Abstract
Shiga toxin-converting bacteriophages (Stx phages) are present as prophages in Shiga toxin-producing Escherichia coli (STEC) strains. Theses phages can be transmitted to previously non-pathogenic E. coli cells making them potential producers of Shiga toxins, as they bear genes for these toxins in their genomes. Therefore, sensitivity of Stx phage virions to various conditions is important in both natural processes of spreading of these viruses and potential prophylactic control of appearance of novel pathogenic E. coli strains. In this report we provide evidence that virions of Stx phages are significantly more sensitive to UV irradiation than bacteriophage λ. Following UV irradiation of Stx virions at the dose of 50 J/m2, their infectivity dropped by 1–3 log10, depending on the kind of phage. Under these conditions, a considerable release of phage DNA from virions was observed, and electron microscopy analyses indicated a large proportion of partially damaged virions. Infection of E. coli cells with UV-irradiated Stx phages resulted in significantly decreased levels of expression of N and cro genes, crucial for lytic development. We conclude that inactivation of Stx virions caused by relatively low dose of UV light is due to damage of capsids that prevents effective infection of the host cells.
Keywords: Shiga toxin-converting bacteriophages, virion stability, UV irradiation
1. Introduction
Virulence of Shiga toxin-producing Escherichia coli (STEC) strains, including their most dangerous subset, enterohemorrhagic E. coli (EHEC), depends on production of Shiga toxins [1,2]. These toxins are deleterious to humans due to strong inhibition of protein synthesis, mediated by specific action of the toxin N-glycosidase activity which leads to modification and inactivation of rRNA [3,4]. Infections by STEC strains are characterized by high morbidity and mortality, mostly as effects of severe complications, including hemolytic uremic syndrome [5]. The high percentage of fatal cases among infected patients take place not only in geographical regions of low levels of medical care, but also in highly developed countries [6,7,8].
In all STEC strains investigated to date, genes coding for Shiga toxins (stx genes) are located on lambdoid prophages, called Shiga toxin-converting prophages or Stx prophages [9]. The Stx bacteriophages belong to the family of lambdoid phages due to similarities of their genomes to that of bacteriophage λ [10]. In lysogenic bacteria, stx genes, like vast majority of phage genes, are silent or expressed at low levels [10,11,12,13,14,15]. Prophage induction, caused by any factors or agents that either provoke the bacterial S.O.S. response or weaken the cI repressor binding to phage promoters [13,16], results in excision of phage DNA from the host chromosome and initiation of the lytic development [12]. At this phase, beside expression of genes coding for phage structural proteins and those involved in phage DNA replication, recombination and regulatory processes, stx genes are also activated. This leads to effective production of Shiga toxins which are released after phage-mediated lysis of the host cell [9,10].
Since lytic development of Stx phages leads not only to production of Shiga toxins, but also to formation of phage progeny and its liberation after cell lysis, it is clear that newly produced virions can infect neighboring E. coli cells. If these cells are not STEC, lysogenization of the new host results in formation of a new STEC strain. Such a scenario is not only theoretical, but it was also documented experimentally [17,18]. In this light, it is important to determine the level of persistence of Stx virions in natural environment, as well as their sensitivity to various factors and agents which could be potentially used for the control of spreading of Stx phages. Both these aspects were investigated previously [19,20,21,22,23,24,25,26], however, while the phenomena were described qualitatively and quantitatively, their specific molecular mechanisms remain largely unknown, particularly at low doses of various factors or agents which correspond to those occurring in the nature. In this work, we have studied sensitivity of virions of five Stx phages to UV irradiation in relation to those of bacteriophage λ, as well as mechanisms of decreased infectivity of Stx virions treated with UV.
2. Results and Discussion
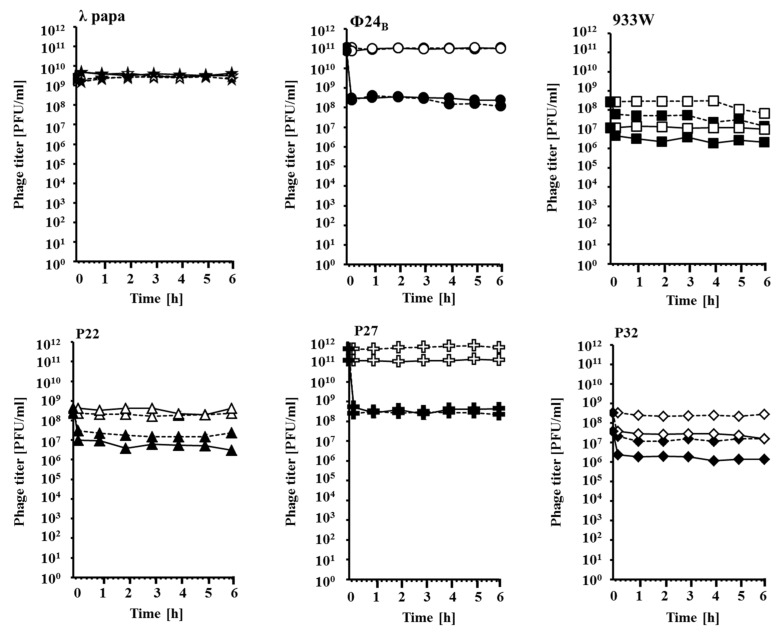
Since the influence of temperature on UV-stimulated spreading of Stx phages has been reported previously [18], we have tested effects of UV irradiation on lambdoid virions at 37 °C (the physiological temperature for E. coli, a host for these phages) and 43 °C (an elevated temperature which might simulate conditions occurring in an infected mammalian organism). When a lysate of bacteriophage λ was irradiated with UV light at the dose of 50 J/m2, no effects on infectivity of virions could be observed at both tested temperatures, 37 °C and 43 °C (Figure 1). However, UV irradiation of virions of five different Shiga toxin-converting bacteriophages (Stx phages) resulted in a significant decrease of the number or plaque forming units (pfu) just after irradiation, and this effect was stable to the end of the experiment, i.e., for 6 h after irradiation (Figure 1). The effect was strongly pronounced in phages Φ24B and P27, where the infectivity dropped by two to three orders of magnitude, while in other tested Stx phages (933W, P22 and P32) number of pfu decreased by one order of magnitude or less, though the decrease was still statistically significant (Figure 1). Therefore, we conclude that virions of Stx phages are sensitive to low doses of UV irradiation, contrary to those of bacteriophage λ. This conclusion has been corroborated by measurement of survival of E. coli cells infected with UV-irradiated and non-irradiated bacteriophages. While no significant differences were observed in the fraction of bacteria surviving both kinds of infection with bacteriophage λ, significantly more cells survived infection with UV-irradiated Stx phages relative to non-treated counterparts (Table 1). Again, the most spectacular differences were observed in phages Φ24B and P27 (Table 1).
Figure 1.
Sensitivity of lamdoid bacteriophages to UV irradiation. Lysates of phages (indicated in each panel) were either non-irradiated (open symbols) or UV-irradiated at 50 J/m2 (closed symbols), and E. coli MG1655 cells were infected at 37 °C (solid lines) or 43 °C (dashed lines). Phage titer was determined at indicated times after irradiation by estimating number of plaque forming units (pfu) per mL. Results are presented as mean values from three independent experiments with error bars indicating SD, however the bars are smaller than the size of symbols. For all experiments but those with bacteriophage λ, statistically significant differences (p < 0.05 in the t-tets) between irradiated and non-irradiated phages were found. The use of phage lysates of different initial titers gave very similar results, thus, the sensitivity to UV irradiation was independent on virion density.
Table 1.
Survival of E. coli MG1655 cells in the liquid culture at 37 °C following infection with lambdoid bacteriophages, either non-irradiated or UV-irradiated, at m.o.i. = 5.
| Bacteriophage | Survival of Cells in Infected Culture (% of Survivors) | |
|---|---|---|
| No UV | 50 J/m2 UV | |
| λ papa | 31 ± 6 | 30 ± 6 |
| Φ24B | 36 ± 8 | 88 ± 7 * |
| 933W | 34 ± 1 | 39 ± 2 |
| P22 | 44 ± 7 | 68 ± 6 * |
| P27 | 43 ± 2 | 93 ± 1 * |
| P32 | 37 ± 7 | 63 ± 9 * |
Asterisks (*) indicate statistically significant differences (p < 0.05 in the t-test) between results obtained with and without UV.
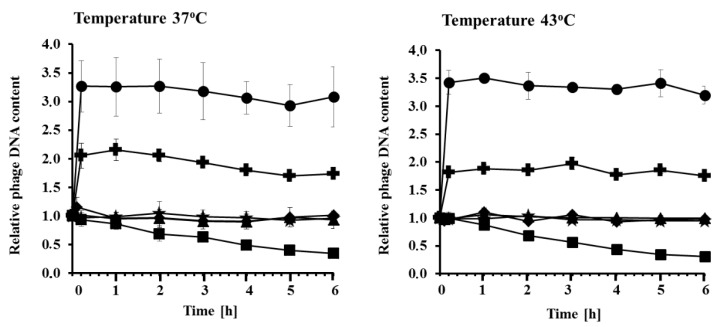
The results presented in Figure 1 and Table 1 raised a question about the mechanism of the decrease in infectivity of virions irradiated with UV light. Damage of DNA appeared an unlikely reason for this effect, as no significant effects of UV irradiation at 50 J/m2 were observed in bacteriophage λ, and there is no reason to suspect significant differences between sensitivity of phages λ and Stx DNAs to UV. Therefore, an alternative hypothesis has been tested, assuming that capsids of Stx virions might be damaged under these conditions. We suspected that severe damage of bacteriophage capsid may result in the release of viral DNA. Therefore, we have determined concentrations of phage DNA in samples of lysates, either untreated or UV irradiated at 50 J/m2, by using a fluorescence assay in which a fluorescent dye emits light only when bound to the target molecule. In these experiments, phage DNA included in the capsid is protected from the dye, and thus undetected, while released DNA can be bound to the dye and visualized by fluorescence. We found that while UV irradiation of λ, 933W, P22, and P32 virions resulted in no increase of the released phage DNA amount (the basal level derived from mechanically damaged virions, always present in each lysate), a significant increase in the level of free phage DNA could be observed in experiments with Φ24B and P27 virions (Figure 2). Note that under these experimental conditions, only severe virion damage, allowing to release of the phage DNA from capsids, might be detected. Therefore, since the effects were most pronounced in the same phages as in the measurement of infectivity (Φ24B and P27), we recognized the results shown in Figure 2 as compatible with those presented in Figure 1 and Table 1. In phages 933W, P22 and P32, the UV-mediated damage of virions might be large enough for the loss of infectivity, but too small to fully open the capsid and allow phage DNA to release. Again, no effects of temperature could be detected (Figure 2). One can assume that changes in a capsid may affect infectivity of virions due to either leakiness of phage DNA or lesions in the protein(s) responsible for effective adsorption of the virus on the host cells, or both.
Figure 2.
Release of phage DNA from capsids after UV irradiation of virions of lambdoid bacteriophages. Lysates of bacteriophage λ (asterisks) and Stx phages Φ24B (circles), 933W (squares), P22 (triangles), P27 (crosses), and P32 (diamonds) were irradiated with UV light at 50 J/m2 at time 0, and incubated at 37 °C or 43 °C as indicated. Amount of released DNA was estimated using Qubit DNA Assay Kit. The results obtained at time 0 are referred to as 1, and other values reflect this value. The presented results are mean values from three experiments with error bars indicating SD (note that in most cases the bars are smaller than the size of symbols).
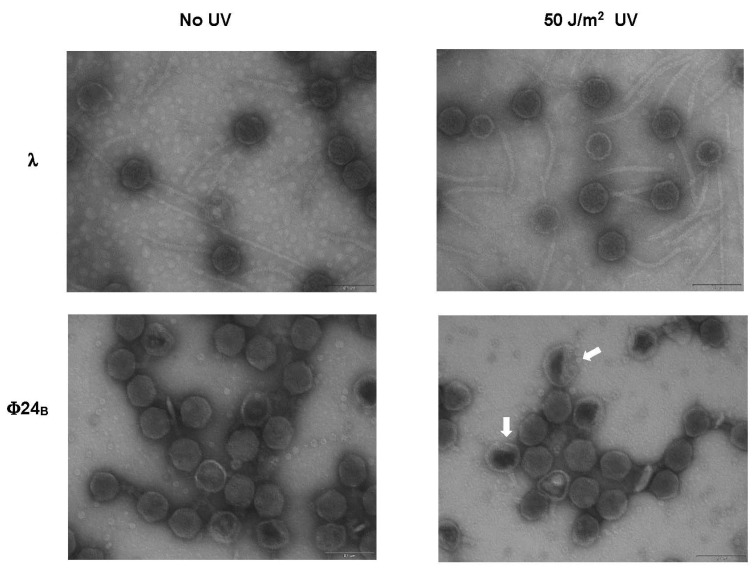
To identify what kind of severe damage of Stx virions can be caused by UV irradiation, we have performed electron microscopic studies. Highly purified (with the cesium chloride ultracentrifugation method) virions were UV irradiated (or not, in control experiments) and subjected to electron microscopic analyses. In the vast majority of lysates of tailed bacteriophages, apart from normal virions, those with empty (devoid of DNA) capsids occur at the levels from a few to several percent [27,28]; such “empty head” virions are called “phage ghosts” [29]. The “empty virions” may appear due to defects in virion assembly or as effects of some environmental conditions. For example, osmotic shock causes a significant increase in the fraction of phage ghosts in virion samples as a result of capsid leakiness and release of phage DNA [29]. We have chosen phages λ and Φ24B, in which no detectable and the most pronounced loss of infectivity and capsid damage were observed, respectively, for electron microscopic studies. In phage λ, UV irradiation at 50 J/m2 resulted in only a slight increase in the fraction of phage ghosts, with no other structural changes detected (Figure 3, Table 2). However, when Φ24B virions were UV irradiated, the fraction of phage ghosts increased considerably relative to non-irradiated samples, and a fraction of untypical virions, with partially destroyed heads, appeared (Figure 3, Table 2). Therefore, we conclude that UV irradiation at the dose of 50 J/m2 caused considerable changes in the structure of Φ24B capsid in a significant fraction of virions.
Figure 3.
Electron micrographs of virions of bacteriophage λ (upper panels) and Stx phage Φ24B (lower panels), either non-irradiated (left panels) or irradiated with UV light at 50 J/m2 (right panels). Untypical Φ24B virions with partially damaged heads are indicated by arrows in the lower right panel. Bars, shown at the lower right corner of each panel, correspond to 100 nm.
Table 2.
Summary of the electron microscopic analysis of λ and Φ24B virions, either non-irradiated or UV-irradiated at 50 J/m2.
| Bacteriophage and Conditions | Fractions of Different Kinds of Virions (%) | ||
|---|---|---|---|
| Normal | Phage Ghosts | Untypical, with Partially Damaged Heads | |
| λ papa (non-irradiated) | 93 | 7 | 0 |
| λ papa (UV-irradiated) | 89 | 11 | 0 |
| Φ24B (non-irradiated) | 91 | 9 | 0 |
| Φ24B (UV-irradiated) | 71 | 23 | 6 |
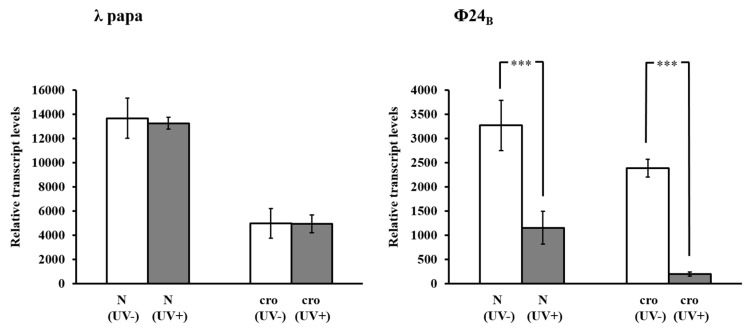
If our conclusion that decreased infectivity of UV-irradiated Stx phages, relative to untreated ones, results from a damage of capsids in a significant proportion of virions is true, one should expect that lower number of phages can introduce their DNA into host cells, and thus, level of expression of phage genes in a population of infected cells should be considerably lower than in E. coli infected with equal number of untreated phages. To test this hypothesis, a sample of the lysate of either λ or Φ24B virions was divided to two halves of equal titer (determined as pfu/mL). One half was UV irradiated at 50 J/m2 while the second was not, and then E. coli cells were infected with either treated or untreated phages. Following infection, total RNA was isolated, and expression of N and cro genes (transcribed from the pL and pR promoter, respectively), crucial for the lytic development [11,12,13,14], was estimated by reverse transcription real-time quantitative PCR (RT-qPCR). Analysis of phage gene expression (RNA level) instead of their copy numbers (DNA level) allowed to be devoid of signal becoming from a fraction of DNA released from capsids in response to UV exposure. In bacteriophage λ, UV irradiation of virions prior to infection did not result in any significant changes in expression of N and cro genes in the population of E. coli cells (Figure 4). On the contrary, in the E. coli population infected with irradiated Φ24B bacteriophage, the level of expression of both genes was significantly lower relative to bacteria infected with non-irradiated viruses (Figure 4).
Figure 4.
Expression of N and cro genes of bacteriophages λ (left panel) and Stx phage Φ24B (right panel) which were either non-irradiated (UV−) or irradiated with UV light at 50 J/m2 (UV+) prior to infection (at m.o.i. = 1). The mRNA levels were estimated in E. coli MG1655 host grown at 37 °C, at 7.5 min or 15 min after infection by λ or Φ24B, respectively. The presented results are mean values from three experiments with error bars indicating SD. Statistically significant differences between results obtained for non-irradiated and UV-irradiated phages (p < 0.001 in the t-test) are indicated by three asterisks.
All the results presented in this report indicate that Stx virions are more sensitive to UV irradiation than those of bacteriophage λ. The process of inactivation of bacteriophages with UV light is a known phenomenon, however, UV doses used by others to decrease infectivity of various phages, including Stx phages, were generally significantly higher than those employed in this work [30,31,32,33,34,35]. When high UV doses were employed, inactivation of bacteriophages was ascribed to DNA lesions caused by the irradiation [36]. However, in this study, at the dose of 50 J/m2, no inactivation of the model virus, bacteriophage λ, could be detected. Therefore, severe DNA damage is unlikely under these conditions, and other mechanisms had to be considered. The release of phage DNA from capsids (determined biochemically), as well as virion dysmorphology (identified in electron microscopic studies), indicated that damage of virions might be responsible for high UV sensitivity of Stx phages. This may have implications for the spreading of these phages and formation of new STEC strains. Although these processes might be effective in habitats devoid of UV, like in human or animal digestive tracts, one can expect their lower efficiency in other environments, outside of mammalian organism. It remains to be elucidated what structural properties of Stx virions make them labile upon UV irradiation at doses which do not cause any considerable damage of bacteriophage λ capsids.
3. Experimental Section
3.1. Bacteria and Bacteriophages
Escherichia coli MG1655 strain [37] and bacteriophages: λ papa (from our collection), Φ24B(∆stx2::cat) [38], 933W∆tox(∆stx2::catGFP), 22∆tox(∆stx2::catGFP), 27∆tox(∆stx2::catGFP), and 32∆tox(∆stx2::catGFP), referred to as Φ24B, 933W, P22, P27 and P32, respectively, in the text, were used in this work. Bacteriophages Φ24B, 933W, P22, P27 and P32 are Shiga toxin-converting phages. Φ24B and 933W have been used for many years as model phages from this group, while P22, P27 and P32 have been described as natural isolates [39]. Derivatives of the Shiga toxin-converting phages 933W, 22, 27, and 32 in which genomes the toxin genes were replaced by cat and/or GFP markers were described previously [39]. Bacteria were routinely cultured in the Luria-Bertani (LB) medium supplemented with 20 μg/mL chloramphenicol (if required) at 37 °C or 43 °C under aerobic conditions. Bacteriophage suspensions were stored in the TM buffer (10 mM Tris-HCl, 10 mM MgSO4, pH 7.2) (Sigma Aldrich, St. Louis, MO, USA) at 4 °C.
3.2. Evaluation of Bacterial Survival after Bacteriophage Infection
The percentage of survived cells after bacteriophage infection was estimated according to the protocol described in details previously [40], with slight modifications. Briefly, host bacteria were grown in LB medium at 37 °C to A600 = 0.2. Sample of 1 mL was withdrawn and centrifuged (2000× g, 10 min, 4 °C). The pellet was suspended in 1 mL of 0.85% NaCl. Then, the sample was centrifuged again and the pellet was suspended in 1.2 mL of LB medium supplemented with MgSO4 and CaCl2 (to a final concentration of 10 mM each). Following incubation for 30 min at 37 °C, the volume of analyzed bacteriophage lysate, corresponding to m.o.i. of 5, untreated or treated with UV light (employing the UV lamp provided by Vilber Lourmat, Marne-la-Vallée, France), at 50 J/m2, (the dose used routinely for lambdoid prophage induction [41,42]) was added, and then mixture was incubated at 37 °C for additional 30 min. Next, TM buffer (10 mM Tris–HCl, 10 mM MgSO4; pH 7.2) was used to prepare serial dilutions. The volume of 40 µL of each dilution was spread on LB agar plates prior to overnight incubation at 37 °C. Percentage of surviving bacteria was calculated relative to the control sample in which TM buffer was added instead of bacteriophage lysate. Each experiment was repeated three times.
3.3. Estimation of Phage Lysate Stability
Phage lysates were obtained after induction of the prophage from the host (E. coli MG1655 strain) as described previously [41], with minor modifications. Briefly, lysogenic bacteria were grown in LB medium at 37 °C, in shake flasks with agitation to an A600 of 0.1. Prophage induction was provoked by mitomycin C which was added to final concentration of 1 μg/mL. After induction of phage lytic development, the culture was incubated for additional 6 to 10 h. Following 15 min agitation with chloroform (added in the volume of 600 μL per 10 mL), the culture was centrifuged (2000× g for 10 min at 4 °C) to remove bacterial debris, and supernatant was filtered using 0.22-μm-pore-size filters (Sigma Aldrich, St. Louis, MO, USA ). Effect of UV irradiation on the phage lysates stability was determined simultaneously at 37 °C or 43 °C, using phage titration method performed according to the standard double agar overlay technique. At times 0 (before UV exposure) and 0.2, 1, 2, 3, 4, 5, 6 h after UV irradiation at 50 J/m2, samples of 0.5 mL were withdrawn, and prepared and titrated as described previously [42]. The phage titer, expressed as plaque forming units (PFU) per mL, was determined. For each phage lysate, three replicates were performed.
3.4. Measurement of Relative DNA Contents
Phage lysates were prepared as described above. Following UV exposure (50 J/m2), one half of the phage lysate was incubated at 37 °C and the second half at 43 °C. Effect of UV irradiation on the amount of DNA released from capsids was determined simultaneously at both temperatures. Before (time zero) and after (the remaining times) exposure of phage lysate to UV light, DNA was quantified by staining with Qubit® dsDNA BR Assay Kit (Invitrogen/Thermo Fisher Scientific, Waltham, MA USA), according to the manufacturer’s protocol. At indicated time points after UV exposure, DNA contents were calculated relative to the DNA amount obtained at time zero which represented value of 1. Each experiment was repeated three times.
3.5. Electron Microscopy
Virions were purified from phage lysates obtained after induction of the prophage from the host E. coli MG1655 strain with mitomycin C, as described above. Purification was performed using cesium chloride density gradient centrifugation method [43]. Electron microscopic analyses of purified phage particles (untreated and treated with UV light at 50 J/m2) were performed employing the Philips CM 100 electron microscope (Philips, Eindhoven, The Netherlands), by using negative staining with uranyl acetate, as described earlier [44].
3.6. Bacteriophage Infection and cDNA Sample Preparation
The experiments were performed according to the protocol described previously [45], with slight modifications. Briefly, E. coli bacteria were cultured to A600 of 0.3 at 37 °C. Then, culture volume of 60 mL was centrifuged and the pallet was washed with 30 mL of 0.85% NaCl. Following additional centrifugation, the sample was suspended in 15 mL of LB medium enriched with MgSO4 and CaCl2 (to a final concentration of 10 mM each). After incubation at 37 °C for 30 min, the sample was chilled on ice. The appropriate volume of analyzed bacteriophage lysate, corresponding to m.o.i of 1 (treated or untreated with UV light at 50 J/m2) was added, and the mixture was incubated on ice. Next, infected bacterial cells were aerated in a water bath shaker at 37 °C. At indicated times (7.5 min for λ and 15 min for Φ24B after phage infection as these phages have various development rates [45]), 1 × 109 cells were treated with NaN3 (Sigma-Aldrich, St. Louis, MO, USA) at final concentration of 10 mM, and harvested. The RNA and cDNA samples for real-time PCR assay were prepared as described previously [40,46] using the High Pure RNA Isolation Kit and Transcriptor Reverse Transcriptase (Roche Applied Science, Basel, Switzerland).
3.7. Real-Time PCR Assay and Data Analysis
The level of N and cro phage genes expression was determined by quantitative real-time reverse transcription-PCR (RT-qPCR) using the LightCycler® 480 Real-Time PCR System (Roche Applied Science) and cDNA samples obtained during bacteriophage infection experiment. According to a procedure described previously [47], transcription rates of tested phage genes were compared in parallel to the icdA bacterial housekeeping gene. Expression of this gene was found to be stable also upon λ or Φ24B phage infection of E. coli MG1655 strain [40,45]. The gene expression analyses were performed using LightCycler® 480 SYBR Green I Master and following primers: pF_λ_N (5′-CTC GTG ATT TCG GTT TGC GA); pR_λ_N (5′-AAG CAG CAA ATC CCC TGT TG); pF_λ_cro (5′-ATG CGG AAG AGG TAA AGC CC); pR_λ_cro (5′-TGG AAT GTG TAA GAG CGG GG); pF_Φ24B_N (5′-AGG CGT TTC GTG AGT ACC TT); pR_Φ24B_N (5′-TTA CAC CGC CCT ACT CTA AGC); pF_Φ24B_cro (5′-CGA AGG CTT GTG GAG TTA GC); pR_ Φ24B_cro (5′-GTC TTA GGG AGG AAG CCG TT); pF_icdA (5′-CGA AGC GGC TGA CCT TAA TTG) and pR_icdA (5′-GTT ACG GTT TTC GCG TTG AT). The reaction components and amplification program were exactly the same as described previously [45]. No template control was included with each run. Gel electrophoresis and melting curve analysis were performed to evaluate the specificity of every amplification reaction. Each experiment was conducted in triplicate. The relative changes in gene expression revealed by quantitative Real-Time PCR experiments were analyzed using the E-Method (with efficiency correction) as described in detail previously [40,45,46].
Acknowledgments
This work was supported by National Science Center, Poland (grant no. 2013/09/B/NZ2/02366 to AW).
Author Contributions
Sylwia Bloch and Bożena Nejman-Faleńczyk have contributed to the conception and the design of the study, they estimated the stability of phage lysates, measured the relative DNA contents, performed RT-qPCR assays after bacteriophage infection, and analyzed and interpreted the results of these experiments. Gracja Topka and Aleksandra Dydecka prepared phage lysates and estimated the percentage of survival of cells after bacteriophage infection. Katarzyna Licznerska and Agnieszka Necel assisted in analyzing the results of experiments and took part in preparation of the phage lysates. Magdalena Narajczyk prepared samples and made microphotographs for electron microscopy analyses. Alicja Węgrzyn was a principal investigator in the project grant, and supervised the study. Grzegorz Węgrzyn participated in planning the experiments and interpretation of their results, and drafted the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gyles C.L. Shiga toxin-producing Escherichia coli: An overview. J. Animal Sci. 2007;6(Suppl. 13):45–62. doi: 10.2527/jas.2006-508. [DOI] [PubMed] [Google Scholar]
- 2.Hunt J.M. Shiga toxin-producing Escherichia coli (STEC) Clin. Lab. Med. 2010;30:21–45. doi: 10.1016/j.cll.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obrig T.G., Moran T.P., Brown J.E. The mode of action of Shiga toxin on peptide elongation of eukaryotic protein synthesis. Biochem. J. 1987;244:287–294. doi: 10.1042/bj2440287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Endo Y., Tsurugi K., Yutsudo T., Takeda Y., Ogasawara T., Igarashi K. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 1988;171:45–50. doi: 10.1111/j.1432-1033.1988.tb13756.x. [DOI] [PubMed] [Google Scholar]
- 5.Razzaq S. Hemolytic uremic syndrome: An emerging health risk. Am. Fam. Physician. 2006;74:991–996. [PubMed] [Google Scholar]
- 6.Bloch S.K., Felczykowska A., Nejman-Faleńczyk B. Escherichia coli O104:H4 outbreak-have we learnt a lesson from it? Acta Biochim. Pol. 2012;59:483–488. [PubMed] [Google Scholar]
- 7.Karch H., Denamur E., Dobrindt U., Finlay B.B., Hengge R., Johannes L., Ron E.Z., Tønjum T., Sansonetti P.J., Vicente M. The enemy within us: Lessons from the 2011 European Escherichia coli O104:H4 outbreak. EMBO Mol. Med. 2012;4:841–848. doi: 10.1002/emmm.201201662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werber D., Krause G., Frank C., Fruth A., Flieger A., Mielke M., Schaade L., Stark K. Outbreaks of virulent diarrheagenic Escherichia coli—Are we in control? BMC Med. 2012;10 doi: 10.1186/1741-7015-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allison H.E. Stx-phages: Drivers and mediators of the evolution of STEC and STEC-like pathogens. Future Microbiol. 2007;2:165–174. doi: 10.2217/17460913.2.2.165. [DOI] [PubMed] [Google Scholar]
- 10.Łoś J.M., Łoś M., Węgrzyn G. Bacteriophages carrying Shiga toxin genes: Genomic variations, detection and potential treatment of pathogenic bacteria. Future Microbiol. 2011;6:909–924. doi: 10.2217/fmb.11.70. [DOI] [PubMed] [Google Scholar]
- 11.Ptashne M. A Genetic Switch: Phage Lambda Revisited. 3rd ed. Cold Spring Harbor Laboratory Press; New York, NY, USA: 2004. pp. 1–154. [Google Scholar]
- 12.Węgrzyn G., Węgrzyn A. Genetic switches during bacteriophage lambda development. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:1–48. doi: 10.1016/S0079-6603(04)79001-7. [DOI] [PubMed] [Google Scholar]
- 13.Węgrzyn G., Licznerska K., Węgrzyn A. Phage λ-new insights into regulatory circuits. Adv. Virus Res. 2012;82:155–178. doi: 10.1016/B978-0-12-394621-8.00016-9. [DOI] [PubMed] [Google Scholar]
- 14.Łoś J.M., Łoś M., Węgrzyn A., Węgrzyn G. Altruism of Shiga toxin-producing Escherichia coli: Recent hypothesis versus experimental results. Front. Cell. Infect. Microbiol. 2013;2 doi: 10.3389/fcimb.2012.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riley L.M., Veses-Garcia M., Hillman J.D., Handfield M., McCarthy A.J., Allison H.E. Identification of genes expressed in cultures of E. coli lysogens carrying the Shiga toxin-encoding prophage Φ24B. BMC Microbiol. 2012;12 doi: 10.1186/1471-2180-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imamovic L., Muniesa M. Characterizing RecA-independent induction of Shiga toxin2-encoding phages by EDTA treatment. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0032393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith D.L., James C.E., Sergeant M.J., Yaxian Y., Saunders J.R., McCarthy A.J., Allison H.E. Short-tailed Stx phages exploit the conserved YaeT protein to disseminate Shiga toxin genes among enterobacteria. J. Bacteriol. 2007;189:7223–7233. doi: 10.1128/JB.00824-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yue W.F., Du M., Zhu M.J. High temperature in combination with UV irradiation enhances horizontal transfer of stx2 gene from E. coli O157:H7 to non-pathogenic E. coli. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0031308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muniesa M., Jofre J. Abundance in sewage of bacteriophages that infect Escherichia coli O157:H7 and that carry the Shiga toxin 2 gene. Appl. Environ. Microbiol. 1998;64:2443–2448. doi: 10.1128/aem.64.7.2443-2448.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muniesa M., Lucena F., Jofre J. Comparative survival of free Shiga toxin 2-encoding phages and Escherichia coli strains outside the gut. Appl. Environ. Microbiol. 1999;65:5615–5618. doi: 10.1128/aem.65.12.5615-5618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johannessen G.S., James C.E., Allison H.E., Smith D.L., Saunders J.R., McCarthy A.J. Survival of a Shiga toxin-encoding bacteriophage in a compost model. FEMS Microbiol. Lett. 2005;245:369–375. doi: 10.1016/j.femsle.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 22.Dumke R., Schröter-Bobsin U., Jacobs E., Röske I. Detection of phages carrying the Shiga toxin 1 and 2 genes in waste water and river water samples. Lett. Appl. Microbiol. 2006;42:48–53. doi: 10.1111/j.1472-765X.2005.01809.x. [DOI] [PubMed] [Google Scholar]
- 23.Imamovic L., Ballesté E., Jofre J., Muniesa M. Quantification of Shiga toxin-converting bacteriophages in wastewater and in fecal samples by real-time quantitative PCR. Appl. Environ. Microbiol. 2010;76:5693–5701. doi: 10.1128/AEM.00107-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rooks D.J., Yan Y., McDonald J.E., Woodward M.J., McCarthy A.J., Allison H.E. Development and validation of a qPCR-based method for quantifying Shiga toxin-encoding and other lambdoid bacteriophages. Environ. Microbiol. 2010;12:1194–1204. doi: 10.1111/j.1462-2920.2010.02162.x. [DOI] [PubMed] [Google Scholar]
- 25.Imamovic L., Muniesa M. Quantification and evaluation of infectivity of Shiga toxin-encoding bacteriophages in beef and salad. Appl. Environ. Microbiol. 2011;77:3536–3540. doi: 10.1128/AEM.02703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imamovic L., Serra-Moreno R., Jofre J., Muniesa M. Quantification of Shiga toxin 2-encoding bacteriophages, by real-time PCR and correlation with phage infectivity. J. Appl. Microbiol. 2010;108:1105–1114. doi: 10.1111/j.1365-2672.2010.04664.x. [DOI] [PubMed] [Google Scholar]
- 27.Ackermann H.W. Bacteriophage electron microscopy. Adv. Virus Res. 2012;82:1–32. doi: 10.1016/B978-0-12-394621-8.00017-0. [DOI] [PubMed] [Google Scholar]
- 28.Ackermann H.W., Prangishvili D. Prokaryote viruses studied by electron microscopy. Arch. Virol. 2012;157:1843–1849. doi: 10.1007/s00705-012-1383-y. [DOI] [PubMed] [Google Scholar]
- 29.Duckworth D.H. Biological activity of bacteriophage ghosts and “take-over” of host functions by bacteriophage. Bacteriol. Rev. 1970;34:344–363. doi: 10.1128/br.34.3.344-363.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allué-Guardia A., Jofre J., Muniesa M. Stability and infectivity of cytolethal distending toxin type V gene-carrying bacteriophages in a water mesocosm and under different inactivation conditions. Appl. Environ. Microbiol. 2012;78:5818–5823. doi: 10.1128/AEM.00997-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark E.M., Wright H., Lennon K.A., Craik V.A., Clark J.R., March J.B. Inactivation of recombinant bacteriophages lambda by use of chemical agents and UV radiation. Appl. Environ. Microbiol. 2012;78:3033–3036. doi: 10.1128/AEM.06800-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Theitler D.J., Nasser A., Gerchman Y., Kribus A., Mamane H. Synergistic effect of heat and solar UV on DNA damage and water disinfection of E. coli and bacteriophages MS2. J. Water Health. 2012;10:605–618. doi: 10.2166/wh.2012.072. [DOI] [PubMed] [Google Scholar]
- 33.Guglielmotti D.M., Mercanti D.J., Reinheimer J.A., Quiberoni Adel L. Review: Efficiency of physical and chemical treatments on the inactivation of dairy bacteriophages. Front. Microbiol. 2012;2 doi: 10.3389/fmicb.2011.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allué-Guardia A., Martínez-Castillo A., Muniesa M. Persistence of infectious Shiga toxin-encoding bacteriophages after disinfection treatments. Appl. Environ. Microbiol. 2014;80:2142–2149. doi: 10.1128/AEM.04006-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diston D., Ebdon J.E., Taylor H.D. Inactivation of bacteriophages infecting Bacteroides strain GB124 using UV-B radiation. Photochem. Photobiol. 2014;90:622–627. doi: 10.1111/php.12223. [DOI] [PubMed] [Google Scholar]
- 36.Wigginton K.R. Virus inactivation mechanisms: Impact of disinfectants on virus function and structural integrity. Environ. Sci. Technol. 2012;46:12069–12078. doi: 10.1021/es3029473. [DOI] [PubMed] [Google Scholar]
- 37.Jensen K.F. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allison H.E., Sergeant M.J., James C.E., Saunders J.R., Smith D.L., Sharp R.J., Marks T.S., McCarthy A.J. Immunity profiles of wild-type and recombinant Shiga-like toxin-encoding bacteriophages and characterization of novel double lysogens. Infect. Immun. 2003;71:3409–3418. doi: 10.1128/IAI.71.6.3409-3418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gamage S.D., Patton A.K., Hanson J.F., Weiss A.A. Diversity and host range of Shiga toxin-encoding phage. Infect. Immun. 2004;72:7131–7139. doi: 10.1128/IAI.72.12.7131-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nejman-Faleńczyk B., Bloch S., Licznerska K., Dydecka A., Felczykowska A., Topka G., Węgrzyn A., Węgrzyn G. A small, microRNA-size, ribonucleic acid regulating gene expression and development of Shiga toxin-converting bacteriophage Φ24Β. Sci. Rep. 2015;5 doi: 10.1038/srep10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nowicki D., Kobiela W., Węgrzyn A., Wegrzyn G., Szalewska-Pałasz A. ppGpp-dependent negative control of DNA replication of Shiga toxin-converting bacteriophages in Escherichia coli. J. Bacteriol. 2013;195:5007–5015. doi: 10.1128/JB.00592-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bloch S., Nejman-Faleńczyk B., Łoś J.M., Barańska S., Łepek K., Felczykowska A., Łoś M., Węgrzyn G., Węgrzyn A. Genes from the exo-xis region of λ and Shiga toxin-converting bacteriophages influence lysogenization and prophage induction. Arch. Microbiol. 2013;195:693–703. doi: 10.1007/s00203-013-0920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J., Russell D.W. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; New York, NY, USA: 2001. [Google Scholar]
- 44.Czajkowski R., Ozymko Z., de Jager V., Siwinska J., Smolarska A., Ossowicki A., Narajczyk M., Łojkowska E. Genomic, proteomic and morphological characterization of two novel broad host lytic bacteriophages ΦPD10.3 and ΦPD23.1 infecting pectinolytic Pectobacterium. spp. and Dickeya. spp. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0119812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bloch S., Nejman-Faleńczyk B., Dydecka A., Łoś J.M., Felczykowska A., Węgrzyn A., Węgrzyn G. Different expression patterns of genes from the exo-xis region of bacteriophage λ and Shiga toxin-converting bacteriophage Ф24B following infection or prophage induction in Escherichia coli. PLoS ONE. 2014;13 doi: 10.1371/journal.pone.0108233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nowicki D., Bloch S., Nejman-Faleńczyk B., Szalewska-Palasz A., Węgrzyn A., Węgrzyn G. Defects in RNA polyadenylation impair both lysogenization by and lytic development of Shiga toxin-converting bacteriophages. J. Gen. Virol. 2015;96:1957–1968. doi: 10.1099/vir.0.000102. [DOI] [PubMed] [Google Scholar]
- 47.Strauch E., Hammerl J.A., Konietzny A., Schneiker-Bekel S., Arnold W., Goesmann A., Pühler A., Beutin L. Bacteriophage 2851 is a prototype phage for dissemination of the Shiga toxin variant gene 2c in Escherichia coli O157:H7. Infect. Immun. 2008;76:5466–5477. doi: 10.1128/IAI.00875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]