Genetic polymorphism may play a role in modulating peripheral arterial disease (PAD) severity when an occlusion is present. With the use of a preclinical model of PAD, we show that a disintegrin and metalloproteinase gene 12, a gene within the limb salvage-associated quantitative trait locus 1, is upregulated in ischemic limbs and sufficient to modify experimental PAD severity in mice, likely through regulation of tyrosine kinase with Ig-like and EGF-like domain 2.
Keywords: peripheral vascular disease, angiogenesis, ischemia, genetics and genes
Abstract
In prior studies from multiple groups, outcomes following experimental peripheral arterial disease (PAD) differed considerably across inbred mouse strains. Similarly, in humans with PAD, disease outcomes differ, even when there are similarities in risk factors, disease anatomy, arteriosclerotic burden, and hemodynamic measures. Previously, we identified a locus on mouse chromosome 7, limb salvage-associated quantitative trait locus 1 (LSq-1), which was sufficient to modify outcomes following experimental PAD. We compared expression of genes within LSq-1 in Balb/c mice, which normally show poor outcomes following experimental PAD, with that in C57Bl/6 mice, which normally show favorable outcomes, and found that a disintegrin and metalloproteinase gene 12 (ADAM12) had the most differential expression. Augmentation of ADAM12 expression in vivo improved outcomes following experimental PAD in Balb/c mice, whereas knockdown of ADAM12 made outcomes worse in C57Bl/6 mice. In vitro, ADAM12 expression modulates endothelial cell proliferation, survival, and angiogenesis in ischemia, and this appeared to be dependent on tyrosine kinase with Ig-like and EGF-like domain 2 (Tie2) activation. ADAM12 is sufficient to modify PAD severity in mice, and this likely occurs through regulation of Tie2.
NEW & NOTEWORTHY
Genetic polymorphism may play a role in modulating peripheral arterial disease (PAD) severity when an occlusion is present. With the use of a preclinical model of PAD, we show that a disintegrin and metalloproteinase gene 12, a gene within the limb salvage-associated quantitative trait locus 1, is upregulated in ischemic limbs and sufficient to modify experimental PAD severity in mice, likely through regulation of tyrosine kinase with Ig-like and EGF-like domain 2.
peripheral arterial disease (PAD), which is caused by atherosclerosis, is a major health care problem. PAD has a prevalence nearly equal to that of ischemic heart disease and morbidity and mortality equivalent to moderate-to-severe heart failure (11, 21). When lower-extremity atherosclerosis is present, patients may be asymptomatic or present with symptoms that fall into one of two major classifications: intermittent claudication (IC) or critical limb ischemia (CLI). IC patients have leg pain (when walking) that is relieved with rest, but the overwhelming majority will not develop an ischemic ulcer nor require amputation. In contrast, CLI patients have leg pain at rest, ulceration, or gangrene; a lower-extremity amputation rate of 20–45%/yr; and an annual mortality as high as 20% (32).
There is growing evidence that genes and pathways that play a role in the development of atherosclerosis also contribute to development of PAD (7, 31). However, given the extensive overlap in risk factors, anatomy, and even hemodynamic measures, observed in patients with IC and those with CLI, there is a strong possibility that genetic polymorphisms may play a role in modulating PAD severity when an occlusion is present (7, 24). Experimental PAD [e.g., hindlimb ischemia (HLI) model detailed in materials and methods] is used widely as a preclinical model for studying the processes that occur following vessel occlusion (5), and outcomes following experimental PAD are influenced by mouse strain backgrounds (13, 35). For example, following experimental PAD, the Balb/c mouse strain had poorer perfusion recovery and typically developed limb necrosis, whereas the C57Bl/6 strain had favorable perfusion recovery without limb necrosis (9, 13, 35). This suggests the existence of a genetic background that affects response to ischemic injury and may affect the PAD severity in the setting of a vessel occlusion. However, to date, no specific genes that modulate PAD severity have been identified.
Previously, we used a preclinical model of PAD and took advantage of the different outcomes in C57Bl/6 and Balb/c mice to map a locus on mouse chromosome 7 that was associated with the favorable outcomes (better perfusion recovery and less tissue loss) in the C57Bl/6 strain and termed it limb salvage-associated quantitative trait locus 1, or LSq-1 (9). Hence, we hypothesized that LSq-1 harbors a gene(s) whose polymorphisms play a role in modulating PAD severity. Therefore, the goal of this study was to refine this locus further, select a candidate gene, determine if its modulation can affect outcomes following experimental PAD, and determine whether polymorphisms of our candidate gene are associated with the severity of PAD in humans.
MATERIALS AND METHODS
Mice.
Twelve- to 14-wk-old male mice (C57Bl/6J, C57BlKS/J, CBA/J, DBA2, Balb/c, 129SI/Svlm, FVB/nJ) were obtained from The Jackson Laboratory (Bar Harbor, ME), either directly or bred internally from parental strains obtained from The Jackson Laboratory. Most of the strains were selected because they were identified previously as having favorable (C57Bl/6J, C57BlKS/J, CBA/J, DBA2, FVB/nJ) or poor (129SI/Svlm) recovery in an ischemic stroke model (22). Twelve- to 18-wk-old male or female C57Bl/6 and Balb/c mice were used in studies assessing a disintegrin and metalloproteinase gene 12 (ADAM12) upregulation in ischemia. Experimental groups were matched for mouse sex and age. Studies were performed under protocols approved by the University of Virginia Animal Care and Use Committee.
Experimental PAD/HLI and perfusion recovery.
Experimental PAD or HLI was achieved by unilateral femoral artery ligation and excision, as described previously (5, 9, 28). Although the HLI model involves more of an acute ischemic injury than is seen in human PAD, conditions that are associated with poor clinical outcome in humans are associated with poorer perfusion recovery and/or increased tissue loss in mice. Nevertheless, this is a widely accepted model of PAD. Blood flow in the ischemic and contralateral, nonischemic limbs was measured by laser Doppler perfusion imaging (LDPI), as described previously (9, 15). Control mice were of the same strain, age, and sex as the experimental mice.
Immunohistochemistry.
In thin sections of ischemic and nonischemic hindlimb muscles [tibialis anterior (TA) or gastrocnemius (GA)], capillaries were identified using a rat anti-mouse CD31 antibody (BD PharMingen, San Diego, CA) and visualized using goat anti-rat conjugated with Alexa Fluor 488 (Molecular Probes, Carlsbad, CA) (14, 39). Three random, high-power (100× magnification) fields were counted, and capillary density was expressed as capillaries/fibers. Muscle fibers were identified using MAb anti-muscle pan-actin clone AC40-Cy3 conjugated (Sigma-Aldrich, St. Louis, MO). Nuclei were stained with 4′,6-diamidino-2-phenylindole (Molecular Probes). Fluorescence microscopy was performed using a Zeiss LSM 700 scanning confocal microscope. Analysis of fluorescence and bright-field microscopy was performed using Image-Pro Plus 3.0 software (Media Cybernetics, Silver Spring, MD).
Cell line and culture.
Human umbilical vein endothelial cells (HUVECs) and endothelial cell (EC) growth medium (ECGM) were obtained from Cell Applications (San Diego, CA). ECGM was supplemented with 10% FBS. In vitro simulation of ischemia was achieved as described previously (30) with modifications as follows: cells were deprived of nutrients and oxygen by growing in endothelial starvation medium (Cell Applications) and cultured in 2% oxygen in a hypoxic chamber fitted with a ProOx model 110 oxygen controller for continuous monitoring and adjustment of the oxygen content of the chamber (BioSpherix, Lacona, NY). In experiments using CD31+ cells from mouse hindlimbs, the cells were isolated, as described previously (19), and detailed below.
Cell isolation from mouse hindlimbs, enrichment for CD31-expressing cells, and flow cytometry.
Cells were isolated from mouse hindlimbs, as described previously (19). CD31-expressing cells were isolated from mouse hindlimbs, as described previously (19). Briefly, GA and TA muscles were extracted from mouse hindlimbs. Muscles were placed in a 50-ml conical tube containing HBSS without calcium and magnesium (Mediatech, Manassas, VA) and minced into ∼1-mm sections. Muscle tissue was digested in 0.2% collagenase type IV (Worthington Biochemical, Lakewood, NJ) for 30 min at 37°C. Digested tissue was passed through a 70-μm strainer (Becton Dickinson, Franklin Lakes, NJ) to remove connective tissues, and cells were collected in the flow-through, which was centrifuged at 300 g for 5 min and resuspended in 30 ml PBS without calcium and magnesium (Invitrogen, Carlsbad, CA) but with 2 mM EDTA (Mediatech) and 0.1% BSA (Sigma-Aldrich). CD31-positive cells were isolated from the cell suspension by incubating the flow-through cells with DSB-X (Invitrogen) biotinylated anti-mouse CD31 (eBioscience, San Diego, CA, and BD Biosciences, San Jose, CA). CD31-expressing cells were then enriched by using FlowComp Dynabeads (Invitrogen), according to the manufacturer's instructions. Briefly, manufacturer-supplied Dynabeads (streptavidin-linked beads) were added to the cell suspension-containing cells treated with biotinylated anti-mouse CD31. A magnet was then applied to the tube containing the cells to pull down Dynabeads bound to CD31-expressing cells, which were then released from the Dynabeads using the manufacturer's supplied release buffer. The fraction of cells from which CD31-expressing cells had been removed was classified as “CD31-depleted fraction,” whereas the flow-through containing CD31-expressing cells was classified as “CD31-enriched fraction.”
Cell staining for flow cytometry and analysis was performed, as described previously (19), with the following antibodies: 7-aminoactinomycin D was used to assess cell viability (Catalog #420403, 1:30 dilution of stock; BioLegend, San Diego, CA), and CD31-expressing cells were detected using anti-CD31 Brilliant Violet (Catalog #102423, 1:500 dilution of stock; BioLegend). ADAM12-expressing cells were stained with anti-ADAM12 antibody (Catalog #39155, 1:500 dilution of stock; Abcam, Cambridge, MA), and secondary antibody for detection of ADAM12+ cells was anti-rabbit IgG phycoerythrin-conjugated goat IgG (Catalog #Fo110, 1:500 dilution of stock; R&D Systems, Minneapolis, MN).
RNA, quantitative PCR, and protein analysis.
Total RNA was isolated from mouse (TA, GA, and ECs) and then used for real-time quantitative (q) RT-PCR using TaqMan assays (Applied Biosystems, Foster City, CA), as described previously (6). The assay identification number for the primer/probe sets used for the amplification of each gene is provided in Table 1. Total hindlimb muscle protein lysates were obtained, as described previously (15). ADAM12 protein was assessed by Western blotting, as described previously (15), using anti-ADAM12 antibody 39155 (Abcam). Total AKT and phosphorylated (p)-AKT were assayed in HUVECs by Western blotting of lysates using anti-AKT (Catalog #9272, 1:1,000 dilution of stock) or anti-p-AKT (S473; Catalog #4060, 1:1,000 dilution of stock; both from Cell Signaling Technology, Danvers, MA). Blots were first probed for p-AKT and then stripped and reprobed for AKT. Secondary antibodies for Western blots were anti-mouse or anti-rabbit IgG, labeled with infrared dye (1:10,000 dilution of stock, 0.5 mg/ml IRDye 800CW; LI-COR Biosciences, Lincoln, NE). Images were saved in fluorescent and white/gray format. Bands from Western blots were quantified using the ImageJ program (34). Cleaved VEGF receptor 2 (VEGFR2) was assayed in cell-culture supernatants using a human VEGFR2 immunoassay (Catalog #DVR200; R&D Systems), per the manufacturer's protocol.
Table 1.
Summary of genes within the 5 haplotype blocks and their mRNA expression profile
| Gene (Assay ID) and Haplotype Block | Expression in Mouse Hindlimbs | Upregulated in Ischemia | C57Bl/6 > Balb/c | Balb/c > C57Bl/6 |
|---|---|---|---|---|
| Block 1 | ||||
| 3100003L05Rik (Mm01277666_m1) | No | No | No | No |
| 4933440M02Rik (Mm01277432_m1) | No | No | No | No |
| 4930571K23Rik (Mm03807449_s1) | No | No | No | No |
| Jmjd5 (Mm00513079_m1) | Yes | No | No | No |
| Nsmce1 (Mm00471126_m1) | Yes | No | No | No |
| Il4ra (Mm01275139_m1) | Yes | Yes | Yes (<2-fold) | No |
| Il21a (Mm00600319_m1) | Yes | Yes | Yes (=2-fold) | No |
| Gtf3c1 (Mm01278522_m1) | Yes | No | No | No |
| D430042009Rik (Mm00525471_m1) | Yes | No | No | No |
| Gsg1l (MM01278522_m1) | No | No | No | No |
| Xpo6 (Mm00503902_m1) | Yes | No | No | No |
| Sbk1 (Mm00455133_m1) | Yes | No | No | No |
| Lat (Mm00456761_m1) | Yes | Yes | No | No |
| Spns1 (Mm00470041_m1) | Yes | No | No | No |
| Nfatc2ip (Mm00803049_m1) | Yes | No | No | No |
| Cd19 (Mm00515420_m1) | No | No | No | No |
| Block 2 | ||||
| Bag3 (Mm00443474_m1) | Yes | No | No | No |
| Inpp5f (Mm00724391_m1) | Yes | Yes | Yes (<2-fold) | No |
| Block 3 | ||||
| ADAM12 (Mm00475719_m1) | Yes | Yes | Yes (>3-fold) | No |
| D7Ertd443e (Mm01131413_m1) | Yes | No | No | No |
| Dock1 (Mm01269874_m1) | Yes | Yes | No | No |
| Block 4 | ||||
| 9430038I01Rik (Mm03991735_g1) | Yes | Yes | No | Yes |
| Block 5 | ||||
| Gpr123 (Mm00623786_m1) | No | No | No | No |
| Kndc1 (Mm006278522_m1) | No | No | No | No |
| Utf1 (Mm00447703_g1) | No | No | No | No |
No expression was defined as no amplification or amplification at comparative threshold > 40.
ADAM12 overexpression and knockdown in mouse hindlimbs.
Full-length murine ADAM12 cDNA was purchased from Open Biosystems (Huntsville, AL). The cDNA was supplied as a Gateway Entry vector (Invitrogen) and was directionally cloned in the p-cytomegalovirus (CMV)-SPORT6 expression vector (Invitrogen) that directs high expression from the CMV promoter upon transfection (or direct injection) into mammalian cells. The cDNA includes the natural stop codon, so there will be no fusion tags or reporter sequences added. For gene transfer into mouse hindlimbs, electric, pulse-mediated gene transfer was performed, as described previously (1). Briefly, under anesthesia (isoflurane), 50 μg (TA) or 100 μg (GA) full-length murine ADAM12 cDNA at a concentration of 2 μg/μl in normal saline was injected in mouse hindlimbs using a 0.5-ml syringe with a 28-gauge needle. Eight electric pulses (100 ms, 1 Hz, and 100 V) were delivered immediately to the injected muscle using a S88K square-pulse stimulator (Grass-Telefactor, West Warwick, RI) through a model 533, 2-Needle Array (BTX Instrument Division, Harvard Apparatus, Holliston, MA), placed on the medial and lateral sides of the muscle so that the electrical field is perpendicular to the long axis of the myofibers. Mice were allowed to recover for 10 days before use in experimental PAD studies. For the in vivo ADAM12 knockdown, we used plasmid that contained small hairpin (sh)RNA, targeting mouse ADAM12, and was coupled to microbubbles. The plasmid (200 μg) was administered to each mouse by retro-orbital injection, and delivery to hindlimbs was achieved through ultrasound-mediated bubble disruption, as described previously (37, 38). Briefly, cationic microbubbles (2 × 108) combined with 200 μg plasmid were infused via retro-orbital injection >1 min. The GA of the left hindlimb were exposed to ultrasound during the injection and for an additional 9 min. Ultrasound exposure was performed using harmonic-power Doppler imaging (Sonos 7500; Philips Ultrasound, Andover, MA) at 1.6 MHz, a pulsing interval of 5 s, a pulse-repetition frequency of 2.5 kHz, and a mechanical index of 0.6, 1.3, or 2.4. Effectiveness of these shRNA in targeting ADAM12 was determined in pilot experiments. Five days postplasmid injection, perfusion recovery by LDPI was initiated, or mice were subjected to experimental PAD. ADAM12 mRNA knockdown in ischemic mouse hindlimbs was determined by qRT-PCR. Effect of shADAM12 on other ADAM isoforms was assessed by qPCR for ADAM15 (Mm00477328) and ADAM17 (Mm00456428) using shADAM12-treated tissue. The human and mouse shRNA kits were obtained from SABiosciences (Qiagen, Valencia, CA; Catalog Numbers KH07647N and KM28766G, respectively). The control shRNA supplied with each kit was used as controls in each experiment.
Transfection of ECs and measurement of in vitro endothelial proliferation, apoptosis, angiogenesis, and cleaved Tie2.
For ADAM12 augmentation and knockdown in vitro, plasmid containing full-length ADAM12 cDNA or shRNA targeting ADAM12 was transfected into HUVECs using a high-efficiency primary cell-transfection reagent (Cytofect EC transfection kit; Cell Applications), following the manufacturer's protocol. Briefly, cells were cultured to ∼75% confluence in a six-well plate. DNA plasmid (3 μg) was mixed with 2.5 μl Cytofect-2 and 2.5 μl polyethylene reagents and incubated at 37°C for 30 min. This plasmid DNA and transfection reagent complex mixture was added to ECs for 1 h and then replaced with ECGM with 10% serum. Cells were used in subsequent experiments, 24 h after transfection. EC proliferation, apoptosis assay, and in vitro angiogenesis were assessed, as described previously (16). Briefly, proliferation was assessed under baseline conditions (ECGM with 10% serum) using tetrazolium dye incorporation (BioVision, Milpitas, CA), 48 h post-transfection, following the manufacturer's protocol. Apoptosis was assessed in transfected cells, 24 h post-transfection, followed by 24 h exposure to simulated ischemia using the terminal deoxynucleotidyltransferase (TdT)-mediated 2′-deoxyuridine 5′-triphosphate nick end-labeling assay (TiterTACS; Trevigen, Gaithersburg, MD), following the manufacturer's protocol. Nuclease-treated wells were used as positive controls, whereas wells without terminal TdT were used as negative controls. For the angiogenesis assay, the cells were plated on growth factor-enriched matrigel (Catalog #356234; BD Biosciences), 48 h post-transfection, and grown in simulated ischemia for 6 h. Each condition was performed in quadruplicate, and representative pictures were taken from each well at 100× magnification. A blinded observer counted the number of complete tubes, which was expressed as tubes/mm2. All experiments described above were repeated at least three times.
For the cleaved/shed tyrosine kinase with Ig-like and EGF-like domain 2 (Tie2) studies, the level of EC Tie2 shedding in ischemia was evaluated by assaying the supernatant of HUVECs exposed to 24 h of simulated ischemia using a human Tie2 ELISA assay (Catalog #DTE200; R&D Systems) and following the manufacturer's protocol. In experiments assessing ADAM12-mediated Tie2 shedding in ischemia, HUVECs, in which ADAM12 had been knocked down (as described above), were exposed to simulated ischemia for 24 h, and the cell culture supernatant was assayed for shed Tie2 by ELISA. Cleaved Tie2 in ischemic and nonischemic mouse hindlimbs was also assessed by Western blotting (15) of GA muscle lysates using anti-Tie2 antibody (Catalog #4224; Cell Signaling Technology). The importance of Tie2 signaling in ADAM12-induced EC proliferation was assessed by cotransfection of ADAM12 cDNA with plasmid containing cDNA for a “kinase dead” Tie2 (lysine to arginine at amino acid 854 or K854R) into HUVECs, 24 h before performing a proliferation assay, as described above. Kinase dead Tie2 was described previously (18) and was provided by Dr. Christopher Kontos (Duke University, Durham, NC).
The knocking down of hypoxia-inducible factor 1-α (HIF-1α) in ECs was achieved by transfection of small interfering (si)RNA, targeting HIF-1α (Catalog #4390824; Ambion, Grand Island, NY). Cell transfection was achieved with siPORT NeoFX transfection agent (Catalog #am4511; Ambion), following the manufacturer's protocol.
Statistical analysis.
All measurements were expressed as means ± SE. Statistical comparisons between two groups (e.g., treated vs. untreated) at a specific time point were performed with the independent Student's t-test. Comparison of more than two groups was performed with ANOVA. In all cases, P < 0.05 was considered statistically significant.
RESULTS
Perfusion recovery across inbred mouse strains and resulting haplotype map refine LSq-1.
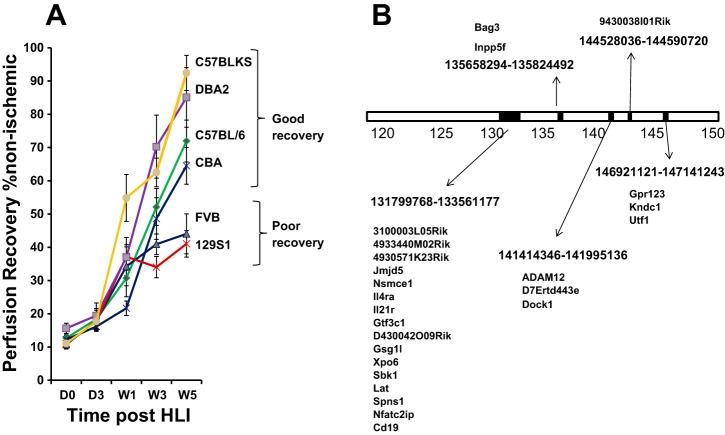
Previously, we showed that LSq-1 defined a region on mouse chromosome 7 that spanned ∼31 mb between markers rs6279696 and rs6216320 and that is estimated to contain 475 genes (9). To narrow genes further within the locus, experimental PAD was performed on six inbred mouse strains, shown previously to demonstrate favorable (C57Bl/6J, C57BlKS/J, CBA/J, DBA2, and FVB/nJ) or poor (129SI/Svlm) recovery in an ischemic stroke model (22), and their perfusion recovery phenotype was determined (Fig. 1A). Based on this phenotype, mouse strains were classified as showing favorable recovery and were aligned with C57Bl/6 or showing poor recovery and were aligned with Balb/c. The results from the strains tested here were combined with that of two other strains tested previously [Balb/c (9) and A/J (9)] to generate haplotype blocks based on single nucleotide polymorphism (SNP) alleles, common in strains that recover well but absent in strains that recover poorly (i.e., C57Bl/6J = C57BlKS/J = CBA/J = DBA2/J but ≠ Balb/cJ, A/J, 129SI/SvlmJ, FVB/nJ). Five haplotype blocks were identified containing 25 genes (Fig. 1B).
Fig. 1.
A: perfusion recovery across inbred mouse strains (6 shown) and resulting haplotype map reduce the number of candidate genes within limb salvage-associated quantitative trait locus 1 (LSq-1). y-Axis shows extent of perfusion in the ischemic limb relative to the nonischemic limb; x-axis shows days (D) and weeks (W) following experimental peripheral arterial disease (PAD; n = 8–10/group). HLI, hindlimb ischemia. B: schematic of haplotype blocks shows location of single nucleotide polymorphisms within LSq-1, where C57Bl/6 = C57BlKS = CBA = DBA2 but ≠ Balb/c, A/J, 129SI/Svlm, FVB/n. Perfusion recovery for Balb/c and A/J was assessed and determined previously to be poor (9). Genes within each haplotype block are shown. ADAM12, a disintegrin and metalloproteinase gene 12.
Gene expression of the 25 genes within the five haplotype blocks in ischemic and nonischemic muscles.
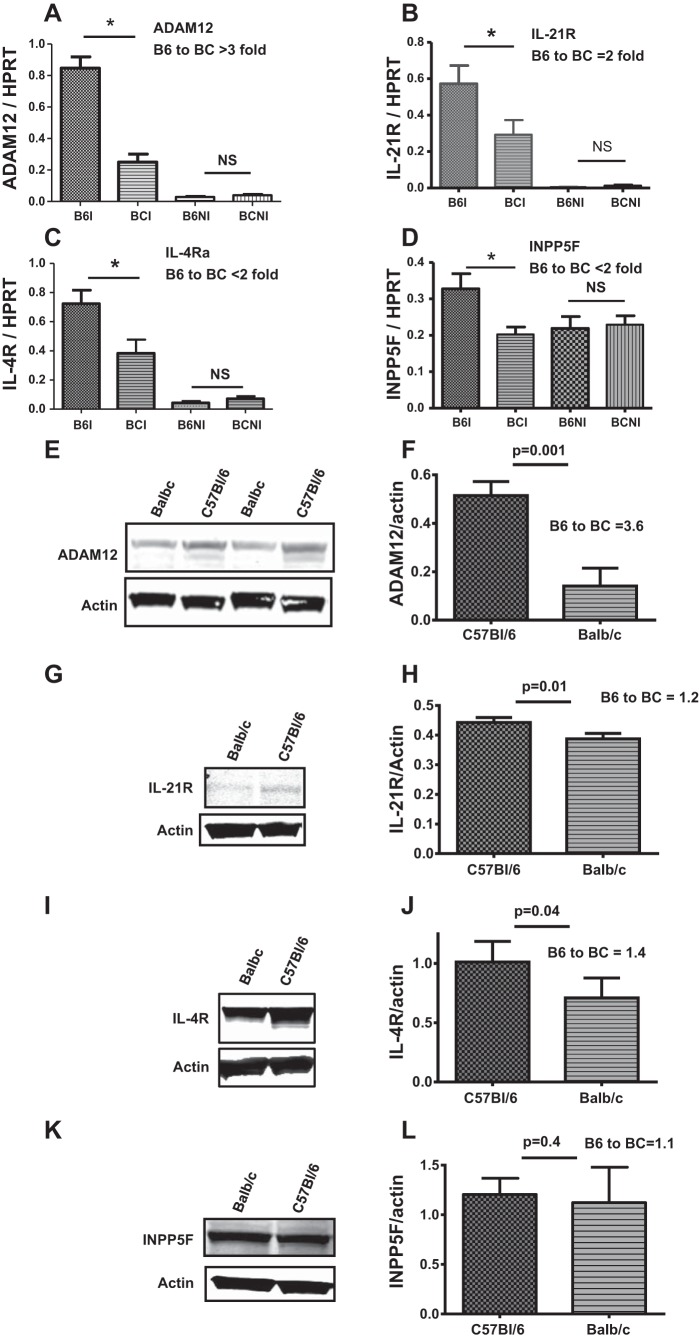
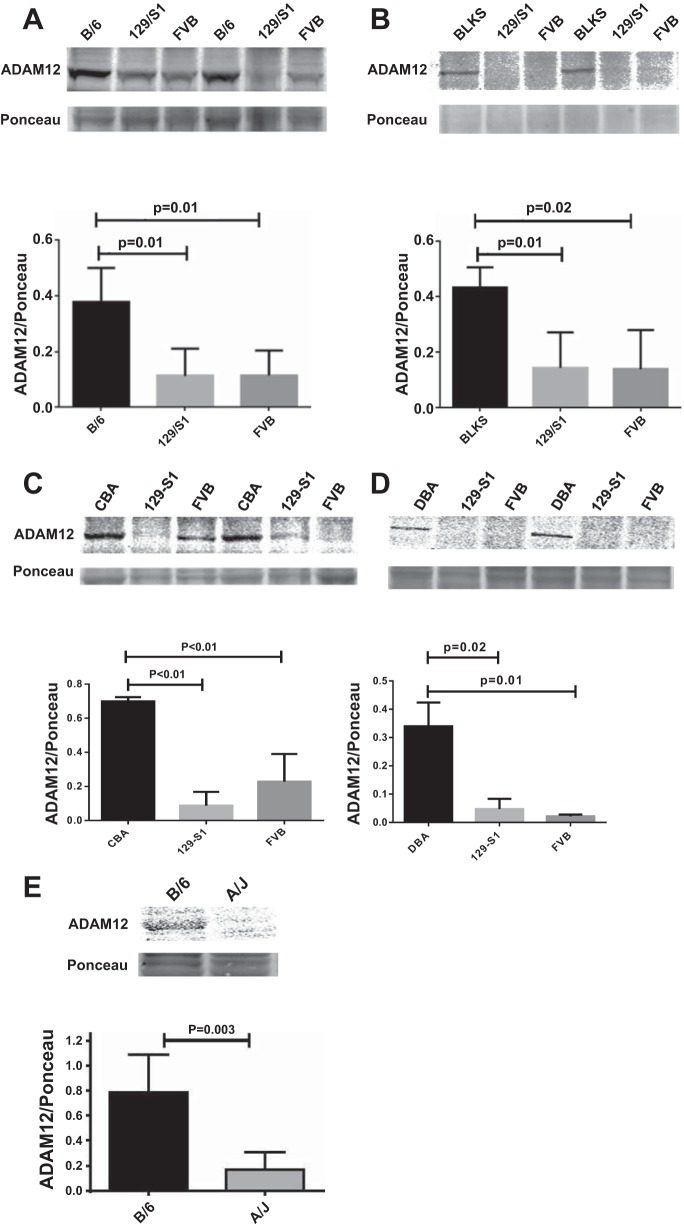
We analyzed the mRNA expression of the above 25 genes in nonischemic and day 3 postischemic hindlimb muscles of C57Bl/6 and Balb/c mice by qRT-PCR. The day 3 postligation time point was used because we had previously shown that perfusion recovery is comparable among these strains then (9). We found eight genes that were not expressed at significant levels [defined as no amplification or amplification at comparative threshold > 40, despite using the same amount of template used for the other detected genes (Table 1)]. Ten genes were expressed but not upregulated in ischemic hindlimbs compared with nonischemic hindlimbs, and seven genes were expressed and upregulated (summarized in Table 1). Of these seven genes, only five showed differential mRNA expression between C57Bl/6 and Balb/c mice. Four of the five genes showed significantly higher mRNA levels in C57Bl/6 than in Balb/c mice (Fig. 2, A–D), whereas one gene showed higher expression in Balb/c (Table 1). We assessed protein expression of the four genes that showed higher mRNA levels in C57Bl/6 compared with Balb/c and found that ADAM12 showed the highest differential expression, and this was more than threefold higher in C57Bl/6 compared with Balb/c (Fig. 2, A, E, and F). The other genes showed less than twofold differential expression between C57Bl/6 and Balb/c (Fig. 2, G–J) or no difference in expression (Fig. 2, K and L). Next, we assessed ADAM12 expression in the mouse strains used in the haplotype block analysis and found the strains that show favorable perfusion recovery also showed higher ADAM12 expression compared with strains that showed poor perfusion recovery (Fig. 3, A–E). Therefore, ADAM12 was chosen as a candidate gene for further testing.
Fig. 2.
ADAM12 showed higher differential expression in C57Bl/6 (B6) compared with Balb/c (BC). ADAM12 (A), IL-21R (B), IL-4Ra (C), and inositol polyphosphate-5-phosphatase F (INPP5F) mRNA (D) are upregulated in ischemic hindlimbs at a higher level in C57Bl/6 than Balb/c. ADAM12 showed the highest difference in expression (>3-fold) in C57Bl/l6 compared with Balb/c [B6I, C57Bl/6 ischemic; BCI, Balb/c ischemic; B6NI, C57Bl/6 nonischemic; BCNI, Balb/c nonischemic; n = 8–10/group, *P < 0.05, not significant (NS) = P > 0.05]. E: higher expression of ADAM12 protein in ischemic hindlimb muscles of C57Bl/6 compared with Balb/c. F: quantification of blot in E (n = 4 for C57Bl/6 and 5 for Balb/c). ADAM12 protein shows the highest differential expression between C57Bl/6 and Balb/c (E and F) compared with the other differentially expressed genes [IL-21R (G and H) and IL-4R (I and J); n = 3–4/group]. INPP5F protein (K and L) did not show significant differential expression (n = 3/group, P = 0.4). HPRT, hypoxanthine guanine phosphoribosyl transferase.
Fig. 3.
Mouse strains with better perfusion recovery (C57Bl/6, C57BlKS, and CBA) show higher ADAM12 protein expression compared with strains with poor perfusion recovery (129-S1, FVB, and A/J). Representative blots showing higher expression of ADAM12 in wk 5 postischemic hindlimb muscles (gastrocnemius) of C57Bl/6 (B/6) compared with 129-S1 and FVB (A); C57BlKS compared with 129-S1 and FVB (B); CBA compared with 129-S1 and FVB (C); DBA compared with 129-S1 and FVB (D); and C57Bl/6 compared with A/J (E). E, top: ADAM12 Western and Ponceau staining; bottom: quantification of bands in top relative to same region on the Ponceau-stained blot (n = 3–4/group).
Augmentation of ADAM12 expression improves perfusion recovery in Balb/c, and the knocking down of ADAM12 impairs perfusion recovery in C57Bl/6.
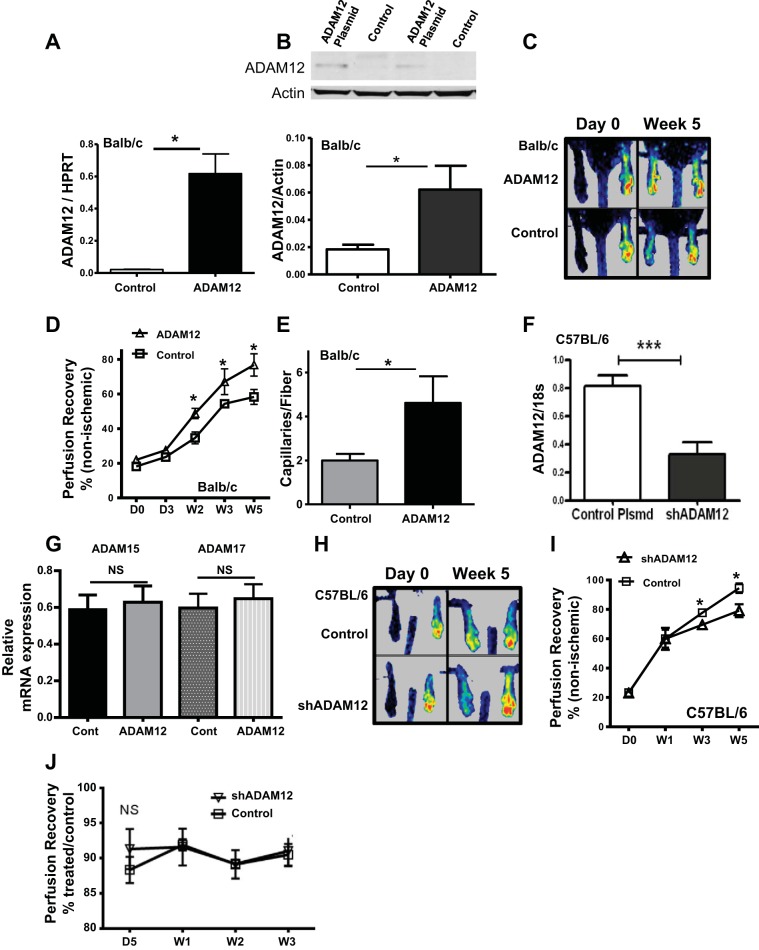
To augment ADAM12 expression, we injected an expression plasmid containing ADAM12 cDNA, or control plasmid, into the hindlimbs of Balb/c mice that normally show poor perfusion recovery and minimal upregulation of ADAM12 in the ischemic hindlimb muscles. ADAM12 mRNA and protein levels were assessed 10 days postplasmid injection. We found increased ADAM12 mRNA [ADAM12/hypoxanthine guanine phosphoribosyl transferase (HPRT) = 0.62 ± 0.10 vs. control/HPRT = 0.02 ± 0.01, n = 6/group, P < 0.05; Fig. 4A] and protein expression (ADAM12/actin = 0.06 ± 0.02 vs. control/actin = 0.02 ± 0.01, n = 3/group, P = 0.04; Fig. 4B) in hindlimbs of mice that received ADAM12 cDNA compared with mice that received control plasmid. Next, we determined the effect of this gene transfer on perfusion recovery following experimental PAD, 10 days postplasmid injection. Balb/c mice receiving ADAM12 cDNA showed better perfusion recovery compared with those receiving control plasmid (wk 5 perfusion recovery as a percent of nonischemic limb = 76.8 ± 6.4 vs. 58.4 ± 4.3, n = 7–9/group, P = 0.02; Fig. 4, C and D). Mice receiving ADAM12 cDNA showed higher capillary density in their hindlimbs at 5 wk postexperimental PAD compared with those receiving control plasmid (4.6 ± 1.2 vs. 2.0 ± 0.3, capillaries/fiber, n = 5–7/group, respectively, P = 0.05; Fig. 4E).
Fig. 4.
Augmentation of ADAM12 expression in Balb/c improved perfusion recovery, whereas knockdown of ADAM12 expression in C57Bl/6 by small hairpin (sh)RNA impaired perfusion recovery following experimental PAD. A: higher ADAM12 mRNA is expressed in hindlimbs that received ADAM12 cDNA compared with those that received control plasmid, 10 days post-treatment (n = 5/group, *P < 0.05). B: ADAM12 Western showing higher expression in muscle lysates from mouse hindlimbs that received ADAM12 cDNA (top); bottom: quantification of bands in top (n = 3/group, *P = 0.04). C: representative laser Doppler perfusion imaging (LDPI) image showing increased perfusion in Balb/c mice that received ADAM12 cDNA. D: LDPI showing perfusion recovery after HLI. y-Axis shows extent of perfusion in the ischemic limb relative to the nonischemic limb; x-axis shows days and weeks following experimental PAD (n = 7–10/group, *P < 0.05). E: anti-CD31 staining of sections of ischemic hindlimbs, 5 wk following experimental PAD, showed a higher number of capillaries/muscle fiber (capillary density) in Balb/c mice that received ADAM12 cDNA (control n = 5, ADAM12 plasmid n = 7, *P = 0.05). F: ischemic hindlimbs from mice treated with shRNA targeting mouse ADAM12 showed decreased ADAM12 mRNA expression compared with hindlimbs treated with control plasmid (n = 6, ***P < 0.01), 10 days postinjection. G: shADAM12-treated hindlimbs show no change in expression of 2 other ADAM isoforms (ADAM15 and ADAM17, n = 6, NS = P > 0.05). H: representative LDPI image showing impaired perfusion in C57Bl/6 mice that received plasmid containing mouse ADAM12 shRNAs. I: y-axis shows extent of perfusion in the ischemic limb relative to the nonischemic limb; x-axis shows days and weeks following experimental PAD (n = 13/group, *P < 0.05). J: perfusion in nonischemic, shADAM12-treated hindlimbs is not different from perfusion in control, nonischemic hindlimbs (NS = P > 0.05, n = 5 for shADAM12, and n = 6 for control).
Next, we assessed the effect of knocking down ADAM12 in C57Bl/6 mice on perfusion recovery following experimental PAD. Mice were injected systemically with microbubbles containing either shRNA, which target ADAM12 expression, or control shRNA, followed by ultrasound disruption of the microbubbles in the hindlimbs (4, 37, 38). Five days after ADAM12 shRNA injection, mice were subjected to experimental PAD. We found significantly decreased ADAM12 mRNA expression in day 3 postischemic limbs of ADAM12 shRNA-treated mice compared with mice that received control plasmid (Fig. 4F). We found no change in expression of two other ADAM isoforms (ADAM15 and ADAM17) in the shADAM12-treated hindlimbs (Fig. 4G). In a different group of mice treated with shRNA, as described, and then subjected to experimental PAD, perfusion recovery was followed by LDPI. ADAM12 shRNA-treated mice showed impaired perfusion recovery following experimental PAD (Fig. 4, H and I). This impairment in recovery was noted starting at 3 wk postexperimental PAD (perfusion recovery as a percent of nonischemic limb = 69.5 ± 0.7 vs. 77.7 ± 1.4, n = 12/group P = 0.04) and was maximal at the end of the experiment at 5 wk (79 ± 4.5 vs. 95 ± 3.3, n = 12/group, P = 0.01; Fig. 4I). The knocking down of ADAM12 in nonischemic hindlimbs showed no difference in limb perfusion compared with perfusion in the control limbs (Fig. 4J).
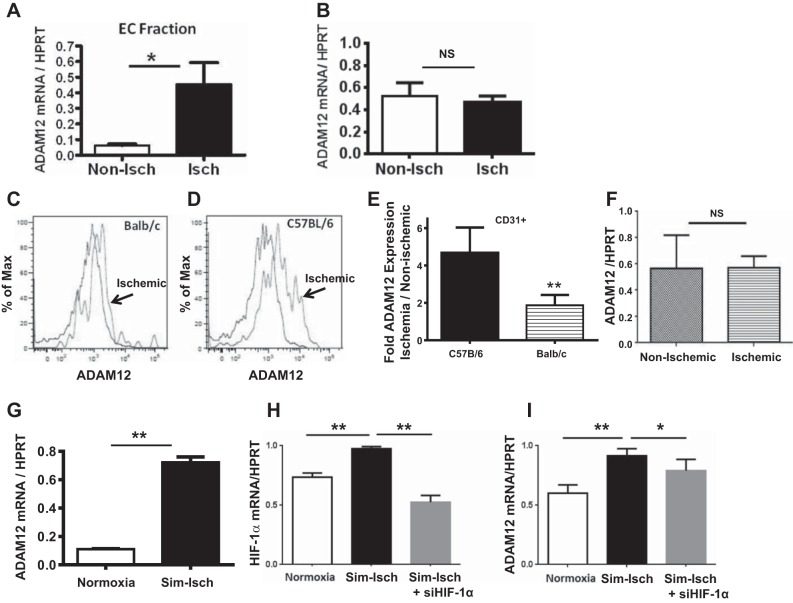
ADAM12 upregulation in ischemia occurs in CD31-expressing cells in vivo and in primary ECs in vitro and is regulated by HIF-1α.
We isolated cells from nonischemic and day 3 postischemic hindlimb muscles (19) and enriched for CD31-expressing cells to determine if ADAM12 upregulation in ischemic hindlimbs was occurring in a cell fraction that was enriched for ECs. We observed higher levels of ADAM12 mRNA expression in the CD31-enriched fraction from ischemic hindlimbs compared with that of nonischemic samples (ADAM12/HPRT = 0.5 ± 0.1 in ischemic cells vs. 0.10 ± 0.01 nonischemic cells, n = 6/group, P = 0.02; Fig. 5A). In contrast, the cells from the CD31-depleted fraction showed no difference in ADAM12 expression between ischemic and nonischemic hindlimbs (ADAM12/HPRT ratio = 0.5 ± 0.1 in ischemic vs. 0.46 ± 0.1 nonischemic, n = 6/group, P = 0.7; Fig. 5B). This suggests that increased expression of ADAM12 in ischemic hindlimbs occurred largely in the CD31-enriched fractions, which includes EC cells. Next, we isolated cells from ischemic and nonischemic hindlimb skeletal muscles from Balb/c and C57Bl/6 mice and analyzed ADAM12 expression on CD31+ cells by flow cytometry. Consistent with our mRNA and Western blot analysis, we found less ADAM12 up-regulation in CD31+ cells isolated from Balb/c mice compared with the cells from C57Bl/6 mice (expression ischemic/nonischemic, Balb/c = 1.9 ± 0.3 vs. C57Bl/6 4.7 ± 0.7; Fig. 5, C–E). To determine whether ADAM12 upregulation occurs in the collateral artery that feeds the ischemic hindlimb, we dissected the profunda femoris arteries from day 3 postischemic and nonischemic C57Bl/6 mouse hindlimbs and assessed ADAM12 mRNA expression by qPCR. We found no upregulation of ADAM12 expression in these collateral vessels (Fig. 5F). Next, we assessed whether primary human ECs would upregulate ADAM12 in ischemia. HUVECs were exposed to simulated ischemia for 24 h (16) in vitro, and then levels of ADAM12 mRNA were determined by qRT-PCR. ADAM12 mRNA is upregulated in HUVECs exposed to simulated ischemia compared with controls cultured in normoxia and standard growth medium (ADAM12/HPRT = 0.70 ± 0.04 vs. 0.10 ± 0.03, respectively, n = 6/group, P < 0.01; Fig. 5G). To determine a possible mechanism regulating ADAM12 upregulation in ischemia, we assessed ADAM12 upregulation in ECs, in which HIF-1α was knocked down by siRNA. We found that the knocking down of HIF-1α (Fig. 5H) resulted in impaired ADAM12 mRNA upregulation in simulated ischemia (Fig. 5I).
Fig. 5.
ADAM12 upregulation in ischemic hindlimb muscles is primarily in CD31-expressing cells, and this upregulation is more robust in CD31+ cells from C57Bl/6 mice compared with CD31+ cells from Balb/c. A: CD31-enriched fraction of cells from ischemic (Isch) hindlimbs showed upregulation of ADAM12 mRNA expression compared with CD31-enriched fraction of cells from nonischemic (Non-Isch) hindlimbs (n = 6/group, *P = 0.02). EC, endothelial cell. B: CD31-depleted cells from ischemic hindlimbs show no upregulation of ADAM12 mRNA expression compared with CD31-depleted cells from nonischemic hindlimbs (n = 6/group, NS = P = 0.7). C and D: representative flow cytometry histograph showing less upregulation of ADAM12 on CD31+ cells isolated from Balb/c mice (C) compared with CD31+ cells from C57Bl/6 mice (D; x-axis shows ADAM12 expression as shown in mean fluorescence). Analysis was gated to exclude dead cells and cell debris and to include only single-cell populations with CD31 expression. E: graph showing ADAM12 expression on CD31+ cells from ischemic and nonischemic hindlimbs from C57Bl/6 and Balb/c mice (n = 4/group, **P < 0.01). F: analysis of ADAM12 mRNA expression in the collateral vessels dissected from C57Bl/6 mice, 3 days post-HLI, shows no upregulation of ADAM12 in vessels from post-HLI limbs compared with vessels from the control limbs (n = 6/group, NS = P > 0.5). G: human umbilical vein ECs (HUVECs) exposed to simulated ischemia (Sim-Isch; hypoxia and nutrient deprivation) for 48 h show robust upregulation of ADAM12 mRNA expression compared with control cells in normoxia and normal growth media (n = 6/group, **P < 0.01). H: simulated ischemia induced increased hypoxia-inducible factor 1-α (HIF-1α) mRNA expression, and this is knocked down by small interfering (si)RNA targeting HIF-1α (n = 4/group, **P < 0.01). I: the knocking down of HIF-1α impairs upregulation of ADAM12 mRNA (n = 4/group, *P < 0.05, **P < 0.01).
Augmentation of ADAM12 expression in vitro increased EC proliferation, whereas shRNA-mediated knockdown of ADAM12 impairs EC proliferation, survival, tube formation, and p-AKT in simulated ischemia.
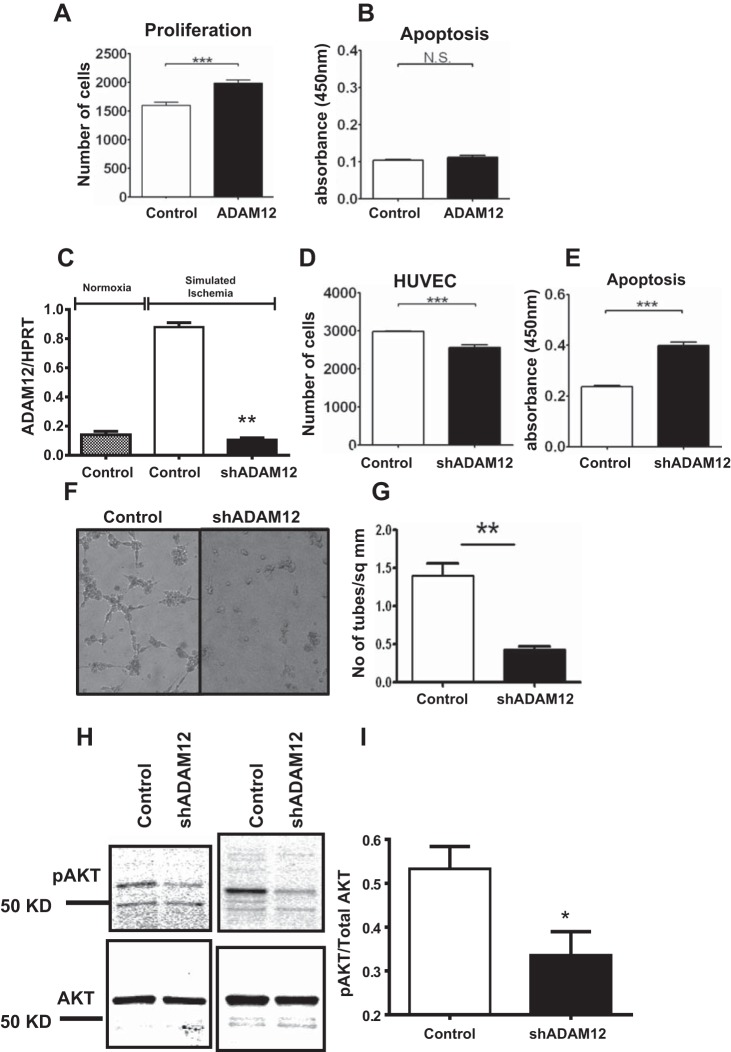
To understand the physiologic mechanism by which ADAM12 improves perfusion recovery, we augmented ADAM12 expression in HUVECs and assessed its effect on cellular proliferation and survival. ECs were transfected with a plasmid containing the ADAM12 cDNA or a control plasmid. We assessed EC proliferation 48 h post-transfection and assessed apoptosis after 24 h of exposure to simulated ischemia. We observed higher EC proliferation in cells transfected with ADAM12 cDNA compared with cells transfected with control plasmid (Fig. 6A). However, there was no difference in the extent of apoptosis in ADAM12 cDNA-transfected ECs compared with controls (Fig. 6B). Hence, augmentation of ADAM12 expression in ECs increased proliferation.
Fig. 6.
Augmentation of ADAM12 increased EC proliferation, whereas knockdown of ADAM12 decreased EC proliferation, survival, angiogenesis, and AKT phosphorylation (p-AKT) in simulated ischemia. A: human ADAM12 cDNA-transfected HUVECs show increased proliferation and no change in apoptosis (B; proliferation and apoptosis assays, n = 6–10/group, ***P < 0.01, NS = P > 0.05). C: transfection of shRNA targeting human ADAM12 results in a significant decrease in ADAM12 mRNA expression (n = 6/group, **P < 0.01). D: shADAM12-transfected HUVECs show decreased proliferation and increased apoptosis in simulated ischemia (E; proliferation and apoptosis assays, n = 6–10/group, ***P < 0.01). F: representative picture showing that shADAM12-transfected HUVECs have decreased tube formation. G: assessment of the number of tubes formed by shADAM12-transfected HUVECs compared with HUVECs transfected with control plasmid (n = 6/group, **P < 0.01). H: representative picture of Western blot from shADAM12-transfected HUVECs probed with anti-AKT and anti-p-AKT. I: quantification of blot in H showing p-AKT:AKT ratio (n = 5/group, *P < 0.05).
A plasmid containing shRNA that targets human ADAM12 expression was transfected into HUVECs that were assayed for proliferation 48 h later in normoxia and normal EC culture medium. We also assessed apoptosis in the transfected cells following 24 h of exposure to simulated ischemia. shRNA transfection resulted in effective knockdown of ADAM12 (Fig. 6C). ECs transfected with shRNA targeting ADAM12 showed decreased proliferation and increased apoptosis in simulated ischemia compared with controls (Fig. 6, D and E). Next, we assayed for the effect of ADAM12 expression on angiogenesis in simulated ischemia in vitro using the matrigel tube-formation assay. Knockdown of ADAM12 resulted in a significantly reduced number of tubes compared with controls (Fig. 6, F and G). Lastly, we assessed the effect of ADAM12 knockdown with shRNA on p-AKT in simulated ischemia. ADAM12 shRNA-transfected HUVECs showed decreased p-AKT (Fig. 6, H and I). Therefore, augmentation of ADAM12 expression in ECs improved proliferation, whereas knockdown of ADAM12 expression impaired proliferation, survival, and angiogenesis and was associated with decreased p-AKT.
ADAM12 modulates Tie2 cleavage in vitro and in vivo.
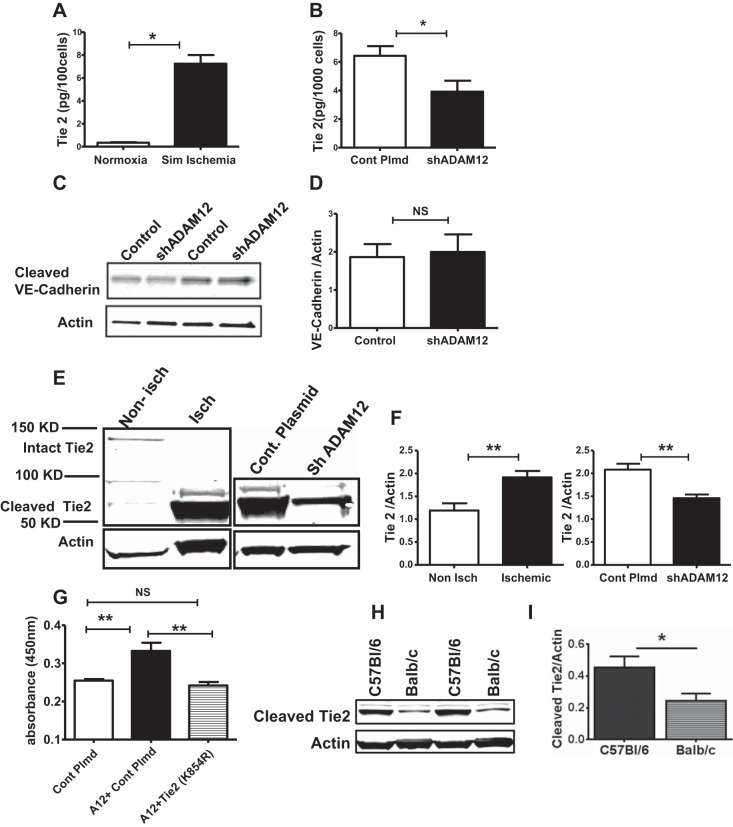
Recent studies by Frohlich et al. (12) showed that ADAM12 is expressed on ECs and can mediate ectodomain shedding/cleavage of a number of EC membrane proteins, including Tie2, vascular endothelial (VE)-cadherin, and VEGFR2, with Tie2 shedding as the most robust of the proteins assessed. Moreover, there is evidence that Tie2 plays a critical role in perfusion recovery following experimental PAD (27), and cleavage of Tie2 leads to Tie2 activation and p-AKT (10, 23). Additionally, in Fig. 6, we show that knocking down ADAM12 results in decreased p-AKT. Therefore, we hypothesized that ADAM12 may modulate perfusion recovery, in part, through shedding of Tie2. We assessed whether there was increased Tie2 shedding under conditions where we observed increased ADAM12 expression in ECs i.e., in simulated ischemia. Our result shows increased shedding of Tie2 by HUVECS exposed to simulated ischemia compared with controls in normoxia (Fig. 7A). Next, we assessed whether ADAM12 is involved in the increased Tie2 shedding by HUVECS in simulated ischemia by knocking down ADAM12 in HUVECS exposed to simulated ischemia. We found decreased Tie2 shedding in the supernatant from HUVECS, in which ADAM12 expression was knocked down (Fig. 7B). We also assessed cleavage of VE-cadherin and VEGFR2 in HUVECs, in which ADAM12 had been knocked down, since both proteins were also previously shown to be cleaved by ADAM12 (12). We found no increase in VE-cadherin cleavage (Fig. 7, C and D), and the VEGFR2 levels were too low to detect in the supernatant tested. We also assessed in vivo whether increased ADAM12 expression in ischemic hindlimbs was associated with increased Tie2 shedding and whether this was mediated by ADAM12. We found, compared with nonischemic mouse hindlimbs, that Tie2 shedding was increased in lysates from day 3 postischemic mouse hindlimbs (Fig. 7, E and F), which we previously showed has increased expression of ADAM12 (Fig. 2A). Next, ADAM12 was knocked down in vivo, as described previously for Fig. 3, and the levels of cleaved Tie2 in lysates from day 3 postischemic hindlimb muscles (GA) were assessed. We found decreased Tie2 shedding in ischemic hindlimb muscles, in which ADAM12 had been knocked down compared with control ischemic hindlimb muscles (Fig. 7, E and F). Next, we assessed whether a physiologic function of ADAM12, i.e., proliferation, is Tie2 dependent. Hence, we cotransfected ADAM12 with a mutant Tie2 (kinase dead) (18) into HUVECS and measured cell proliferation. As expected, we found that augmentation of ADAM12 expression increased proliferation of HUVECS; however, cotransfection with mutant Tie2 abolished the increased proliferation (Fig. 7G). Lastly, we compared cleaved Tie2 levels in lysates from ischemic hindlimb muscles (GA) of C57Bl/6 with that in ischemic hindlimb muscles of Balb/c mice (24 h post-HLI). We found higher levels of cleaved Tie2 (Fig. 7, H and I) in ischemic C57Bl/6 hindlimb muscles compared with that of Balb/c. This is also consistent with the higher ischemia-induced ADAM12 expression that we observed in C57Bl/6 compared with Balb/c mice. Taken together, these data suggest that ADAM12 regulates ischemia-induced Tie2 shedding in vitro and in vivo and mediates endothelial proliferation through Tie2.
Fig. 7.
ADAM12 regulates tyrosine kinase with Ig-like and EGF-like domain 2 (Tie2) shedding, and ADAM12-induced EC proliferation is Tie2 dependent. A: exposure of HUVECs to simulated ischemia increased Tie2 shedding (n = 4/group, *P < 0.01). B: ADAM12 “knock down” (shADAM12) decreased HUVEC Tie2 shedding in simulated ischemia (n = 6/group, *P < 0.05). C: the levels of the cleaved vascular endothelial (VE)-cadherin were not different between control and ADAM12 knockdown HUVECS (n = 6/group, NS = P > 0.05). D: quantification of the bands in C. E, left: lysates from ischemic mouse hindlimbs show higher levels of cleaved Tie2 compared with nonischemic hindlimbs; right: lysates from ischemic hindlimbs, in which ADAM12 was “knocked down” (shADAM12), show decreased cleaved Tie2 compared with ischemic hindlimbs that received control plasmid (Cont Plmd). F, left: quantification of blot represented in E, left; right, quantification of the blot represented in E, right (n = 4 for nonischemic, n = 5 for ischemic, **P < 0.01). G: kinase dead Tie2 (lysine to arginine at amino acid 854 or K854R) abolishes ADAM12 (A12)-induced HUVEC proliferation (n = 7–8/group, **P < 0.01, NS = P > 0.05). H: lysates from ischemic C57Bl/6 hindlimb muscles show higher levels of cleaved Tie2 compared with that from Balb/c. I: quantification of band in H (n = 4/group, *P < 0.05).
DISCUSSION
Our study demonstrates the first identification of a gene (ADAM12) that is sufficient to modify the severity of PAD in mice. Differences in the extent of recovery across inbred mouse strains following experimental PAD have been known for >1 decade (13, 17, 35). To date, no single gene has been identified that modulates the severity of PAD. We began this study following our identification of a LSq-1, which contains genetic sequences from C57Bl/6 mice that conferred relatively favorable outcomes (perfusion recovery and extent of tissue loss), following experimental PAD, but relatively poor outcomes when absent (9). By the combination of ancestral haplotype-sharing patterns from several inbred mouse strains with analysis of gene-expression studies, we identified ADAM12 as a lead candidate gene. In this study, we did not assess whether the differences in perfusion by the different strains of mice had an impact on exercise performance, since the goal of the study was to identify genes within the LSq-1 locus involved in perfusion recovery. Additionally, although we did not assess blood pressure in the different mouse strains studied here, prior studies have shown no significant difference in blood pressure among the C57Bl/6, Balb/c, and A/J strains (29, 33), yet our data show that Balb/c and A/J show poor perfusion recovery compared with C57Bl/6 (9). Moreover, the 129s strain has higher blood pressure than the C57Bl/6 strain (33) yet has poorer perfusion recovery and impaired ADAM12 expression. Additionally, whereas the baseline blood flow in the different strains was not assessed, the blood flow measured on day 0, immediately after the HLI surgery, was not different among the strains, suggesting that the degree of ischemia induced in the mice is comparable. Taken together, these findings suggest that differences in blood pressure or ischemia induced by the surgery are not likely to explain the difference in perfusion recovery observed among the strains tested.
C57Bl/6 postischemic hindlimbs upregulated ADAM12 to a higher extent than Balb/c postischemic hindlimbs. Augmentation of ADAM12 expression resulted in “gain of function” with improved perfusion recovery in an inbred mouse strain (Balb/c) that normally displays poor perfusion recovery. Similarly, the impairment of ADAM12 expression in an inbred mouse strain that normally displays favorable perfusion recovery (C57Bl/6) resulted in “loss of function” with poorer perfusion recovery.
We identified ECs as one of the cell types in which ADAM12 is upregulated in ischemia. CD31+ cells from C57Bl/6 mice upregulate ADAM12 protein expression to a higher extent than Balb/c mice. Moreover, augmentation of ADAM12 expression in ECs improved proliferation, whereas knockdown of ADAM12 impaired EC proliferation, survival, tube formation, and AKT activation in simulated ischemia. ADAM12-induced EC proliferation was Tie2 dependent, and C57Bl/6 mice, which upregulate ADAM12 to a higher extent than Balb/c, also showed higher levels of cleaved Tie2 in ischemic muscles than did Balb/c.
ADAM12 is a member of the ADAM family of proteins, and up to 40 members of this gene family have been described to date (http://people.virginia.edu/%7Ejw7g/Table_of_the_ADAMs.html). Mouse ADAM12 was originally identified in 1995 as a protein involved in muscle fusion but has since been implicated in the regulation of cell proliferation and differentiation (25). Some of the functions of ADAM12 are thought to be due to its ability to regulate membrane-bound proteins through ectodomain shedding (2, 20). More recently, ADAM12 has been implicated in processes that involve excessive growth, including cardiac hypertrophy and cancer (12, 25). Nevertheless, our present study is the first to investigate the role of ADAM12 in ischemia-induced angiogenesis. Previous studies by our lab (8, 15) and others (5, 26) have shown impaired perfusion recovery following HLI in mice with a PAD risk factor, such as diabetes or atherosclerosis. Although it is beyond the scope of this particular study, we speculate that risk factors for PAD may, in fact, alter regulation of ADAM12 expression in ischemia and therefore, would be an interesting subject for future studies.
Previously, we found that the LSq-1 locus did not contribute to wound healing, which occurred in a nonischemic setting (9), suggesting that the effect of the locus may be important in the response to ischemia rather than for generalized tissue regeneration. The extent of the native collateral circulation in healthy tissues and the capacity of collaterals to enlarge/remodel have also been implicated as possible mechanisms that may account for the difference in perfusion recovery among mouse strains (3). Moreover, a locus mapping to mouse chromosome 7 but different from the LSq-1 locus was shown to be associated with the extent of pre-existing collaterals (36). We found no ADAM12 upregulation in the collateral vessels proximal to the site of vessel ligation in C57Bl/6 ischemic hindlimbs. Differential ADAM12 expression between C57Bl/6 and Balb/c mice was only in ischemic hindlimbs. Moreover, ADAM12 upregulation in ECs appears to be regulated by HIF-1α. Our findings do not rule out the possibility that other genes within or outside of LSq-1 may contribute to perfusion recovery or limb preservation following experimental PAD. We also cannot rule out the possibility that other cell types other than ECs may upregulate ADAM12 in ischemia and contribute to perfusion recovery.
A comparison of the C57Bl/6 ADAM12 exon sequences with that of Balb/c, using the SNP Query form (http://www.informatics.jax.org/javawi2/servlet/WIFetch?page=snpQF) at The Jackson Laboratory, did not reveal any differences. This suggests that sequences that control the difference in ADAM12 expression in ischemia between the two strains lie in the noncoding regions.
GRANTS
Support for this work was provided by a Robert Wood Johnson Foundation, Harold Amos Medical Faculty Development Program (Award GF12160), and a diversity supplement (3R01 HL101200-01S1; to A. O. Dokun). Support for the work was also provided by the National Heart, Lung, and Blood Institute (R01HL116455 and R01HL121635; to B. H. Annex).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: A.O.D. conception and design of research; A.O.D., L.C., M.O., C.R.F., S.H., and R.J.L. performed experiments; A.O.D., L.C., M.O., C.R.F., W.S.J., and D.C. analyzed data; A.O.D. and S.H.S. interpreted results of experiments; A.O.D. prepared figures; A.O.D. drafted manuscript; A.O.D., W.S.J., D.A.M., R.J.L., S.H.S., and B.H.A. edited and revised manuscript; A.O.D., L.C., W.S.J., D.A.M., R.J.L., and B.H.A. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors acknowledge technical assistance with in vivo microbubble delivery by Dr. Alexander Klibanov and other technical assistance provided by Rebecca Maddux.
REFERENCES
- 1.Akimoto T, Sorg BS, Yan Z. Real-time imaging of peroxisome proliferator-activated receptor-gamma coactivator-1alpha promoter activity in skeletal muscles of living mice. Am J Physiol Cell Physiol 287: C790–C796, 2004. [DOI] [PubMed] [Google Scholar]
- 2.Asakura M, Kitakaze M, Takashima S, Liao Y, Ishikura F, Yoshinaka T, Ohmoto H, Node K, Yoshino K, Ishiguro H, Asanuma H, Sanada S, Matsumura Y, Takeda H, Beppu S, Tada M, Hori M, Higashiyama S. Cardiac hypertrophy is inhibited by antagonism of ADAM12 processing of HB-EGF: metalloproteinase inhibitors as a new therapy. Nat Med 8: 35–40, 2002. [DOI] [PubMed] [Google Scholar]
- 3.Chalothorn D, Faber JE. Strain-dependent variation in collateral circulatory function in mouse hindlimb. Physiol Genomics 42: 469–479, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christiansen JP, French BA, Klibanov AL, Kaul S, Lindner JR. Targeted tissue transfection with ultrasound destruction of plasmid-bearing cationic microbubbles. Ultrasound Med Biol 29: 1759–1767, 2003. [DOI] [PubMed] [Google Scholar]
- 5.Couffinhal T, Silver M, Zheng L, Kearney M, Witzenbichler B, Isner J. Mouse model of angiogenesis. Am J Pathol 152: 1667–1679, 1998. [PMC free article] [PubMed] [Google Scholar]
- 6.Dai Q, Huang J, Klitzman B, Dong C, Goldschmidt-Clermont PJ, March KL, Rokovich J, Johnstone B, Rebar EJ, Spratt SK, Case CC, Kontos CD, Annex BH. Engineered zinc finger-activating vascular endothelial growth factor transcription factor plasmid DNA induces therapeutic angiogenesis in rabbits with hindlimb ischemia. Circulation 110: 2467–2475, 2004. [DOI] [PubMed] [Google Scholar]
- 7.Dokun AO, Annex B, Ginsburg HW. Genetic polymorphisms in peripheral arterial disease role of genomic methodologies. In: Genomic and Personalized Medicine. New York: Elsevier, 2008, p. 1–2. [Google Scholar]
- 8.Dokun AO, Chen L, Lanjewar SS, Lye RJ, Annex BH. Glycaemic control improves perfusion recovery and VEGFR2 protein expression in diabetic mice following experimental PAD. Cardiovasc Res 101: 364–372, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dokun AO, Keum S, Hazarika S, Li Y, Lamonte GM, Wheeler F, Marchuk DA, Annex BH. A quantitative trait locus (LSq-1) on mouse chromosome 7 is linked to the absence of tissue loss after surgical hindlimb ischemia. Circulation 117: 1207–1215, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Findley CM, Cudmore MJ, Ahmed A, Kontos CD. VEGF induces Tie2 shedding via a phosphoinositide 3-kinase/Akt dependent pathway to modulate Tie2 signaling. Arterioscler Thromb Vasc Biol 27: 2619–2626, 2007. [DOI] [PubMed] [Google Scholar]
- 11.Fowkes FG. Peripheral vascular disease: a public health perspective. J Public Health Med 12: 152–159, 1990. [DOI] [PubMed] [Google Scholar]
- 12.Frohlich C, Klitgaard M, Noer JB, Kotzsch A, Nehammer C, Kronqvist P, Berthelsen J, Blobel C, Kveiborg M, Albrechtsen R, Wewer UM. ADAM12 is expressed in the tumour vasculature and mediates ectodomain shedding of several membrane-anchored endothelial proteins. Biochem J 452: 97–109, 2013. [DOI] [PubMed] [Google Scholar]
- 13.Fukino K, Sata M, Seko Y, Hirata Y, Nagai R. Genetic background influences therapeutic effectiveness of VEGF. Biochem Biophys Res Commun 310: 143–147, 2003. [DOI] [PubMed] [Google Scholar]
- 14.Hazarika S, Angelo M, Li Y, Aldrich AJ, Odronic SI, Yan Z, Stamler JS, Annex BH. Myocyte specific overexpression of myoglobin impairs angiogenesis after hind-limb ischemia. Arterioscler Thromb Vasc Biol 28: 2144–2150, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazarika S, Dokun AO, Li Y, Popel AS, Kontos CD, Annex BH. Impaired angiogenesis after hindlimb ischemia in Type 2 diabetes mellitus: differential regulation of vascular endothelial growth factor receptor 1 and soluble vascular endothelial growth factor receptor 1. Circ Res 101: 948–956, 2007. [DOI] [PubMed] [Google Scholar]
- 16.Hazarika S, Farber CR, Dokun AO, Pitsillides AN, Wang T, Lye RJ, Annex BH. MicroRNA-93 controls perfusion recovery after hindlimb ischemia by modulating expression of multiple genes in the cell cycle pathway. Circulation 127: 1818–1828, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helisch A, Wagner S, Khan N, Drinane M, Wolfram S, Heil M, Ziegelhoeffer T, Brandt U, Pearlman JD, Swartz HM, Schaper W. Impact of mouse strain differences in innate hindlimb collateral vasculature. Arterioscler Thromb Vasc Biol 26: 520–526, 2006. [DOI] [PubMed] [Google Scholar]
- 18.Huang LT, Rao P, Peters KG. GRB2 and SH-PTP2: potentially important endothelial signaling molecules downstream of the TEK/TIE2 receptor tyrosine kinase. Oncogene 11: 2097–2103, 1995. [PubMed] [Google Scholar]
- 19.Imoukhuede PI, Dokun AO, Annex BH, Popel AS. Endothelial cell-by-cell profiling reveals the temporal dynamics of VEGFR1 and VEGFR2 membrane localization after murine hindlimb ischemia. Am J Physiol Heart Circ Physiol 304: H1085–H1093, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito N, Nomura S, Iwase A, Ito T, Kikkawa F, Tsujimoto M, Ishiura S, Mizutani S. ADAMs, a disintegrin and metalloproteinases, mediate shedding of oxytocinase. Biochem Biophys Res Commun 314: 1008–1013, 2004. [DOI] [PubMed] [Google Scholar]
- 21.Kannel WB. Update on some epidemiologic features of intermittent claudication: the Framingham Study. J Am Geriatr Soc 33: 13–18, 1985. [DOI] [PubMed] [Google Scholar]
- 22.Keum S, Marchuk DA. A locus mapping to mouse chromosome 7 determines infarct volume in a mouse model of ischemic stroke/clinical perspective. Circ Cardiovasc Genet 2: 591–598, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim C, Seon Lee H, Lee D, Lee SD, Cho EG, Yang SJ, Kim SB, Park D, Kim MG. Epithin/PRSS14 proteolytically regulates angiopoietin receptor Tie2 during transendothelial migration. Blood 117: 1415–1424, 2010. [DOI] [PubMed] [Google Scholar]
- 24.Knowles JW, Assimes TL, Li J, Quertermous T, Cooke JP. Genetic susceptibility to peripheral arterial disease: a dark corner in vascular biology. Arterioscler Thromb Vasc Biol 27: 2068–2078, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kveiborg M, Albrechtsen R, Couchman JR, Wewer UM. Cellular roles of ADAM12 in health and disease. Int J Biochem Cell Biol 40: 1685–1702, 2008. [DOI] [PubMed] [Google Scholar]
- 26.Lefèvre J, Michaud SÉ, Haddad P, Dussault S, Ménard C, Groleau J, Turgeon J, Rivard A. Moderate consumption of red wine (cabernet sauvignon) improves ischemia-induced neovascularization in ApoE-deficient mice: effect on endothelial progenitor cells and nitric oxide. FASEB J 21: 3845–3852, 2007. [DOI] [PubMed] [Google Scholar]
- 27.Lekas M, Lekas P, Mei SHJ, Deng Y, Dumont DJ, Stewart DJ. Tie2-dependent neovascularization of the ischemic hindlimb is mediated by angiopoietin-2. PLoS One 7: e43568, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Hazarika S, Xie D, Pippen AM, Kontos CD, Annex BH. In mice with Type 2 diabetes, a vascular endothelial growth factor (VEGF)-activating transcription factor modulates VEGF signaling and induces therapeutic angiogenesis after hindlimb ischemia. Diabetes 56: 656–665, 2007. [DOI] [PubMed] [Google Scholar]
- 29.Mattson DL. Comparison of arterial blood pressure in different strains of mice. Am J Hypertens 14: 405–408, 2001. [DOI] [PubMed] [Google Scholar]
- 30.McClung JM, McCord TJ, Keum S, Johnson S, Annex BH, Marchuk DA, Kontos CD. Skeletal muscle specific genetic determinants contribute to the differential strain-dependent effects of hindlimb ischemia in mice. Am J Pathol 180: 2156–2169, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDermott MM, Lloyd-Jones DM. The role of biomarkers and genetics in peripheral arterial disease. J Am Coll Cardiol 54: 1228–1237, 2009. [DOI] [PubMed] [Google Scholar]
- 32.Norgren L, Hiatt WR, Dormandy JA, Nehler MR, Harris KA, Fowkes FG. Inter-society consensus for the management of peripheral arterial disease (TASC II). J Vasc Surg 45: S5–S67, 2007. [DOI] [PubMed] [Google Scholar]
- 33.Ryan MJ, Didion SP, Davis DR, Faraci FM, Sigmund CD. Endothelial dysfunction and blood pressure variability in selected inbred mouse strains. Arterioscler Thromb Vasc Biol 22: 42–48, 2002. [DOI] [PubMed] [Google Scholar]
- 34.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scholz D, Ziegelhoeffer T, Helisch A, Wagner S, Friedrich C, Podzuweit T, Schaper W. Contribution of arteriogenesis and angiogenesis to postocclusive hindlimb perfusion in mice. J Mol Cell Cardiol 34: 775–787, 2002. [DOI] [PubMed] [Google Scholar]
- 36.Sealock R, Zhang H, Lucitti JL, Moore SM, Faber JE. Congenic fine-mapping identifies a major causal locus for variation in the native collateral circulation and ischemic injury in brain and lower extremity. Circ Res 114: 660–671, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tlaxca JL, Rychak JJ, Ernst PB, Konkalmatt PR, Shevchenko TI, Pizzaro TT, Rivera-Nieves J, Klibanov AL, Lawrence MB. Ultrasound-based molecular imaging and specific gene delivery to mesenteric vasculature by endothelial adhesion molecule targeted microbubbles in a mouse model of Crohn's disease. J Control Release 165: 216–225, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie A, Belcik T, Qi Y, Morgan TK, Champaneri SA, Taylor S, Davidson BP, Zhao Y, Klibanov AL, Kuliszewski MA, Leong-Poi H, Ammi A, Lindner JR. Ultrasound-mediated vascular gene transfection by cavitation of endothelial-targeted cationic microbubbles. JACC Cardiovasc Imaging 5: 1253–1262, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie D, Li Y, Reed EA, Odronic SI, Kontos CD, Annex BH. An engineered vascular endothelial growth factor-activating transcription factor induces therapeutic angiogenesis in ApoE knockout mice with hindlimb ischemia. J Vasc Surg 44: 166–175, 2006. [DOI] [PubMed] [Google Scholar]