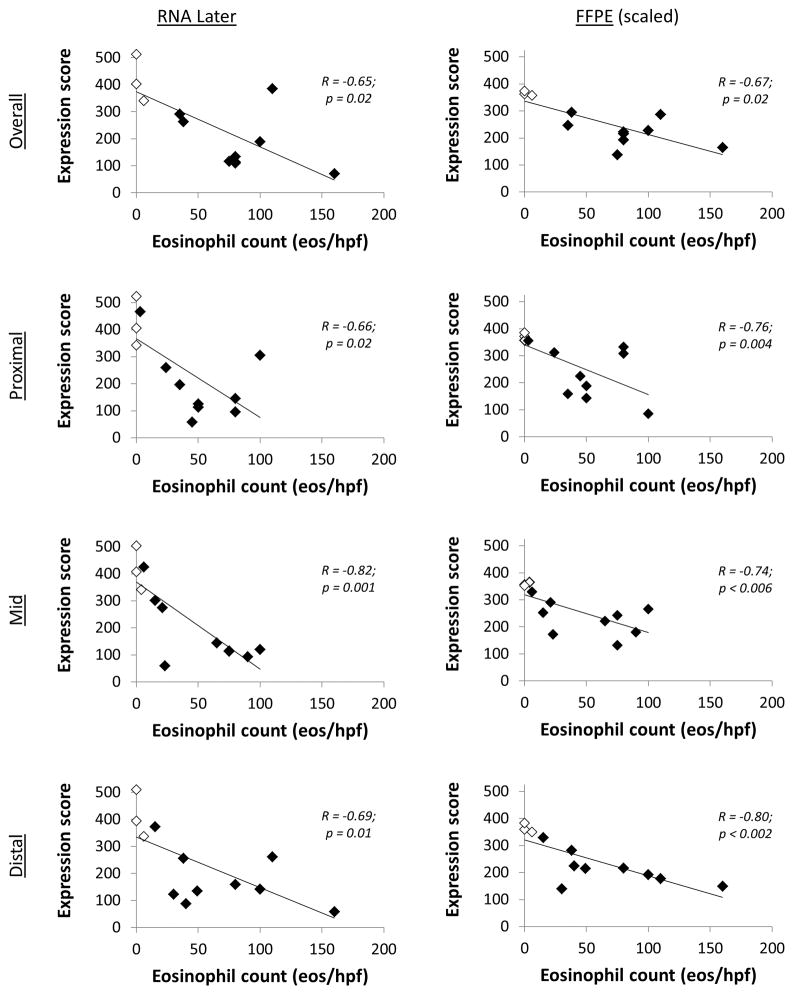
There were also significant inverse correlations between the esophageal eosinophil count and the gene score for both RNAL and FFPE, both overall and at all esophageal levels (Figure 4). Specifically, higher eosinophilic counts were associated with lower gene scores, and this association held regardless of case or control status. For example, there was one EoE subject with low proximal and mid esophageal eosinophil counts, but high distal counts (3, 6, and 110 eos/hpf, respectively). This subject had correspondingly higher gene scores at the proximal and mid- levels for both RNAL (468 and 426, respectively) and FFPE (358 and 330), but lower scores distally (262 for RNAL; 177 for FFPE).
Figure 4.
Correlation between gene expression score, by preservation method, and the peak eosinophil count, both for the overall scores and for three esophageal levels. For all graphs, the white diamonds are the controls, and the black diamonds are the EoE cases.
Discussion
The current diagnostic algorithm for EoE requires that both clinical and histologic features are present and that competing causes of esophageal eosinophilia are excluded [1–3]. However, it remains difficult to distinguish EoE and GERD clinically [4–12], and few techniques have been validated to do so [20]. The recent development of a gene expression profile test, however, holds great promise for doing just that [29], but whether it was optimal to use RNAL- or FFPE-preserved tissue, and whether there was a difference in expression by level of the esophagus, was not known. The goals of this study were to determine whether gene scores for FFPE specimens were equivalent to those from RNAL, and whether there was variability in gene expression scores based on biopsy location. The results were strong and consistent. First, RNAL and FFPE gene scores were not statistically different, either overall or by biopsy level, and the scores were highly correlated in individual patients, regardless of biopsy location. We also did not detect any differences in individual gene expression by level. Second, the gene scores almost perfectly distinguished the EoE cases from the controls. This has clinical implications for this methodology: one biopsy, regardless of the tissue preservation method or the esophageal location, can potentially be used to help diagnose EoE.
The genetic expression profile of patients with EoE, subsequently termed the EoE transcriptome, was first described by Blanchard and colleagues in a study of children with either active EoE, reflux esophagitis, or normal controls [30]. They demonstrated that there were approximately 340 upregulated and 230 downregulated genes characteristic of EoE. Additionally, using high-throughput whole transcriptome RNA sequencing techniques, Sherrill et al recently expanded the genetic signature of EoE, identifying 1607 differentially expressed genes [37]. Identification of differential gene expression led to the development of a molecular diagnostic approach for EoE, recently published by Wen et al [29]. In this landmark study, the technique was developed in 15 pediatric patients with active EoE and 14 normal controls, and then confirmed in an independent population of 18 EoE cases and 14 controls. Additional analysis showed similar results in 12 adults with EoE, and examined diagnostic utility in an additional 50 controls and 82 EoE cases. While the majority of samples assessed in this study were in RNAL or were fresh tissue, there were a subset tested in FFPE that yielded similar results. However, there was no assessment of expression at different levels of the esophagus. Our study assessed the same 96 gene panel in an adult population, using a similar but commercialized platform. As with the data from the Wen study, the panel was nearly perfect in its ability to discriminate EoE cases from controls, and we have expanded on their findings with our analysis throughout different levels of the esophagus.
This study has some potential limitations, as well as notable strengths, to address. First, this was conducted at a single center and included only adults, so we are unable to comment on whether the same findings would be seen in other settings or in children. However, the study by Wen et al presented comparable results for RNAL and FFPE in a pediatric population [29]. Our cases and controls are also not well-matched regarding age, and further validation will be needed in an age-matched population. Second, it is possible that the gene score is a marker of inflammation and may not be disease specific, but the study design does not allow us to comment on the specificity of the gene panel in non-EoE inflammatory conditions, including proton pump inhibitor-responsive esophageal eosinophilia. Third, while the number of subjects included in this study was small, there were a large number of specimens analyzed and the results were robust and consistent across several analysis techniques. Moreover, this study utilized prospectively collected and banked tissue samples, with standardized protocols and storage methods, from well-characterized EoE cases and GERD controls. This is a clinically-relevant population in whom this gene panel could potentially be applicable, and allowed for a unique comparison between paired specimens from multiple esophageal locations with the two tissue preservation techniques. We were also able to perform analyses correlating gene scores with eosinophil counts, both overall and by esophageal level. We feel that these strengths outweigh the possible limitations of the study. Additionally, while EoE has been shown to be a patchy disease histologically, with wide variations in eosinophil counts throughout the esophagus [38], it appears that the gene expression may be more consistent. An assessment of gene expression related to variations in esophageal eosinophilia should be explored in future studies.
In conclusion, this analysis of prospectively collected and banked esophageal biopsy samples showed that gene expression scores in EoE cases and GERD controls were equivalent in FFPE and RNAL tissue. Further, gene expression scores were similar in the three esophageal locations tested, and correlated strongly with the esophageal eosinophil counts. This implies that a single biopsy in either FFPE or RNAL from anywhere in the esophagus may have the potential to be used for genetic diagnosis of EoE.
Supplementary Material
Acknowledgments
Financial support: This research was conducted with support, in part, by an education grant from Diagnovus and by NIH award K23DK090073 (ESD).
Footnotes
Competing interests and disclosures: Dr. Dellon has received an educational grant from Diagnovus. Drs. Yellore, Andreatta, and Stover are employees of Diagnovus and hold interest in the company.
References
- 1.Dellon ES, Liacouras CA. Advances in Clinical Management of Eosinophilic Esophagitis. Gastroenterology. 2014;147:1238–1254. doi: 10.1053/j.gastro.2014.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liacouras CA, Furuta GT, Hirano I, et al. Eosinophilic esophagitis: Updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20. e6. doi: 10.1016/j.jaci.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 3.Dellon ES, Gonsalves N, Hirano I, et al. ACG Clinical Guideline: Evidence based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis. Am J Gastroenterol. 2013;108:679–92. doi: 10.1038/ajg.2013.71. [DOI] [PubMed] [Google Scholar]
- 4.Ngo P, Furuta GT, Antonioli DA, Fox VL. Eosinophils in the esophagus--peptic or allergic eosinophilic esophagitis? Case series of three patients with esophageal eosinophilia. Am J Gastroenterol. 2006;101:1666–70. doi: 10.1111/j.1572-0241.2006.00562.x. [DOI] [PubMed] [Google Scholar]
- 5.Dellon ES, Farrell TM, Bozymski EM, Shaheen NJ. Diagnosis of eosinophilic esophagitis after fundoplication for ‘refractory reflux’: implications for preoperative evaluation. Dis Esophagus. 2010;23:191–5. doi: 10.1111/j.1442-2050.2009.01019.x. [DOI] [PubMed] [Google Scholar]
- 6.Rodrigo S, Abboud G, Oh D, et al. High intraepithelial eosinophil counts in esophageal squamous epithelium are not specific for eosinophilic esophagitis in adults. Am J Gastroenterol. 2008;103:435–42. doi: 10.1111/j.1572-0241.2007.01594.x. [DOI] [PubMed] [Google Scholar]
- 7.Aceves SS, Newbury RO, Dohil R, Schwimmer J, Bastian JF. Distinguishing eosinophilic esophagitis in pediatric patients: clinical, endoscopic, and histologic features of an emerging disorder. J Clin Gastroenterol. 2007;41:252–6. doi: 10.1097/01.mcg.0000212639.52359.f1. [DOI] [PubMed] [Google Scholar]
- 8.Steiner SJ, Kernek KM, Fitzgerald JF. Severity of Basal Cell Hyperplasia Differs in Reflux Versus Eosinophilic Esophagitis. J Pediatr Gastroenterol Nutr. 2006;42:506–509. doi: 10.1097/01.mpg.0000221906.06899.1b. [DOI] [PubMed] [Google Scholar]
- 9.Dellon ES, Gibbs WB, Fritchie KJ, et al. Clinical, endoscopic, and histologic findings distinguish eosinophilic esophagitis from gastroesophageal reflux disease. Clin Gastroenterol Hepatol. 2009;7:1305–1313. doi: 10.1016/j.cgh.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller S, Neureiter D, Aigner T, Stolte M. Comparison of histological parameters for the diagnosis of eosinophilic oesophagitis versus gastro-oesophageal reflux disease on oesophageal biopsy material. Histopathology. 2008;53:676–84. doi: 10.1111/j.1365-2559.2008.03187.x. [DOI] [PubMed] [Google Scholar]
- 11.Parfitt JR, Gregor JC, Suskin NG, Jawa HA, Driman DK. Eosinophilic esophagitis in adults: distinguishing features from gastroesophageal reflux disease: a study of 41 patients. Mod Pathol. 2006;19:90–6. doi: 10.1038/modpathol.3800498. [DOI] [PubMed] [Google Scholar]
- 12.Molina-Infante J, Ferrando-Lamana L, Mateos-Rodriguez JM, Perez-Gallardo B, Prieto-Bermejo AB. Overlap of reflux and eosinophilic esophagitis in two patients requiring different therapies: A review of the literature. World J Gastroenterol. 2008;14:1463–6. doi: 10.3748/wjg.14.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spechler SJ, Genta RM, Souza RF. Thoughts on the complex relationship between gastroesophageal reflux disease and eosinophilic esophagitis. Am J Gastroenterol. 2007;102:1301–6. doi: 10.1111/j.1572-0241.2007.01179.x. [DOI] [PubMed] [Google Scholar]
- 14.Aceves SS, Newbury RO, Dohil MA, Bastian JF, Dohil R. A symptom scoring tool for identifying pediatric patients with eosinophilic esophagitis and correlating symptoms with inflammation. Ann Allergy Asthma Immunol. 2009;103:401–6. doi: 10.1016/S1081-1206(10)60359-6. [DOI] [PubMed] [Google Scholar]
- 15.von Arnim U, Wex T, Rohl FW, et al. Identification of clinical and laboratory markers for predicting eosinophilic esophagitis in adults. Digestion. 2011;84:323–7. doi: 10.1159/000331142. [DOI] [PubMed] [Google Scholar]
- 16.Mulder DJ, Hurlbut DJ, Noble AJ, Justinich CJ. Clinical features distinguish eosinophilic and reflux-induced esophagitis. J Pediatr Gastroenterol Nutr. 2013;56:263–70. doi: 10.1097/MPG.0b013e3182794466. [DOI] [PubMed] [Google Scholar]
- 17.Protheroe C, Woodruff SA, de Petris G, et al. A novel histologic scoring system to evaluate mucosal biopsies from patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2009;7:749–755. e11. doi: 10.1016/j.cgh.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dellon ES, Chen X, Miller CR, et al. Tryptase staining of mast cells may differentiate eosinophilic esophagitis from gastroesophageal reflux disease. Am J Gastroenterol. 2011;106:264–71. doi: 10.1038/ajg.2010.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dellon ES, Chen X, Miller CR, Woosley JT, Shaheen NJ. Diagnostic utility of major basic protein, eotaxin-3, and leukotriene enzyme staining in eosinophilic esophagitis. Am J Gastroenterol. 2012;107:1503–11. doi: 10.1038/ajg.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dellon ES, Speck O, Woodward K, et al. Markers of Eosinophilic Inflammation for Diagnosis of Eosinophilic Esophagitis and Proton Pump Inhibitor-Responsive Esophageal Eosinophilia: A Prospective Study. Clin Gastroenterol Hepatol. 2014;12:2015–22. doi: 10.1016/j.cgh.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanchard C, Stucke EM, Rodriguez-Jimenez B, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127:208–17. 217 e1–7. doi: 10.1016/j.jaci.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lucendo AJ, Navarro M, Comas C, et al. Immunophenotypic characterization and quantification of the epithelial inflammatory infiltrate in eosinophilic esophagitis through stereology: an analysis of the cellular mechanisms of the disease and the immunologic capacity of the esophagus. Am J Surg Pathol. 2007;31:598–606. doi: 10.1097/01.pas.0000213392.49698.8c. [DOI] [PubMed] [Google Scholar]
- 23.Kirsch R, Bokhary R, Marcon MA, Cutz E. Activated mucosal mast cells differentiate eosinophilic (allergic) esophagitis from gastroesophageal reflux disease. J Pediatr Gastroenterol Nutr. 2007;44:20–6. doi: 10.1097/MPG.0b013e31802c0d06. [DOI] [PubMed] [Google Scholar]
- 24.Gupta SK, Fitzgerald JF, Kondratyuk T, HogenEsch H. Cytokine expression in normal and inflamed esophageal mucosa: a study into the pathogenesis of allergic eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2006;42:22–6. doi: 10.1097/01.mpg.0000188740.38757.d2. [DOI] [PubMed] [Google Scholar]
- 25.Konikoff MR, Blanchard C, Kirby C, et al. Potential of blood eosinophils, eosinophil-derived neurotoxin, and eotaxin-3 as biomarkers of eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2006;4:1328–36. doi: 10.1016/j.cgh.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Huang JJ, Joh JW, Fuentebella J, et al. Eotaxin and FGF enhance signaling through an Extracellular signal-related kinase (ERK)-dependent pathway in the pathogenesis of Eosinophilic Esophagitis. Allergy Asthma Clin Immunol. 2010;6:25. doi: 10.1186/1710-1492-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnsson M, Bove M, Bergquist H, et al. Distinctive blood eosinophilic phenotypes and cytokine patterns in eosinophilic esophagitis, inflammatory bowel disease and airway allergy. J Innate Immun. 2011;3:594–604. doi: 10.1159/000331326. [DOI] [PubMed] [Google Scholar]
- 28.Subbarao G, Rosenman MB, Ohnuki L, et al. Exploring potential noninvasive biomarkers in eosinophilic esophagitis in children. J Pediatr Gastroenterol Nutr. 2011;53:651–8. doi: 10.1097/MPG.0b013e318228cee6. [DOI] [PubMed] [Google Scholar]
- 29.Wen T, Stucke EM, Grotjan TM, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145:1289–99. doi: 10.1053/j.gastro.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanchard C, Wang N, Stringer KF, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dellon ES, Speck O, Woodward K, et al. Clinical and Endoscopic Characteristics do Not Reliably Differentiate PPI-Responsive Esophageal Eosinophilia and Eosinophilic Esophagitis in Patients Undergoing Upper Endoscopy: A Prospective Cohort Study. Am J Gastroenterol. 2013;108:1854–60. doi: 10.1038/ajg.2013.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dellon ES, Rusin S, Gebhart JH, et al. Utility of a non-invasive serum biomarker panel for diagnosis and monitoring of EoE: A prospective study. Am J Gastroenterol. 2015 doi: 10.1038/ajg.2015.57. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burk CM, Beitia R, Lund PK, Dellon ES. High rate of galactose-alpha-1,3-galactose sensitization in both eosinophilic esophagitis and patients undergoing upper endoscopy. Dis Esoph. 2015 doi: 10.1111/dote.12356. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dellon ES, Fritchie KJ, Rubinas TC, Woosley JT, Shaheen NJ. Inter- and intraobserver reliability and validation of a new method for determination of eosinophil counts in patients with esophageal eosinophilia. Dig Dis Sci. 2010;55:1940–9. doi: 10.1007/s10620-009-1005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dellon ES, Aderoju A, Woosley JT, Sandler RS, Shaheen NJ. Variability in diagnostic criteria for eosinophilic esophagitis: A systematic review. Am J Gastroenterol. 2007;102:2300–13. doi: 10.1111/j.1572-0241.2007.01396.x. [DOI] [PubMed] [Google Scholar]
- 36.Gusnanto A, Calza S, Pawitan Y. Identification of differentially expressed genes and false discovery rate in microarray studies. Curr Opin Lipidol. 2007;18:187–93. doi: 10.1097/MOL.0b013e3280895d6f. [DOI] [PubMed] [Google Scholar]
- 37.Sherrill JD, Kiran KC, Blanchard C, et al. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun. 2014;15:361–9. doi: 10.1038/gene.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dellon ES, Speck O, Woodward K, et al. Distribution and variability of esophageal eosinophilia in patients undergoing upper endoscopy. Mod Pathol. 2015;28:383–90. doi: 10.1038/modpathol.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.