Abstract
Succinate dehydrogenase (SDH) is a hetero-tetrameric nuclear-encoded complex responsible for the oxidation of succinate to fumarate in the tricarboxylic acid (TCA) cycle. Loss-of-function mutations in any of the SDH genes are associated with cancer formation. However, the impact of SDH loss on cell metabolism and the mechanisms enabling growth of SDH-defective cells are largely unknown. Here, we generated Sdhb-ablated kidney mouse cells and employed comparative metabolomics and stable isotope-labelling approaches to identify nutritional requirements and metabolic adaptations to SDH loss. We found that lack of SDH activity commits cells to consume extracellular pyruvate, which sustains Warburg-like bioenergetic features. We further demonstrated that pyruvate carboxylation diverts glucose-derived carbons into aspartate biosynthesis, thus sustaining cell growth. By identifying pyruvate carboxylase as an essential gene for the proliferation and tumorigenic capacity of SDH-deficient cells, this study revealed a metabolic vulnerability for potential future treatment of SDH-associated malignancies.
INTRODUCTION
TCA cycle is a pivotal source for mitochondrial NADH and the core metabolic route for production of many biosynthetic precursors. Over the last fifteen years, oncogenic mutations in three TCA cycle-related enzymes, SDH, fumarate hydratase (FH) and isocitrate dehydrogenase (IDH) have been identified1. While these discoveries may implicate altered metabolism as an underlying hallmark of neoplastic transformation, they left the question how these cancer cells fulfil bioenergetic and anabolic demands largely unresolved.
SDH is a hetero-tetrameric nuclear-encoded protein complex responsible for oxidation of succinate to fumarate in the TCA cycle and for feeding electrons into the mitochondrial respiratory chain for ATP production (complex II). Inactivating mutations in the human genes for any of the SDH subunits, or the SDH complex assembly factor (SDHAF2), are associated with susceptibility to develop neuroendrocrine neoplasms, gastrointestinal stromal tumours as well as renal cell carcinoma (RCC)2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13. Loss of SDH causes succinate accumulation in cells, which activates hypoxia-inducible factors at normal oxygen tension and inhibits α-ketoglutarate-dependent histone and DNA demethylases, thereby establishing, respectively, a pseudohypoxic and hypermethylator phenotype in tumours14, 15, 16.
Although SDH is the first discovered TCA cycle enzyme with tumour suppressor properties, the metabolic consequences of SDH disabling and the molecular mechanisms enabling survival and growth of SDH-defective cells remain largely unexplored. Knowledge of metabolic profile of SDH-mutated tumours is limited and largely inferred from few steady-state metabolic changes observed in situ in human studies. However, whereas pan-metabolomic analyses are useful to identify biochemical signatures for tumour stratification, they do not allow for the understanding of biochemical fluxes necessary to disclose targetable cancer metabolic vulnerabilities. Considering this limitation, the RNA interference-mediated silencing of SDH subunits in cancer cell allowed a partial understanding of cellular effects of mitochondrial complex II deficiency14, 17, 18. However, as SDH levels are never completely depleted by RNAi, the residual SDH activity might still play a role in succinate oxidation in mitochondria, thereby masking the effective rewiring of metabolic networks in tumours devoid of functional SDH. To overcome this limitation, we generated Sdhb-ablated immortalized kidney mouse cells and by unsupervised metabolomic screenings and isotope-labelling approaches we identified metabolic pathways essential to support their proliferation. We found that SDHB loss is sufficient to ensure a complete block of the TCA cycle and to drive Warburg-like bioenergetic features of aerobic glycolysis in proliferating cells. We demonstrated that ablation of SDH activity commits cells to consume extracellular pyruvate needed to sustain maximal glycolytic flux and support the diversion of glucose-derived carbons into aspartate biosynthesis via pyruvate carboxylase (PCX for mouse and PC for human). By identifying Pcx as an essential gene for SDH-deficient but dispensable for normal cells, this study unveils a metabolic vulnerability for potential treatment of SDH-associated neoplasms.
RESULTS
Sdhb deletion induces complete truncation of the TCA cycle and commits cells to fulfill energetic needs through glycolysis
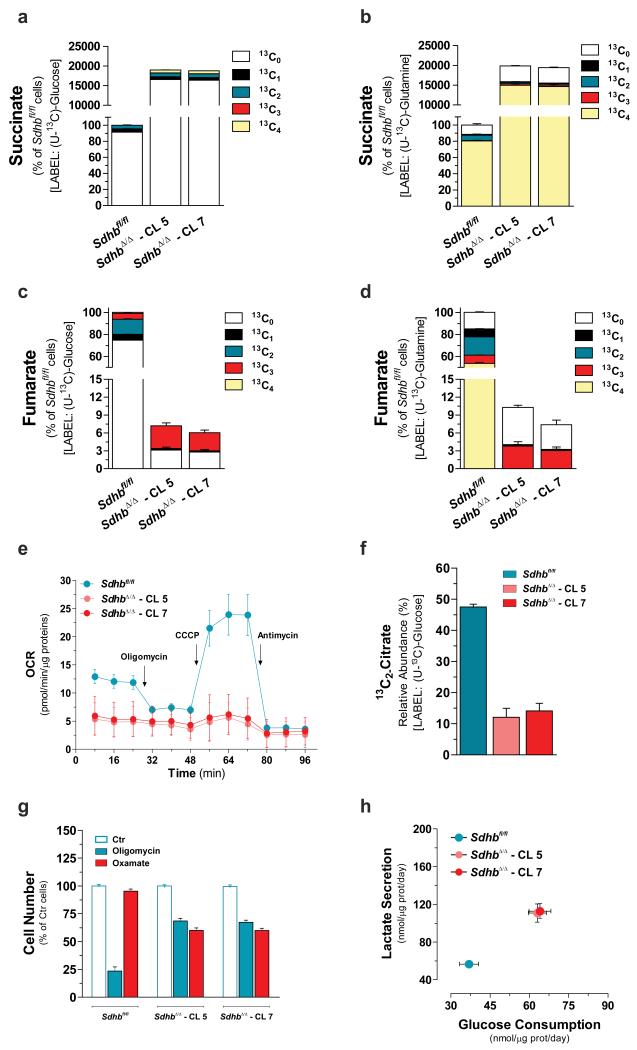
To predict and validate metabolic alterations induced by FH loss, we previously used genetically modified kidney mouse cells in which Fh1 has been deleted19, 20, 21. Similarly, to disclose metabolic rewiring induced by SDH loss, we first produced genetically modified mice containing LoxP sites flanking exon 3 of the endogenous Sdhb gene (Supplementary Fig. 1a) and then immortalized primary kidney epithelial cells isolated from these mice (Sdhbfl/fl). To generate Sdhb knockout cells (SdhbΔ/Δ), Sdhbfl/fl cells were infected with recombinant adenovirus expressing Cre recombinase. Two clones (SdhbΔ/Δ - CL 5 and SdhbΔ/Δ - CL 7) were selected from the infected pool and genetically confirmed to contain homozygous Sdhb-deleted alleles (Supplementary Fig. 1b, c). Suppression of exon 3 leads to a premature stop codon and, as a result, SdhbΔ/Δ cells presented with a complete loss of SDHB protein production and complete impairment of the overall SDH complex activity (Supplementary Fig. 1d, e). Carbon supply to the TCA cycle is achieved mainly through the catabolism of glucose and glutamine. Therefore, to reveal the effects of SDHB loss on TCA cycle function, cells were cultured in medium containing uniformly labelled U-13C-glucose or U-13C-glutamine, and the 13C-labelling of succinate and fumarate was analysed by liquid chromatography-mass spectrometry (LC-MS). SDHB loss gave rise to a build-up of intracellular succinate, which reached levels approximately 200-fold higher than that of Sdhbfl/fl cells, and a concomitant decrease of fumarate (Fig. 1a-d). When U-13C-glucose was used, less than 15% of cellular succinate was labelled (Fig. 1a). However, over 80% of the succinate was fully labelled (13C4) when cells were cultured with U-13C-glutamine (Fig. 1b), indicating that glutamine is a major source of carbons for the TCA cycle in both Sdhbfl/fl and SdhbΔ/Δ cells. Importantly, the fumarate pool of the Sdhbfl/fl cells fed with either 13C6-labelled glucose or 13C5-labelled glutamine contained considerable fractions of isotopologues with 2 and 4 13C atoms respectively, due to the processing of succinate in and beyond the SDH step (Fig. 1c, d). The absence of these isotopologues in SdhbΔ/Δ cells demonstrates that loss of SDHB is sufficient for blocking the TCA cycle (Fig. 1c, d). FADH2, generated during SDH catalysis and NADH, produced mainly in the mitochondria by other dehydrogenases, feed the respiratory chain for oxygen consumption and ATP production. Therefore, the effects of complex II deficiency and TCA cycle truncation on the oxygen consumption rate (OCR) of SDH-null cells were investigated. Sdhb-deficient clones display lower basal mitochondrial respiration with respect to SDH-proficient cells (Fig. 1e). This feature was strongly associated with a reduced entry of glucose carbons into the TCA cycle via pyruvate dehydrogenase as indicated by the diminished pool of citrate containing two 13C atoms in SDHB-null cells fed with U-13C-glucose with respect to normal counterparts (Fig. 1f). In line with this finding, lower labelling of lipogenic acetyl-CoA (AcCoA) from glucose was observed in SDH-null cells compared to their normal counterparts (Supplementary Fig. 1f). On the contrary, glutamine represents the main source of labelled lipogenic AcCoA when SDHB is lost (Supplementary Fig. 1f). In-depth analysis of the respiratory profile indicated that whereas under basal conditions Sdhbfl/fl cells consume molecular oxygen at a sub-maximal capacity, both the maximal OCR and the reserve capacity are reduced upon SDH loss, indicating that SdhbΔ/Δ cells respire at a rate close to their bioenergetic limit (Fig. 1e and Supplementary Fig. 2a). Importantly, the near complete loss of oxygen consumption is not due to a reduction in the number of mitochondria. Indeed, as observed in SDHB-associated renal cell tumours22, SDH-null cells display an increase in mitochondrial mass (Supplementary Fig. 2b). Importantly, the OCR decline was also associated with the specific decrease of mitochondrial complex I proteins, as previously reported for SDH-defective neuroendocrine tumours23, and with the resulting drop of its catalytic activity (Supplementary Fig. 2c, d). It is worth-noting that, approximately 60% of the mitochondrial respiration is coupled to oxidative phosphorylation in Sdhbfl/fl cells while SdhbΔ/Δ cells show a reduced coupling efficiency (Supplementary Fig. 2a). This indicates that oxidative phosphorylation contributes only marginally to ATP synthesis when SDH activity is lost. In line with these respiratory adjustments, the ATP-synthase inhibitor oligomycin induced energetic stress in Sdhbfl/fl cells only, preferentially limiting their proliferation (Fig. 1g and Supplementary Fig. 2e, f). The respiratory features of SDH-null cells imply that their ATP requirements must be fulfilled mainly by glycolysis. In line with this deduction, the reduced OCR of SdhbΔ/Δ cells is associated with an increase in the glycolytic flux, as indicated by the augmented extracellular acidification rate (Supplementary Fig. 2g) and confirmed by the increase in glucose consumption and lactate production rates (Fig. 1h). Importantly, whereas glycolysis inhibition elicited by the lactate dehydrogenase (LDH) inhibitor oxamate did not affect the energetic status of Sdhbfl/fl cells, it raised the AMP/ATP ratio in SDH-null cells and selectively impaired their proliferative potential (Fig. 1g and Supplementary Fig. 2h-j, e, m).
Figure 1. Loss of Sdhb is sufficient to truncate TCA cycle and disable mitochondrial respiration.
Isotopologues distribution of intracellular succinate and fumarate after incubation for 24h with either U-13C-glucose (a, c) or U-13C-glutamine (b, d). All data are presented as mean±s.e.m of one representative experiment (n=3 wells) independently replicated twice (c, d) and three times (a, b). (e) Respiratory profile of the specified cells. Arrows indicate incubation of cells with the indicated compounds. Data are presented as mean±s.e.m of n=27 (Sdhbfl/fl), n=31 (SdhbΔ/Δ – CL 5) and n=29 (SdhbΔ/Δ – CL 7) wells pooled from four independent experiments. (f) Fraction of intracellular citrate pool containing two 13C atoms as result of pyruvate dehydrogenase activity. Data are presented as mean±s.e.m of one representative experiment (n=3 wells) independently replicated twice. (g) Cell number of the indicated cells measured after 96h of treatment with 0.5 μM oligomycin or 5 mM oxamate. Data are presented as mean±s.e.m of n=12 wells pooled from four independent experiments. (h) Rates of the indicated metabolites upon 48h of incubation of Sdhbfl/fl and SdhbΔ/Δ cells with U-13C-glucose. The sum of all isotopologues is reported for clarity. Data are presented as mean±s.e.m of n=18 wells pooled from three independent experiments. In this and all other figures, “wells” represent technical replicate samples set up and assessed under identical conditions and in parallel within a single experiment. Raw data of independently repeated experiments are provided in Supplementary Table 3.
Extracellular pyruvate is required to maintain maximal glycolytic flux and to enable proliferation of Sdhb-null cells
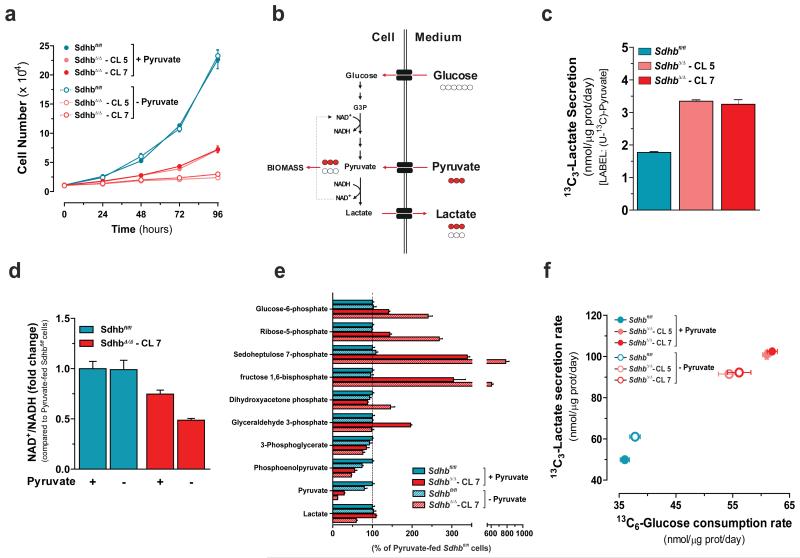
In order to study the changes in cell metabolism upon SDHB loss, the exchange rate of metabolites between cells and the extracellular medium was profiled by LC-MS. In accordance with being the major carbon source for the TCA cycle, glutamine was the second most consumed metabolite by all cell types, but with different rates (Supplementary Fig. 3a). Interestingly, despite the presence of pyruvate in the medium, a net secretion of this organic acid by Sdhbfl/fl cells was observed. On the contrary, pyruvate ranks as the third most-consumed metabolite in the SDH-null clones (Supplementary Fig. 3a). In agreement with this metabolic feature, pyruvate deprivation selectively impaired the proliferation of SdhbΔ/Δ cells, sparing their normal counterparts (Fig. 2a). Whereas SdhbΔ/Δ cells consumed pyruvate, this exogenous metabolite contributed only marginally to its intracellular pool, as found by incubating cells in U-13C-pyruvate (Supplementary Fig. 3b). Moreover, as it is mainly synthesized de novo through glycolysis, the large majority of intracellular pyruvate was fully labelled (13C3) in cells cultured with U-13C-glucose (Supplementary Fig. 3c). Surprisingly, when pyruvate was deprived from the medium, intracellular steady-state levels of this metabolite were dramatically decreased in SdhbΔ/Δ clones, suggesting that the demand for pyruvate exceeded the cells’ biosynthetic capacity (Supplementary Fig. 3c). By converting pyruvate to lactate, LDH restores the NAD+ pool which is rate-limiting for glycolytic flux. Similarly, an extra pool of NAD+ derived from the synthesis of lactate from extracellular pyruvate, may allow SDH-null cells to maximize their glycolytic capacity and divert some glucose-derived carbons for biomass generation (Fig. 2b). In agreement with this hypothesis, the conversion rate of exogenous pyruvate into lactate (13C3-Lactate) was almost 2.5 fold higher in SdhbΔ/Δ than in Sdhbfl/fl cells (Fig. 2c) and the NAD+/NADH ratio was decreased in pyruvate-deprived SdhbΔ/Δ cells, as well as in LDH-inhibited cells (Fig. 2d and Supplementary Fig. 2k). As a result of the NAD+/NADH unbalance, a marked accumulation of pentose phosphate pathway and glycolytic intermediates upstream of the NAD+-dependent oxidation of glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate, was observed in pyruvate-deprived SDH-deficient cells (Fig. 2e). In line with these observations, a consequent decrease in glucose-derived lactate (13C3-Lactate) secretion and glucose (13C6-glucose) consumption rates were measured upon pyruvate depletion (Fig. 2f), demonstrating that SDHB loss constrains cells to the uptake of extracellular pyruvate to sustain maximal glycolytic flux.
Figure 2. SDH deficiency commits cells to consume extracellular pyruvate to maintain maximal glycolytic flux and cell growth.
(a) Number of cells cultured in presence (+) or absence (−) of pyruvate measured at the indicated time points. Data are presented as mean±s.e.m of one representative experiment (n=4 wells), independently replicated three times. (b) Schematic representation of effects of pyruvate consumption on maintenance of NAD+/NADH redox state and glycolytic flux. Red and white circles indicate 13C and 12C carbon unit, respectively. (c) Secretion rate of exogenous pyruvate-derived lactate (13C3-lactate) of Sdhbfl/fl and SdhbΔ/Δ cells cultured for 48h with U-13C-Pyruvate. Data are presented as mean±s.e.m of n=6 wells pooled from two independent experiments. (d) Measurement of NAD+/NADH ratio expressed as fold change compared to pyruvate-fed Sdhbfl/fl cells (e) Pentose phosphate pathway and glycolytic intermediates in cells cultured for 24h in presence (+) or absence (−) of pyruvate. Data in d, e are presented as mean±s.e.m of one representative experiment (n=3 wells), independently replicated twice. (f) Exchange rates of the indicated labeled metabolites upon 48h of incubation of Sdhbfl/fl and SdhbΔ/Δ cells with U-13C-glucose in presence (+) or absence (−) of pyruvate. Data are presented as mean±s.e.m of n=18 wells pooled from three independent experiments. Raw data of independently repeated experiments are provided in Supplementary Table 3.
Glucose-dependent aspartate biosynthesis dictates pyruvate dependency of Sdhb-deficient cells
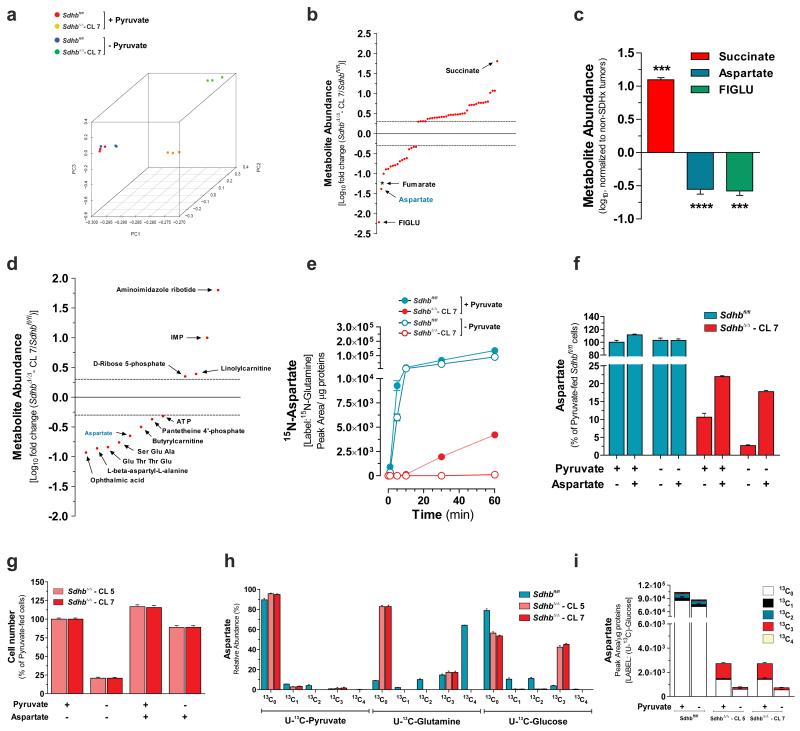
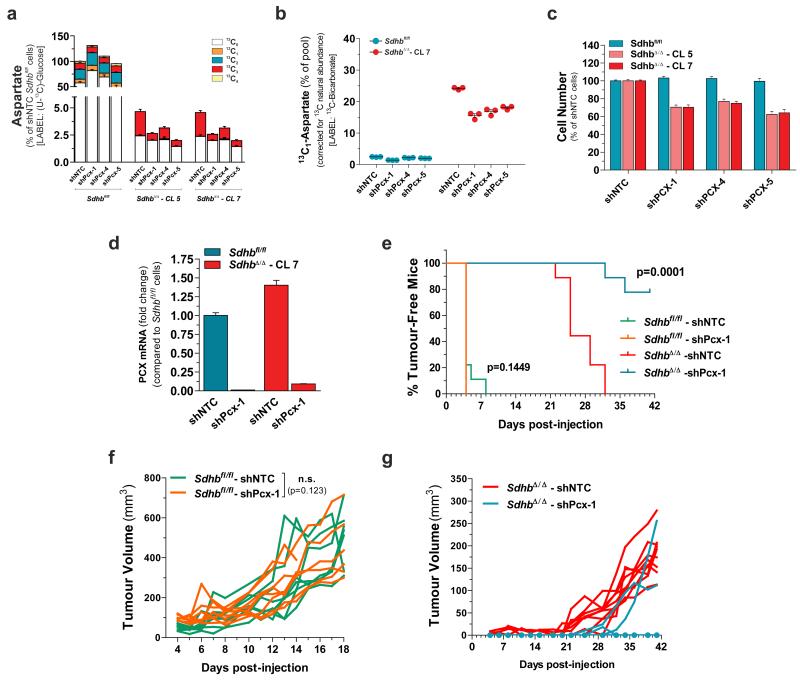
To obtain further insights on metabolic changes due to SDH loss, an untargeted endo-metabolomic profiling coupled with multivariate statistics was performed (Supplementary Table 1). The principal component analysis (PCA) showed a clear separation between SDH-null and SDH-containing cells. Moreover, the occurrence of sub-clustering of pyruvate-fed and pyruvate-deprived SdhbΔ/Δ cells, but not Sdhbfl/fl cells, indicates that the removal of pyruvate from the medium generates a metabolic signature only when SDHB is deleted (Fig. 3a). Metabolite profiling (in the presence of pyruvate) revealed that, as expected, succinate is the most increased metabolite in the SDH-deficient cells as compared to control cells. Interestingly however, formiminoglutamic acid (FIGLU), a catabolic product of histidine, and aspartate, a non-essential amino acid synthesized de novo from oxaloacetate, ranked as the first and second decreased metabolites in SDH-null cells (Fig. 3b). Furthermore, in agreement with this in vitro observation, these metabolic changes were also detected in human SDH-mutated paragangliomas and pheochromocytomas with respect to SDH-unrelated tumours (Fig. 3c and Supplementary Table 2 and 3), thus validating our in vitro observations. Furthermore, the steady state levels of aspartate, but not FIGLU, were further dampened by about 4-fold upon pyruvate withdrawal in SdhbΔ/Δ cells, whereas it remained unchanged in SDHB-proficient cells (Fig. 3d and Supplementary Table 1). Supporting these observations, de novo biosynthesis of aspartate (15N-aspartate) was decreased in SdhbΔ/Δ cells cultured with α-15N-glutamine and was further diminished upon pyruvate deprivation (Fig. 3e), suggesting that reduced aspartate availability for biomass generation may underlie the pyruvate-dependency of SDH-deficient cells. In line with this hypothesis, supra-physiological aspartate supplementation elevated its intracellular steady-state levels (Fig. 3f) and almost completely relieved the pyruvate-addiction of SdhbΔ/Δ cells (Fig. 3g). To further explore the influence of exogenous pyruvate availability on aspartate biosynthesis, we analysed the pattern of fractional 13C-enrichment of this amino acid after culturing cells in the presence of U-13C-pyruvate, U-13C-glutamine or U-13C-glucose (Fig. 3h). The 13C-enrichment profiles revealed that less than 10% of the total aspartate pool contained isotopologues derived from extracellular pyruvate, regardless of cell genotype. Rather, approximately 80% of aspartate was derived from glutamine in Sdhbfl/fl cells whereas, due to TCA cycle truncation, glutamine carbons contributed to less than 20% of aspartate production via reductive carboxylation (13C3) in SdhbΔ/Δ cells. Surprisingly, whereas glucose supplied only up to 20% of the aspartate pool in Sdhbfl/fl cells, approximately 45% of aspartate carbons were derived from glucose in SDH-null cells. Therefore, as pyruvate accelerates the glycolytic flux in SdhbΔ/Δ cells, we postulated that it would also sustain the diversion of glucose-derived carbons into the aspartate biosynthetic pathway. Accordingly, the relative contribution of glucose carbons to the aspartate pool dropped from 45% to 15% in SdhbΔ/Δ cells upon pyruvate withdrawal, whereas it remained unaffected in SDH-proficient counterparts (Fig. 3i).
Figure 3. Glucose-dependent aspartate biosynthesis dictates pyruvate dependency of Sdhb-deficient cells.
(a) PCA of the untargeted endo-metabolomic screening of Sdhbfl/fl and SdhbΔ/Δ cells cultured for 24h in presence (+) or absence (−) of pyruvate. (b) Untargeted metabolic profiling of Sdhb-null cells. Each dot represents a metabolite with a fold change >2 or <0.5 and a FDR-corrected p-value <0.0001. Only metabolites that meet the above-mentioned criteria in two out of three independent experiments (n=3 wells per experiment) are presented. * Since in this untargeted technique fumarate could not be measured, the fold change of fumarate are derived from results presented in Fig.1 c and d. FIGLU, Formiminoglutamic acid. (c) Targeted metabolites profiling of n=4 human SDHx-mutated paraganglioma/pheochromocytoma specimens. Data are expressed as log10 fold change with respect to n=18 non-SDHx-mutated tumours. ***P < 0.001, ****P < 0.0001, two-tailed Student’s t-test. (d) Untargeted metabolic profiling of pyruvate-deprived SdhbΔ/Δ cells. Each point represents a metabolite with a fold change > 2 or < 0.5 and a FDR-corrected p-value <0.0001. Only known endo-metabolites that meet the above-mentioned criteria in two out of three independent experiments (n=3 wells each) are presented. (e) Accumulation of 15N-aspartate in cells incubated with α-15N-glutamine for the indicated time in presence/absence of pyruvate. Data are presented as mean±s.e.m (n=3 wells) of one experiment, performed once. (f) Aspartate levels in cells cultured for 24h in presence or absence of pyruvate and/or 2.5 mM aspartate. Data are presented as mean±s.e.m (n=3 wells) of one representative experiment, independently replicated twice. (g) Cell number measured after 96h of culture in presence/absence of pyruvate and/or 2.5 mM aspartate. Data are presented as mean±s.e.m of n=16 wells pooled from four independent experiments. (h) Isotopologues labelling profile (mean±s.e.m.) of aspartate in cells cultured for 24h in presence of U-13C-pyruvate, U-13C-glutamine (n=6 wells for both conditions, pooled from two independent experiments) or U-13C-glucose (n=9 wells pooled from three independent experiments). (i) Aspartate levels in cells cultured for 24h with U-13C-glucose in presence/absence of pyruvate. Data are presented as mean±s.e.m (n=3 wells) of one representative experiment, independently replicated twice. Raw data of independently repeated experiments are provided in Supplementary Table 3.
Pyruvate carboxylase expression is induced in SDH-deficient cells and SDHx-mutated human tumours
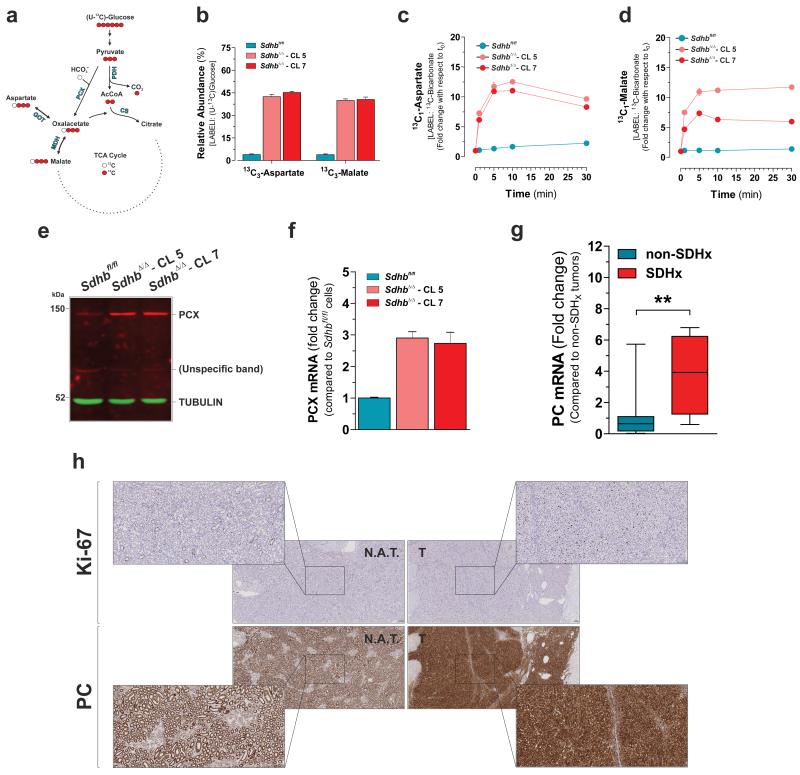
In Sdhbfl/fl cells glucose mainly provides two carbons (13C2-aspartate) to the aspartate skeleton by pyruvate oxidation in the TCA cycle via pyruvate dehydrogenase. On the contrary, the U-13C-glucose-derived aspartate pool of SdhbΔ/Δ cells contained exclusively isotopologues with three 13C atoms. This indicated that SDH loss primed cells to engage pyruvate carboxylase to generate oxaloacetate (Fig. 3i and 4a). This was further supported by the observation that, in SdhbΔ/Δ cells cultured with U-13C-glucose, the fractional 13C-enrichment of malate and fumarate recapitulated that of aspartate (Fig. 4a, b and Fig. 1c). Potential increased PC activity (based on the labelling pattern of glucose-derived TCA cycle metabolites) was recently observed also in patient-derived FH-deficient RCC24, 25. Furthermore it was previously suggested that PC may become essential for SDH- and FH-deficient cells26. PCX fixates bicarbonate into pyruvate to generate oxaloacetate. Therefore, as an additional test of PCX activity, cells were cultured in the presence of 13C-bicarbonate and the kinetics of synthesis of 13C1-aspartate and 13C1-malate were monitored. In agreement with our related findings, both 13C-labelled metabolites were enriched faster in SDH-null cells compared to normal counterparts (Fig. 4c, d). Furthermore, PCX protein level was up-regulated in SdhbΔ/Δ cells (Fig. 4e) in association with an enhancement in its gene expression (Fig. 4f), substantiating the increase in enzyme activity. To extend the impact of these findings and to gain insights into whether pyruvate carboxylase is related to SDH-deficient cancer pathophysiology, we analysed PC expression in SDH-defective human tumours. PC was found induced in SDHx-mutated paragangliomas and pheochromocytomas with respect to non-SDHx-mutated tumours (Fig. 4g). In addition, stronger granular cytoplasmic immunostaining of PC was revealed in tumour epithelial cells of human SDHB-mutated RCC compared to their normal adjacent renal tissues (Fig. 4h and Supplementary Fig. 4a).
Figure 4. Pyruvate carboxylase expression is induced in SDH-defective cells and SDHx-mutated human tumours.
(a) Predicted labeling pattern of the indicated metabolites in cells cultured with U-13C-glucose. Red and white circles indicate 13C and 12C carbon unit, respectively. PDH, Pyruvate Dehydrogenase; PCX, Pyruvate Carboxylase; CS, Citrate Synthase; MDH, Malate Dehydrogenase; GOT, Glutamate-Oxalacetate Transaminase. (b) Abundance of 13C3-Aspartate and 13C3-malate in U-13C-glucose-cultured cells. Data in b are presented as mean±s.e.m of n=9 wells pooled from three independent experiments. Accumulation of 13C1-aspartate (c) and 13C1-Malate (d) in cells incubated with 13C-bicarbonate for the indicated time. Data in c, d are presented as mean±s.e.m of n=3 wells of one representative experiment, independently replicated twice. (e) Western blot analysis of PCX protein levels in the indicated cells. Image representative of two independent experiments. (f) qPCR analysis of PCX mRNA levels in the indicated cells. Data are presented as mean±s.e.m of n=9 wells pooled from three independent experiments. (g) qPCR analysis of PC mRNA levels in the n=4 human SDHx-mutated paraganglioma/pheochromocytoma specimens. Data are expressed as fold change with respect to n=18 non-SDHx mutated tumours. **P < 0.01. two-tailed Student’s t-test. (h) Immuhistochemical assessment of PC levels in a human SDHB-mutated RCC (Tumour ID 21542, details are reported in Supplementary Table 2). The cell proliferation marker Ki-67 identifies iperproliferative neoplastic areas of the section. N.A.T., normal adjacent tissue; T., tumour. Scale bars = 100 μM. Raw data of independently repeated experiments are provided in Supplementary Table 3.
Pyruvate carboxylase sustains proliferation and tumorigenic capacity of transformed Sdhb-null cells
Our findings prompted us to investigate the essentiality of PCX for growth of SDH-deficient cells. To this aim, Pcx expression was stably silenced by three independent shRNAs via lentiviral infection (Supplementary Fig. 4b). This reduced the incorporation of 13C carbons derived from U-13C-glucose (Fig. 5a and Supplementary Fig. 4c) or from 13C-bicarbonate (Fig. 5b and Supplementary Fig. 4d) into both malate and aspartate, without eliciting appreciable changes in the contribution of glutamine carbons to the pool of this amino acid (Supplementary Fig. 4e). Furthermore, silencing of Pcx significantly reduced the proliferation of SDH-null cells in vitro, sparing the normal counterparts (Fig. 5c). We then evaluated the role of PCX in vivo by xenografting H-RASV12-transformed Sdhbfl/fl and SdhbΔ/Δ cells in immuno-deficient mice after silencing PCX expression by two different shRNAs (Fig. 5d and Supplementary Fig. 4f). PCX down-regulation in SDH-proficient cells did not result in a significant alteration in tumour initiation and growth (Fig. 5e-g and Supplementary Fig. 4g-i). On the contrary, PCX was required to support tumorigenesis in SDHB-deficient cells. Indeed, mice xenografted with SdhbΔ/Δ-shPcx cells gave rise to tumours with a significantly delayed onset in both experiments. It’s worth noting that whereas tumour formation was observed only in two out of nine mice xenografted with SdhbΔ/Δ-shPcx-1 cells, tumours emerged in all mice injected with SdhbΔ/Δ-shPcx-5 cells, but exhibited a significantly reduced growth with respect to their PCX expressing counterparts (Fig. 5e-g and Supplementary Fig. 4g-i).
Figure 5. Pyruvate carboxylation is a vulnerable adaptation to Sdhb loss.
(a) Aspartate abundance in PCX-silenced cells cultured for 24h with U-13C-glucose. Data are presented as mean±s.e.m of n=9 wells pooled from three independent experiments. (b) Relative abundance of 13C1-aspartate in PCX-silenced cells incubated with 13C-bicarbonate for 10 min. Data are presented as mean±s.e.m of n=3 wells of one representative experiment, independently replicated twice. (c) Data are presented as mean±s.e.m of n=12 wells pooled from three independent experiments.. (d) PCX expression in H-RasV12-transformed cells infected with lentiviruses expressing either a shNTC or shPcx-1 sequence prior to injection into nude mice. Data are presented as mean±s.e.m (n=3 wells) of one experiment, performed once. (e-g) In vivo growth of cells described in (d) xenografted in athymic nude mice. The % of tumour-free mice over time (e) and the tumour volumes of each xenografted mouse (f, g) are presented (n=8 Sdhbfl/fl-shNTC, n=8 Sdhbfl/fl-shPcx1, n=9 SdhbΔ/Δ-shNTC, n=9 SdhbΔ/Δ-shPcx1). The Log-rank (Mantel-Cox) test was used to calculate the statistical significance between curves in (e). A statistical permutation test was used to compare the statistical significance between curves of the selected genotypes in (f), as described in Methods. Data derive from one experiment, performed once. Raw data of independently repeated experiments are provided in Supplementary Table 3.
Pyruvate carboxylase enables growth of Sdhb-null cells by supporting aspartate biosynthesis
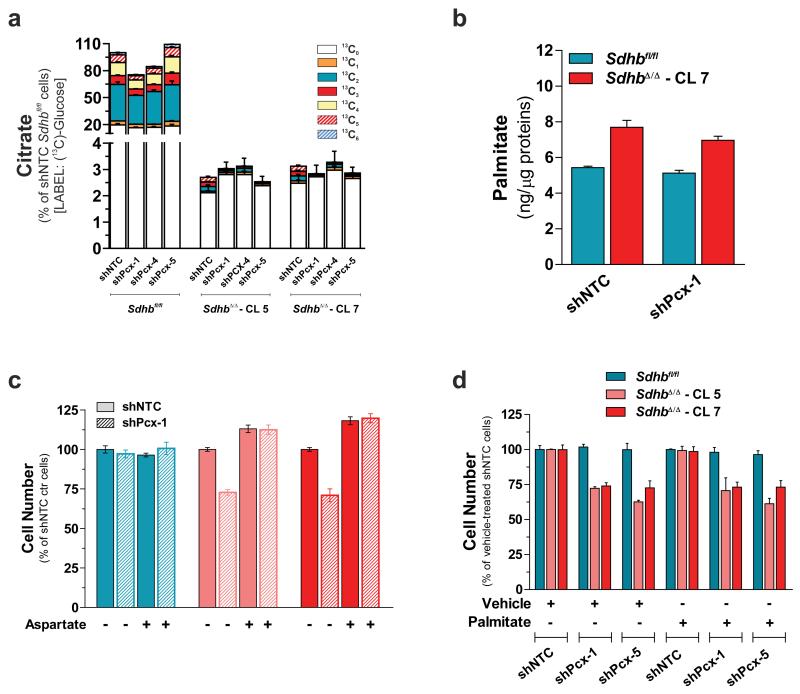
PC is required for the growth of cancer cells under in vitro glutamine-deprived conditions and allows cells to use glucose-derived pyruvate, rather than glutamine, for replenishing TCA cycle intermediates which are used in cataplerotic reactions, such as fatty acids (palmitate) biosynthesis27. However, our results suggest that the engagement of PCX provides SDH-deficient cells with a metabolic adaptation that mainly supports aspartate production. Indeed, compared to aspartate, PCX contributes marginally to citrate labelling (13C3-citrate and 13C5-citrate) (Supplementary Fig. 5a). Moreover, citrate and palmitate steady-state levels remained unchanged in PCX-silenced cells (Fig. 6a, b), and whereas their proliferative capacity was completely rescued by aspartate supplementation, providing excess palmitate to PCX-silenced SDH-deficient cells did not support in vitro proliferation (Fig. 6c, d and Supplementary Fig. 5b, c). The rescue of the growth of PCX-silenced SDH-deficient cells by aspartate also indicates that the effect of both shRNAs used to silence PCX is on target.
Figure 6. Pyruvate carboxylase supports growth of Sdhb-null cells by sustaining aspartate biosynthesis.
(a) Citrate abundance in PCX-silenced cells cultured for 24h with U-13C-glucose. Data are presented as mean±s.e.m of n=9 wells pooled from three independent experiments. (b) Effect of PCX silencing on the total intracellular palmitate levels. Data are presented as mean±s.e.m (n=3 wells) of one experiment, performed once. (c) Number of PCX-silenced cells in the presence (+) or absence (−) of 2.5 mM aspartate measured after 96h of culture. Data are presented as mean±s.e.m (n=4 wells) of one experiment, performed once. (d) Number of control and PCX-silenced cells supplemented with 50 μM palmitate measured after 96h of culture. Data are presented as mean±s.e.m (n=4 wells) of one experiment, performed once. Raw data of independently repeated experiments are provided in Supplementary Table 3.
DISCUSSION
Cancer cell metabolism is shaped by changing fluxes along key metabolic pathways and altering nutrient utilization in order to support biomass accumulation required for continuous mitotic divisions. Here, we show that the impairment of mitochondrial bioenergetics due to SDHB deficiency results in dependency on extracellular pyruvate to sustain the glycolytic flux and to direct glucose-derived carbons into aspartate biosynthesis via pyruvate carboxylase. Whereas the anaplerotic oxidative metabolism of glutamine into the TCA cycle allows SDH-proficient cells to replenish TCA cycle intermediates, which are required for the production of cellular building blocks, the TCA cycle truncation forces SDH-null cells to rely on PCX to generate oxaloacetate.
The impact of these findings is strengthened by the histopathological observation that high-proliferative regions of SDHB-mutated RCC are characterized by higher PC expression in comparison to low-proliferative areas in the same tumour and in normal adjacent tubules (Fig. 4h). However, although our results suggest a direct link between PC levels and SDHB loss in renal cancers and neuroendocrine tumours, induction of PC gene expression may be dispensable, as metabolic fluxes through this enzyme can be shaped by allosteric regulation of PC function28 and by post-transcriptional modulation of PC protein abundance27. In fact, the extent of the induction of Pcx RNA in SDH-deficient cells was inconsistent (compare Fig. 4f to Suppl. Fig. 4b), nevertheless, the induction of PCX activity in these cells, as measured by bicarbonate-derived 13C incorporation into aspartate, was equivalently high (compare Fig. 4c to Fig. 5b). It is noteworthy that PC was recently found to be essential for non-small-cell lung cancer proliferation29. Furthermore, FH deficiency24, 25 or the expression of oncogenic IDH1 mutation30, increased anaplerotic contribution of pyruvate carboxylation, although no essentiality of PC in these cells was reported.
Beyond the direct contribution to protein biosynthesis, aspartate is the precursor for asparagine as well as pyrimidine and purine nucleotides, and since aspartate is not abundant in human plasma, its de novo synthesis is required for cell growth and proliferation. Therefore, by relating glycolytic-dependency of SDH-null cells to aspartate biosynthesis via PC, this study uncovers a metabolic vulnerability that may be exploited for treatments of malignancies arising as a direct consequence of SDH loss-of-function and, possibly, of other TCA cycle defects. Furthermore, the evidence that aspartate levels are dramatically depleted in SDHB-deficient cells and tumours raises the possibility that a mild inhibition of aspartate amino-transferases could limit preferentially their proliferative potential as well. Similarly, because PC fixates bicarbonate into pyruvate, inhibition of bicarbonate uptake or synthesis could limit substrate availability for this enzyme, thereby offering an additional therapeutic window for the treatment of SDH-mutated tumours.
METHODS
Generation of conditional Sdhb mice and immortalized Sdhb-null mouse kidney cells
A DNA fragment comprising 240bp homology arms flanking an F3-KAN-F3-LoxP cassette was recombineered into intron 3-4 of the Sdhb gene of a pool of three BAC clones in the EL250 strain of E.coli31. Subsequent expression of Flp in these cells allowed excision of the Kanamycin-resistance gene. The same cells were then subjected to recombineering with a DNA fragment comprising 120bp and 400bp homology arms flanking an FRT-HYG-FRT-LoxP cassette into intron 2-3.The plasmid pFlexDTA, a modified version of pFlexible32, was linearised by NotI-AscI and a retrieval fragment cloned into these sites. This comprised 1140bp of homology sequence bisected by a SwaI site to allow for subsequent linearization. The homology arms of this plasmid are approximately 5kb distal to the Hygro cassette on the BAC DNA. The resultant SwaI-linearised vector was used to retrieve DNA sequence from the modified Sdhb locus by recombineering to generate the targeting vector transfected in mid-log phase G4 mESCs (129S6/SvEvTac × C57BL/6Ncr F1)33 by electroporation. Electroporation was performed under standard conditions (250V; 500μF; infinite resistance; cuvette width: 4mm) in a Biorad GenePulser XCell with capacitance extender. After plating onto DR4 irradiated MEF monolayers34, cells were placed under Hyg selection for 6-7 days. Individual colonies were picked on to 96-well plates for cryopreservation and further analysis. Correct targeting of the vector to the Sdhb locus on both the 5′ and 3′ sides was confirmed using PCR on genomic DNA prepared from Hyg-resistant colonies. PCR genotyping was done using Expand Long Template (Roche) according to the manufacturer’s recommendations. The presence of the isolated LoxP/F3 sequence in intron 3-4 was confirmed separately. Following confirmation of targeting by PCR, clones were recovered and expanded under Hyg-ESC medium prior to re-confirmation by PCR. Following identification of correctly targeted clones, mouse lines were derived by injection of Sdhbwt/fl ES cells into C57BL/6J blastocysts according to standard protocols35. Following breeding of high percentage male chimeras, germline offspring were identified by coat colour and the presence of the modified allele was confirmed by PCR across the isolated LoxP/F3 sequence as described above. Sdhbwt/fl mice were then intercrossed to obtain Sdhbfl/fl mice that were used to isolate primary epithelial kidney cells following the method described previously36. Sdhbfl/fl cells were immortalized by infection with the pBABE-Hygro Retroviral Vector carrying the SV40 T-Ag gene, kindly provided by Dr. Peter D. Adams (Cancer Research UK, The Beatson Institute for Cancer Research). 1× 105 Sdhbfl/fl cells were then either left untreated or infected with 200 p.f.u/cell of a Cre recombinase-encoding adenovirus (Ad5-CMV-Cre-GFP, Vector Development Laboratory). After 3 days of infection, a clonal approach by limiting dilution was used to obtain homogenous Sdhb-deleted (SdhbΔ/Δ) cells. The cells were negative for Mycoplasma infection and were maintained at 37°C in a 5% CO2 atmosphere in air using 25 mM glucose and 1 mM pyruvate-containing DMEM (21969-035, Gibco, Lyfe Technologies) supplemented with 10% FBS and 2 mM glutamine. All cell lines used in this paper are not listed in the database of commonly misidentified cell lines maintained by ICLAC and NCBI Biosample.
DNA and RNA extraction
Genomic DNA (gDNA) and RNA were extracted using, respectively, the QIAamp® DNA Blood Mini Kit (51104, Qiagen) and RNeasy Mini Kit (74104, Qiagen) following manufacturer’s instructions. All nucleic acids were quantified by the Eppendorf Biophotometer using Eppendorf single sealed cuvettes, UVette (952010051, Eppendorf UK Limited). gDNA was assessed to determine the genotypes of the different cell lines using the primers: 3LoxP U1 (5′-CATTGTGGTCCTTGGCACAG-3′), 3LoxP D1 (5′-AAGGGCGTTGTCTTCTCACC-3′) and Hyg U1 (5′-CTTGTATGGAGCAGCAGACG-3′). The PCR of gDNA was performed using GoTaq DNA polymerase (Promega) and 0.5 μM primers following the manufacturer’s indications. The PCR program was: 120 s at 95 °C followed by 33 cycles of 30 s at 95 °C, 60 s at 63 °C and 60 s at 72 °C.
qPCR analyses
For Real-Time (qPCR) analysis, 2 μg of total RNA was retro-transcribed into cDNA using SuperScript® VILO™ reverse transcriptase (11755-050, Invitrogen, Life Technologies). cDNAs were mixed with the Fast SYBR® Green Master Mix (4385612, Applied Biosystems) according to the manufacturer instruction and 4 pmol of both forward and reverse primers for: Pcx (forw. 5′-GGCGCATGAGTTCTCCAACA-3′; rev. 5′-GTAGGCCCGGGTGTAATTCTC-3′) or PC (forw. 5′CCAGAGGCAGGTCTTCTTTG-3′; rev. 5′- GGCCCTTCACGTCCTTTAG-3′). qPCR was performed by the 7500 Fast Real-Time PCR System (Applied Biosystems) and the amplification steps were 95°C for 20 s, followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. The relative quantitation of each mRNA was performed by the comparative ΔΔCt method using β-actin for normalization.
Cell number assessment
Cells were plated at 1×104 cells/well in 24-well plates. At the specified experimental time, each well was washed once with PBS, trypsinized, re-suspended in a Casyton solution and counted with a Casy cell counter.
Measurement of OCR and ECAR rate
2×104 Sdhbfl/fl cells and 3×104 SdhbΔ/Δ cells were plated onto XF96 plates (09814, Seahorse Bioscience) and incubated at 37 °C, 5% CO2 for 24 h. The medium was then replaced with 150 μl of unbuffered assay media (Seahorse Bioscience) supplemented with 25 mM glucose, 2% FBS, 1mM pyruvate (pH 7.4) and cells were then placed at 37 °C in a CO2-free incubator for 45 minutes. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were then recorded using the XF96 Extracellular Flux Analyzer (Seahorse Bioscience). OCR and ECAR were normalized to the protein content in each well calculated at the end of the experiments by the Lowry assay. One μM oligomycin A, 0.75 μM CCCP and 1 μM antimycin A were added to measure ATP-coupled, maximal, and mitochondrial-dependent basal OCR, respectively as previously reported37.
Measurement of SDH activity
The activity of SDH (succinate:coenzyme Q oxidoreductase activity) was measured by a spectrophotometric assay as described previously38.
Measurement of the mitochondrial mass
Mitochondrial mass was assessed cytofluorimetrically by measuring the incorporation of 100 nM the mitochondrial potential-insensitive dye MitoTracker Green FM (Life Technologies) as described previously39.
Mitochondria isolation and measurement of Complex I activity
Mitochondria were isolated from Sdhbfl/fl and SdhbΔ/Δ cells as previously described40 and used to determine levels of several mitochondrial proteins by immunoblotting and to assess the activity of Complex I by the dipstick assay kit (Abcam, ab109720), according to manufacturer instructions.
Immunoblotting
Cells were washed twice with cold PBS and lysed in RIPA buffer supplemented with protein and phosphatase inhibitor cocktails. Protein concentration was determined by the Bicinchoninic Acid Assay (Thermo scientific) using BSA as a standard. Equal amounts of protein were mixed with Laemmli buffer 2x warmed at 95°C for 5 min, and loaded on 4-12% gels for SDS-PAGE. After electrophoretic separation, proteins were blotted onto 0.22 mm nitrocellulose (Millipore), blocked with 10% non-fat milk in TBS-Tween, and incubated at 4°C overnight with the following antibodies: anti-succinate dehydrogenase b (goat polyclonal Santa Cruz Biotech sc-34150, 1:500), anti-β-tubulin (mouse monoclonal Sigma-Aldrich, T5201, 1:10000), anti-pyruvate carboxylase (rabbit monoclonal Abcam, ab126707, 1:1000), anti-NDUFS4 (mouse monoclonal, Abcam, Ab87399, 1:1000), anti-MTND6 (mouse monoclonal, Invitrogen, A31857, 1:1000), anti-COX IV (rabbit polyclonal, Abcam, Ab16056, 1:2000), anti-membrane integrity WB cocktail (mouse monoclonal, Abcam, Ab110414, 1:1000). Membranes were then washed and incubated with secondary anti-mouse (926-32212 or 926-32222, LI-COR), anti-rabbit (926-68073, LI-COR) or anti-goat (926-32214, LI-COR) at 1:10000 dilution. The IR scanning was performed using Licor Odyssey scanner (channel, 700 and 800) and acquired using Image Studio 2.0.
Metabolites Extraction and LC-MS
Sdhbfl/fl and SdhbΔ/Δ cells were plated in 6-well plates in 25 mM glucose and 1 mM pyruvate-containing DMEM (21969-035, Gibco, Lyfe Technologies) supplemented with 10% FBS and 2 mM glutamine at 2×105 and 3×105 cells/well, respectively. After 24h the medium was replaced with DMEM (11966-025, Gibco, Lyfe Technologies) supplemented with 10% FBS, 25 mM uniformly labeled (U-13C) or unlabelled glucose in presence of 1 mM U-13C or unlabelled pyruvate. For pyruvate-deprivation experiments, the above medium devoid of pyruvate was used. For U-13C-glutamine-tracing experiments, complete DMEM (21969-035, Gibco, Lyfe Technologies) supplemented with 10% FBS and 4 mM U-13C-glutamine was used. For α-15N-glutamine-tracing assays, cells were plated in 25 mM glucose and 4 mM glutamine-containing custom made DMEM supplemented with 10% FBS with or without 1 mM pyruvate. After 24h, media were replaced with the same medium containing both 2 mM unlabelled and 2 mM α-15N-glutamine in presence or absence of 1 mM pyruvate. For 13C-bicarbonate-tracing experiments, 24h after cell plating, media were replaced with DMEM (12800-017, Gibco, Lyfe Technologies) supplemented with 10% FBS and 20 mM sodium bicarbonate. After 8 hours, cells were incubated with 13C-labelled or unlabelled 24 mM bicarbonate.
Cells were cultured in the above specified conditions for 24h (if not otherwise indicated), for intracellular metabolites (endo-metabolites) determination, or for 48h, to analyze metabolites present in the medium (exo-metabolites). At the end of the incubations, monolayers were rapidly washed 3 times with ice-cold PBS and extracted with 600 μl of ice-cold extraction solution, composed of methanol, acetonitrile, and water (5:3:2), for endo-metabolites determination. Alternatively, an aliquot of the media was diluted in extraction solution for exo-metabolites analyses. Media derived from wells lacking of cells but incubated in the same conditions were used as a reference to quantify the exchange rate (consumption/secretion) of exo-metabolites. Both media and cell extracts were centrifuged at 16,000g for 30 min at 4°C and the supernatants were analyzed by LC-MS. For exo-metabolites quantification, aliquots of DMEM media were processed similarly and supplemented with known concentrations of either U-13C labelled metabolites or essential and non-essential amino acids to prepare calibration curves. Endo-metabolites were normalized to the protein content in each well calculated, at the end of the experiment, by the Lowry assay. The exchange rates were obtained normalizing the variation in consumption or secretion of each exo-metabolite to the average protein content of each samples since the beginning of the experiment (t = 0h) to its end (t = 48h). For the measurement of fumarate and its isotopologues, a Q-Exactive Orbitrap mass spectrometer (Thermo Scientific, Bremen, Germany) was used together with a Thermo Scientific Dionex Ultimate 3000 HPLC system. The HPLC setup consisted of a ZIC-HILIC column (150mm × 4.6mm, 5μm, SeQuant, Merck KGaA, Darmstadt, Germany), preceded with a guard column SeQuant ZIC-HILIC (20mm × 2.1mm). The aqueous mobile phase solvent is 0.1% formic acid in water and the organic mobile phase is 0.1% formic acid in acetonitrile. The mobile phase gradient runs from 80% organic mobile phase to 50% over the first 12 minutes, is then maintained at 50% organic mobile phase for the next 14 minutes, increased and maintained at 20% organic mobile phase for 8 minutes, and then equilibrated back to 80% organic mobile phase. The flow rate used is 300μl/min and the column temperature is maintained at 30°C. Samples are maintained at 4°C in a chilled autosampler, prior to injection. The total analysis time is 45min and 5μl of sample is injected. The Q-Exactive mass spectrometer was operated in full scan mode, with electrospray (ESI) ionization and polarity switching, over a mass range of 75-1000 m/z at a resolution of 35,000 (at 200m/z). The Q-Exactive was also used in SIM (Selected Ion Monitoring) mode, for improving sensitivity, where only fumarate and its isotopologues were monitored by the mass spectrometer, using negative ion mode and a resolution of 70,000 (at 200m/z). The mass accuracy for both methods was below 5ppm. Data were acquired with Thermo Xcalibur software. The peak areas of fumarate and its isotopologues were determined using Thermo LCquan software, identified by the exact mass of each singly charged ion and by known retention time on the HPLC column. For the unbiased metabolomics analysis and the targeted analysis of all the other metabolites, cell and medium extracts were injected on a ZIC-pHILIC column (SeQuant, 150 × 2.1mm, 5μm). Mobile phase C: 20 mM ammonium carbonate, pH 9.4. Mobile phase D: acetonitrile. The flow rate was 100 μL/min over the gradient as follows: 0 minutes 80% D to 30 minutes 20% D and the column was then re-equilibrated until 38 minutes at 80%D. All metabolites were detected over a mass range of 75-1000 m/z using the Exactive Orbitrap mass spectrometer (Thermo Scientific, Waltham, MA, USA) with electrospray (ESI) ionization and polarity switching.
Determination of lipogenic AcCoA
Isotopic enrichment in palmitate in cells incubated for 72h with U-13C-glucose or U-13C-glutamine were quantified by LC-MS and the fraction of labelling of lipogenic AcCoA was inferred from that of palmitate, using the bionomial model introduced by Kamphorst et al41. The synthesis of palmitate results in the incorporation of 8 acetyl groups from AcCoA into palmitate. Each acetyl unit can be unlabelled (M+0) or fully labelled (M+2). If p is the M+2 fraction then the expected M+2x (x=1,…,8) fractions (P) of palmitate follow the bionomial model (eq. 1).
| (eq.1) |
We determined p by fitting the value resulting in the minimum mean squared deviation between the predicted (eq. 1) and the observed values (fraction at each isotopologues).
Palmitate quantification and GS-MS
For quantification of total intracellular palmitate level, the cells were cultured in 6-well plates. At time of extraction, medium was aspirated, cells rinsed twice with 1 mL room temperature PBS, 0.75 mL of methanol/PBS (1:1, v/v) solution at −20°C added, and the plates kept at −20°C for 10 min. Cells were then scraped into a glass tube (Fisher Scientific), chloroform (0.5 ml) and internal standard mixture (20 μL of 50 μg/mL TAG 14:0/17:1/18:1 and PC 17:0/17:0 (Avanti Polar Lipids)) added, the mixture vortexed for 1 min and then centrifuged at 500 g for 10 min, and the chloroform layer transferred to a new glass vial. The extract was dried under N2, followed by trans-esterification and formation of fatty acid methyl esters (FAMEs) by heating the lipid extract at 100°C with 80 μL of toluene, 600 μL of methanol and 120 μL of methanolic-HCl for 60 min. Thereafter, 400 μL of water was added to the reaction mixture and FAMEs were extracted with 300 μL of hexane and analysed by GC-MS. FAMEs were separated on an Agilent 7890B GC system with a Phenomenex ZB-1701 column (30m × 0.25mm × 0.25 μm), coupled to an Agilent 7000 Triple Quadrupole GC/MS system. GC-MS parameters and temperature program were as previously described42. Peak integration was performed with Agilent Mass Hunter B.06.00 software. Peak height of methyl palmitate (C16:0) was normalized to both methyl heptadecanoate (C17:0) and heptadecenoate (C17:1), and calculated value was used to determining the amount of palmitate (in ng) in sample by employing calibration curve.
Short hairpin (shRNA) interference of mouse Pcx
The lentiviral non-targeted shNTC and shRNA plasmids against Pcx were purchased from Sigma-Aldrich and identified as follows: shPcx-1, TRCN0000112425 (CCGGCCCTTCAGCTATTTGTCCTTTCTCGAGAAAGGACAAATAGCTGAAGGGTTTTTG) shPcx-4, TRCN0000112428 (CCGGGCACTACTTCATCGAGGTCAACTCGAGTTGACCTCGATGAAGTAGTGCTTTTTG) shPcx-5, TRCN0000112429 (CCGGCTTTCGCTCTAAGGTGCTAAACTCGAGTTTAGCACCTTAGAGCGAAAGTTTTTG). Lentiviral infections were carried out as previously described19. Infected cells were incubated with medium containing 4 μg/ml puromycin (selection medium) for additional 48h before being used for further experiments.
Immunohystochemistry (IHC) of human SDHB-associated RCC
Six microns cut paraffin embeded tissue slices of SDHB-associated RCC and non-neoplastic adjacent kidney tissue were immunostained by the Dako Autostainer for PC (Sigma-Aldrich, HPA043922) and Ki-67 (Dako, M7240). Epitope retrieval was performed at 98°C in Tris-EDTA retrieval buffer, pH8. Samples were counterstained with haematoxylin, dehydrated and coverslipped. The diagnostic mutational analysis was performed by the East of Scotland Regional Genetics service by using NHS standardised mutation screening procedures. PCR amplification was carried out on Hamilton Starlet. Bidirectional sequencing of all exons was performed by the ABI Prism 3730 and Mutation Surveyor.
Analyses of human paragangliomas and pheochromocytomas
Pheochromocytoma/Paraganglioma susceptibility genes (SDHA, B, C, D, AF2, RET, VHL, MAX and TMEM127) have been analysed for point mutations and small deletions/insertions in 22 samples available for this study. Bidirectional sequencing of the coding regions and splice sites was performed on a ABI3130xl Genetic Analyzer and analysed by SeqScape Software. MLPA analysis (VHL p016c2; SDHB,C,D,AF2 p226b2; MAX,SDHA p429b1) and semiquantitative multiplex-PCR (TMEM127) have been performed to evaluate for a deletion/duplication of one or more exons of these genes and analysed with Coffalyser.net software. RNA was isolated by TRIzol ® Reagent (Ambion) following manufacturer’s indications and further processed as described above. Tumour metabolites were extracted by incubating 40 mg of tumour tissue in 1 ml of ice-cold extraction solution and further processed as described above.
Tumour xenografts
Sdhbfl/fl and SdhbΔ/Δ - CL 7 cells were transformed by pWZL retroviral vector encoding H-RasV12 oncogene34 kindly provided by Prof. Karen Vousden (Cancer Research UK, Beatson Institute), infected with a non-targeted shRNA (shNTC) or shRNA against Pcx and selected with 4 μg/ml puromycin for 7 days. In a first experiment, 2 × 106 transformed shNTC and shPcx-5 cells were subcutaneously injected in the rear flanks of 10 female 6-week old Balb\c Nu nude mice. In a second experiment, 1 × 106 transformed shNTC and shPcx-1 cells were subcutaneously injected in the rear flanks of 8 (for Sdhbfl/fl cells) and 9 (for SdhbΔ/Δ cells) male 6-week old Balb\c Nu nude mice. No statistical method was used to predetermine sample size. Tumour lengths (L) and widths (W) of sagittal sections were measured 3 times per week by manual caliper and tumour volumes were calculated by the formula tumour volume = 0.5×L×W2. Tumour onset was defined by the presence of a tumour mass with a volume ≥ 40 mm3 recorded in at least two consecutive measurements. The fractions of tumour-free mice were determined by GraphPad Prism and their statistical analyses were performed by using the Log-rank (Mantel-Cox) Test. Where applicable, a statistical permutation test (http://bioinf.wehi.edu.au/software/compareCurves/) was used to compare the statistical significance between curves of the selected genotypes. Ten thousands permutations were used in each analysis. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Ethical approval of human and animal studies
The study of human samples included in this manuscript was approved by the Tayside Tissue Bank research ethics committee (TR000106 and TR000341), Ethics Committee of the COR (Regional Cancer Center) and of the Veneto Institute of Oncology Padova (Italy). Informed consent was obtained from all subjects. Animal work was carried out with ethical approval from University of Glasgow under the Animal (Scientific Procedures) Act 1986 and the EU Directive 2010 (PPL 60/4181). Animals were housed in individual ventilated cages in a barrier facility proactive in environmental enrichment.
Reagents
U-13C-glutamine (CLM-1822), U-13C-Glucose (CLM-1396-5), U-13C-Pyruvate (CLM-2440) and α-15N-glutamine (NLM-1016) were from Cambridge Isotopes Laboratories; 13C-bicarbonate (372382), L-aspartate (A8949) and Sodium Oxamate (O2751) and all remaining reagents were obtained from Sigma-Aldrich.
Statistical Analyses
Two-tailed Student’s t were performed with Graph Pad Prism 5.01 software (GraphPad Software Inc). When unequal variances between experimental groups were computated, Welch’s correction was applied. Raw data of independently repeated experiments are provided in Supplementary Table 3. Statistical analyses were performed using the number of wells as the sample size (n). Wells represent technical replicate samples set up and assessed in parallel within a single experiment using identical conditions. Details on the numbers of wells assessed and the number of times experiments were performed independently are provided in every figure legend.
Bioinformatic processing and statistical analysis of the untargeted metabolomic data
Log transformed metabolic intensities were analysed in R using Limma package implementing moderated t-statistic with Empirical Bayes correction and Benjamini and Hochberg adjusted p-values. Three dominant principal components analysis43, built on metabolites displaying a maximal coefficient of variance <0.1, depicts the effect of genotype and treatment in the data.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge Saverio Tardito and Zachary T. Schug for comments and interpretation of the results, Ayala King for editorial work, Umasuthan Srirangalingam for help in human specimen collection and the Beatson Institute mouse facility staff for housing of mice and xenografts measurements.
GRANT SUPPORT
This work was funded by Cancer Research UK. S.C. is recipient of a FEBS long-term fellowship.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
Eyal Gottlieb is a shareholder and a consultant of MetaboMed Israel, Ltd.
REFERENCES
- 1.Gaude E, Frezza C. Defects in mitochondrial metabolism and cancer. Cancer Metab. 2014;2:10. doi: 10.1186/2049-3002-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baysal BE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 3.Niemann S, Müller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nature Genet. 2000;26:268–270. doi: 10.1038/81551. [DOI] [PubMed] [Google Scholar]
- 4.Astuti D, et al. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am. J. Hum. Genet. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baysal BE, et al. Prevalence of SDHB, SDHC, and SDHD germline mutations in clinic patients with head and neck paragangliomas. J. Med. Genet. 2002;39:178–183. doi: 10.1136/jmg.39.3.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ni Y, et al. Germline mutations and variants in the succinate dehydrogenase genes in Cowden and Cowden-like syndromes. Am. J. Hum. Genet. 2008;83:261–268. doi: 10.1016/j.ajhg.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnichon N, et al. SDHA is a tumour suppressor gene causing paraganglioma. Hum. Mol. Genet. 2010;19:3011–3020. doi: 10.1093/hmg/ddq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frezza C, Pollard PJ, Gottlieb E. Inborn and acquired metabolic defects in cancer. J. Mol. Med. 2011;89:213–220. doi: 10.1007/s00109-011-0728-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill AJ, et al. Renal tumours and hereditary pheochromocytoma-paraganglioma syndrome type 4. N. Engl. J. Med. 2011;364:885–886. doi: 10.1056/NEJMc1012357. [DOI] [PubMed] [Google Scholar]
- 10.Ricketts CJ, et al. Succinate dehydrogenase kidney cancer: an aggressive example of the Warburg effect in cancer. J. Urol. 2012;188:2063–2071. doi: 10.1016/j.juro.2012.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williamson SR, et al. Succinate dehydrogenase-deficient renal cell carcinoma: detailed characterization of 11 tumours defining a unique subtype of renal cell carcinoma. Mod. Pathol. 2015;28:80–94. doi: 10.1038/modpathol.2014.86. [DOI] [PubMed] [Google Scholar]
- 12.Vanharanta S, et al. Early-onset renal cell carcinoma as a novel extraparaganglial component of SDHB-associated heritable paraganglioma. Am. J. Hum. Genet. 2004;74:153–159. doi: 10.1086/381054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ricketts C, Woodward ER, Killick P, Morris MR, Astuti D, Latif F, Maher ER. Germline SDHB mutations and familial renal cell carcinoma. J. Natl. Cancer Inst. 2008;100:1260–1262. doi: 10.1093/jnci/djn254. [DOI] [PubMed] [Google Scholar]
- 14.Selak MA, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 15.Xiao M, et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumour suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letouzé E, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;10:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 17.Guzy RD, Sharma B, Bell E, Chandel NS, Schumacker PT. Loss of the SdhB, but not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxiainducible factor activation and tumourigenesis. Mol. Cell. Biol. 2008;28:718–731. doi: 10.1128/MCB.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao M, et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumour suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frezza C, et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011;17:225–228. doi: 10.1038/nature10363. [DOI] [PubMed] [Google Scholar]
- 20.Zheng L, et al. Reversed argininosuccinate lyase activity in fumarate hydratase-deficient cancer cells. Cancer Metab. 2013;1:1–12. doi: 10.1186/2049-3002-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng L, et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat. Communn. 2015;6:6001. doi: 10.1038/ncomms7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Housley SL, et al. Renal carcinoma with giant mitochondria associated with germ-line mutation and somatic loss of the succinate dehydrogenase B gene. Histopathology. 2010;56:405–408. doi: 10.1111/j.1365-2559.2010.03482.x. [DOI] [PubMed] [Google Scholar]
- 23.Favier J, et al. Warburg effect is genetically determined in inherited pheochromocytomas. PLoS One. 2009;4:e7094. doi: 10.1371/journal.pone.0007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullen AR, et al. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 2014;7:1679–1690. doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang C, et al. Metabolic reprogramming for producing energy and reducing power in fumarate hydratase null cells from hereditary leiomyomatosis renal cell carcinoma. PLoS One. 2013;8:e72179. doi: 10.1371/journal.pone.0072179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Folger O, Jerby L, Frezza C, Gottlieb E, Ruppin E, Shlomi T. Predicting selective drug targets in cancer through metabolic networks. Mol Syst Biol. 2011;7:501. doi: 10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng T, et al. Pyruvate carboxylase is required for glutamine-independent growth of tumour cells. Proc. Natl. Acad. Sci. USA. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jitrapakdee S, Wallace JC. Structure, function and regulation of pyruvate carboxylase. Biochem. J. 1999;340:1–16. doi: 10.1042/bj3400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sellers K, et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J. Clin. Invest. 2015;125:687–698. doi: 10.1172/JCI72873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izquierdo-Garcia JL, Cai LM, Chaumeil MM, Eriksson P, Robinson AE, Pieper RO, Phillips JJ, Ronen SM. Glioma Cells with the IDH1 Mutation Modulate Metabolic Fractional Flux through Pyruvate Carboxylase. PLoS One. 2014;22:9. doi: 10.1371/journal.pone.0108289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.