Abstract
Termination of protein synthesis occurs when a translating ribosome encounters one of three universally conserved stop codons: UGA, UAA, or UAG. Release factors recognise stop codons in the ribosomal A site to mediate release of the nascent chain and recycling of the ribosome. Bacteria decode stop codons using two separate release factors with differing specificities for the second and third bases1. By contrast, eukaryotes rely on an evolutionarily unrelated omnipotent release factor (eRF1) to recognise all three stop codons2. The molecular basis of eRF1 discrimination for stop codons over sense codons is not known. Here, we present electron cryo-microscopy (cryo-EM) structures at 3.5 – 3.8 Å resolution of mammalian ribosomal complexes containing eRF1 interacting with each of the three stop codons in the A site. Binding of eRF1 flips nucleotide A1825 of 18S rRNA so that it stacks on the second and third stop codon bases. This configuration pulls the fourth position base into the A site, where it is stabilised by stacking against G626 of 18S rRNA. Thus, eRF1 exploits two rRNA nucleotides also used during tRNA selection to drive mRNA compaction. Stop codons are favoured in this compacted mRNA conformation by a hydrogen-bonding network with essential eRF1 residues that constrains the identity of the bases. These results provide a molecular framework for eukaryotic stop codon recognition and have implications for future studies on the mechanisms of canonical and premature translation termination3,4.
Termination of translation in eukaryotes is initiated when a ternary complex of eRF1-eRF3-GTP binds to a stop codon in the ribosomal A site5,6. GTP hydrolysis by eRF3 induces a conformational change that leads to its dissociation, permitting eRF1 to accommodate fully in the A site. This change is thought to bring a universally conserved GGQ motif close to the ester bond between the nascent polypeptide and the tRNA, stimulating its hydrolysis. Concomitant with these events, the ATPase ABCE1 is recruited to the ribosome after eRF3 dissociation and, together with eRF1, catalyses splitting of the ribosomal subunits to recycle post-termination complexes3,4,7,8.
We reasoned that a catalytically inactive eRF1 mutant may trap a pre-hydrolysis termination complex with two key features. First, eRF1 would be in complex with the stop codon it had recognised. Second, the unreleased nascent polypeptide would provide a unique affinity handle to enrich this species for structural analysis. Therefore, we substituted the glycines of the GGQ motif with alanines (eRF1AAQ)9 and added this mutant to in vitro translation reactions in rabbit reticulocyte lysate. Peptide release at all three stop codons was inhibited by eRF1AAQ as judged by persistence of a peptidyl-tRNA (Extended Data Fig. 1a, b). Affinity purification of these ribosome-nascent chain complexes (RNCs) via the nascent chain recovered both eRF1AAQ and ABCE1 (Extended Data Fig. 1c, d), suggesting that eRF1AAQ was trapped on the RNCs in its accommodated state. Association of ABCE1 was enhanced with eRF1AAQ-stalled RNCs relative to RNCs stalled with a truncated mRNA (Extended Data Fig. 1c), consistent with a report that the function of ABCE1 in post-termination recycling requires peptidyl-tRNA hydrolysis7.
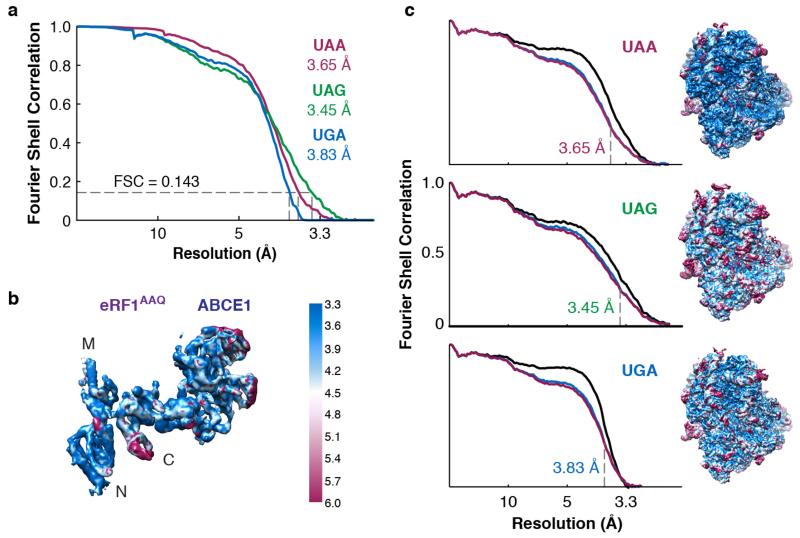
Purified RNCs stalled with eRF1AAQ at each stop codon (Extended Data Fig. 1d) were directly utilised for cryo-EM. Multiple rounds of three-dimensional classification in silico revealed that ~10% of the particles contained eRF1AAQ-ABCE1 (Extended Data Fig. 2). Datasets of between 20,000 and 50,000 particles for the three stop codons yielded maps with overall resolutions of 3.45 Å (UAG), 3.65 Å (UAA) and 3.83 Å (UGA), against which the models were refined (Fig. 1, Extended Data Fig. 3, and Extended Data Table 1).
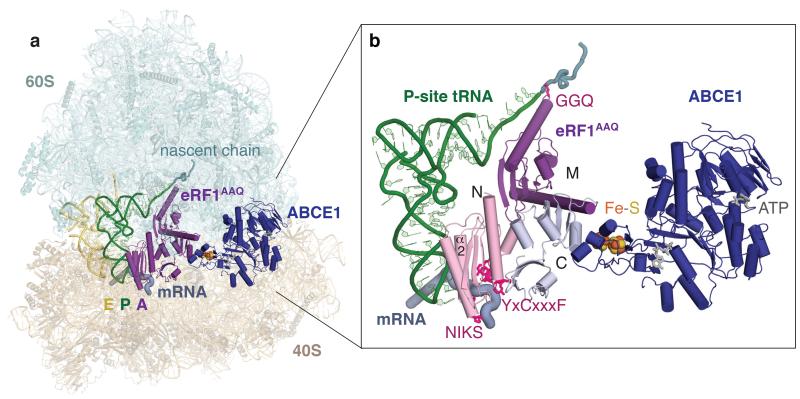
Figure 1. Overall structure of a eukaryotic translation termination complex.
a, Overview of the structure of an eRF1AAQ-stalled mammalian ribosome-nascent chain complex containing the UAG stop codon showing the 40S and 60S ribosomal subunits, E- (yellow) and P-site (green) tRNAs, eRF1AAQ (purple) occupying the A site, and ABCE1 (blue) occupying the GTPase centre. b, Close-up view of eRF1AAQ coloured by domain (N, M, C) with the GGQ, NIKS and YxCxxxF motifs highlighted (pink). Also shown are the P-site tRNA (green), nascent polypeptide (teal), the mRNA containing the UAG stop codon (slate), and ABCE1 (blue) with its iron-sulfur clusters (orange/yellow) and nucleotide binding sites (gray) indicated.
In each reconstruction, eRF1 is in its extended conformation10, and ABCE1 occupies the GTPase centre (Fig. 1a). The three domains of eRF1 (N, M and C) have moved relative to one another compared to the crystal structure11 (Extended Data Fig. 4a) and are each well resolved (Extended Data Fig. 3b). Direct interactions of the N domain with the codon deep in the decoding centre argues against an earlier suggestion that eRF1 disengages from the stop codon in the presence of ABCE110.
The N and M domains of eRF1 independently contact the P-site tRNA and together structurally resemble a tRNA in the A site (Fig. 1b). Helix α2 of the N domain runs parallel to, and interacts with, the anticodon stem-loop of the P-site tRNA (Extended Data Fig. 4b). The M domain is functionally analogous to the tRNA acceptor stem11, and positions the GGQ motif9 in the peptidyl transferase centre (Fig. 1b). To occupy a similar position as the 3’ end of an A-site tRNA, the GGQ-loop is shifted by 10 Å compared to the crystal structure11 (Extended Data Fig. 4c). This positions the alanine residues of the mutated GGQ motif directly opposite the terminal 3’ adenosine of the P-site tRNA and the glutamine (Gln185) in proximity of the ester bond between the nascent polypeptide and the tRNA. This conformation closely resembles that of the GGQ-loop in bacterial release factors12-14 (Fig. 2a).
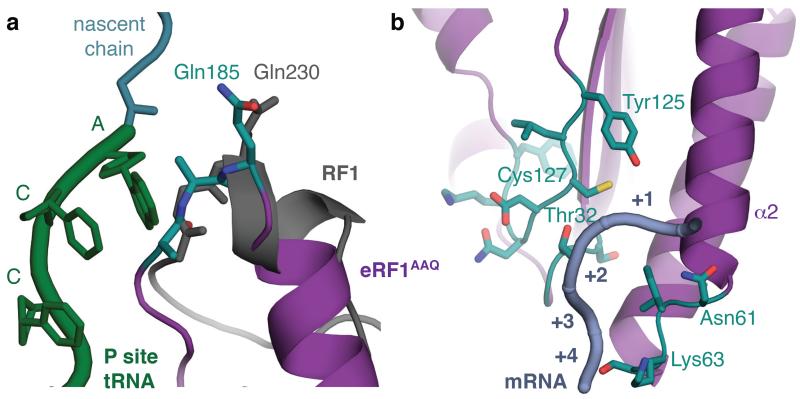
Figure 2. Conformation of essential eRF1 motifs.
a, Conformation of the GGQ-loop (teal) of eRF1AAQ (purple) within the peptidyl transferase centre. The AAQ tripeptide, positioned next to the CCA end of the P-site tRNA (green), closely resembles the conformation adopted by GGQ of a bacterial release factor (RF1, grey) bound to the ribosome. b, Positions of NIKS, YxCxxxF, and GTS (teal) motifs in the N domain of eRF1AAQ (purple) relative to the mRNA (slate). The positions of the stop codon (+1 to +3) and the following base (+4) are indicated.
At the decoding centre, the interactions between the N domain of eRF1 and the stop codon are well resolved (Fig. 2b and Extended Data Fig. 5). The most striking feature is a compact configuration of mRNA that accommodates four nucleotides in the A site instead of three (Fig. 3a, b). mRNA compaction depends crucially on the ‘flipping out’ of A1825 (A1493 in bacteria) in helix 44 (h44) of 18S rRNA, forming a stacking interaction with the +2 nucleotide of the stop codon, on which the +3 nucleotide stacks. This configuration allows the +4 nucleotide to stack with base G626 (G530 in bacteria) of 18S rRNA (Fig. 3b). Stacking with G626 would be more stable for purines, explaining their statistical bias at the +4 position in eukaryotes15. Compaction of mRNA in the A site probably results in pulling downstream mRNA further into the mRNA channel. This is consistent with protection of two additional nucleotides of 3’ mRNA upon eRF1 binding to the ribosome16,17.
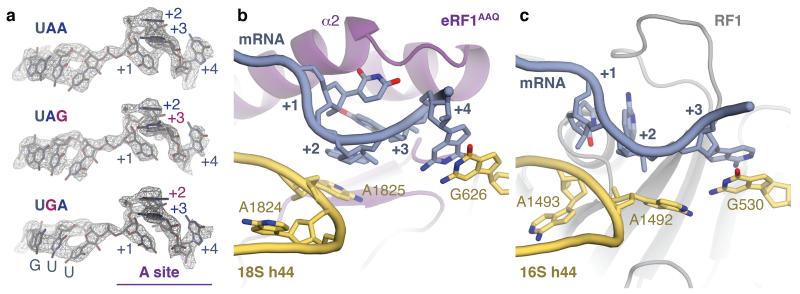
Figure 3. Stop codon configuration in the eukaryotic decoding centre.
a, EM map densities of the mRNA in the termination complexes containing the UAA, UAG and UGA stop codons reveal that they adopt the same compacted conformation. The ValGUU codon in the P site and the stop codon (+1 to +3) and following (+4) bases in the A site (purple) are indicated. b, The core termination signal recognised by eRF1AAQ (purple) is formed by four mRNA bases (+1 to +4, slate) that occupy the A site. Bases +2 and +3 stack on A1825, which is flipped out of helix 44 (h44), and base +4 on G626 of 18S rRNA (yellow). c, In bacteria, RF1 (grey) recognises a more extended stop codon configuration where the +3 base stacks on G530 (the equivalent of G626) of 16S rRNA.
The structures of the bacterial release factor complexes are qualitatively distinct (Fig. 3c). Instead of A1493, the adjacent A1492 is flipped out12-14. However, this flipped base does not stack with any of the stop codon bases and does not lead to mRNA compaction. G530 therefore stacks with the +3 base of the stop codon12-14 instead of the +4 base. While the +4 base also impacts stop codon recognition in bacteria, the greatest preference appears to be for uridine rather than for purines18. Thus, eukaryotes appear to exploit the +4 base to stabilise mRNA compaction. The unique stacking of the +2 and +3 bases in this compacted state is an important element for stop codon recognition by eRF1 (see below).
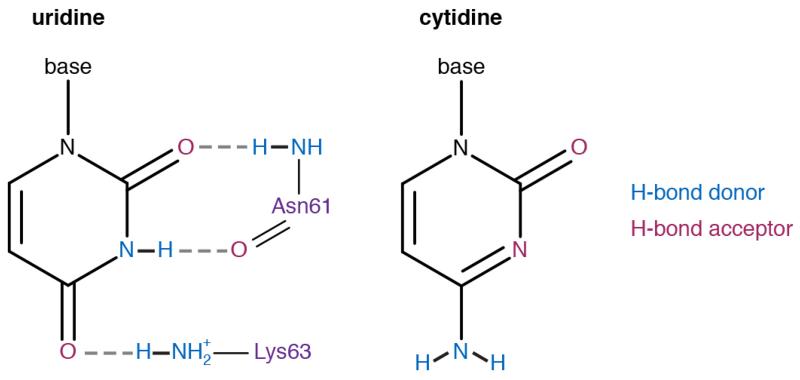
The conserved NIKS, YxCxxxF, and GTS motifs in eRF1 have been crosslinked to stop codon bases19,20. In the structures here, the essential NIKS sequence (residues 61-64)21, located at the end of helix α2, imposes a requirement for uridine in the first (+1) position via interactions with its Watson-Crick edge. A local distortion in helix α2 and the subsequent loop helps to optimise hydrogen bond interactions with the nucleotide (Fig. 4a). Notably, the side chains of Asn61 and Lys63 form hydrogen bonds with the uracil carbonyl groups. A further hydrogen bond forms with the main chain carbonyl of Asn61. Hydroxylation of Lys63 (at the Cδ position) reduces stop codon read-through and promotes peptide release in vivo22. The hydroxyl group may allow an additional interaction with the phosphate backbone of the mRNA and help position the ε-amino group for optimal hydrogen-bonding.
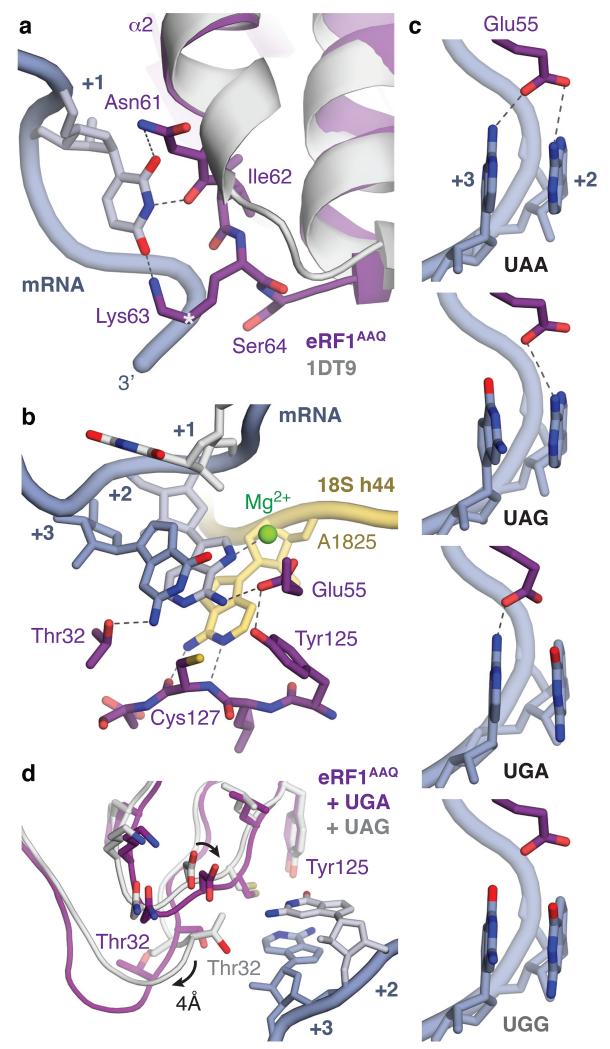
Figure 4. Molecular basis of stop codon recognition by eRF1.
a, The conformation of the NIKS motif (purple) at the end of helix α2 of ribosome-bound eRF1AAQ compared to the eRF1 crystal structure (PDB ID: 1DT9, light grey) allows it to form hydrogen bonds with the uridine in the +1 position of stop codons (slate; see also Extended Data Fig. 6). The hydroxyl groups of Ser64 and Cδ–hydroxylated (*) Lys63 help to position the NIKS motif by interacting with the mRNA phosphate backbone. b, Detailed interactions between the UAG stop codon (slate), eRF1AAQ (purple), and A1825 (yellow), depicting (i) stacking of the +2 and +3 bases with A1825, (ii) stabilisation of the flipped out position of A1825 by hydrogen bonds with the main chain of Cys127, (iii) a hydrogen bonding network involving Thr32, Glu55 and Tyr125 that specifies stop codon selectivity, and (iv) coordination of a Mg2+ ion (green) by the +2 adenosine. c, Model for stop codon discrimination by eRF1. Stacking of the +2 and +3 bases (slate) and possible hydrogen bonding interactions with Glu55 of eRF1 (purple) are shown for the three stop codons (UAA, UAG, UGA) and the sense codon UGG, which codes for tryptophan. d, The UGA stop codon induces a conformational change in the YxCxxxF and GTS motifs of eRF1AAQ (purple) compared to UAG- (white) and UAA-bound eRF1AAQ and eRF1 crystal structures.
Purines would be disfavoured at the +1 position by steric hindrance with eRF1 while cytidine would be incompatible with the hydrogen bonding requirements of the NIKS motif (Extended Data Fig. 6). Thus, the universality of uridine in the first position of stop codons is defined by extensive hydrogen bonding with eRF1. In bacteria, RF1 and RF2 also utilise hydrogen-bonding networks to specify uridine in the +1 position, but with different interactions12, highlighting how independent solutions have evolved for the same specificity problem.
Interactions of the YxCxxxF motif and Glu55 of eRF1 with the +2 and +3 purines provide a basis for stop codon discrimination from sense codons (Fig. 4b, c). The main chain of Cys127 from the YxCxxxF motif forms two hydrogen bonds with the Watson-Crick edge of A1825 to stabilise its flipped orientation, facilitating stacking of the +2 and +3 bases (Fig. 4b). The invariant residues Glu55 from helix α2 and Tyr125 act as a pair23 to position the glutamate side chain so that it can hydrogen bond with the N6 atoms of adenosines at the +2 and/or +3 positions. These interactions are only possible with purines, excluding pyrimidines from the +2 and +3 positions in stop codons. In the case of UGG, which codes for tryptophan, the two O6 atoms of consecutive stacked guanosines would not satisfy the hydrogen-bonding requirements, and their increased repulsion with each other and with the negatively-charged Glu55 would also disfavor the formation of the conformation that allows these interactions (Fig. 4c).
The conserved GTS motif, located towards the sugar edge of the +3 base, adopts two different conformations that are interdependent with the position of the YxCxxxF motif24. With adenosine in the +2 position, Thr32 faces the +3 base and can hydrogen bond with the N2 atom of the guanosine in UAG (Fig. 4b). A guanosine at the +2 position is accommodated by a movement of the YxCxxxF motif towards the stacked pair by ~1 Å, which propagates a 4 Å movement of the GTS motif so that Thr32 now faces away from the stop codon (Fig. 4d). Consistent with these observations, perturbation of any residue contributing to this network has substantial effects on stop codon recognition and specificity17,23,25,26.
In conclusion, these structures show how stop codons are specifically selected by eRF1. At the +1 position, only uridine can form the network of interactions with the NIKS motif. The flipping of A1825 results in its stacking onto the +2 and +3 bases of a distorted mRNA so that they are decoded as a single unit (Fig. 4b). This solves the puzzle of how guanosine can occur at either the +2 or +3 position but not at both: two successive guanosines would lead to repulsion between their O6 atoms and with Glu55 (Fig. 4c). By requiring two purines while specifically excluding consecutive guanosines, the selectivity of eRF1 at the +2 and +3 positions is logically equivalent to a NAND gate.
This mechanism for stop codon recognition by eRF1 is distinct from that used by the evolutionarily and structurally unrelated bacterial release factors, even though they share some common strategies including the extensive use of hydrogen bonding for decoding specificity and an invariant GGQ motif to catalyse peptide hydrolysis. The work here paves the way for studies on the mechanism of termination and ribosome recycling, including the roles of eRF3 and ABCE1, as well as the interaction of termination factors with quality control pathways such as nonsense-mediated mRNA decay27. Finally, insight into stop codon recognition can provide a framework for the development of inhibitors of termination that may be useful to treat the ~11% of hereditary diseases caused by premature termination28,29.
Methods
Plasmids and antibodies
An SP64-based plasmid encoding 3× Flag-tagged Sec61β containing the autonomously-folding villin headpiece (VHP) domain30 was modified by individually inserting each stop codon (TAA, TAG, TGA) after the valine at position 68 of unmodified Sec61β. The remaining C-terminal portion of the protein was deleted using Phusion mutagenesis (Thermo Scientific). In vitro transcription reactions were performed using PCR products generated with primers flanking the SP6 promoter and the 3’UTR of the SP64 vector as previously described31. The cDNA encoding human eRF1 (Origene) was subcloned into a pRSETA expression vector after an N-terminal 6× His tag and TEV cleavage site using standard procedures. The eRF1AAQ mutant was generated via Phusion mutagenesis. Antibodies against Hbs1 and ABCE1 have been described32; the eRF1 antibody was from New England Biolabs.
Release factor purification
Wild-type and mutant eRF1 (eRF1AAQ) were expressed in and purified from Escherichia coli BL21(DE3) cells grown under antibiotic selection in LB. Transformed cells were induced at A600 = 0.4-0.6 with 0.2 mM IPTG for 2 hr at 37°C and lysed with a microfluidiser in lysis buffer (1× PBS, pH 7.5, 250 mM NaCl, 10 mM imidazole, 1 mM DTT) containing 1× protease inhibitor cocktail (Roche). Lysates were clarified by centrifugation and the supernatant passed over a NiNTA column. After washing with 25 column volumes of lysis buffer, elutions were carried out with 250 mM imidazole in lysis buffer. Peak fractions were pooled, dialysed overnight in the presence of TEV protease against 50 mM Hepes, pH 7.4, 150 mM KAc, 5 mM MgAc2, 10 mM imidazole, 10% glycerol, 1 mM DTT. The TEV protease and cleaved His tag were removed by passage over a NiNTA column.
In vitro translations and sample preparation
In vitro translations in a rabbit reticulocyte (RRL) system were for 25 min at 32°C as before30,31. Where indicated, 0.5 μM eRF1AAQ was included to trap termination complexes. 4 mL translation reactions were directly incubated with 100 μL (packed volume) of anti-Flag M2 beads (Sigma) for 1-1.5 hr at 4°C with gentle mixing. The beads were washed sequentially with 6 mL 50 mM Hepes, pH 7.4, 100 mM KAc, 5 mM MgAc2, 0.1% Triton X-100, 1 mM DTT, 6 mL 50 mM Hepes, pH 7.4, 250 mM KAc, 5 mM MgAc2, 0.5% Triton X-100, 1 mM DTT and 6 mL RNC buffer (50 mM Hepes, pH 7.4, 100 mM KAc, 5 mM MgAc2, 1 mM DTT). Two sequential elutions were carried out with 100 μL 0.1 mg/mL 3× Flag peptide (Sigma) in RNC buffer at room temperature for 25 min. The elutions were combined and centrifuged at 100,000 rpm at 4°C for 40 min in a TLA120.2 rotor (Beckman Coulter) before resuspension of the ribosomal pellet in RNC buffer.
Electron microscopy
3 μL aliquots of purified ribosome complexes at a concentration of 120 nM were applied onto Quantifoil R2/2 cryo-EM grids covered with continuous carbon (estimated to be 50 Å thick) at 4°C and 100% ambient humidity. After 30 s incubation, the grids were blotted for 3 s and vitrified in liquid ethane using a Vitrobot MKIII (FEI).
Automated data collection (EPU software, FEI) was conducted on a Titan Krios microscope equipped with a XFEG electron source using 300 kV acceleration voltage. For each 1.1 s exposure, 17 movie frames were recorded on a Falcon II direct electron detector (FEI) at a calibrated magnification of 104,478, resulting in a pixel size of 1.34 Å33. A dose rate of ~30 electrons per Å2 per second was used. Defocus values ranged from −1.1 to −5.9 μm for the UAA-eRF1AAQ dataset, −0.7 to −4.1 μm for the UAG-eRF1AAQ dataset, and −0.7 to −3.8 μm for the UGA-eRF1AAQ dataset (Extended Data Table 1), as more images were collected closer to focus on the latter two datasets.
Image Processing
The 17 frames were aligned using whole-image motion correction34 to reduce beam-induced blurring of the images. Parameters of the contrast transfer function for each motion-corrected micrograph were obtained using Gctf (K. Zhang, MRC-LMB, in development). Ribosome particles were selected using semi-automated particle picking implemented in RELION35. All two- and three-dimensional classifications and refinements were performed using RELION36. We used reference-free two-dimensional class averaging to discard non-ribosomal particles and three-dimensional classification to sort different compositions and conformations of the ribosome complexes. After two-dimensional classification, the UAA-eRF1AAQ dataset contained 556,994 particles from two independent data acquisitions, while the UAG-eRF1AAQ and UGA-eRF1AAQ datasets contained 216,276 and 250,705 particles, respectively, both from a single data acquisition.
A 40 Å low-pass filtered cryo-EM reconstruction of the rabbit 80S ribosome (Electron Microscopy Data Bank (EMDB) accession 2704)32 was used as an initial model for the three-dimensional refinement. In a subsequent three-dimensional classification run with 12 classes, an angular sampling of 1.8° was combined with local angular searches, and the refined model from the first refinement was used as a starting model. A further round of refinement and three-dimensional classification into 6 classes using an angular sampling of 0.9° combined with local angular searches was necessary to isolate a homogeneous class of ribosomes bound with eRF1-ABCE1 (see Extended Data Fig. 2). This resulted in 49,979 particles for the UAA-eRF1AAQ dataset, 20,515 particles for the UAG-eRF1AAQ dataset, and 22,058 particles for the UGA-eRF1AAQ dataset.
Prior to a final round of refinement and classification with classes combined from different datasets, statistical particle-based movie correction was performed in RELION-1.4. For these calculations we used running averages of five movie frames, and a standard deviation of one pixel for the translational alignment. In addition, we used a resolution and dose-dependent model for the radiation damage, in which each frame is B-factor weighted as estimated from single-frame reconstructions37.
Reported resolutions are based on the Fourier shell correlation (FSC) 0.143 criterion. High-resolution noise substitution was used to correct for the effects of a soft mask on FSC curves38. Before visualization, density maps were corrected for the modulation transfer function of the Falcon II detector and then sharpened by applying a negative B-factor that was estimated using automated procedures39 (Extended Data Table 1). Local resolution was quantified using ResMap40.
Model building
The reconstruction was initially interpreted by docking the large and small subunits of the mammalian 80S ribosome, with respective Protein Data Bank (PDB) accession codes 3J92 and 3J7P41,42, into the map using Chimera43. The crystal structures of human eRF1 (PDB accession code 1DT9)11 and Pyrococcus abysii ABCE1 (PDB accession code 3BK7)44 were docked into the A site and GTPase center, respectively, before being subjected to a Jiggle Fit in Coot45. To model the bound tRNAs, bacterial tRNA (PDB accession code 4V51)46 was used as a template and modified to the sequence of the most prevalent tRNA for the particular codon from the genomic tRNA database47. The atomic models were then modified in Coot v0.8 to agree with the rabbit sequences and optimised for fit to density.
Model refinement and validation
Reciprocal space refinement was carried out in REFMAC v5.8 optimised for EM maps utilizing external restraints generated by ProSMART and LIBG45. The model was refined against amplitudes and phases from the experimental map that were unchanged during the course of refinement. FSCaverage was monitored during refinement to follow the fit-to-density, and the final model was validated using MolProbity48 (Extended Data Table 1). The Ramachandran statistics for eRF1 are 93.0% favoured, 1.5% outliers and for ABCE1, 91.1% favoured, 1.9 % outliers. Cross-validation against over-fitting was calculated as previously described45,49 (Extended Data Figure 3c).
Extended Data
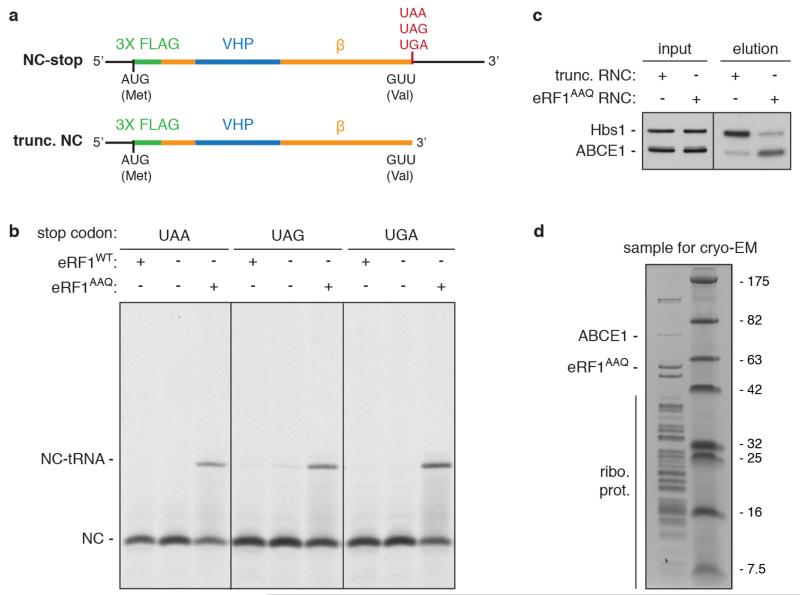
Extended Data Figure 1. eRF1AAQ stalls ribosomes at stop codons.
a, Line diagrams of mRNA encoding nascent chain (NC) substrates used in this study. The cytosolic portion of human Sec61β (residues 1-68, orange) was modified to contain an N-terminal 3× Flag tag (green) for affinity purification and the autonomously-folding villin headpiece (VHP, blue) domain. The three stop codons were individually inserted after Val68 of Sec61β to generate substrates for eRF1AAQ-mediated stalling, or the mRNA was truncated after the same residue to generate an independently-stalling substrate. b, In vitro translation reactions of NC-stop substrates containing the indicated stop codon (see panel a) in the presence of 35S-methionine without or with excess eRF1WT or eRF1AAQ. Reactions were for 25 min at 32°C and directly analyzed by SDS-PAGE and auto-radiography. The terminated (NC) and tRNA-associated (NC-tRNA) nascent chain products are indicated. Addition of eRF1AAQ selectively prevents peptide hydrolysis when the stop codon is reached. c, Anti-Flag affinity purifications of ribosome-nascent chains (RNCs) stalled either by mRNA truncation or at the UAA stop codon with eRF1AAQ (see panel a) were immunoblotted for the splitting factors Hbs1 and ABCE1. The different amounts of Hbs1 and ABCE1 co-purified despite identical nascent chain sequences in each RNC complex suggest that eRF1AAQ selectively traps ABCE1 on pre-termination complexes. d, SDS-PAGE and Coomassie staining of affinity-purified eRF1AAQ-stalled ribosome-nascent chains containing the UGA stop codon utilised for cryo-EM analysis. Bands corresponding to ribosomal proteins, ABCE1, and eRF1AAQ, which were verified by immunoblotting and mass spectrometry (data not shown), are indicated.
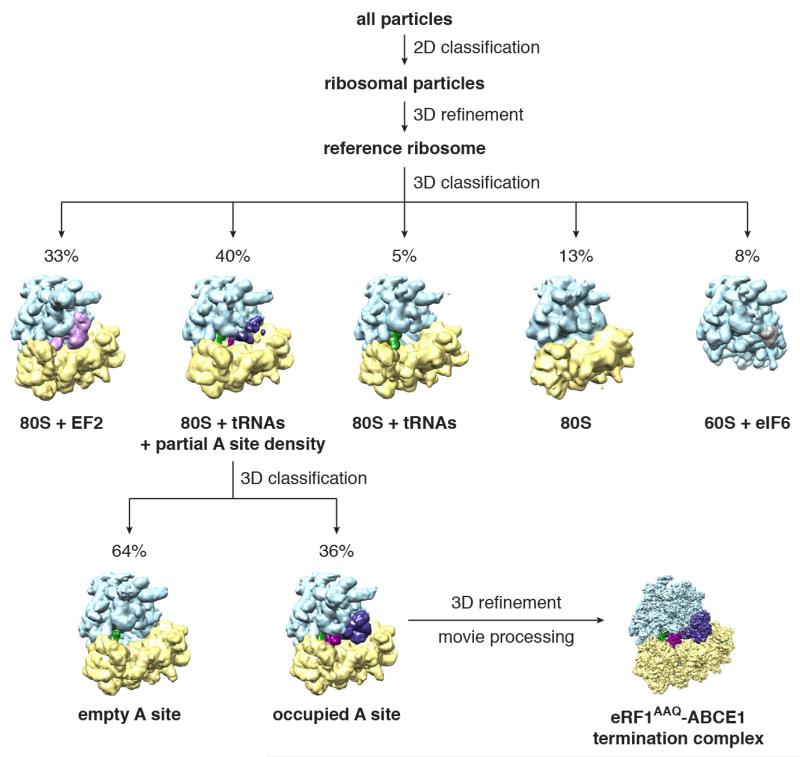
Extended Data Figure 2. In silico 3D classification scheme for cryo-EM datasets.
Particles extracted from automated particle picking in Relion were subjected to 2D classification. Non-ribosomal particles were discarded and the remaining particles were combined for a 3D refinement. The resulting map was used as a reference for 3D classification, which typically isolated 5 distinct classes of ribosomal complexes with the indicated distributions. Classes containing 80S ribosomes with canonical P- and E-site tRNAs and weak factor density in the A site (~40%) were combined and subjected to another round of 3D classification for A site occupancy. Approximately 1/3 of this population contained strong density for eRF1AAQ and ABCE1. These particles were combined for subsequent 3D refinement and movie processing. All four datasets (two for the UAA stop codon and one each for the UAG and UGA stop codons) were processed similarly. The eRF1AAQ-ABCE1-containing particles of the two UAA datasets after the two rounds of classification were combined for refinement to yield the final map.
Extended Data Figure 3. Quality of maps and models.
a, Fourier shell correlation (FSC) curves for the EM maps of each termination complex containing the indicated stop codon. b, Isolated eRF1AAQ-ABCE1 density from the UAA termination complex map coloured by local resolution. c, Fit of models to maps. FSC curves calculated between the refined model and the final map (black), and with the self- (blue) and cross-validated (magenta) correlations for each stop codon complex. The EM map of each termination complex coloured by local resolution (as in panel b) is displayed next to the corresponding curves.
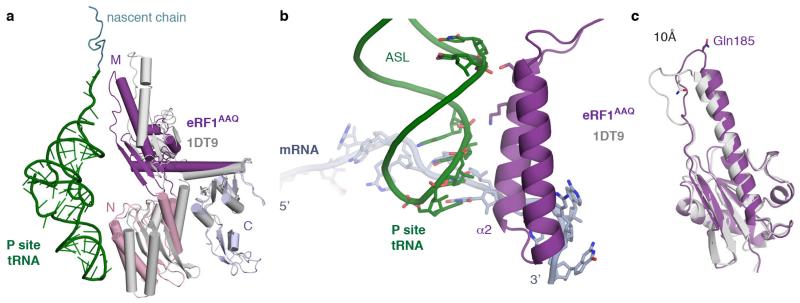
Extended Data Figure 4. eRF1AAQ interactions within the termination complex.
a, Comparison of ribosome-bound eRF1AAQ (coloured by domain) with the crystal structure of eRF1 (PDB ID: 1DT9, grey) superposed on the C domain. Both the N and M domains of eRF1 rotate upon stop codon recognition on the ribosome. The P-site tRNA (green) and nascent chain (teal) are shown for orientation. b, Interaction of helix α2 of the N domain of eRF1AAQ (purple) with the anticodon stem loop (ASL) of the P-site tRNA (green). c, Superposition of the eRF1AAQ M domain (purple) with the eRF1 crystal structure (PDB ID: 1DT9) showing a 10 Å movement of the GGQ-loop to accommodate within the peptidyl transferase centre.
Extended Data Figure 5. Examples of map densities.
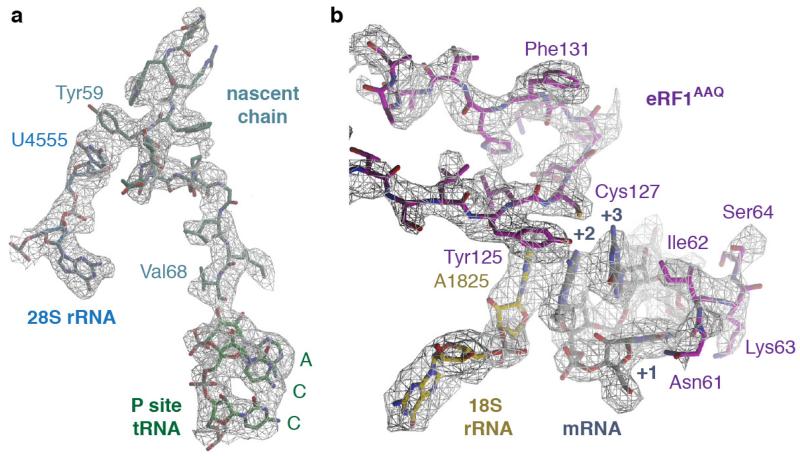
a, Density (from the UAG-containing termination complex) for the nascent chain (teal) attached to the CCA end of the P-site tRNA (green) is of sufficient resolution to model the defined sequence of the C-terminal end of the programmed nascent chain. This provides additional verification that the termination complexes are stalled at Val68 of Sec61β (human numbering) with the stop codon in the A site (see also Extended Data Fig. 1a). A stacking interaction between an aromatic residue of the nascent chain and U4555 (blue) lining the ribosomal exit tunnel can also be observed. b, Densities for the interactions between the UAG stop codon (grey), a portion of h44 of 18S rRNA (yellow) and the YxCxxxF and NIKS motifs of eRF1AAQ (purple). The invariant isoleucine of the NIKS motif provides a hydrophobic base for the stacking of the +2 and +3 bases of the stop codon with A1825. Unlike the tyrosine and cysteine residues of the YxCxxxF motif, the phenylalanine does not contribute to stop codon recognition, but to the hydrophobic packing of the eRF1 N domain.
Extended Data Figure 6. Hydrogen bonds specify for uridine at the +1 position.
Chemical diagrams of uridine and cytidine with hydrogen bond donors (blue) and acceptors (magenta) indicated. Two of the three hydrogen bonds that uridine forms with Asn61 and Lys63 of the NIKS motif of eRF1AAQ (purple) are not possible with cytidine (see also Fig. 4a).
Extended Data Table 1.
Refinement and model statistics.
| UAA | UAG | UGA | |
|---|---|---|---|
| Data Collection | |||
| Particles | 49,979 | 20,515 | 22,058 |
| Pixel size (Å) | 1.34 | 1.34 | 1.34 |
| Defocus range (μm) | 1.1-5.9 | 0.7-4.1 | 0.7-3.8 |
| Defocus mean (μm) | 3.2 | 2.4 | 2.3 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron dose (e- Å−2) | 30 | 30 | 30 |
| Model composition | |||
| Non-hydrogen atoms | 226,532 | 226,533 | 226,533 |
| Protein residues | 12,676 | 12,676 | 12,676 |
| RNA bases | 5,820 | 5,820 | 5,820 |
| Ligands (Zn2+/Mg2+/ADP) | 8/197/2 | 8/197/2 | 8/197/2 |
| Refinement | |||
| Resolution (Å) | 3.65 | 3.45 | 3.83 |
| Map sharpening B-factor (Å2) | −81.7 | −50.6 | −82.7 |
| Average B factor (Å2) | 105.8 | 87.4 | 93.5 |
| FSCaverage | 0.85 | 0.84 | 0.88 |
| FSCaverage (eRF1) | 0.70 | 0.64 | 0.74 |
| FSCaverage (ABCE1) | 0.71 | 0.62 | 0.75 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.006 | 0.006 | 0.008 |
| Bond angles (°) | 1.19 | 1.22 | 1.40 |
| Validation | |||
| Molprobity score | 2.7 (93rd percentile) | 2.8 (89th percentile) | 3.0 (89th percentile) |
| Clashscore, all atoms | 5.2 (100th percentile) | 6.2 (97th percentile) | 8.2 (97th percentile) |
| Good rotamers (%) | 88.2 | 87.5 | 86.4 |
| Ramachandran plot | |||
| Favored (%) | 87.0 | 85.8 | 83.5 |
| Outliers (%) | 3.3 | 3.5 | 4.2 |
| Validation (RNA) | |||
| Correct sugar puckers (%) | 96.5 | 93.4 | 96.0 |
| Good backbone conformations (%) | 68.2 | 66.8 | 65.8 |
Acknowledgements
We thank C. Savva, F. de Haas, and S. Welsch for assisting with cryo-EM data collection, J. Grimmett and T. Darling for computing support, D. Barford for critically reading the manuscript, and I. Fernández, J. Llácer, G. Murshudov, S. Scheres, and R. Voorhees for useful discussions. This work was supported by the UK Medical Research Council (MC_UP_A022_1007 to R.S.H. and MC_U105184332 to V.R.). A.B. was supported by a Career Development Fellowship. S.S. was supported by a St John’s College Title A fellowship. J.M. thanks T. Dever, NICHD, and the NIH Oxford-Cambridge Scholars’ Program for support. V.R. was supported by a Wellcome Trust Senior Investigator award (WT096570), the Agouron Institute, and the Jeantet Foundation.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Author information
Maps have been deposited with the EMDB under accession codes XXX, YYY and ZZZ. Atomic coordinates have been deposited with the Protein Data Bank under accession codes XXX, YYY and ZZZ.
The authors declare no competing financial interests.
References
- 1.Scolnick E, Tompkins R, Caskey T, Nirenberg M. Release factors differing in specificity for terminator codons. Proc. Natl. Acad. Sci. U.S.A. 1968;61:768–774. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frolova L, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 3.Dever TE, Green R. The Elongation, Termination, and Recycling Phases of Translation in Eukaryotes. Cold Spring Harbor Perspectives in Biology. 2012;4:a013706–a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson RJ, Hellen CUT, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 5.Muhs M, et al. Cryo-EM of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus IRES. Molecular Cell. 2015;57:422–432. doi: 10.1016/j.molcel.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor D, et al. Cryo-EM structure of the mammalian eukaryotic release factor eRF1-eRF3-associated termination complex. Proc. Natl. Acad. Sci. U.S.A. 2012;109:18413–18418. doi: 10.1073/pnas.1216730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pisarev AV, et al. The Role of ABCE1 in Eukaryotic Posttermination Ribosomal Recycling. Molecular Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E1392–E1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frolova LY, et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preis A, et al. Cryoelectron Microscopic Structures of Eukaryotic Translation Termination Complexes Containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep. 2014;8:59–65. doi: 10.1016/j.celrep.2014.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song H, et al. The Crystal Structure of Human Eukaryotic Release Factor eRF1— Mechanism of Stop Codon Recognition and Peptidyl-tRNA Hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 12.Laurberg M, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 13.Weixlbaumer A, et al. Insights into Translational Termination from the Structure of RF2 Bound to the Ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korostelev A, et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown CM, Stockwell PA, Trotman CN, Tate WP. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Research. 1990;18:6339–6345. doi: 10.1093/nar/18.21.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shirokikh NE, et al. Quantitative analysis of ribosome-mRNA complexes at different translation stages. Nucleic Acids Research. 2010;38:e15–e15. doi: 10.1093/nar/gkp1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kryuchkova P, et al. Two-step model of stop codon recognition by eukaryotic release factor eRF1. Nucleic Acids Research. 2013;41:4573–4586. doi: 10.1093/nar/gkt113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poole ES, Brown CM, Tate WP. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995;14:151–158. doi: 10.1002/j.1460-2075.1995.tb06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chavatte L, Seit-Nebi A, Dubovaya V, Favre A. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 2002;21:5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bulygin KN, et al. Three distinct peptides from the N domain of translation termination factor eRF1 surround stop codon in the ribosome. RNA. 2010;16:1902–1914. doi: 10.1261/rna.2066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frolova L, Seit-Nebi A, Kisselev L. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA. 2002;8:129–136. doi: 10.1017/s1355838202013262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng T, et al. Optimal Translational Termination Requires C4 Lysyl Hydroxylation of eRF1. Molecular Cell. 2014;53:645–654. doi: 10.1016/j.molcel.2013.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolosov P, et al. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Research. 2005;33:6418–6425. doi: 10.1093/nar/gki927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong LE, Li Y, Pillay S, Frolova L, Pervushin K. Selectivity of stop codon recognition in translation termination is modulated by multiple conformations of GTS loop in eRF1. Nucleic Acids Research. 2012;40:5751–5765. doi: 10.1093/nar/gks192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Z, et al. Structural insights into eRF3 and stop codon recognition by eRF1. Genes & Development. 2009;23:1106–1118. doi: 10.1101/gad.1770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seit-Nebi A, Frolova L, Kisselev L. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002;3:881–886. doi: 10.1093/embo-reports/kvf178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czaplinski K, et al. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes & Development. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keeling KM, Xue X, Gunn G, Bedwell DM. Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet. 2014;15:371–394. doi: 10.1146/annurev-genom-091212-153527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mort M, Ivanov D, Cooper DN, Chuzhanova NA. A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 2008;29:1037–1047. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 30.Shao S, Malsburg, von der K, Hegde RS. Listerin-Dependent Nascent Protein Ubiquitination Relies on Ribosome Subunit Dissociation. Molecular Cell. 2013;50:637–648. doi: 10.1016/j.molcel.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma A, Mariappan M, Appathurai S, Hegde RS. Protein Secretion. Vol. 619. Humana Press; 2010. pp. 339–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao S, Hegde RS. Reconstitution of a Minimal Ribosome-Associated Ubiquitination Pathway with Purified Factors. Molecular Cell. 2014;55:880–890. doi: 10.1016/j.molcel.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai X-C, Fernandez IS, McMullan G, Scheres SHW. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. elife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheres SHW. Semi-automated selection of cryo-EM particles in RELION-1.3. Journal of Structural Biology. 2015;189:114–122. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. elife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenthal PB, Henderson R. Optimal Determination of Particle Orientation, Absolute Hand, and Contrast Loss in Single-particle Electron Cryomicroscopy. Journal of Molecular Biology. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 40.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2013;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voorhees RM, Fernandez IS, Scheres SHW, Hegde RS. Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell. 2014;157:1632–1643. doi: 10.1016/j.cell.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shao S, Brown A, Santhanam B, Hegde RS. Structure and Assembly Pathway of the Ribosome Quality Control Complex. Molecular Cell. 2015;57:433–444. doi: 10.1016/j.molcel.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Karcher A, Schele A, Hopfner KP. X-ray Structure of the Complete ABC Enzyme ABCE1 from Pyrococcus abyssi. J. Biol. Chem. 2008;283:7962–7971. doi: 10.1074/jbc.M707347200. [DOI] [PubMed] [Google Scholar]
- 45.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selmer M. Structure of the 70S Ribosome Complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 47.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Research. 2009;37:D93–7. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2009;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]