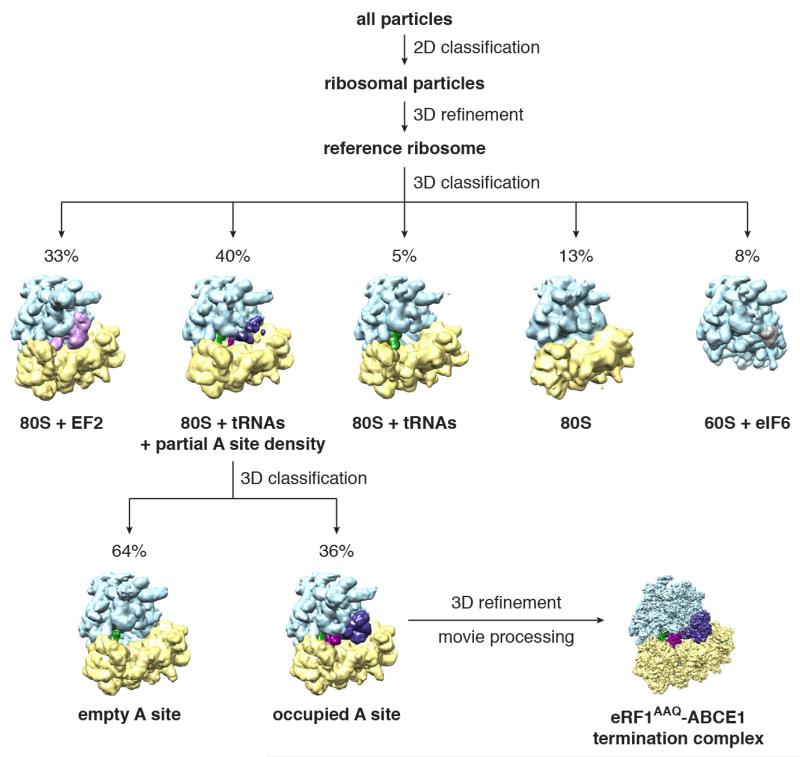
Extended Data Figure 2. In silico 3D classification scheme for cryo-EM datasets.
Particles extracted from automated particle picking in Relion were subjected to 2D classification. Non-ribosomal particles were discarded and the remaining particles were combined for a 3D refinement. The resulting map was used as a reference for 3D classification, which typically isolated 5 distinct classes of ribosomal complexes with the indicated distributions. Classes containing 80S ribosomes with canonical P- and E-site tRNAs and weak factor density in the A site (~40%) were combined and subjected to another round of 3D classification for A site occupancy. Approximately 1/3 of this population contained strong density for eRF1AAQ and ABCE1. These particles were combined for subsequent 3D refinement and movie processing. All four datasets (two for the UAA stop codon and one each for the UAG and UGA stop codons) were processed similarly. The eRF1AAQ-ABCE1-containing particles of the two UAA datasets after the two rounds of classification were combined for refinement to yield the final map.