Abstract
All-trans retinoic acid (ATRA) is instrumental to male germ cell differentiation, but its mechanism of action remains elusive. To address this question, we have analyzed the phenotypes of mice lacking, in spermatogonia, all rexinoid receptors (RXRA, RXRB and RXRG) or all ATRA receptors (RARA, RARB and RARG). We demonstrate that the combined ablation of RXRA and RXRB in spermatogonia recapitulates the set of defects observed both upon ablation of RAR in spermatogonia. We also show that ATRA activates RAR and RXR bound to a conserved regulatory region to increase expression of the SALL4A transcription factor in spermatogonia. Our results reveal that this major pluripotency gene is a target of ATRA signaling and that RAR/RXR heterodimers are the functional units driving its expression in spermatogonia. They add to the mechanisms through which ATRA promote expression of the KIT tyrosine kinase receptor to trigger a critical step in spermatogonia differentiation. Importantly, they indicate also that meiosis eventually occurs in the absence of a RAR/RXR pathway within germ cells and suggest that instructing this process is either ATRA-independent or requires an ATRA signal originating from Sertoli cells.
Author Summary
Differentiation of spermatozoa from immature germ cells, called spermatogonia, critically depends on retinoic acid (ATRA), the active metabolite of vitamin A that acts though binding to nuclear receptors called RXR and RAR. To understand the mechanism by which ATRA control germ cell differentiation, we generated mice simultaneously lacking all RXR or all RAR specifically in spermatogonia. From their phenotypic analysis, we demonstrate that meiosis does not require a RAR/RXR-dependent pathway in germ cells and propose that this process is either ATRA-independent or requires an ATRA signal originating from somatic cells. We also show that RXR, in the form of dimers with RAR, can drive spermatogonia differentiation through binding to a regulatory region located in the Sall4 gene. This finding is significant, as the transcription factor encoded by Sall4 is known to regulate the expression of KIT, a key tyrosine kinase receptor which is frequently deregulated in testicular cancer.
Introduction
Spermatogenesis is a tightly regulated, cyclical, cell differentiation process, taking place in the seminiferous epithelium of the testis and yielding mature spermatozoa from stem cells. Spermatogonia in the single cell state, known as A single (As) spermatogonia, have traditionally be considered as the main spermatogonia stem cells in the mouse. Upon division, As spermatogonia give rise either to two new single cells or to a pair of daughter cells called A paired (Apr) spermatogonia that do not complete cytokinesis and remain connected through an intercellular bridge. The Apr spermatogonia divide further to form syncytial chains of 4 to 16 A aligned (Aal) spermatogonia [1]. Collectively, As, Apr and Aal (referred to as “undifferentiated spermatogonia”) are present throughout the seminiferous epithelial cycle and retain stem cell properties. Subsequently, Aal cells differentiate without mitotic division into A1 spermatogonia. Five cell divisions follow A1 formation, forming successively A2, A3 A4, In (intermediate) and B spermatogonia. Collectively, A1 to B spermatogonia (referred to as “differentiating spermatogonia”) express the prototypic marker KIT [2,3] and differentiate at given stages of the seminiferous epithelium cycle, each step of differentiation being associated with a mitotic division [4].
In rodent, all-trans retinoic acid (ATRA), the biologically active form of vitamin A (retinol) is instrumental to spermatogonia differentiation as assessed from vitamin A deficiency studies. In mice fed a vitamin A-deficient (VAD) diet from weaning onwards, all spermatogonia progressively arrest at the Aal-A1 transition, yielding seminiferous tubules that contain only Aal spermatogonia and Sertoli cells. Systemic administration of ATRA to VAD mice reinitiates spermatogenesis from mitotically-arrested Aal spermatogonia, resulting in their massive differentiation into spermatogonia expressing KIT, the marker of the Aal-A1 transition [2], and resuming their proliferation/differentiation [5,6]. The molecular mechanism through which ATRA controls Kit expression is however not yet fully elucidated. Characterizing this mechanism is important not only in the field of reproduction, but also for a better understanding of the biology of testicular germ cell tumors as KIT is also frequently deregulated in seminomas [7].
In cells, ATRA binds to and activates nuclear receptors (RARA, RARB and RARG), which are ligand-dependent transcriptional regulators. They usually function in the form of heterodimers with rexinoid receptors (RXRA, RXRB and RXRG) to control expression of ATRA-target genes through binding to specific sites located in genomic regulatory regions and called retinoic acid response elements (RARE) [8]. In the adult mouse testis, RARG cell-autonomously transduces an ATRA signal required for spermatogonia differentiation. Accordingly, the testes abnormalities observed upon deletion of Rarg either in the whole organism or specifically in spermatogonia in sexually mature males are similar to those present in VAD males [9]. As to RXR isotypes, the situation is contrasted. Our in situ hybridization (ISH) analyses failed to detect any of them in spermatogonia in the normal mouse testis [10], while another study evidenced RXRA in spermatogonia by immunohistochemistry (IHC) [11]. Thus, either RXR is absent and therefore dispensable for RAR functioning in spermatogonia, as it is the case in Sertoli cells [12], or RXRA is required but its expression level in spermatogonia too low to be detected by ISH.
To discriminate between these two possibilities, we have generated mice lacking all RXR isotypes specifically in spermatogonia from PN5 onwards and analyzed their phenotype. We demonstrate that ablation of all Rxr genes arrests differentiation of some spermatogonia at the Aal-A1 transition and recapitulates the full set of defects characteristic of the vitamin A deficiency-induced testis degeneration. We further show that efficient ablation of the 3 Rar genes in spermatogonia using the same genetic approach resembles ablation of the 3 Rxr genes. Importantly, some A1 spermatogonia still differentiate in these mutants, indicating the existence of a mechanism allowing the Aal to A1 transition independently of RAR/RXR in germ cells. Along these lines, both meiotic and post-meiotic cells devoid of RAR or RXR are also produced, in contrast to the situation when ATRA synthesis is impaired [13]. We propose that paracrine signals emanating from and transduced in Sertoli cells by ATRA-activated RARA stimulate some Aal spermatogonia to become A1 and trigger entry into meiosis. We finally provide evidence that RXR and RAR bind to the same Sall4 regulatory region to control ATRA-dependent expression of SALL4A in the RAR/RXR-dependent spermatogonia. As SALL4A is known to impair ZBTB16-mediated Kit repression [14], our study provides novel insights into the molecular mechanism by which ATRA could control KIT expression, and thereby the differentiation of Aal into A1 spermatogonia in vivo.
Results and Discussion
The ATRA signal is generally transduced by RAR/RXR heterodimers, notably during mouse embryonic development [15]. However, it can also be relayed by RAR independently of RXR, as it is the case in Sertoli cells [12]. Analysis by reverse transcription (RT) coupled to quantitative PCR (qPCR) of mRNA extracted from fluorescent-activated cell sorting (FACS) purified spermatogonia [13] showed that Rxra and Rxrb mRNA were present in these cells. Thus, contrary to our previous thoughts [10] but in agreement with IHC analyses [11], RXR are actually present in spermatogonia and could well be necessary for their ATRA-induced differentiation. To test for this possibility, we generated mutant mice lacking all three RXR isotypes in undifferentiated spermatogonia and their descendants (hereafter called Rxra;b;g Spg–/– mutants) using Tg(Stra8-cre) 1Reb transgene [16]. In this transgenic line, Cre-mediated ablation occurs as early as post-natal day 3 (PN3), as assessed using a reporter assay (S1 Fig) and in agreement with previous reports [14,16]. The crosses also generated control males devoid of the Cre transgene and carrying loxP-flanked alleles of Rxra, Rxrb and Rxrg. They did not display testis defects and were hereafter referred to as control mice.
Loss of RXR in undifferentiated spermatogonia yields age-related testis degeneration
The different generations of germ cells form cellular associations of fixed composition called epithelial stages. In control testes only the twelve normal epithelial stages (I–XII) [17] were identified (Fig 1A). In contrast, analysis of 12-week-old Rxra;b;g Spg–/– mutant testes (n = 5), revealed that, aside from normal epithelial stages (Fig 1C), 36.3 ± 9.6% of the tubule sections exhibited a degenerated seminiferous epithelium (Fig 1B) either lacking a large proportion of germ cells (T3) or containing only spermatogonia and Sertoli cells (T4). In addition, 17.8 ± 3.4% of the tubule sections lacked, around their entire circumference, either one or two generations of germ cells, yielding abnormal variants of the epithelial stages (T2). The missing germ cell layers included: preleptotene spermatocytes (Fig 1E and 1G), pachytene spermatocytes (Fig 1D and 1H), and/or round spermatids (Fig 1F and 1H). Thus, germ cell differentiation appeared altered in Rxra;b;g Spg–/– mutants. Analysis of other combinations of compound mutants at the age of 12 weeks revealed that the pathological phenotype was generated solely upon the simultaneous ablation of Rxra and Rxrb (Fig 2). This indicates that both RXRA and RXRB exert redundant functions in spermatogonia, while RXRG is dispensable. One year-old controls (n = 4) displayed only normal germ cell associations, whereas mutants (n = 4) displayed tubule sections containing only Sertoli cells and spermatogonia (Fig 1I and 1J). The latter expressed molecular markers of undifferentiated spermatogonia such as Gfra1 and Zbtb16 [3,18], but not of differentiating spermatogonia such as Kit and Stra8 [2,19] (Fig 3A–3H).
Fig 1. Ablation of RXR in spermatogonia induces age-related testis degeneration.
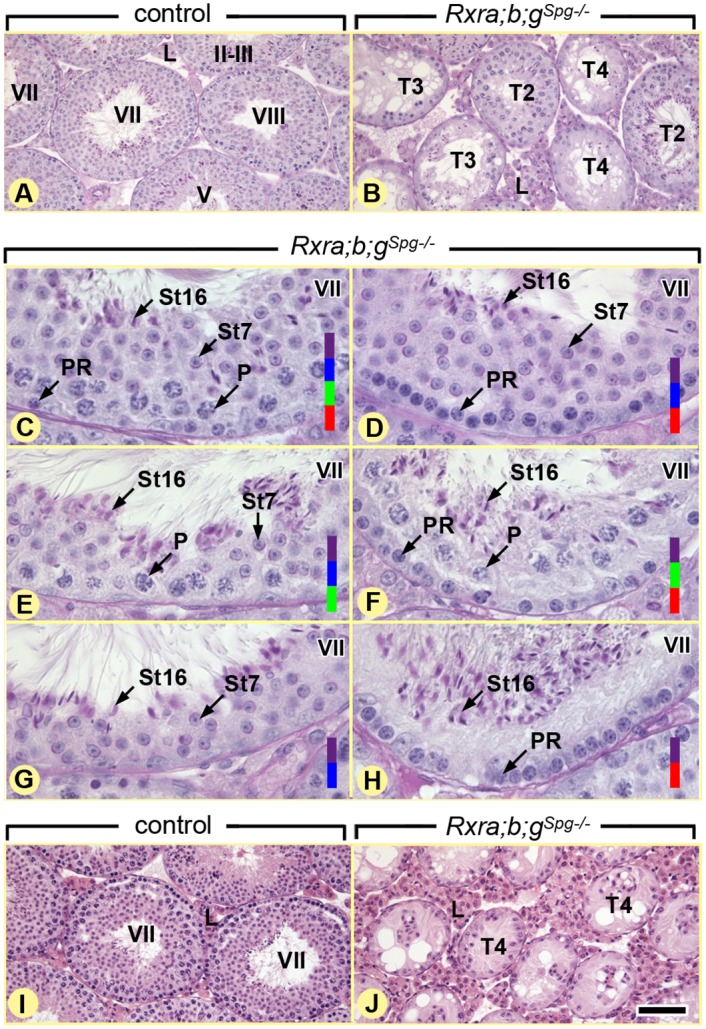
(A,B) Periodic acid-Schiff stains illustrating overviews and (C-H) details of germ cell associations in the seminiferous epithelium of 12 week-old control and Rxra;b;g Sgp–/– testes, as indicated. Normal gem cell associations at epithelial stage VII (C) coexist with abnormal associations mimicking, to some extent, this epithelial stage, but lacking: pachytene spermatocytes (D,H), preleptotene spermatocytes (E,G) and round spermatids (F,H). (I,J) Hematoxylin and eosin stain showing overviews of 12 month-old control and Rxra;b;g Sgp–/– testes: seminiferous tubules containing only spermatogonia and Sertoli cells represent the end-stage of degeneration in the mutant testes. PR and P, preleptotene and pachytene spermatocytes, respectively; St7 and St16, step 7 and 16 spermatids, respectively; T2, tubule sections lacking generation(s) of germ cells around their entire circumference; T3, tubule sections with disorganization of the germ cell layer; T4, tubule sections containing only spermatogonia and Sertoli cells. Germ cell populations present in a given seminiferous tubule cross-section are highlighted by colored bars: red, preleptotene spermatocytes; green, pachytene spermatocytes; blue, step 7 (round) spermatids; purple, step 16 (elongated, mature) spermatids. Roman numerals indicate the stages of the seminiferous epithelium cycle. Scale bar, 80 μm (A,B and I,J) and 30 μm (C-H).
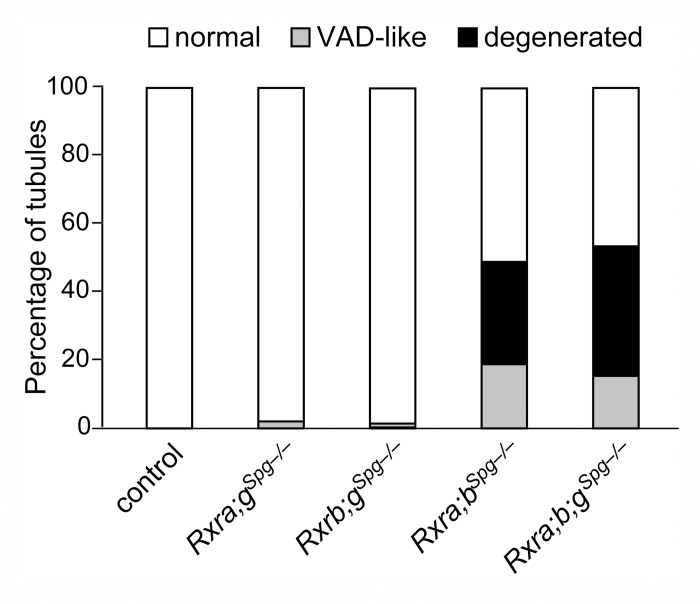
Fig 2. RXRA and RXRB are both instrumental to spermatogonia differentiation.
Mean percentages of tubule sections showing normal cellular associations (white bars), abnormal associations resembling the VAD situation with either one or two generations of germ cells lacking (grey bars), and degenerated epithelium containing only spermatogonia and Sertoli cells (black bars) in testes of 12 month-old mice (n = 5) with the indicated genotype. Mice lacking Rxrg and either Rxra (Rxra;g Sgp–/– mutants) or Rxrb (Rxrb;g Sgp–/– mutants) are marginally affected. In contrast, mice simultaneously lacking Rxra and Rxrb (Rxra;b Sgp–/– mutant) displayed a high proportion of affected tubule sections. Additional ablation of Rxrg does not worsen the pathological phenotype (Rxra;b Sgp–/– mutant). This indicates that RXRG is dispensable, whereas RXRA and RXRB are both required and exert redundant functions in spermatogonia.
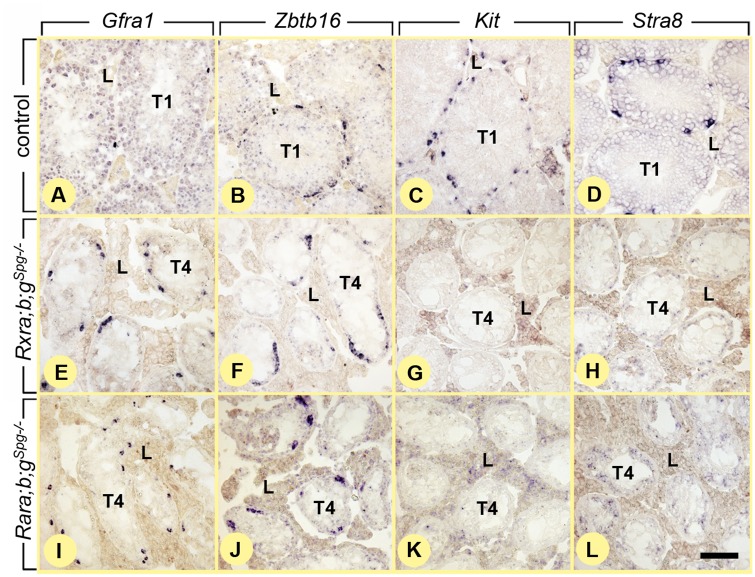
Fig 3. Ablation of RXR blocks spermatogonia are at the undifferentiated stage.
In situ hybridization with anti-sense probes for Gfra1, Zbtb16, Kit and Stra8 on histological sections of 12 month-old control (A-D), Rxra;b;g Spg–/–(E-H) and Rara;b;g Spg–/–(I-L) testes. Undifferentiated spermatogonia expressing Gfra1 and Zbtb16 are found in both control and mutant testes. In contrast differentiating spermatogonia expressing Kit and Stra8 are found in control (C,D) but not in mutant (G,H,K,L) testes. T1, tubule sections showing normal germ cell associations; T4, tubule sections containing only spermatogonia and Sertoli cells; L, Leydig cells. Scale bar: 80 μm.
Ablation of all Rar in spermatogonia yields age-related testis degeneration resembling that induced by loss of RXR
The histological defects displayed by Rxra;b;g Spg–/– mutants appeared to be much more severe than those observed when Rarg and Rara genes are deleted using the Tg(Neurog3-cre) 24Syos transgene [9]. This raised the possibility that RXR isotypes could be instrumental to some aspects of spermatogonia differentiation, independently of RARG and RARA. To test for this hypothesis, we analyzed the outcome of deleting Rar genes in spermatogonia by means of the same Tg(Stra8-cre) 1Reb transgene. Accordingly, mice carrying loxP-flanked alleles of Rara, Rarb and Rarg were crossed with Tg(Stra8-cre) 1Reb mice to generate Rara;b;g Spg–/– mutants and their controls.
In 12 week-old Rara;b;g Spg–/– mutants (n = 5), 24.5 ± 10.1% of the seminiferous tubule sections were abnormal, amongst which 11.5 ± 5%, identified as variants of the normal epithelial stages, lacked one or two generations of germ cells and 12.9 ± 7.3% exhibited a seminiferous epithelium either with a complete disorganization of the germ cell layers or with spermatogonia and Sertoli cells only (S2 Fig). In one-year-old mutants (n = 3), the seminiferous epithelium consisted only in Sertoli cells and spermatogonia, which expressed genes that are typical of undifferentiated spermatogonia (i.e., Gfra1 and Zbtb16), but not of differentiating spermatogonia (i.e., Kit and Stra8) (Fig 3I–3L). Altogether, these data indicate that age-matched Rara;b;g Spg–/– and Rxra;b;g Spg–/– mutants display similar, if not identical, phenotypes, including a slow and progressive loss of differentiating germ cells and the presence of spermatogonia blocked at an undifferentiated, Aal, stage (i.e., ZBTB16-positive, KIT-negative [2,3]) in aged mutants, both of which are features of the VAD testis [20].
To further document the similarities between the phenotypes induced by Rxr and Rar loss-of-functions, we examined the effect of Rxr ablation on germ cell apoptosis. Terminal deoxynucleotidyl-transferase dUTP nick end-labeling (TUNEL) assays indicated that apoptosis of preleptotene spermatocytes was not increased in testes of 8 week-old Rxra;b;g Spg–/– mutants, relative to age-matched controls (Fig 4A and 4B). Actually, we did not detect a single TUNEL-positive preleptotene spermatocyte in controls and in Rxra;b;g Spg–/– mutants (n = 3 males for each genotype; n > 200 preleptotene spermatocytes per testis). Therefore, similarly to the situation in mice lacking Rara and Rarg in spermatogonia [9], cell-death cannot account for the missing germ cell layers observed in Rxra;b;g Spg–/– mutant testes.
Fig 4. Ablation of RXR in spermatogonia blocks their division, but does not affect meiosis.
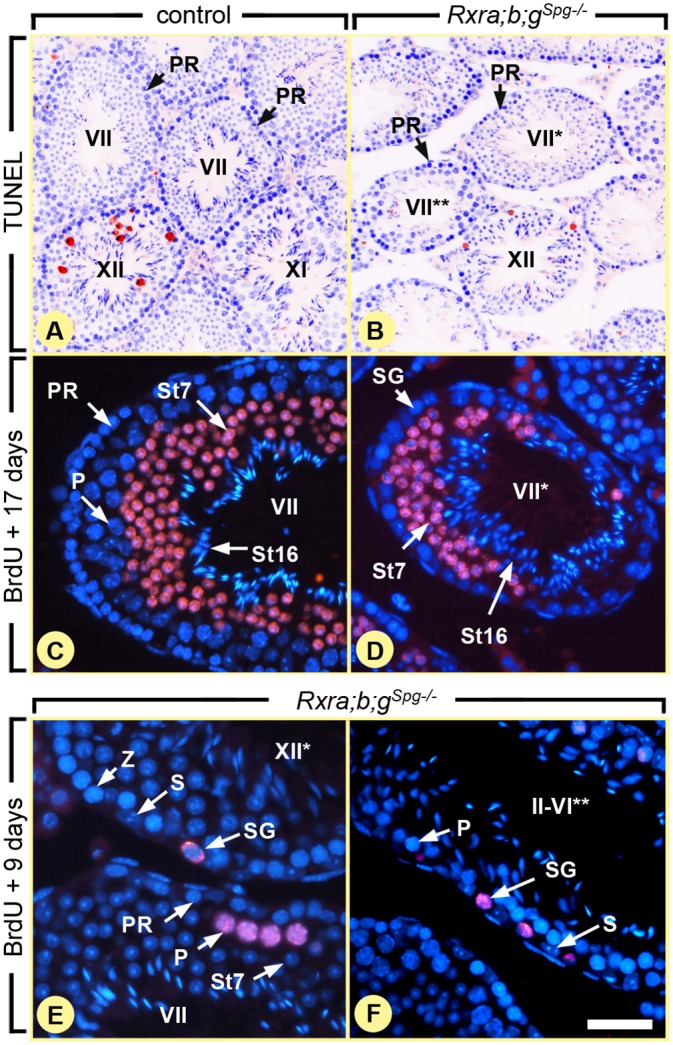
(A,B) TUNEL assays on histological sections from 8 week-old control and Rxra;b;g Spg–/– testis as indicated. Red signals correspond to apoptotic cells and nuclei are counterstained with DAPI (in blue). (C-F) Immunohistochemical detection of BrdU (red signals). After administration, incorporated BrdU has been similarly transferred to spermatids at 17 days (C,D) or to pachytene spermatocytes at 9 days (E,F) both in control and mutant seminiferous tubules. In contrast, spermatogonia retaining BrdU are observed only in mutants (E,F). PR and P, preleptotene and pachytene spermatocytes, respectively; S, Sertoli cells; SG, spermatogonia; St7 and St16, step 7 and 16 spermatids, respectively; Z, zygotene spermatocytes. Roman numerals refer to the stages of the seminiferous epithelium cycle. In mutant testes, one asterisk and two asterisks indicate tubule sections without pachytene spermatocytes and without round spermatids, respectively. Scale bar: 160 μm (A,B), 40 μm (C,D) and 25 μm (E,F).
We next examined the effect of Rxr ablation on the pace of preleptotene spermatocyte differentiation, because any delay or an arrest of this process may lead: (i) to the disappearance of pachytene spermatocytes through their normal differentiation into round, step 7, spermatids after one cycle of the seminiferous epithelium (i.e., 8.6 days), then (ii) to the disappearance of step 7 spermatids through their normal transformation into mature, step 16, spermatids after completion of a second cycle. Thus, we evaluated the duration of meiotic phase of spermatogenesis after 5-bromo-2'-deoxyuridine (BrdU) incorporation into S-phase nuclei. In adult testis, BrdU is mainly incorporated into B spermatogonia and preleptotene spermatocytes [20]. We fate-mapped the BrdU-labeled descendants of these cells 9 and 17 days after injection of the tracer. At the latter time-point, the most advanced, BrdU-positive, cell-type was step 7 spermatids in both control and Rxra;b;g Spg–/– testes (n = 3 for each genotype; 8 week-old) and there was no retained labeling in any spermatocyte (Fig 4C and 4D). Thus, similarly to the situation in mice lacking Rara and Rarg in spermatogonia [9], the duration of meiosis is not altered in Rxra;b;g Spg–/– mutant testes. In this context, it seems logical to assume that ablation of Rxr genes induces germ cell depletion solely through altering the spermatogonia proliferation/differentiation process. Accordingly, BrdU was detected in Rxra;b;g Spg–/– testes, 9 days after its incorporation, in cells displaying histological features of spermatogonia (Fig 4E and 4F). BrdU was never detected in spermatogonia of control testes at this time-point after injection because its amount becomes diminished by half in each daughter cell upon cell-division, yielding a progressive decrease of the signal over time and its absence 9 days after BrdU incorporation. This observation indicates that some of the spermatogonia that had incorporated BrdU in the Rxra;b;g Spg–/– testes did not divide further or divided more slowly than in control testes, as it is the case for spermatogonia lacking Rar [9] and in VAD testis [20]. Our study shows therefore that the outcomes of ablating RAR or RXR in spermatogonia are identical at the histological level.
Differentiation of A1 spermatogonia and meiosis still can occur when RAR and RXR are absent in germ cells
Given the central role assigned to ATRA in spermatogonia differentiation and in male meiosis [13,21], it was surprising that differentiation of only some spermatogonia was impaired, and that meiosis always proceeded normally in the absence of either Rar or Rxr. To exclude the possibility that Cre-mediated excision was mosaic, thereby resulting in the absence of RAR or RXR in some, but not all, differentiating germ cells, we analyzed their expression in Rara;b;g Spg–/– and Rxra;b;g Spg–/– testes. RT-qPCR analysis of whole testis RNA showed that the amount of Rarg transcripts was markedly reduced (4-fold) in Rara;b;g Spg–/– mutants, as early as PN5 (S3 Fig). Consistent with this finding, IHC analyses indicated a total absence of RARG in germ cells of Rara;b;g Spg–/– mutants, at PN5 and at PN60 (S3 Fig). In fact 100% of spermatogonia and of preleptotene spermatocytes were devoid of RARG in Rara;b;g Spg–/– mutants. Efficient loss of RARA and RARB in these mutants can be assessed neither by RT-qPCR because they are not, or only weakly, expressed in spermatogonia nor by IHC analyses because reliable antibodies are not available [9,10]. However, efficient ablation of loxP-flanked Rara and Rarb alleles was assessed at the genomic level, using FACS-purified germ cell populations [22]. PCR analysis demonstrated that excised (null, L–), but not conditional (L2) alleles, were detected in genomic DNA isolated from spermatogonia, spermatocytes and spermatids of PN60 mice bearing the Tg(Stra8-cre) 1Reb transgene (S3 Fig). Together, these data indicate that ablation of all 3 Rar genes was efficient in all germ cells, as early as PN5. As the outcome of ablating all RAR in spermatogonia appears very close to that induced upon Rarg knockout (Rarg –/– mutants) [9], our data suggest that RARG is the major functional RAR isotype in spermatogonia. Similar results were obtained in Rxra;b;g Spg–/– mutants, with Rxra transcript amounts markedly reduced (4- to 5-fold) as early as PN5 (S3 Fig).
Assuming that the excision of Rar and Rxr genes was complete from PN5 onwards, the impact of RAR or RXR loss-of-functions during the pubertal development of the testis was evaluated at PN20, i.e., when the first post-meiotic cells appear. At this developmental stage, control, Rxra;b;g Spg–/– and Rara;b;g Spg–/– mutant testes (n = 4 for each genotype) were indistinguishable: in both situations, late pachytene and diplotene spermatocytes represented the most advanced germ cell in the vast majority of tubule sections (Fig 5A). These results indicate that the spermatocytes present at PN20 in the mutants testes, appeared in due time, likely because the spermatogonia from which they derived started to differentiate before PN3, at a time when Rar or Rxr genes were not yet knocked out. More importantly, they also indicate that all preleptotene spermatocytes initiated meiosis normally (around PN8), at a time when they were devoid of RAR or RXR since ablation was obvious from PN5 in their precursors (see above). Analyzing the seminiferous epithelium later during pubertal development revealed the occurrence of abnormal cellular associations at PN25: few tubule sections in mutant testes displayed spermatogonia associated with round spermatids but without the intervening layers of preleptotene and pachytene spermatocytes (Fig 5B and 5C). This observation confirms the initial wave of A1 spermatogonia differentiation was not affected (yielding step 7 spermatids at PN25), and suggests the second wave was arrested (or delayed) in few tubules at some point before meiosis, leading to the absence of spermatocytes at PN25. However, the presence of normal cellular associations in the majority of tubule sections indicates that both A1 spermatogonia differentiation and meiosis occurred, despite absence of RAR or RXR in germ cells (see above). In keeping with this, KIT-positive A1 spermatogonia were found at stages VII-VIII of the seminiferous epithelium cycle in Rara;b;g Spg–/– mutants at PN60, similarly to the situation in control mice (Fig 5E and 5F).
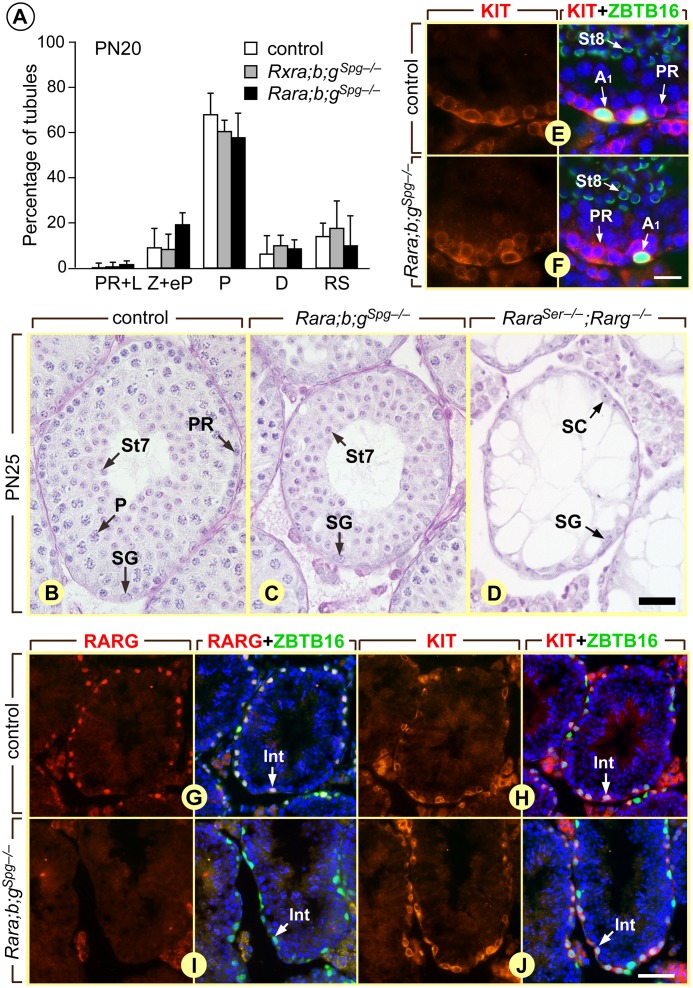
Fig 5. Ablation of either RXR or RAR in spermatogonia does not alter the first round of spermatogenesis.
(A) Percentages of seminiferous tubule cross-sections in which preleptotene/leptotene (PR+L), zygotene and early pachytene (Z+eP), late pachytene (P) and diplotene (D) spermatocytes or round spermatids (RS) represent the most advanced germ cell-types in control (white bars), Rxra/b/g Spg–/–(black bars) and Rara/b/g Spg–/–(grey bars) testes at post-natal day 20 (PN20). The bars represent mean ± s.e.m. (n = 4–5). (B-D) Histological sections of seminiferous from post-natal day 25 (PN25) control, Rara/b/g Spg–/– and Rara Ser–/–/Rarg –/– mice stained with hematoxylin and eosin. Note the absence of spermatocytes (C) or of all meiotic and post meiotic germ cells (D) in the mutant testes. (E-J) Detection of spermatogonia expressing KIT (red signal in E, F, H and J) and RARG (red signal in G and I) in control and Rara/b/g Spg–/– testes at 6 weeks of age. ZBTB16 (green nuclear signal in E, F, H and J) identifies spermatogonia. Alexa Fluor 488-conjugated peanut agglutinin (in E and F) labels the acrosomal system of spermatids, allowing precise identification of the stage of the seminiferous epithelium cycle. (G and H) and (I and J) represent consecutive histological sections. All sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to label nuclei (blue signal). A1, A1 spermatogonia, based on their presence in seminiferous tubule sections that contain both preleptotene spermatocytes (KIT-positive and ZBTB16-negative) and step8 spermatids. Int, intermediate spermatogonia, based on cell density co-expression of KIT and ZBTB16, and peanut hemagglutinin staining of acrosomes on consecutive sections. PR, P, preleptotene and pachytene spermatocytes, respectively; SG, spermatogonia, St7 and St8, step 7 and step8 round spermatids. Scale bars: 30 μm (B-D), 20 μm (E and F) and 55 μm (G-J).
The reason why ablation of RAR or RXR in germ cells affects only a fraction of A1 spermatogonia is unclear. In some cases the A1 transition appears to take place in due time, yielding normal cell associations, while in other instances no A1 are formed and Aal spermatogonia have to wait one (or several) epithelial cycles to become A1, yielding seminiferous tubule segments with missing generations of spermatocytes and spermatids. Moreover, spermatogonia do not transition at random because, as a result of stage-dependent cell divisions [4], extensive rows of differentiating, KIT-positive, RARG-negative, spermatogonia were observed at stages I-VI of the seminiferous epithelium cycle in Rara;b;g Spg–/– mutants, as it is the case in control males (Fig 5G–5I). An unknown signal, distinct from ATRA, may operate as a backup only in the context of Rara;b;g Spg–/– and Rxra;b;g Spg–/– mutants to promote spermatogonia differentiation. Alternatively, two populations of Aal spermatogonia may normally exist in the seminiferous epithelium at stages VII-VIII, one requiring RAR/RXR to transition, the other one being committed to become A1, independently of RAR/RXR heterodimers. As A1 spermatogonia were less and less often generated with aging and no longer observed in old Rara;b;g Spg–/– and Rxra;b;g Spg–/– mutants, the second population may become depleted with time resulting in a complete arrest of spermatogenesis. Given the pivotal role of ATRA and the precise timing in spermatogonia differentiation at stage VII-VIII of the seminiferous epithelium cycle [4], instructing the transition in this second population may also require ATRA. We propose that an ATRA signal transduced by RARA in Sertoli cells stimulates some Aal spermatogonia to become A1 even though no RAR/RXR pathway is functional within these latter cells. In keeping with this proposal, it worth noting that the seminiferous epithelium of mice lacking both RARA in Sertoli cells and RARG in spermatogonia (Rara Ser–/–;Rarg –/– mutants) was found to consist only in Sertoli cells and undifferentiated spermatogonia (Fig 5D), as it is the case when ATRA synthesis is specifically abolished in Sertoli cells [13].
More surprisingly, following the Aal to A1 transition, germ cell differentiation progressed at a normal pace despite the lack of RAR or RXR inside these cells. This indicates that initiation (and progression) of meiosis can proceed even in the absence of a functional ATRA signaling pathway in spermatocytes. If one considers that ATRA is as a mandatory meiosis-inducing substance in vertebrates [23], then our finding necessarily implies that the ATRA-dependent pathway instructing preleptotene spermatocytes is not autocrine in nature, as previously proposed [13], but instead operates in Sertoli cells. In this context, ATRA would control the synthesis by Sertoli cells of yet unknown, intermediate, secreted factor(s) acting on spermatocytes to trigger meiosis. Alternatively, the possibility exists that meiotic initiation does not require ATRA in male germ cells, as it was shown to be the case in female germ cells [24]. Discriminating between these two possibilities awaits further investigations. Regardless of the scenario, our findings indicate that expression of Stra8 does not require a RAR/RXR-dependent signaling pathway in preleptotene spermatocytes, even though this receptor heterodimer can efficiently bind to RARE in the Stra8 promoter [13,24].
RARG/RXR heterodimers bind to Sall4 and induce expression of SALL4A in spermatogonia
The similarities between Rar and Rxr gene ablations indicate that RAR and RXR exert convergent functions in spermatogonia, and support the possibility that ATRA signaling in these cells involves RAR/RXR heterodimers. They also indicate that RXR in spermatogonia are unlikely to play a role other than controlling differentiation in conjunction with RAR. To test whether RAR and RXR were actually recruited to an endogenous gene promoter in spermatogonia in vivo, we performed immunoprecipitation (IP) of these nuclear receptors using chromatin from PN5 wild-type testes, followed by qPCR analysis of the recovered DNA fragments, and assessed binding to Stra8, which is proposed as a RAR target-gene in spermatogonia [21]. Both anti-RAR and anti-RXR antibodies were able to precipitate, with similar efficiencies, the DNA sequences containing the RAR binding sites of the Stra8 promoter (S4 Fig). These data further support the notion that RAR/RXR heterodimers can be the functional units transducing the ATRA-signal in spermatogonia. However, although STRA8 promotes spermatogonia differentiation, it is not strictly required for this process [19,25,26] and its expression does not appears to be dependent upon RAR/RXR-signaling (see above). Thus, effectors acting downstream of ATRA and distinct from STRA8 likely account for the Aal to A1 spermatogonia transition.
To gain insights into the genetic cascade controlled by RAR/RXR heterodimers and aside from STRA8, we set up an experiment aimed at identifying ATRA-controlled genes in spermatogonia. To this purpose, we used Aldh1a1-3 Ser−/− mutants as a model, in which all retinaldehyde dehydrogenase activity is ablated in Sertoli cells. These mutants were chosen because (i) their spermatogonia differentiation is blocked at the Aal stage, (ii) Aal spermatogonia express RARG [9] and (iii) Aal spermatogonia are poised to differentiate into A1 spermatogonia upon activation of ATRA signaling [13]. We treated organotypic cultures of Aldh1a1-3 Ser−/− testes with the RARG-selective agonist BMS961 (n = 5) or with its vehicle (n = 5) for 6 hours and extracted mRNA. Microarray expression profiling identified only a few transcripts that were differentially expressed upon activation of RARG, amongst which Sall4. This gene encodes two isoforms named SALL4A and SALL4B [27]. They are zinc-finger transcription factors, which participate in regulatory networks and are critical for cell fate decisions and lineage specification [28,29]. In the mouse testis, their expression is restricted to spermatogonia [30,31] and mice deficient for Sall4 in these cell-type display testis defects that resemble those observed in Rxra;b;g Spg–/– and Rara;b;g Spg–/– mutants, namely loss of differentiating, KIT-positive, spermatogonia and of meiotic cells [14]. Thus Sall4 gene appears particularly relevant to ATRA-induced spermatogonia differentiation.
We confirmed by RT-qPCR that Sall4a mRNA steady state level was increased upon BMS961 administration in Aldh1a1-3 Ser−/− testes, without the need for intermediate protein synthesis as this increase occurred in the presence of cycloheximide (Fig 6A). Western-blot analysis of protein extracts from Aldh1a1-3 Ser−/− testes revealed that SALL4A protein level was increased by BMS961-activated RARG (Fig 6B, compare lane 1 to 2); this increase was prevented in mice additionally carrying a Rarg knock-out (Fig 6B, compare lane 3 to 4) and was not observed in BMS961-treated Rara;b;g Spg–/– mutants (Fig 6B, compare lane 5 to 6). In addition, Sall4a mRNA levels were significantly decreased in whole testis of Rara;b;g Spg–/– and Rxra;b;g Spg–/– mutants at PN60, while Sall4b and Zbtb16 mRNA levels were unchanged (Fig 6C). The finding that Sall4b mRNA level was not altered is in keeping with previous reports showing that SALL4B is expressed at a constant level in spermatogonia [31,32]. Altogether our results indicate that (i) Sall4a expression is decreased in testes of mice lacking RAR or RXR in spermatogonia (Rara;b;g Spg–/– and Rxra;b;g Spg–/– testes); (ii) SALL4A is detected at a low level in the seminiferous epithelium of mice deficient in ATRA (Aldh1a1-3 Ser−/− testes), but at a high level when RARG is activated by BMS961 in these mice (Aldh1a1-3 Ser−/− testes treated with BMS961), except when RARG is lacking (Aldh1a1-3 Ser−/−;Rarg −/− testes, treated with BMS961); and (iii) SALL4A is not detected in testes of adult mice lacking RAR in spermatogonia even in the presence of the RARG agonist (Rara;b;g Spg–/– testes, treated with BMS961). Altogether, these data indicate that Sall4a expression is controlled by ATRA-activated RARG in spermatogonia.
Fig 6. Sall4a expression in undifferentiated spermatogonia is controlled by ligand-activated RARG.
(A) Relative expression of Sall4a mRNA quantified by RT-qPCR in Aldh1a1-3 Ser−/− testes cultured in the absence (−) or in the presence (+) of cycloheximide (CHX) and treated for 6 hours with vehicle (white bar) and BM961 (grey bars). Error bars represent s.e.m. (n = 5); * p < 0.05. (B) Western blot analysis of protein extracts from testes of mutants as indicated treated with BMS961 (+) or with vehicle (−), using anti-SALL4 or anti-ACTIN antibodies. NS points to an unspecific signal. (C) Relative expression of Sall4a, Sall4b and Zbtb16 mRNA quantified by RT-qPCR in whole testes from control (white bars), Rara;b;g Spg−/− (grey bars) and Rxra;b;g Spg−/− (black bars) mice at PN60. Error bars represent s.e.m. (n = 5); * p < 0.05.
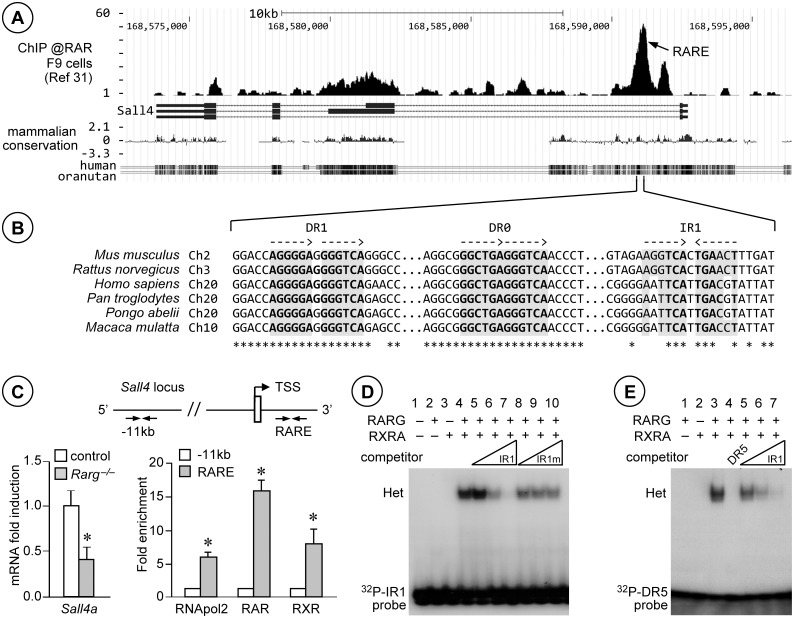
Using data sets locating RAR-occupied sites genome-wide in several cell-types [33,34], we identified a 700 bp-long RAR-binding region located in the first intron of Sall4 (RARE, Fig 7A). This DNA fragment contained a RAR binding sequence called IR1, consisting of inverted repeats (two core motifs 5’-RGKTSA-3’ oriented head-to-tail) separated by 1 bp (Fig 7B), as well as two additional sites called DR1 and DR0 (direct repeats of the core motif separated by 1 and 0 bp, respectively). We performed triplicate IP experiments with anti-RAR and anti-RXR antibodies using chromatin extracted from PN5 wild-type mouse testes as substrate. At this developmental stage, Sall4a expression was dependent upon RARG (Fig 7C, left panel). We analyzed the immuno-precipitated chromatin fragments by qPCR and evidenced robust binding of both RAR and RXR in vivo, in a 106bp-long region restricted to chr2:168,591,142–168,591,247 (NCBI37/mm9) in Sall4 (RARE, Fig 7C, right panel). To further confirm interaction of IR1 with RAR/RXR heterodimers, we performed electrophoretic mobility shift assays (EMSA) (Fig 7D). They revealed that RARG isotype in combination with RXRA isotype (lane 4), but neither RARG nor RXRA alone (lanes 2 and 3, respectively), bound the radiolabelled IR1 sequence. Binding was competed by increasing amounts of unlabeled IR1 (lanes 5–7), but not by IR1m bearing point-mutations in the first core motif (lanes 8–10). They also showed that unlabeled IR1 efficiently competed binding of RARG/RXRA heterodimers to the radiolabeled, canonical, RAR binding site of Rarb gene (called DR5, Fig 7E). The data suggested therefore that RARG/RXRA heterodimers could enhance expression of SALL4A through binding to an IR1 motif located in Sall4 intron. This motif appeared moderately well-conserved in mouse, rat, human and primate genomes (Fig 7B), but single mismatches do not necessarily abrogate RAR/RXR binding, even when located at highly conserved positions [33]. The DR1 and DR0 were also able to bind RARG/RXRA heterodimers (S5 Fig). Their sequences were even well-conserved across the species than that of IR1 (Fig 7B). Thus the RAR binding region in Sall4 belongs to the category of “composite elements”, the functionality of which has already been demonstrated [34]. Interestingly, SALL4A is also expressed in human spermatogonia [35]. Moreover, Fertilysin (N,N’-1,8-octanediylbis[2,2-dichloro-acetamide], also called WIN 18,446), which acts by inhibiting ATRA synthesis [36], reversibly inhibits spermatogenesis in men by inducing an arrest of germ cell differentiation at the spermatogonia stage [37,38], which resembles the phenotype we describe here in the mouse. Thus it is possible that RAR/RXR heterodimers also drive SALL4A expression in human spermatogonia.
Fig 7. RARG/RXRA heterodimers bind to Sall4 in testis chromatin, on an IR1 motif located in the first intron.
(A) UCSC Genome Browser snapshot of the Sall4 locus in NCBI37/mm9 assembly, including tracks for anti-RAR ChIP-seq [34], RefSeq genes and mammalian conservation (from top to bottom). RARE points to the RAR-binding region identified according to Moutier et al. [34]. (B) Alignments of the DNA sequences from the indicated species and corresponding to the region containing the RAR-binding region in mouse Sall4. The dotted arrows indicate orientations of the core motifs. Stars and grey boxes highlight the DNA residues and the RAR binding sequences that are conserved in all 6 species, respectively. (C) Left panel: relative expression of Sall4a mRNA quantified by RT-qPCR in whole testes from control (white bars) and Rarg −/− (grey bars) mice at PN5. Error bars represent s.e.m. (n = 9); * p < 0.05. Right panel: schematic representation of Sall4 locus and analysis of DNA recovered from testis chromatin immunoprecipitated using antibodies directed against RNApol2, all RAR or all RXR isotypes (RAR or RXR, respectively). The untranslated exon and the transcription start site (TSS) are depicted by an open box and a broken arrow, respectively. The locations of primers used are indicated at −11 kb and in the RAR-binding region (RARE). Mean fold enrichment of three experiments at RARE binding site (grey bars) is relative to the amount of DNA recovered at −11 kb (set at 1, white bars). Error bars represent s.e.m. (n = 4 to 5); * p < 0.05. (D) EMSA showing that RARG/RXRA heterodimers (Het) bind to the 32P-labelled IR1 of Sall4 (lane 4). Binding is competed when increasing amounts of unlabeled IR1 are added to reaction (lanes 5–7), but not when a mutated form IR1 is added (IR1m, lanes 8–10). 32P-IR1 probe indicates unbound DNA. (E) EMSA showing that RARG/RXRA heterodimers (Het) bound to the DR5 of Rarb (lane 3) are competed both when unlabeled DR5 (lane 4) or increasing amounts of IR1 (lanes 5–7) are added to reaction. 32P-DR5 probe indicates unbound DNA.
A model for the effects of ATRA on KIT expression
A hallmark of the transition to a differentiating state in spermatogonia is the expression of KIT receptor at the surface of A1 spermatogonia [5]. However, it is unclear what regulatory steps control the expression of this crucial cell surface receptor. Several studies have shown that undifferentiated spermatogonia are primed to turns on KIT and initiate differentiation upon activation of an ATRA signal [2]. This ATRA signal acts indirectly on Kit because its mRNA is not induced by BMS961, as assessed from our microarray expression profiling (see above) and no RAR binding site is found in Kit [39]. From our present study, we propose that ATRA enhances the level of Sall4a mRNA, allowing thereby an increase of the amount of SALL4A in spermatogonia. In agreement with this proposal, SALL4A appears at PN3-PN4 in wild-type spermatogonia [31,32], coinciding with the onset of endogenous ATRA signaling [40] and differentiation of the first KIT-positive spermatogonia [41]. SALL4A in high amount could (i) sequester ZBTB16, resulting in the release of the ZBTB16-mediated repression of Kit expression [14]; (ii) interact with DNMT3A and/or DNMT3B [42], allowing the epigenetic shift which is instrumental to A1 transition to take place properly [43]; and (iii) act on yet unknown other components of the differentiation program required in spermatogonia to transition to the A1 state.
In agreement with our proposal, Hobbs et al. [14] reported that the total amount of SALL4 protein detected in spermatogonia is higher at the A1 stage (i.e., in the ZBTB16-high, KIT-positive cell population) than at the Aal stage (i.e., in the ZBTB16-high, KIT-negative cell population). It is however not possible to show by IHC that Aal spermatogonia expressing SALL4A upon ATRA signaling activation differentiate into A1 spermatogonia and express KIT because, contrary to what has been stated in a previous report [31], antibodies to SALL4 do not distinguish between SALL4A and SALL4B. As SALL4B is expressed in spermatogonia and in their precursors from embryonic day 17.5 onwards [31,32], these antibodies are unsuitable to detect a specific increase in the expression of the sole SALL4Aisoform.
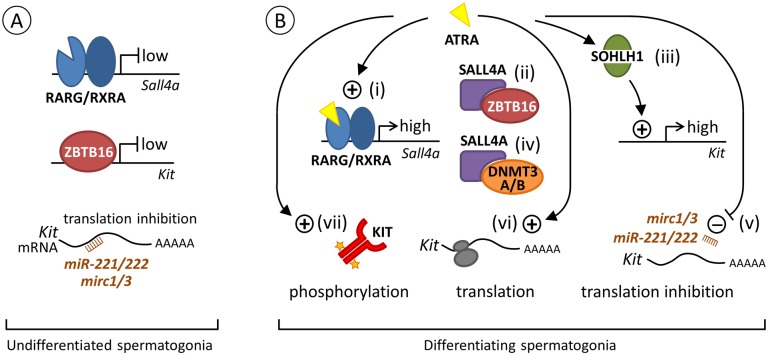
A comprehensive model summarizing the combination of transcriptional, post-transcriptional and non-genomic effects of ATRA pathways possibly controlling KIT expression and the commitment of Aal spermatogonia towards the A1 fate is proposed (Fig 8). The interest of better understanding the control of KIT expression in spermatogonia is not restricted to gametogenesis, but extends to testicular cancer. In fact, seminoma cells frequently bear somatic mutations activating KIT, or overexpress KIT or SALL4 [7,44,45]. Therefore, pharmacological modulation of mechanisms that regulate KIT expression in spermatogonia, such as antagonizing ATRA action, might have important applications for future therapeutic strategies.
Fig 8. Proposed model for the regulation of Kit expression by ATRA during the transition from Aal to A1 spermatogonia.
(A) Spermatogonia at an undifferentiated state. ATRA is not available to activate RARG/RXRA heterodimer and transcription of Sall4 is low. Transcription of Kit is also low because ZBTB16 is bound to its promoter [48]. In addition, translation of Kit mRNA already present in cells [2,49,50] is prevented by the Mirc1, Mirc3 and miR221/222 small interfering RNAs (brown comb) [50,51]. (B) Spermatogonia at a differentiating state upon ATRA action. (i) At the Aal-A1 transition, one possible way for ATRA (yellow triangle) to regulate Kit expression is to activate RARG/RXRA heterodimer, which increases Sall4a expression (high, our study). (ii) SALL4A in large amount can then sequesters ZBTB16 [14], clearing Kit promoter and relieving the repression of Kit transcription normally exerted by ZBTB16 [48]. (iii) ATRA is also proposed to increase the level of SOHLH1, which can replace ZBTB16 on regulatory regions to increase Kit expression (high) [52]. Sohlh1 is however not a direct target of RARG as RAR-binding sites are not found in this gene [34] and its expression is not induced by BMS961 (our study). (iv) Alternatively, SALL4A can also interact with DNMT3A/B to facilitate the epigenetic shift required for A1 differentiation [42,43]. (v) In parallel, ATRA can further induce KIT protein through decreasing expression of microRNA such as Mirc1, Mirc3 and miR221/222 that prevent Kit mRNA translation. How ATRA regulate miRNA expression is however unknown, as RARE have not been identified in the vicinity of their promoters [34,50,51]. ATRA can also function as a rapid, non-genomic, agent by (vi) increasing the loading of Kit mRNA on polysomes (grey ovals) and its translation [41] and (vii) inducing phosphorylation of KIT (orange stars) and of downstream KIT-effectors, reinforcing commitment towards the A1 spermatogonia fate [53].
Materials and Methods
Mice and treatments
Mice were on a mixed C57BL/6-129/Sv (50–50%) genetic background. They were housed in a licensed animal facility (agreement #A67-218-37). All experiments were approved by the local ethical committee (Com’Eth, accreditations #2012–080 and #2012–081), and were supervised by N.B.G. or M.M. who are qualified in compliance with the European Community guidelines for laboratory animal care and use (2010/63/UE). To inactivate Rar- or Rxr-coding genes in spermatogonia, mice carrying loxP-flanked alleles (L2) of Rara, Rarb and Rarg or of Rxra, Rxrb and Rxrg [12, and references therein] were crossed with mice bearing the Tg(Stra8-cre) 1Reb transgene [16]. In F1, Rxra L2/L2;Rxrb L2/L2;Rxrg L2/L2 females were crossed with males bearing one copy of the transgene (Stra8 tg/0). The resulting males (Stra8-Cre tg/0;Rxra +/L2;Rxrb +/L2;Rxrg +/L2) were backcrossed on Rxra L2/L2;Rxrb L2/L2;Rxrg L2/L2 females to generate mutant males in F2 (Stra8-Cre tg/0;Rxra L2/L2;RxrbL2/L2;Rarg L2/L2), and their control littermates (Rxra +/L2;Rxrb +/L2;Rxrg +/L2 and Rxra L2/L2;Rxrb L2/L2;Rxrg L2/L2 males). The same approach was used to inactivate Rara, Rarb and Rarg in spermatogonia. Aldh1a1-3 ser-/- mutants were generated as described previously [13]. Mice lacking RARA in Sertoli cells in a RARG-null genetic background were obtained by crossing Rara Ser–/–[12] and Rarg –/– mice [9] together. BMS961 (50 mg/kg body weight, Tocris Bioscience) dissolved in dimethylsulfoxide was administered to the mice by intra peritoneal injections. BrdU (Sigma-Aldrich) was dissolved in phosphate buffered saline and injected by intra peritoneal at 50 mg/kg body weight.
Isolation of germ cell populations by flow cytometry and organotypic cultures
Germ cell populations were purified from testes of Aldh1a1-3 ser-/-, Rara;b;g Spg–/– and Rxra;b;g Spg–/– mice by FACS and characterized as described previously [13,22]. Organotypic cultures of testes from Aldh1a1-3 ser-/- mice were also as described previously, except that the RARG-selective agonist BMS961 at 10–7M (Tocris Bioscience) was used to activate RAR signaling instead of BMS753 [13].
Histology, detection of apoptotic and proliferating cells, immunohistochemistry (IHC) and in situ hybridization (ISH)
For histology, testis samples were fixed in Bouin’s fluid for 16 hours and embedded in paraffin. Histological sections (5 μm-thick) were stained with hematoxylin and eosin or with periodic acid-Shiff (PAS). The percentage of affected seminiferous tubules was established on PAS-stained histological sections by counting cross-sections of tubules (n > 400 per testis). For all other methods, testes were fixed for 16 hours in 4% (wt/vol) buffered paraformaldehyde (PFA). For detection of apoptotic cells, TUNEL assays were performed using the In Situ Cell-Death Detection kit, Fluorescein (Roche Diagnostics). BrdU incorporation was detected by using an anti-BrdU antibody (Roche Molecular Biochemicals) and immunofluorescence labeling as described [9]. For IHC, 10 μm-thick frozen sections were incubated overnight at 4°C with rabbit anti-STRA8 (Ab49602, Abcam), rabbit anti-RARG1 (D3A4 #8965, Cell Signaling Technology), goat anti-ZBTB16 (AF2944, R&D Systems) and rabbit anti-KIT (D13A2 #3074, Cell Signaling Technology) antibodies diluted 1:200 to 1:500. Detection of bound primary antibodies was achieved by incubating the section with Cy3-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) or Alexa Fluor 488-conjugated donkey anti-goat IgG (Life Technologies). ISH using digoxigenin-labeled probes for detection of Gfra1, Kit, Stra8 and Zbtb16 expression was performed as described [9,10,20]. The sections were all counterstained with 0.001% (vol/vol) 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) and mounted in Vectashield (Vector Laboratories). The pattern of Cre expression driven by Tg(Stra8-cre) 1Reb [16] was assessed through testing excision in mice carrying the Gt(ROSA) 26Sortm1Sor reporter transgene [46]. In these mice, E. coli beta-galactosidase is synthesized only in cells that have experienced Cre-mediated deletion of an intervening stop sequence. Analysis of beta-galactosidase activity was as described [10].
Analysis of RNA and chromatin
Total RNA was prepared using TRIzol reagent (Life Technologies). Reverse transcription of total RNA followed by PCR amplification of cDNA was performed using QuantiTect Reverse Transcription (Qiagen) and LightCycler 480 SYBR Green I Master (Roche Diagnostics) kits, respectively. Primers were as indicated in Table 1. Triplicates of at least three samples were used in each experimental condition. The transcript levels were normalized relative to that of Rplp0 or Gapdh transcripts, whose expressions are not changed by retinoid administration. Data were expressed as fold induction relative to vehicle or control conditions. To prepare chromatin, PN5 testes were fixed with 0.4% PFA (wt/vol) for 15 minutes, before being sonicated to shear DNA to an average size of 500 bp. For each reaction, 100 μg of chromatin was first incubated with 18 μg of ChIP grade anti-RAR (sc-773; Santa Cruz biotechnology), anti-RXR (sc-774; Santa Cruz biotechnology) or anti-RNA polymerase II (RNApol2; sc-9001; Santa Cruz biotechnology) antibodies and then with protein G-Sepharose. Beads were washed, and eluted DNA–protein complexes were reverse cross-linked and purified. ChIP was performed in triplicate, using distinct chromatin extracts. The recovered immuno-precipitated DNA was analyzed by triplicate qPCR and was compared with input DNA. Quantitation was determined by the enrichment of the binding site compared with a site located upstream the TSS (–11kb), and were expressed as mean fold-enrichment (n = 3). The sequences of the oligonucleotides used are indicated in Table 2. Statistical significance was assessed by Student t tests or by one-way ANOVA followed by the post hoc Newman-Keuls test for comparison by pairs.
Table 1. Primers used in quantitative RT-PCR.
| Gene | Accession no. | Primers | Position (nt) | Size (nt) |
|---|---|---|---|---|
| Gapdh | NM_001289726.1 | 5’-AAGGTCATCCATGACAACTT-3’ | 570–657 | 88 |
| 5’-GGCCATCCACAGTCTTCTGG-3’ | ||||
| Rara | NM_001177 | 5’-AGCACCAGCTTCCAGTCAGT-3’ | 569–733 | 165 |
| 5’-AGTGGTAGCCGGATGATTTG-3’ | ||||
| Rarb | NM_011243 | 5’-GCTGGGTCGTCGTTTTCTAA-3’ | 1167–1282 | 135 |
| 5’-GAAACAGGCCTTCTCAGTGC-3’ | ||||
| Rarg | NM_011244.4 | 5’-CTCGGGTCTATAAGCCATGC-3’ | 746–805 | 60 |
| 5’-CCCCATAGTGGTAGCCAGAA-3’ | ||||
| Rplp0 | NM_007475 | 5’-ACCCTGAAGTGCTCGACATC-3’ | 720–908 | 208 |
| 5’-AGGAAGGCCTTGACCTTTTC-3’ | ||||
| Rxra | NM_001290481.1 | 5’-GATATCAAGCCGCCACTAGG-3’ | 384–534 | 151 |
| 5’-TTGCAGCCCTCACAACTGTA-3’ | ||||
| Rxrb | NM_001205 | 5’-GGGCTGCAAGGGTTTCTTCA-3’ | 186–348 | 163 |
| 5’-CTCCTGAACCGCCTCCCTTT-3’ | ||||
| Rxrg | NM_009107 | 5’-TGTGGTCAACAGTGTCAGCA-3’ | 6–190 | 185 |
| 5’-AGAAGCCTTTGCAACCTTCA-3’ | ||||
| Sall4a | NM_175303.4 | 5’-AGTGTCACCTGCCAATAGCC-3’ | 2560–2726 | 167 |
| 5’-TGCCAGGCACTTCAACTTT-3’ | ||||
| Sall4b | NM_201395.3 | 5’-CTCGACCAGTCCAAGAAAGG-3’ | 1232–1394 | 163 |
| 5’-TGCCAGGCACTTCAACTTT-3’ | ||||
| Zbtb16 | NM_001033324.2 | 5’-AACGGTTCCTGGACAGTTTG-3’ | 1563–1734 | 172 |
| 5’-CCACACAGCAGACAGAAGA-3’ |
Gene names, accession numbers, forward (upper line) and reverse (lower line) primers, their positions in the sequences and sizes of the amplified fragments are indicated. nt: nucleotide.
Table 2. Primers used for ChIP and EMSA assays.
| Gene | Primer | Assay |
|---|---|---|
| IR1 | 5’-ATGAGGTAGAAGGTCACTGAACTTTGATAAACTCG-3’ | EMSA |
| 5’-CGAGTTTATCAAAGTTCAGTGACCTTCTACCTCAT-3’ | ||
| mutated IR1 | 5’-ATGAGGTAGAAAGGCGCTGAACTTTGATAAACTCG-3’ | EMSA |
| 5’-CGAGTTTATCAAAGTTCAGCGCCTTTCTACCTCAT-3’ | ||
| DR0 | 5’-GCAGGCGGGCTGAGGGTTAACCCTTTTGTT-3’ | EMSA |
| 5’-AACAAAAGGGTTAACCCTCAGCCCGCCTGC-3’ | ||
| DR1 | 5’-CATCGCAGGACCAGGGGAGGGGTCAGGGCC-3’ | EMSA |
| 5’-GGCCCTGACCCCTCCCCTGGTCCTGCGATG-3’ | ||
| Rarb DR5 | 5’-CGGGTAGGGTTCACCGAAAGTTCACTCGCA-3’ | EMSA |
| 5’-TGCGAGTGAACTTTCGGTGAACCCTACCCG-3’ | ||
| Sall4 | 5’-AGCAATGACCTTCCAGTTGC-3’ | ChIP |
| 5’-TGGGATCCTACTTTTCCCAAA-3’ | ||
| Sall4 –11kb | 5’-TTGATCGGACAGCTTTTGTG-3’ | ChIP |
| 5’-GGGACTGGAGGGAGAAAAAG-3’ | ||
| Stra8 –3kb | 5’-GGCAGCAGGCCACCAATAAA-3’ | ChIP |
| 5’-TAGGCTTGGTTCCCCGTGTG-3’ | ||
| Stra8 DR2 | 5’-AGGTCATCTTGCTCCTTCCA-3’ | ChIP |
| 5’-ATCACAGCCCTGTCACTGC-3’ | ||
| Stra8 DR4 | 5’-GTGAGGTAGATCCCGGATTG-3’ | ChIP |
| 5’-GACCTGAGGTGAGCTGCTTC-3’ |
Gene, forward (upper line) and reverse (lower line) primers and their use are indicated. IR0, inverted repeats separated by 0 nucleotide; DR0, DR1, DR2, DR4 and DR5, direct repeats separated by 0, 1, 2, 4 and 5 nucleotides, respectively.
Electrophoretic mobility shift assays
They were performed as described previously [47]. Briefly, the oligonucleotides were annealed and labeled with [γ-32P]ATP (Amersham Bioscience). For competition assays, unlabeled oligonucleotides were added in the incubation mixture in 1- to 1000-fold molar excess. The sequences of the oligonucleotides used are described in Table 2.
Western blotting
Protein extracts were prepared in 50 mM Tris-HCl (pH7.5) buffer containing 150 mM NaCl, 0.5% (wt/vol) sodium deoxycholate, 1% (vol/vol) NP40, 0.2% (vol/vol) sodium dodecyl sulfate (SDS), and protease inhibitor mixture (Roche diagnostics). They were resolved by 4–16% (wt/vol) gradient SDS polyacrylamide gel electrophoresis (Expedeon) and transferred to nitrocellulose membranes (Protran) using standard protocols. The membranes were incubated with anti-SALL4 antibodies (Ab29112; Abcam) diluted 1:500 and IgG were detected using goat anti-mouse coupled to horseradish peroxidase as secondary antibodies (diluted 1:5000) followed by chemiluminescence according to the manufacturer’s protocol (GE Healthcare). The blots were subsequently incubated 2 times for 10 minutes at room temperature in 0.2M glycine pH 2.2 containing 0.1% (wt/vol) SDS and 0.1% (wt/vol) Tween 20, and were further probed with anti-actin antibodies (sc-58673; Santa Cruz biotechnology) diluted 1:500 to verify for equivalent loading in all the lanes.
Supporting Information
The pattern of Cre-mediated gene excision was assessed through testing DNA excision in mice also carrying the Gt(ROSA) 26Sortm1Sor reporter transgene. In these mice, E. coli beta-galactosidase is synthesized only in cells that have experienced Cre-mediated deletion of an intervening stop sequence (Soriano, 1999). (A-C,E,F) X-Gal staining and (D) STRA8 IHC on testis sections in mice bearing both Tg(Stra8-cre) 1Reb and Gt(ROSA) 26Sortm1Sor transgenes. At PN3 (i.e., shortly after the onset of spermatogenesis), beta-galactosidase activity (red signal) is already initiated in about 50% of the spermatogonia and in all seminiferous cord (A,B), whereas spermatogonia expressing endogenous STRA8 are exceptional. At PN5, beta-galactosidase-positive spermatogonia become widespread (C), and staining for STRA8 on consecutive sections (D) shows that its activity is initiated within a subset of spermatogonia that outnumbers those expressing the endogenous Stra8. At PN20, beta-galactosidase activity is observed in all spermatocytes and spermatids (E,F). These data indicate that excision of the reporter transgene occurs in undifferentiated spermatogonia as early as PN3 and is complete in all germ cells from PN5 onwards, in accordance with previous reports [14,16]. The Tg(Stra8-cre) 1Reb thus appears to be a suitable tool to study the role of RXR and RAR in spermatogonia. Note that (i) both the cytosolic and the juxta-nuclear, punctate (green arrowheads), beta-galactosidase activities indicate Cre-mediated excision of LacZ; (ii) in A, B and F, the staining resulting from beta-galactosidase activity was converted to a red false color, and the DAPI nuclear stain was converted to a bright-field image and then to a blue false color using Photoshop software; (iii) a processing artifact of consecutive sections has induced a distortion making the distances between the seminiferous cords wider in panel D than in panel C; and (iv) X-Gal-positive cells are never detected in testes of age-matched Gt(ROSA) 26Sortm1Sor mice that do not harbor the Tg(Stra8-cre) 1Reb transgene. The box in A indicates the region displayed at a higher magnification in B. Legend: C, seminiferous cords; G spermatogonia; PR and P, preleptotene and pachytene spermatocytes, respectively; R, round spermatids. Scale bar: 80 μm (A and C-F) and 25 μm (B).
(TIF)
(A-B) Section from 12 week-old control and Rara;b;g Spg–/– testes. (C-H) Germ cell associations in the seminiferous epithelium of Rara;b;g Spg–/– mutants. Normal gem cell associations at epithelial stage VII (C) coexist with abnormal associations lacking: pachytene spermatocytes (D,H), preleptotene spermatocytes (E,G) and round spermatids (F,H). (I,J) Section from 12 month-old control and Rara;b;g Spg–/– testes: seminiferous tubules containing only spermatogonia and Sertoli cells represent the end-stage of degeneration in the mutant testes (J). PR and P, preleptotene and pachytene spermatocytes, respectively; St7 and St16, step 7 and 16 spermatids, respectively; T2, tubules sections lacking generation(s) of germ cells around their entire circumference; T3, tubules sections with complete disorganization of the germ cell layer; T4, tubules sections containing only spermatogonia and Sertoli cells. Germ cell populations present in a given tubule cross-section are highlighted by colored bars: red, preleptotene spermatocytes; green, pachytene spermatocytes; blue, step 7 (round) spermatids; purple, step 16 (elongated, mature) spermatids. Roman numerals indicate the stages of the seminiferous epithelium cycle. Periodic acid-Schiff (A-H) and hematoxylin and eosin (I-J) stains. Scale bar, 80 μm (A-B and I-J) and 30 μm (C-H).
(TIF)
(A) Relative expression of Rarg and Rxra mRNA quantified by RT-qPCR in whole testes from control (white bars), Rara;b;g Spg−/− (grey bars) and Rxra;b;g Spg−/− (black bars) mice at PN5 (upper panel) and PN60 (lower panel). Error bars represent s.e.m. (n = 5); * p < 0.05. (B) PCR analysis of genomic DNA extracted from FACS-purified germ cells in heterozygote control mice (left panel) and Rara;b;g Spg−/− mutant mice (right panel) at PN60. This experiment proves efficient excision of the Rara, Rarb and Rarg alleles in all germ cells populations isolated from the mutant testes, as assessed by the absence of Rara L2 alleles and the trace amounts of Rarb or Rarg L2 alleles, which might be attributed to a low, contaminating, number of somatic cells. SG, PR, Z/P, P and RS, purified germ cell populations containing spermatogonia, preleptotene/leptotene spermatocytes, zygotene/early pachytene spermatocytes, late pachytene/diplotene spermatocytes and round spermatids, respectively. L2 and L–, conditional (loxP-flanked) and null (excised) alleles, respectively. CTL– and CTL+, negative (no DNA added) and positive (L2/L2 or L–/L–DNA added) control PCR reactions, respectively. (C-F) Immunohistochemical detection of spermatogonia expressing RARG (red nuclear signal) in control and Rara;b;g Spg–/– testes at PN5 and PN60. ZBTB16 expression (green nuclear signal) identifies spermatogonia. Spermatogonia nuclei co-expressing RARG and ZBTB16 appear in yellow. VII, stage VII of the seminiferous epithelium cycle; SG, spermatogonia. PR, preleptotene spermatocytes; asterisks indicate non-specific fluorescence in Leydig cells. Scale bars: 55 μm (C and D) and 40 μm (E and F).
(TIF)
Schematic representation of Stra8 locus and analysis by qPCR of DNA recovered from PN5 wild-type testis chromatin immunoprecipitated using antibodies directed against RNApol2, all RAR isotypes (RAR) or all RXR isotypes (RXR) at the Stra8 locus. The untranslated exons and the two transcription start sites (TSS1 and TSS2) are depicted by open boxes and broken arrows, respectively. The locations of primers used for qPCR are indicated at −3 kb and in Stra8. Mean fold enrichment of three experiments at DR4 (grey bars) and DR2 (black bars) binding sites is relative to the amount of DNA recovered at −3 kb (set at 1, white bars). Error bars represent s.e.m (n = 5); * p < 0.05.
(TIF)
EMSA showing that RARG/RXRA heterodimers (Het) bound to the DR5 of Rarb (lane 3) are competed both when unlabeled DR5 (lane 5) or increasing amounts of DR1 (lanes 6–8), DR0 (lanes 9–11) and IR1 (lanes 12–14) are added to reaction. 32P-DR5 probe indicates unbound DNA.
(TIF)
Acknowledgments
We thank common services and platforms of IGBMC, and Cathie ERB for helpful advices on EMSA.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from CNRS, INSERM, UNISTRA, Agence Nationale pour la Recherche (ANR 09-BLAN-0282; 10-BLAN-1239; 13-BSV2-0017), Fondation pour la Recherche Médicale (FDT20110922849; DEQ20071210544) and European community (FP7-PEOPLE-2012-IEF Marie Curie action, project #331687). It was also supported in part by the grant ANR-10-LABX-0030-INRT under the frame program Investissements d'Avenir labeled ANR-10-IDEX-0002-02. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. de Rooij DG. Proliferation and differentiation of spermatogonial stem cells. Reproduction 2001; 121:347–354. [DOI] [PubMed] [Google Scholar]
- 2. Schrans-Stassen BH, van de Kant HJ, de Rooij DG, van Pelt AM. Differential expression of c-kit in mouse undifferentiated and differentiating type A spermatogonia. Endocrinology 1999; 140:5894–5900. [DOI] [PubMed] [Google Scholar]
- 3. Suzuki H, Sada A, Yoshida S, Saga Y. The heterogeneity of spermatogonia is revealed by their topology and expression of marker proteins including the germ cell-specific proteins Nanos2 and Nanos3. Dev Biol. 2009; 336:222–231. 10.1016/j.ydbio.2009.10.002 [DOI] [PubMed] [Google Scholar]
- 4. de Rooij DG. Stem cells in the testis. Int J Exp Pathol. 1998; 79:67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Pelt AM, de Rooij DG. Retinoic acid is able to reinitiate spermatogenesis in vitamin A-deficient rats and high replicate doses support the full development of spermatogenic cells. Endocrinology. 1991; 128:697–704. [DOI] [PubMed] [Google Scholar]
- 6. de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl. 2000; 21:776–798. [PubMed] [Google Scholar]
- 7. Rossi P. Transcriptional control of KIT gene expression during germ cell development. Int J Dev Biol. 2013; 57:179–184. 10.1387/ijdb.130014pr [DOI] [PubMed] [Google Scholar]
- 8. Chambon P. The nuclear receptor superfamily: a personal retrospect on the first two decades. Mol Endocrinol. 2005; 19:1418–1428. [DOI] [PubMed] [Google Scholar]
- 9. Gely-Pernot A, Raverdeau M, Célébi C, Dennefeld C, Feret B, Klopfenstein M, et al. Spermatogonia differentiation requires retinoic acid receptor γ. Endocrinology 2012; 153:438–449. 10.1210/en.2011-1102 [DOI] [PubMed] [Google Scholar]
- 10. Vernet N, Dennefeld C, Rochette-Egly C, Oulad-Abdelghani M, Chambon P, Ghyselinck NB, et al. Retinoic acid metabolism and signaling pathways in the adult and developing mouse testis. Endocrinology 2006; 147:96–110. [DOI] [PubMed] [Google Scholar]
- 11. Gaemers IC, van Pelt AM, van der Saag PT, Hoogerbrugge JW, Themmen AP, de Rooij DG. Differential expression pattern of retinoid X receptors in adult murine testicular cells implies varying roles for these receptors in spermatogenesis. Biol Reprod. 1998; 58:1351–1356. [DOI] [PubMed] [Google Scholar]
- 12. Vernet N, Dennefeld C, Guillou F, Chambon P, Ghyselinck NB, Mark M. Prepubertal testis development relies on retinoic acid but not rexinoid receptors in Sertoli cells. EMBO J. 2006; 25:5816–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raverdeau M, Gely-Pernot A, Féret B, Dennefeld C, Benoit G, Davidson I, et al. Retinoic acid induces Sertoli cell paracrine signals for spermatogonia differentiation but cell autonomously drives spermatocyte meiosis. Proc Natl Acad Sci USA 2012; 109:16582–16587. 10.1073/pnas.1214936109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hobbs RM, Fagoonee S, Papa A, Webster K, Altruda F, Nishinakamura R, et al. Functional antagonism between Sall4 and Plzf defines germline progenitors. Cell Stem Cell 2012; 10:284–298. 10.1016/j.stem.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mark M, Ghyselinck NB, Chambon P. Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu Rev Pharmacol Toxicol. 2006; 46:451–480. [DOI] [PubMed] [Google Scholar]
- 16. Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 2008; 46:738–742. 10.1002/dvg.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Russell LD, Ettlin RA, SinhaHikim AP, Clegg ED. Histological and histopathological evaluation of the testis. Clearwater, FL: Cache River Press, 1990. [Google Scholar]
- 18. Naughton CK, Jain S, Strickland AM, Gupta A, Milbrandt J. Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate. Biol Reprod. 2006; 74:314–321. [DOI] [PubMed] [Google Scholar]
- 19. Mark M, Jacobs H, Oulad-Abdelghani M, Dennefeld C, Féret B, Vernet N, et al. STRA8-deficient spermatocytes initiate, but fail to complete, meiosis and undergo premature chromosome condensation. J Cell Sci. 2008; 121:3233–3242. 10.1242/jcs.035071 [DOI] [PubMed] [Google Scholar]
- 20. Ghyselinck NB, Vernet N, Dennefeld C, Giese N, Nau H, Chambon P, et al. Retinoids and spermatogenesis: lessons from mutant mice lacking the plasma retinol binding protein. Dev Dyn. 2006; 235:1608–1622. [DOI] [PubMed] [Google Scholar]
- 21. Hogarth CA, Griswold MD. The key role of vitamin A in spermatogenesis. J Clin Invest. 2010; 120:956–962. 10.1172/JCI41303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Getun IV, Torres B, Bois PR. Flow cytometry purification of mouse meiotic cells. J Vis Exp. 2011; 50:e2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Griswold MD, Hogarth CA, Bowles J, Koopman P. Initiating meiosis: the case for retinoic acid. Biol Reprod. 2012; 86:35 10.1095/biolreprod.111.096610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kumar S, Chatzi C, Brade T, Cunningham TJ, Zhao X, Duester G. Sex-specific timing of meiotic initiation is regulated by Cyp26b1 independent of retinoic acid signalling. Nat Commun. 2011; 2:151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AM, Page DC. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci USA 2008; 105:14976–14980. 10.1073/pnas.0807297105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Endo T, Romer KA, Anderson EL, Baltus AE, de Rooij DG, Page DC. Periodic retinoic acid-STRA8 signaling intersects with periodic germ-cell competencies to regulate spermatogenesis. Proc Natl Acad Sci USA 2015; 112:E2347–E2356. 10.1073/pnas.1505683112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rao S, Zhen S, Roumiantsev S, McDonald LT, Yuan GC, Orkin SH. Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol Cell Biol. 2010; 30:5364–5380. 10.1128/MCB.00419-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elling U, Klasen C, Eisenberger T, Anlag K, Treier M. Murine inner cell mass-derived lineages depend on Sall4 function. Proc Natl Acad Sci USA 2006; 103:16319–21634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sakaki-Yumoto M, Kobayashi C, Sato A, Fujimura S, Matsumoto Y, Takasato M, et al. The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development 2006; 133:3005–3013. [DOI] [PubMed] [Google Scholar]
- 30. Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004; 71:319–330. [DOI] [PubMed] [Google Scholar]
- 31. Gassei K, Orwig KE. SALL4 expression in gonocytes and spermatogonial clones of postnatal mouse testes. PLoS One 2013; 8:e53976 10.1371/journal.pone.0053976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liao HF, Chen WS, Chen YH, Kao TH, Tseng YT, Lee CY, et al. DNMT3L promotes quiescence in postnatal spermatogonial progenitor cells. Development 2014; 141:2402–2413. 10.1242/dev.105130 [DOI] [PubMed] [Google Scholar]
- 33. Delacroix L, Moutier E, Altobelli G, Legras S, Poch O, Choukrallah MA, et al. Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol Cell Biol. 2010; 30:231–244. 10.1128/MCB.00756-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moutier E, Ye T, Choukrallah MA, Urban S, Osz J, Chatagnon A, et al. Retinoic acid receptors recognize the mouse genome through binding elements with diverse spacing and topology. J Biol Chem. 2012; 287:26328–265341. 10.1074/jbc.M112.361790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eildermann K, Aeckerle N, Debowski K, Godmann M, Christiansen H, Heistermann M, et al. Developmental expression of the pluripotency factor sal-like protein 4 in the monkey, human and mouse testis: restriction to premeiotic germ cells. Cells Tissues Organs 2012; 196:206–220. 10.1159/000335031 [DOI] [PubMed] [Google Scholar]
- 36. Paik J, Haenisch M, Muller CH, Goldstein AS, Arnold S, Isoherranen N, et al. Inhibition of retinoic acid biosynthesis by the bisdichloroacetyldiamine WIN 18,446 markedly suppresses spermatogenesis and alters retinoid metabolism in mice. J Biol Chem 2014; 289:15104–15117. 10.1074/jbc.M113.540211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heller CG, Moore DJ, Paulsen CA. Suppression of spermatogenesis and chronic toxicity in men by a new series of bis(dichloroacetyl) diamines. Toxicol Appl Pharmacol. 1961; 3:1–11. [DOI] [PubMed] [Google Scholar]
- 38. Heller CG, Flageolle BY, Matson LJ. Histopathology of the human testes as affected by bis(dichloroacetyl)diamines. Exp Mol Pathol Suppl. 1963; 2:107–114. [PubMed] [Google Scholar]
- 39. Hayashi S, Kunisada T, Ogawa M, Nishikawa S. Identification of the control regions for mouse c-kit gene transcription induced by retinoic acid. DNA Res. 1995; 2:211–218. [DOI] [PubMed] [Google Scholar]
- 40. Snyder EM, Small C, Griswold MD. Retinoic acid availability drives the asynchronous initiation of spermatogonial differentiation in the mouse. Biol Reprod. 2010; 83:783–790. 10.1095/biolreprod.110.085811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Busada JT, Chappell VA, Niedenberger BA, Kaye EP, Keiper BD, Hogarth CA, et al. Retinoic acid regulates Kit translation during spermatogonial differentiation in the mouse. Dev Biol. 2015; 397:140–149. 10.1016/j.ydbio.2014.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang J, Corsello TR, Ma Y. Stem cell gene SALL4 suppresses transcription through recruitment of DNA methyltransferases. J Biol Chem. 2012; 287:1996–2005. 10.1074/jbc.M111.308734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shirakawa T, Yaman-Deveci R, Tomizawa S, Kamizato Y, Nakajima K, Sone H, et al. An epigenetic switch is crucial for spermatogonia to exit the undifferentiated state toward a Kit-positive identity. Development 2013; 140:3565–3576. 10.1242/dev.094045 [DOI] [PubMed] [Google Scholar]
- 44. Coffey J, Linger R, Pugh J, Dudakia D, Sokal M, Easton DF, et al. Somatic KIT mutations occur predominantly in seminoma germ cell tumors and are not predictive of bilateral disease: report of 220 tumors and review of literature. Genes Chromosomes Cancer 2008; 47:34–42. [DOI] [PubMed] [Google Scholar]
- 45. Cao D, Li J, Guo CC, Allan RW, Humphrey PA. SALL4 is a novel diagnostic marker for testicular germ cell tumors. Am J Surg Pathol. 2009; 33:1065–1077. 10.1097/PAS.0b013e3181a13eef [DOI] [PubMed] [Google Scholar]
- 46. Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999; 21:70–71. [DOI] [PubMed] [Google Scholar]
- 47. Hwang JJ, Chambon P, Davidson I. Characterization of the transcription activation function and the DNA binding domain of transcriptional enhancer factor-1. EMBO J. 1993; 12:2337–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Filipponi D, Hobbs RM, Ottolenghi S, Rossi P, Jannini EA, Pandolfi PP, et al. Repression of kit expression by Plzf in germ cells. Mol Cell. Biol. 2007; 27:6770–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Prabhu SM, Meistrich ML, McLaughlin EA, Roman SD, Warne S, Mendis S, Itman C, Loveland KL. Expression of c-Kit receptor mRNA and protein in the developing, adult and irradiated rodent testis. Reproduction. 2006; 131:489–499. [DOI] [PubMed] [Google Scholar]
- 50. Yang QE, Racicot KE, Kaucher AV, Oatley MJ, Oatley JM. MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development 2013; 140:280–290. 10.1242/dev.087403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tong MH, Mitchell DA, McGowan SD, Evanoff R, Griswold MD. Two miRNA clusters, Mir-17-92 (Mirc1) and Mir-106b-25 (Mirc3), are involved in the regulation of spermatogonial differentiation in mice. Biol Reprod. 2012; 86:72 10.1095/biolreprod.111.096313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Barrios F, Filipponi D, Campolo F, Gori M, Bramucci F, Pellegrini M, et al. SOHLH1 and SOHLH2 control Kit expression during postnatal male germ cell development. J Cell Sci. 2012; 125:1455–1464. 10.1242/jcs.092593 [DOI] [PubMed] [Google Scholar]
- 53. Pellegrini M, Filipponi D, Gori M, Barrios F, Lolicato F, Grimaldi P, et al. ATRA and KL promote differentiation toward the meiotic program of male germ cells. Cell Cycle 2008; 7:3878–3888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The pattern of Cre-mediated gene excision was assessed through testing DNA excision in mice also carrying the Gt(ROSA) 26Sortm1Sor reporter transgene. In these mice, E. coli beta-galactosidase is synthesized only in cells that have experienced Cre-mediated deletion of an intervening stop sequence (Soriano, 1999). (A-C,E,F) X-Gal staining and (D) STRA8 IHC on testis sections in mice bearing both Tg(Stra8-cre) 1Reb and Gt(ROSA) 26Sortm1Sor transgenes. At PN3 (i.e., shortly after the onset of spermatogenesis), beta-galactosidase activity (red signal) is already initiated in about 50% of the spermatogonia and in all seminiferous cord (A,B), whereas spermatogonia expressing endogenous STRA8 are exceptional. At PN5, beta-galactosidase-positive spermatogonia become widespread (C), and staining for STRA8 on consecutive sections (D) shows that its activity is initiated within a subset of spermatogonia that outnumbers those expressing the endogenous Stra8. At PN20, beta-galactosidase activity is observed in all spermatocytes and spermatids (E,F). These data indicate that excision of the reporter transgene occurs in undifferentiated spermatogonia as early as PN3 and is complete in all germ cells from PN5 onwards, in accordance with previous reports [14,16]. The Tg(Stra8-cre) 1Reb thus appears to be a suitable tool to study the role of RXR and RAR in spermatogonia. Note that (i) both the cytosolic and the juxta-nuclear, punctate (green arrowheads), beta-galactosidase activities indicate Cre-mediated excision of LacZ; (ii) in A, B and F, the staining resulting from beta-galactosidase activity was converted to a red false color, and the DAPI nuclear stain was converted to a bright-field image and then to a blue false color using Photoshop software; (iii) a processing artifact of consecutive sections has induced a distortion making the distances between the seminiferous cords wider in panel D than in panel C; and (iv) X-Gal-positive cells are never detected in testes of age-matched Gt(ROSA) 26Sortm1Sor mice that do not harbor the Tg(Stra8-cre) 1Reb transgene. The box in A indicates the region displayed at a higher magnification in B. Legend: C, seminiferous cords; G spermatogonia; PR and P, preleptotene and pachytene spermatocytes, respectively; R, round spermatids. Scale bar: 80 μm (A and C-F) and 25 μm (B).
(TIF)
(A-B) Section from 12 week-old control and Rara;b;g Spg–/– testes. (C-H) Germ cell associations in the seminiferous epithelium of Rara;b;g Spg–/– mutants. Normal gem cell associations at epithelial stage VII (C) coexist with abnormal associations lacking: pachytene spermatocytes (D,H), preleptotene spermatocytes (E,G) and round spermatids (F,H). (I,J) Section from 12 month-old control and Rara;b;g Spg–/– testes: seminiferous tubules containing only spermatogonia and Sertoli cells represent the end-stage of degeneration in the mutant testes (J). PR and P, preleptotene and pachytene spermatocytes, respectively; St7 and St16, step 7 and 16 spermatids, respectively; T2, tubules sections lacking generation(s) of germ cells around their entire circumference; T3, tubules sections with complete disorganization of the germ cell layer; T4, tubules sections containing only spermatogonia and Sertoli cells. Germ cell populations present in a given tubule cross-section are highlighted by colored bars: red, preleptotene spermatocytes; green, pachytene spermatocytes; blue, step 7 (round) spermatids; purple, step 16 (elongated, mature) spermatids. Roman numerals indicate the stages of the seminiferous epithelium cycle. Periodic acid-Schiff (A-H) and hematoxylin and eosin (I-J) stains. Scale bar, 80 μm (A-B and I-J) and 30 μm (C-H).
(TIF)
(A) Relative expression of Rarg and Rxra mRNA quantified by RT-qPCR in whole testes from control (white bars), Rara;b;g Spg−/− (grey bars) and Rxra;b;g Spg−/− (black bars) mice at PN5 (upper panel) and PN60 (lower panel). Error bars represent s.e.m. (n = 5); * p < 0.05. (B) PCR analysis of genomic DNA extracted from FACS-purified germ cells in heterozygote control mice (left panel) and Rara;b;g Spg−/− mutant mice (right panel) at PN60. This experiment proves efficient excision of the Rara, Rarb and Rarg alleles in all germ cells populations isolated from the mutant testes, as assessed by the absence of Rara L2 alleles and the trace amounts of Rarb or Rarg L2 alleles, which might be attributed to a low, contaminating, number of somatic cells. SG, PR, Z/P, P and RS, purified germ cell populations containing spermatogonia, preleptotene/leptotene spermatocytes, zygotene/early pachytene spermatocytes, late pachytene/diplotene spermatocytes and round spermatids, respectively. L2 and L–, conditional (loxP-flanked) and null (excised) alleles, respectively. CTL– and CTL+, negative (no DNA added) and positive (L2/L2 or L–/L–DNA added) control PCR reactions, respectively. (C-F) Immunohistochemical detection of spermatogonia expressing RARG (red nuclear signal) in control and Rara;b;g Spg–/– testes at PN5 and PN60. ZBTB16 expression (green nuclear signal) identifies spermatogonia. Spermatogonia nuclei co-expressing RARG and ZBTB16 appear in yellow. VII, stage VII of the seminiferous epithelium cycle; SG, spermatogonia. PR, preleptotene spermatocytes; asterisks indicate non-specific fluorescence in Leydig cells. Scale bars: 55 μm (C and D) and 40 μm (E and F).
(TIF)
Schematic representation of Stra8 locus and analysis by qPCR of DNA recovered from PN5 wild-type testis chromatin immunoprecipitated using antibodies directed against RNApol2, all RAR isotypes (RAR) or all RXR isotypes (RXR) at the Stra8 locus. The untranslated exons and the two transcription start sites (TSS1 and TSS2) are depicted by open boxes and broken arrows, respectively. The locations of primers used for qPCR are indicated at −3 kb and in Stra8. Mean fold enrichment of three experiments at DR4 (grey bars) and DR2 (black bars) binding sites is relative to the amount of DNA recovered at −3 kb (set at 1, white bars). Error bars represent s.e.m (n = 5); * p < 0.05.
(TIF)
EMSA showing that RARG/RXRA heterodimers (Het) bound to the DR5 of Rarb (lane 3) are competed both when unlabeled DR5 (lane 5) or increasing amounts of DR1 (lanes 6–8), DR0 (lanes 9–11) and IR1 (lanes 12–14) are added to reaction. 32P-DR5 probe indicates unbound DNA.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.