Abstract
Fungal polyketides are a diverse class of natural products, or secondary metabolites (SMs), with a wide range of bioactivities often associated with toxicity. Here, we focus on a group of non-reducing polyketide synthases (NR-PKSs) in the fungal phylum Ascomycota that lack a thioesterase domain for product release, group V. Although widespread in ascomycete taxa, this group of NR-PKSs is notably absent in the mycotoxigenic genus Fusarium and, surprisingly, found in genera not known for their secondary metabolite production (e.g., the mycorrhizal genus Oidiodendron, the powdery mildew genus Blumeria, and the causative agent of white-nose syndrome in bats, Pseudogymnoascus destructans). This group of NR-PKSs, in association with the other enzymes encoded by their gene clusters, produces a variety of different chemical classes including naphthacenediones, anthraquinones, benzophenones, grisandienes, and diphenyl ethers. We discuss the modification of and transitions between these chemical classes, the requisite enzymes, and the evolution of the SM gene clusters that encode them. Integrating this information, we predict the likely products of related but uncharacterized SM clusters, and we speculate upon the utility of these classes of SMs as virulence factors or chemical defenses to various plant, animal, and insect pathogens, as well as mutualistic fungi.
Keywords: mycotoxin, gene cluster, evolution, polyketide synthase
1. Introduction
The Kingdom Fungi is well known for its ability to synthesize bioactive secondary metabolites (SMs), also known as natural products. The dominant taxa producing SMs belong to several filamentous ascomycete genera, many of which are renowned plant, insect, and/or human pathogens. Virulence is often associated with the production of toxic SMs in these fungi [1]. In nature, where studied, SMs afford various fitness advantages to the producing species ranging from protection from fungivory and physical insults (e.g., UV light) to competition with other microbes for niche securement [2,3,4].
SMs can be classified according to chemical types. For example, polyketides are derived from acetyl/malonyl-coenzyme A (CoA), non-ribosomal peptides from amino acids, and terpenes from isoprene. The reader is referred to several recent reviews for in-depth coverage of each chemical class [5,6,7]. Each class is associated with a defining synthase, polyketide synthase (PKS), non-ribosomal peptide synthetase (NRPS), and terpene cyclase/synthase, respectively. Ribosomal peptides [8] and hybrid synthases, commonly hybrid PKS-NRPS enzymes, have also been described. The synthases contain conserved catalytic domains easily detectable by statistical analysis, i.e., hidden Markov models, thus making them highly suitable for phylogenetic analyses and evolutionary inferences [9,10,11]. Typically, the genes encoding these synthases are physically clustered with additional enzymatic genes required to form the end metabolite; these are termed SM gene clusters.
Polyketides in particular have drawn considerable interest due to their impact on human and plant health both positively (e.g., lovastatin) and detrimentally (e.g., aflatoxin). The first fungal PKS to be identified and characterized was 6-methylsalicylic acid synthase, found in several Penicillium and Aspergillus spp. [12,13,14]. Shortly thereafter, PKSs required for spore pigmentation as well as the mycotoxins aflatoxin and sterigmatocystin were characterized in several Aspergillus spp. [15,16,17,18]. Additional PKSs involved in toxin synthesis include Pks1 and Pks2 (T toxin in Cochliobolus heterostrophus, [19,20]), FusA (fusarin production in Fusarium fujikuroi, [21] and F. verticillioides, F. venenatum [22]), Zea1 and Zea2 (zearalenone production in F. graminearum, [23,24]), Fum1 (fumonisin production in F. verticillioides, F. fujikuroi, [25,26]), Pks-CT (citrinin production in Monascus purpureus, [27]), and NhPKS1 (bostrycoidin and fusarubin in F. solani, [28]). PKSs and hybrid PKS-NRPSs have also been associated with the development of fungal spores and overwintering structures such as sclerotia [29,30,31].
Most fungal PKSs are multi-functional enzymes known as iterative type I PKSs, where each catalytic domain is encoded in one gene, though a few fungal PKSs are of type III [32]. There are two main classes of type I PKS known as non-reducing (NR) and highly reducing (HR). All PKSs harbor three essential domains—the β-ketoacyl synthase (KS), malonyl-CoA:acyl carrier protein transacylase (MAT), and acyl carrier protein (ACP) domains—however, NR- and HR-PKSs vary in their catalytic domains that impact the reduction or dehydration steps of the growing carbon chain (Figure 1). The minimal architecture of HR-PKSs is typically composed of ketoreductase (KR), dehydratase (DH), and enoyl reductase (ER) domains, thereby resembling fatty acid synthases (FASs). Another key difference is the presence of a C-methyltransferase (CMeT) domain found in most HR-PKSs and only in one subset of NR-PKSs (e.g., A. nidulans AfoE, [33]). However, despite the presence of CMeT domains in HR-PKSs, analysis in Fusarium spp. suggests that the domain can be non-functional [34].
Figure 1.
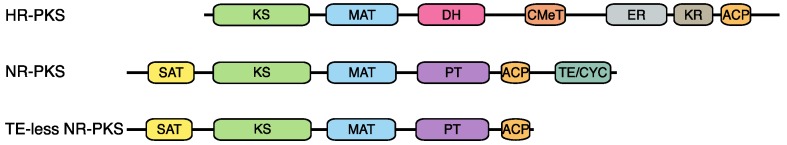
A diagram showing the domains of typical highly reducing (HR), non-reducing (NR), and group V NR type I fungal polyketide synthases. SAT = starter-unit:ACP transacylase, KS = β-ketoacyl synthase, MAT = malonyl-CoA:ACP transacylase, DH = dehydratase, CMeT = C-methyltransferase, ER = enoyl reductase, KR = ketoreductase, PT = product template, ACP = acyl carrier protein, TE = thioesterase, CYC = Claisen cyclase.
NR-PKSs are characterized by the absence of KR, DH, and ER domains and the presence of starter-unit:ACP transacylase (SAT) and product template (PT) domains. While N-terminal SAT domains of most NR-PKSs accept acetyl-CoA as starter unit [35] there are certain examples where the starter unit is either provided by dedicated FAS-like proteins [36] or HR-PKSs [37] which are usually encoded by genes located within the co-regulated cluster region. While the KS domain largely controls chain length [38], the PT domain, which is always located between the MAT and ACP domains, determines the cyclization pattern of the polyketide product [39]. The generated cyclized products of NR-PKSs are released by a variety of mechanisms, that in some cases contribute to cyclization patterns as well [40]. The thioesterase (TE)-mediated product release by a canonical TE domain is the most common mechanism (e.g., orsellinic acid/F9775 [41,42,43]). This domain often extends to a domain capable of C–C Claisen cyclization (a TE/CYC domain) (e.g., aflatoxin and sterigmatocystin PKSs and many pigment PKSs, [44,45]). In some cases, NR-PKSs contain a reductase-releasing (R) domain (e.g., asperfuranone [33]); in others, e.g., the PKS involved in asperthecin biosynthesis in A. nidulans, there is no releasing domain. These latter NR-PKSs are usually coupled with metallo-β-lactamase-type TE proteins (MβL-TE) that allow for release of the nascent polyketide chain. These MβL-TEs are encoded by distinct genes located within their respective gene clusters, as illustrated for asperthecin (AptB), endocrocin (EncB), viridicatumtoxin (VrtG), TAN-1612 (AdaB), geodin (GedB), monodictyphenone (MdpF), pestheic acid (PtaB), trypacidin (TpcB), and neosartoricin (NscB) [46,47,48,49,50,51,52,53]. Here we conducted a phylogenetic analysis of PKS genes from currently available fungal genomes, with a focus on the TE-less NR-PKSs, group V. Our data strengthen predictions of subdivisions within this group, and, moreover, we expand upon the predictive power of this analysis to suggest directions for future research.
2. Group V NR-PKS Phylogeny
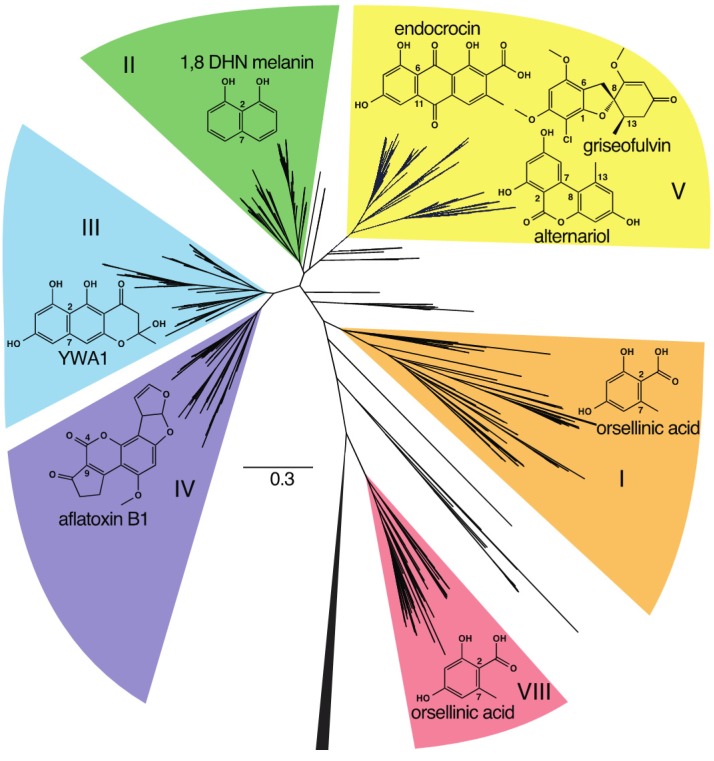
Previous phylogenetic analyses have used the whole PKS, the KS domain, or the PT domain for comparison, but these have been noted to reflect one another, indicating their coevolution [54]. These studies classified the NR-PKSs first into three subclades [55], then further into five [56], seven [54], and most recently eight groups [57]. These NR-PKSs are present in many ascomycetes and some basidiomycetes, though only members of group VIII have been identified in basidiomycetes [57] with only one characterized example to date [58]. These groups generally represent unique combinations of product length and cyclization register. Of the eight current groups, we focus on NR-PKSs belonging to the TE-less group V in this study (Figure 2). Group V has thirteen characterized gene clusters with eleven described products including endocrocin, monodictyphenone, trypacidin, geodin, pestheic acid, asperthecin, TAN-1612, neosartoricin, viridicatumtoxin, griseofulvin, and alternariol (Table 1, Figure 3). All group V PKSs lack a domain for product release, but they vary in the length (hepta- to decaketide) of their products; most make C6–C11 connections, but a couple of exceptional cases make C1–C6 or C2–C7 connections. Generally, the thioesterase activity is encoded in a separate but adjacent MβL-TE. Based on our analysis of the KS domains of 908 fungal PKSs identified through the NCBI’s BLAST utility and manually using AspGD (Figure S1), we have identified 188 PKSs belonging to group V (Figure S2). These PKSs are derived from 88 species of 39 genera distributed across five classes of ascomycetes (Table 1). Notably, no group V PKSs were identified in the well characterized Fusarium spp. [59]. This phylogenetic tree allowed us to visualize relationships between these unknown PKSs and the thirteen examples, thus defining the best contexts in which to deduce the functions of the putative clusters to which these PKSs belong.
Figure 2.
A maximum likelihood phylogenetic tree constructed with FastTree [60] using the KS domains of 908 NR-PKSs. Clades corresponding to characterized groups I–VIII are highlighted and labeled. No members of groups VI and VII were identified by our methods. Examples of structures produced by each group are shown in their respective highlighted regions and the numbering of the carbon-carbon bonds indicates the mode of cyclization of the PKSs that produce these compounds. The outgroup, consisting of the KS domains of 70 HR- and hybrid PKS/NRPSs, as well as the human fatty acid synthase, FASN, is collapsed.
Table 1.
List of the classes, genera, and species with PKSs belonging to group V, their accession numbers, their subgroup, characterized products, and predicted products. These predictions are based on the presence or absence of homologs of the requisite genes to produce these compounds as determined by previous studies of group V SM gene clusters. These requisite genes are discussed in the sections corresponding to either the relevant group (e.g., V1, V2, and V3) or compound (e.g., alternariol or viridicatumtoxin).
| Class | Genus | Species | PKS | Sub-group | Characterized product | Predicted product | Reference |
|---|---|---|---|---|---|---|---|
| Dothidiomycete | Alternaria | alternata | AFN68300 | V3 | Alternariol | Characterized | [61] |
| Aureobasidium | melanogenum | KEQ66138 | V1 | None | Emodin anthrone | This study | |
| Bipolaris | maydis | EMD89515 | V3 | None | No data | This study | |
| ENH99769 | V3 | None | No data | This study | |||
| ENI07798 | V1 | None | Other anthrone | This study | |||
| AAR90273 | V1 | None | No data | This study | |||
| AAR90274 | V3 | None | No data | This study | |||
| oryzae | EUC41199 | V1 | None | Benzophenone | This study | ||
| EUC45162 | V1 | None | Other anthrone | This study | |||
| sorokiniana | EMD62925 | V1 | None | Other anthrone | This study | ||
| EMD66882 | V1 | None | Benzophenone | This study | |||
| victoriae | EUN25734 | V1 | None | Other anthrone | This study | ||
| zeicola | EUC29913 | V1 | None | Other anthrone | This study | ||
| Macrophomina | phaseolina | EKG11397 | V1 | None | No product | This study | |
| EKG18431 | V1 | None | No data | This study | |||
| Phaeosphaeria | nodorum | EAT76667 | V3 | Alternariol | Characterized | [62] | |
| Pseudocercospora | fijiensis | EME79056 | V1 | None | Emodin | This study | |
| Pseudogymnoascus | destructans | ELR08155 | V1 | None | Endocrocin anthrone | This study | |
| pannorum | KFY01830 | V1 | None | Endocrocin anthrone | This study | ||
| KFY04191 | V1 | None | Other anthrone | This study | |||
| KFY04767 | V1 | None | Endocrocin anthrone | This study | |||
| KFY14212 | V1 | None | Emodin anthrone | This study | |||
| KFY24933 | V1 | None | Endocrocin anthrone | This study | |||
| KFY28347 | V1 | None | Endocrocin anthrone | This study | |||
| KFY28376 | V1 | None | Endocrocin anthrone | This study | |||
| KFY41668 | V1 | None | Emodin anthrone | This study | |||
| KFY50141 | V1 | None | Grisandiene | This study | |||
| KFY61750 | V1 | None | Endocrocin anthrone | This study | |||
| KFY73941 | V1 | None | Endocrocin anthrone | This study | |||
| KFY81274 | V1 | None | Other anthrone | This study | |||
| KFY90954 | V1 | None | Other anthrone | This study | |||
| KFY97098 | V1 | None | Endocrocin anthrone | This study | |||
| KFZ02364 | V1 | None | Endocrocin anthrone | This study | |||
| KFZ03027 | V1 | None | Other anthrone | This study | |||
| KFZ03785 | V1 | None | Endocrocin anthrone | This study | |||
| KFZ09857 | V1 | None | Endocrocin anthrone | This study | |||
| Pyrenophora | teres | EFQ95560 | V1 | None | Endocrocin anthrone | This study | |
| tritici | EDU45231 | V1 | None | Endocrocin anthrone | This study | ||
| Setosphaeria | turcica | EOA88807 | V1 | None | Other anthrone | This study | |
| Sphaerulina | musiva | EMF17386 | V1 | None | Other anthrone | This study | |
| Verruconis | gallopava | KIW05310 | V1 | None | No product | This study | |
| Eurotiomycete | Arthroderma | benhamiae | EFE32713 | V2 | None | Neosartoricin, fumicycline | This study |
| otae | EEQ30779 | V2 | None | Neosartoricin, fumicycline | This study | ||
| EEQ31623 | V3 | None | Alternariol | This study | |||
| Aspergillus | acidus | Aspfo1_0068040 | V2 | None | TAN-1612 | This study | |
| Aspfo1_0069798 | V1 | None | No data | This study | |||
| aculeatus | Aacu16872_063282 | V1 | None | No data | This study | ||
| Aacu16872_063333 | V1 | None | No data | This study | |||
| brasiliensis | Aspbr1_0070836 | V1 | None | No data | This study | ||
| Aspbr1_0071307 | V2 | None | TAN-1612 | This study | |||
| clavatus | EAW13612 | V1 | None | No data | This study | ||
| flavus | EED53479 | V1 | None | No data | This study | ||
| KJJ30826 | V3 | None | No data | This study | |||
| fumigatus | EAL84397 | V1 | Endocrocin | Characterized | [47] | ||
| EAL84875 | V2 | Neosartoricin, fumicycline | Characterized | [49,63] | |||
| EAL89339 | V1 | Trypacidin | Characterized | [53] | |||
| EDP47078 | V1 | Endocrocin | Characterized | [47] | |||
| EDP47964 | V2 | None | Neosartoricin, fumicycline | This study | |||
| EDP50840 | V1 | None | Trypacidin | This study | |||
| KEY78897 | V1 | None | Endocrocin | This study | |||
| KEY82310 | V1 | None | Other anthraquinone | This study | |||
| KEY82351 | V2 | None | Neosartoricin, fumicycline | This study | |||
| glaucus | Aspgl1_0045725 | V1 | None | No data | This study | ||
| kawachii | GAA85937 | V2 | None | TAN-1612 | This study | ||
| GAA88581 | V1 | None | No data | This study | |||
| nidulans | CBF70387 | V2 | Asperthecin | Characterized | [46] | ||
| CBF79143 | V3 | Alternariol | Characterized | [54] | |||
| CBF90097 | V1 | Monodictyphenone | Characterized | [51] | |||
| niger | EHA20150 | V1 | None | No data | This study | ||
| XP_001402309 | V1 | None | No product | This study | |||
| AEN83889 | V2 | TAN-1612 | Characterized | [64] | |||
| CAK40778 | V2 | None | TAN-1612 | This study | |||
| CAK47960 | V1 | None | No data | This study | |||
| ochraceoroseus | KKK15179 | V1 | None | Emodin | This study | ||
| KKK17199 | V1 | None | No data | This study | |||
| oryzae | BAE58990 | V1 | None | No data | This study | ||
| BAE62229 | V3 | None | Alternariol | This study | |||
| KDE80734 | V3 | None | Alternariol | This study | |||
| KDE81226 | V1 | None | No data | This study | |||
| XP_001820992 | V1 | None | No data | This study | |||
| parasiticus | KJK64046 | V3 | None | No data | This study | ||
| rambellii | KKK15908 | V1 | None | No data | This study | ||
| KKK27047 | V1 | None | Other anthraquinone | This study | |||
| ruber | EYE98259 | V1 | None | Endocrocin anthrone | This study | ||
| sydowii | Aspsy1_0090693 | V1 | None | No data | This study | ||
| Aspsy1_0144848 | V1 | None | No data | This study | |||
| Aspsy1_0151845 | V2 | None | Asperthecin | This study | |||
| Aspsy1_0157033 | V2 | None | Neosartoricin, fumicycline | This study | |||
| Aspsy1_1049255 | V1 | None | No data | This study | |||
| terreus | EAU31624 | V1 | Geodin | Characterized | [65,66] | ||
| EAU37396 | V1 | None | No data | This study | |||
| BAB88752 | V1 | None | No data | This study | |||
| tubingensis | Asptu1_0059858 | V2 | None | No data | This study | ||
| Asptu1_0123892 | V2 | None | TAN-1612 | This study | |||
| ustus | KIA75323 | V1 | None | Benzophenone | This study | ||
| KIA75530 | V2 | None | Asperthecin | This study | |||
| KIA75835 | V2 | None | Viridicatumtoxin-like naphthacenedione | This study | |||
| versicolor | Aspve1_0089706 | V2 | None | Neosartoricin, fumicycline | This study | ||
| Aspve1_0122449 | V1 | None | No data | This study | |||
| Aspve1_0886277 | V2 | None | Asperthecin | This study | |||
| wentii | Aspwe1_0034272 | V1 | None | No data | This study | ||
| zonatus | Aspzo1_2112764 | V1 | None | No data | This study | ||
| Capronia | epimyces | EXJ89638 | V1 | None | Other anthrone | This study | |
| Cladophialophora | carrionii | ETI19899 | V1 | None | Benzophenone | This study | |
| yegresii | EXJ61970 | V1 | None | Benzophenone | This study | ||
| Endocarpon | pusillum | ERF75912 | V1 | None | No product | This study | |
| Microsporum | gypseum | EFR03594 | V2 | None | Neosartoricin, fumicycline | This study | |
| Neosartorya | fischeri | EAW20700 | V2 | Neosartoricin, fumicycline | Characterized | [49,63] | |
| EAW24682 | V1 | None | Benzophenone | This study | |||
| EAW24697 | V1 | None | Grisandiene | This study | |||
| EAW25724 | V1 | None | No data | This study | |||
| Penicillium | aethiopicum | ADI24926 | V2 | Viridicatumtoxin | Characterized | [48] | |
| ADI24953 | V3 | Griseofulvin | Characterized | [48] | |||
| expansum | KGO43750 | V1 | None | Benzophenone | This study | ||
| KGO65172 | V1 | None | Emodin | This study | |||
| oxalicum | EPS34273 | V1 | None | Benzophenone | This study | ||
| Talaromyces | cellulolyticu | GAM33809 | V1 | None | No data | This study | |
| GAM37897 | V1 | None | Grisandiene | This study | |||
| GAM40075 | V3 | None | No data | This study | |||
| GAM42425 | V1 | None | No data | This study | |||
| GAM43179 | V1 | None | No data | This study | |||
| islandicus | CRG83532 | V1 | None | No data | This study | ||
| CRG86674 | V3 | None | No data | This study | |||
| CRG92129 | V1 | None | No data | This study | |||
| marneffei | KFX46552 | V1 | None | Grisandiene | This study | ||
| KFX52365 | V1 | None | Other anthrone | This study | |||
| ADH01670 | V1 | None | No data | This study | |||
| ADH01674 | V1 | None | No data | This study | |||
| stipitatus | EED18910 | V1 | None | Emodin anthrone | This study | ||
| EED18976 | V1 | None | Benzophenone | This study | |||
| Trichophyton | equinum | EGE06343 | V2 | None | Neosartoricin, fumicycline | This study | |
| interdigitale | KDB28089 | V2 | None | Neosartoricin, fumicycline | This study | ||
| rubrum | EZG11077 | V2 | None | Neosartoricin, fumicycline | This study | ||
| KDB38561 | V2 | None | Neosartoricin, fumicycline | This study | |||
| soudanense | EZF78756 | V2 | None | Neosartoricin, fumicycline | This study | ||
| tonsurans | EGD99348 | V2 | None | Neosartoricin, fumicycline | This study | ||
| verrucosum | EFE44835 | V2 | None | Neosartoricin, fumicycline | This study | ||
| Lecanoromycete | Usnea | longissima | AGI60157 | V1 | None | No data | This study |
| Leotiomycete | Blumeria | graminis | CCU75801 | V3 | None | No data | This study |
| EPQ66189 | V3 | None | No data | This study | |||
| Botrytis | cinerea | AAR90250 | V3 | None | No data | This study | |
| EMR83380 | V3 | None | No data | This study | |||
| XP_001553397 | V3 | None | No data | This study | |||
| Oidiodendron | maius | KIM92894 | V1 | None | Grisandiene | This study | |
| KIM93459 | V2 | None | No data | This study | |||
| KIM96903 | V2 | None | No data | This study | |||
| KIM99919 | V3 | None | No data | This study | |||
| Sclerotinia | borealis | ESZ98980 | V1 | None | Benzophenone | This study | |
| Sordariomycete | Claviceps | purpurea | CCE31584 | V1 | None | Benzophenone | This study |
| Colletotrichum | graminicola | EFQ33703 | V3 | None | Alternariol | This study | |
| sublineola | KDN60962 | V3 | None | No data | This study | ||
| KDN62802 | V3 | None | No product | This study | |||
| Diaporthe | ampelina | KKY32371 | V1 | None | Benzophenone | This study | |
| KKY34489 | V1 | None | Endocrocin anthrone | This study | |||
| KKY37364 | V1 | None | No product | This study | |||
| Eutypa | lata | EMR67234 | V1 | None | Benzophenone | This study | |
| Gaeumannomyces | graminis | EJT69423 | V3 | None | Alternariol | This study | |
| Grosmannia | clavigera | EFX02748 | V1 | None | Benzophenone | This study | |
| EFX04268 | V3 | None | No product | This study | |||
| Metarhizium | acridum | EFY89907 | V2 | None | Viridicatumtoxin-like naphthacenedione | This study | |
| album | KHN97625 | V2 | None | Viridicatumtoxin-like naphthacenedione | This study | ||
| anisopliae | KID64953 | V2 | None | Viridicatumtoxin-like naphthacenedione | This study | ||
| KID65337 | V1 | None | Grisandiene | This study | |||
| KJK89454 | V2 | None | Viridicatumtoxin-like naphthacenedione | This study | |||
| KJK95825 | V1 | None | Grisandiene | This study | |||
| brunneum | KID65403 | V1 | None | Grisandiene | This study | ||
| KID75074 | V2 | None | Viridicatumtoxin-like naphthacenedione | This study | |||
| guizhouense | KID84413 | V1 | None | Grisandiene | This study | ||
| KID87428 | V2 | None | Viridicatumtoxin-like naphthacenedione | This study | |||
| majus | KID93731 | V1 | None | Grisandiene | This study | ||
| KID97686 | V2 | None | Viridicatumtoxin-like naphthacenedione | This study | |||
| robertsii | EXU97310 | V2 | None | Viridicatumtoxin-like naphthacenedione | This study | ||
| EXV00352 | V1 | None | Grisandiene | This study | |||
| Myceliophthora | thermophila | AEO58356 | V1 | None | Benzophenone | This study | |
| Pestalotiopsis | fici | AGO59040 | V1 | Pestheic acid | Characterized | [52] | |
| ETS76950 | V1 | Pestheic acid | Characterized | This study | |||
| Phaeomoniella | chlamydospora | KKY14863 | V3 | None | No data | This study | |
| Stachybotrys | chartarum | KEY72288 | V1 | None | Benzophenone | This study | |
| KFA45466 | V3 | None | No data | This study | |||
| KFA54249 | V1 | None | Benzophenone | This study | |||
| KFA70942 | V3 | None | No data | This study | |||
| KFA71118 | V1 | None | Benzophenone | This study | |||
| Thielavia | terrestris | AEO66245 | V1 | None | Grisandiene | This study | |
| Trichoderma | atroviride | EHK46042 | V1 | None | No data | This study | |
| harzianum | KKO99957 | V1 | None | Other anthraquinone | This study | ||
| KKP03797 | V3 | None | No data | This study | |||
| reesei | ETR97748 | V3 | None | Alternariol | This study | ||
| virens | EHK20655 | V3 | None | Alternariol | This study |
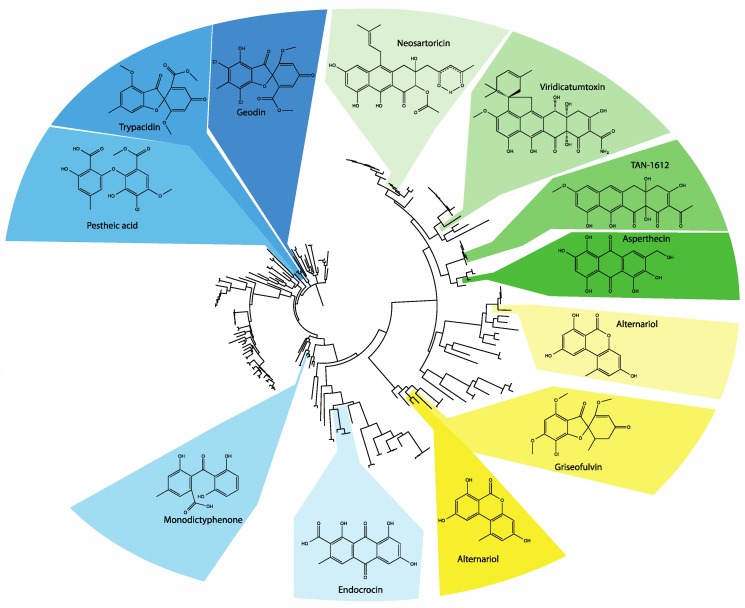
Figure 3.
A maximum likelihood phylogenetic tree constructed with FastTree [60] using the KS domains of a subset of 188 group V NR-PKSs extracted from the broader set used above. The leaves corresponding to characterized PKSs are highlighted in shades of blue for group V1, green for group V2, and yellow for group V3. The structures produced by these PKSs and their associated decorating enzymes are shown adjacently.
3. Phylogenetics Directs Product Prediction
Directed by our phylogenetic analysis, we sought to predict the products of the uncharacterized members of group V by comparison to the characterized gene clusters. Coevolution of the genes in SM gene clusters allows that phylogenetic analysis of a constituent gene, or the protein it encodes, can be used to inform the search for clusters with interesting similarities or differences to known clusters. We applied the existing knowledge of the unique attributes of group V gene clusters to the clusters identified in our phylogenetic tree, and discuss examples from groups of gene clusters that are potentially of interest or that might be insignificantly different from known clusters. These results should aid in identifying interesting targets for future study and in avoiding duplication of efforts in the SM research community.
3.1. Group V1
Within the group V PKSs, a subset known as group V1 consists of octaketide synthases with C6–C11 cyclization. Five clusters from this subgroup have been characterized and their products determined. These include endocrocin, monodictyphenone, trypacidin, geodin, and pestheic acid, produced by A. fumigatus, A. nidulans, A. fumigatus, A. terreus, and Pestalotiopsis fici, respectively [47,51,52,53,66]. Many of the initial studies characterizing these clusters relied on earlier biochemical characterization of the geodin and aflatoxin biosynthetic pathways [50,65,67,68,69,70,71,72]. Group V1 is notable for the large number of aflatoxin homologs its clusters contain. Some clusters belonging to groups V2 and V3 have homologs of aflL (vrtK) and alfO (gsfD), but the characterized clusters in group V1 collectively contain homologs of as many as seven afl or stc (sterigmatocystin) cluster genes. These include homologs of aflR, aflS, aflX, aflY, aflM, hypC, and stcT (Figure 4A). The trypacidin and geodin clusters have previously been noted to contain partial aflatoxin clusters [73], but this is true of group V1 clusters in general. Manual analysis of our MultiGeneBLAST (MGB) [74] results additionally revealed a gene with significant similarity to versicolorin B-synthase (Vbs, AflK) associated with several uncharacterized clusters (Figure S2). Study of clusters containing vbs homologs might reveal an interesting role for this addition afl cluster homolog in group V. We speculate that the high number of afl/stc gene homologs reflects that group V1 clusters share a common ancestor with the afl/stc clusters.
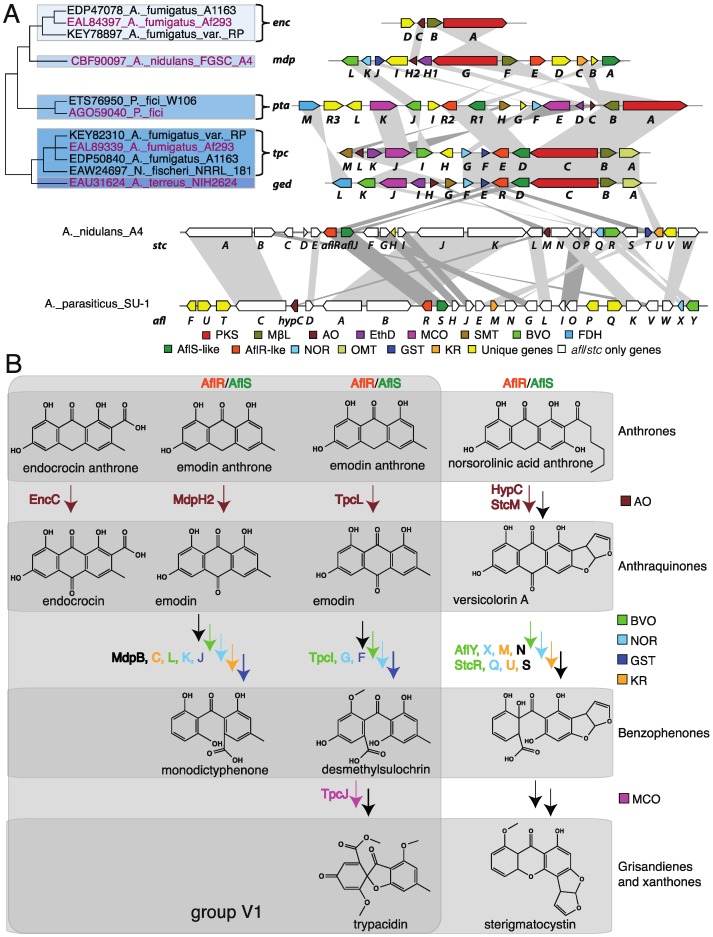
Figure 4.
(A) A phylogenetic tree created from the group V maximum likelihood tree (Figure S2) showing just the relationships between the characterized group V1 PKSs. The gene cluster diagrams next to brackets depict the cluster corresponding to the PKS with its accession number highlighted in red, but all of the bracketed PKSs belong to clusters which are identical in terms of the presence and synteny of their group V-cluster homologs. Genes are represented as arrows with a color corresponding to their ortholog group and these are connected by shaded regions. Genes colored in yellow are unique among clusters shown here. Genes with no color in the afl and stc clusters do not have a homolog in the group V1 clusters shown; (B) A comparison of the analogous reactions catalyzed by the enzymes encoded by homologs of afl cluster genes in the endocrocin, monodictyphenone, trypacidin, and aflatoxin pathways. The reactions of the trypacidin pathway are representative of the geodin and pestheic acid biosynthetic pathways. Pathways of group V1 clusters are enclosed in a grey box. The enzymes catalyzing each reaction are shown to the left of the arrows and the color of the text and arrows matches Figure 4A. Arrows in black represent reactions not shown or reactions for which the enzymes, also labeled in black, are not homologous, except for AflN and StcS, which are homologous. PKS = Polyketide synthase, MβL = Metallo-β-lactamase-type thioesterase, AO = Anthrone oxidase, EthD = EthD domain-containing protein, a putative decarboxylase [51,53], MCO = multicopper oxidase, SMT = S-adenosylmethionine-dependent methyltransferase, BVO = Baeyer-Villiger oxidase, FDH = Flavin-dependent halogenase, AflS = Transcriptional co-regulator of the aflatoxin biosynthetic gene cluster [76], AflR = Transcriptional regulator of the aflatoxin biosynthetic gene cluster, NOR = NADH-dependent oxidoreductase, OMT = O-methyltransferase, GST = Glutathione S-transferase, KR = Ver-1-like ketoreductase [77,78].
All characterized group V1 clusters produce metabolites with an anthraquinone skeleton, such as endocrocin, emodin, and versicolorin A, by action of anthrone oxidases, e.g., HypC, StcM, EncC, MdpH2, and TpcL. Except for endocrocin which represents an end-product, a subset may then be processed into an open-ringed benzophenone structure, like monodictyphenone, by a Baeyer-Villiger oxidase (BVO), an NADH-dependent oxidoreductase (NOR), and potentially a glutathione S-transferase (GST). These reactions are similar to early steps in aflatoxin biosynthesis and this is reflected in the conservation of homologs for these key enzymes in group V1 clusters, though the sets of enzymes required between the aflatoxin/sterigmatocystin and group V1 cluster pathways are only partially overlapping (Figure 4B). Benzophenones may further be converted into closed-ring structures by spontaneous dehydration to xanthones, such as sterigmatocystin, or enzymatically to grisandienes, like trypacidin and geodin, by a multicopper oxidase (MCO), e.g., TpcJ. We leverage the knowledge of these conserved enzymes that catalyze the conversions between these chemical classes to make predictions about the products of related but uncharacterized clusters in this group (Table 1).
3.1.1. Endocrocin-Like Clusters
The smallest characterized cluster from group V1, the four-gene endocrocin cluster was characterized as a virulence factor in A. fumigatus [47,75] and is also found in Neosartorya fischeri. A similar PKS is present in A. terreus (EAU37396), but appears to not be surrounded by any decorating genes, suggesting that the cluster might be a remnant of an endocrocin-like cluster. The endocrocin cluster encodes only two other enzymes in addition to the PKS (EncA) and MβL-TE (EncB) characteristic of group V clusters, an anthrone oxidase (EncC) and a 2-oxoglutarate-Fe(II)-type oxidoreductase (EncD). This minimal complement of decorating enzymes enables the production of the simple anthraquinone endocrocin. For our predictions of the products of uncharacterized group V1 clusters, the presence of genes encoding a PKS and an MβL-TE was considered sufficient to produce anthrones, with the additional presence of an anthrone oxidase-encoding gene required to predict an anthraquinone product (Table 1). Closely related PKSs in a series of other aspergilli are associated with homologs of encB, but no homologs of encC or encD were found in clusters that were available for analysis by MGB or AspGD. Similar to what was observed in A. terreus, it appears that the clusters in this clade may be remnants of endocrocin-like clusters (Figure S2). It is intriguing to speculate that cluster duplications from more complex clusters present in group V1 (see below) and subsequent deterioration led to this sub-group. Recent evidence for chemical redundancy between endocrocin and intermediates from the trypacidin cluster in A. fumigatus could explain the potential decay of the endocrocin and endocrocin-like clusters [53].
3.1.2. Monodictyphenone-Like Clusters
The monodictyphenone-producing cluster in A. nidulans consists of 12 genes, notably including homologs of the three genes required for endocrocin production in the enc cluster. These encode a PKS (MdpG), an MβL-TE (MdpF), and an anthrone oxidase, MdpH2. Though the latter is annotated as part of a larger gene, mdpH, studies of the trypacidin- and geodin-producing clusters [53,66] suggest that mdpH is actually two separate genes, herein referred to as mdpH1 and mdpH2. The mdp cluster is capable of producing endocrocin, but only in the absence of mdpH [51], suggesting that the other half of this gene, mdpH1, encodes a decarboxylase. This cluster additionally produces prenylated xanthones with the activity of prenyltransferases encoded outside of the cluster itself [79]. Among the proteins encoded by characterized group V cluster genes, MdpB, MdpC, MdpD, and MdpI are unique to the mdp cluster. The presence of the corresponding genes can be used to differentiate mdp-like clusters from clusters more similar to the other members of group V1.
Although not products of a group V PKS, the polyketide mycotoxins aflatoxin and sterigmatocystin share decorating enzymes to all group V1 pathways. Biosynthesis of aflatoxin and sterigmatocystin involves the conversion of the anthraquinone precursor versicolorin A to the xanthone demethylsterigmatocystin through a benzophenone-like intermediate and involves the actions of a cytochrome P450 monooxygenase (P450), AflN/StcS [67,68,80], a ketoreductase, AflM/StcU [77], an NOR, AflX/StcQ [70,77], and a BVO, AflY/StcR [71]. In the biosynthesis of monodictyphenone and prenyl xanthones in A. nidulans, a similar ring-opening reaction involving the conversion of the anthraquinone chrysophanol to the benzophenone aldehyde arugosins was recently proposed to be carried out by a glutathione S-transferase (GST), MdpJ, an NOR, MdpK, and a BVO, MdpL [51,78]. The genes encoding these enzymes bear significant similarity to StcT, AflX, and AflY, respectively (Figure 4). No known role is proposed for StcT in sterigmatocystin biosynthesis and no homolog of stcT is present in the aflatoxin gene cluster. Interestingly, action of only the BVO MdpL followed by hydrolysis is sufficient to produce the benzophenone carboxylic acid monodictyphenone as a shunt product. The mdp cluster also contains an aflM homolog in mdpC, but MdpC has been speculated to catalyze the conversion of emodin to chrysophanol in combination with MdpB and not to be involved in the ring-opening step [78]. The biosyntheses of the related compounds trypacidin and geodin in A. fumigatus and A. terreus, respectively, involve ring-opening conversion of the anthraquinone questin to the benzophenone desmethylsulochrin speculated to be catalyzed by a BVO (TpcI/GedK) and potentially an NOR (TpcG/GedF) and GST (TpcF/GedE) [53,66,67,68,78]. In pestheic acid biosynthesis in P. fici, the ring-opening of the anthraquinone physcion to the benzophenone desmethylisosulochrin is similarly proposed to be mediated by a BVO, PtaJ, and an NOR, PtaF, but this cluster encodes no GST, suggesting that this enzymatic activity might not be required for this transition [52].
In summary, these anthraquinone ring-opening reactions to form benzophenones all involve BVOs, NORs, and potentially GSTs. This is a similar but distinct set of enzymes required for the analogous chemical reactions in the biosynthesis of aflatoxin and sterigmatocystin. The presence of genes encoding these conserved anthraquinone ring-opening enzymes, a BVO and a NOR, in addition to the basic enzymes required to produce an anthraquinone, a PKS and MβL-TE, was used as the criterion for prediction of benzophenones as the products of many uncharacterized group V1 clusters (Table 1). Taken together, it is remarkable that, despite the obvious differences between the aflatoxin/sterigmatocystin PKSs (belonging to group IV) and the PKSs present in group V, this subset of enzymatic genes catalyzing ring-opening reactions is shared. Similar to this set of enzymatic genes, homologs of the two regulatory proteins AflR/S are also conserved in group V1 (Figure 4). It is noteworthy that, unlike the ring-opening enzymes, genes encoding AflR/S homologs can be found in other group IV clusters, i.e., the fusarubin cluster [81], and even in group III clusters, i.e., the bikaverin cluster [82].
One cluster from A. ustus, speculated to produce monodictyphenone in a recent study [83] due to its close phylogenetic relationship with that cluster, also has an MCO similar to that of the trypacidin, geodin, and pestheic acid clusters (see below). This suggests that this cluster might produce a chemical structure more similar to these latter clusters than to monodictyphenone and thereby exemplifies the need for a close evaluation of the whole cluster architecture.
3.1.3. Trypacidin, Geodin, and Pestheic Acid-Like Clusters
As noted above, the monodictyphenone cluster shares many similarities with the trypacidin and geodin biosynthetic pathways in A. fumigatus and A. terreus, respectively, and pestheic acid biosynthesis in P. fici. All of these clusters catalyze anthraquinone to benzophenone ring-opening reactions using a BVO and an NOR. In the biosyntheses of trypacidin and geodin, the transition from the open benzophenone to closed grisandiene is catalyzed by a MCO. Specifically, in trypacidin biosynthesis TpcJ converts monomethylsulochrin to trypacidin, and, in geodin biosynthesis, GedJ converts dihydrogeodin to geodin [48,84]. The presence of a gene encoding an MCO in addition to the enzymatic machinery required to produce a benzophenone, a PKS, MβL-TE, BVO, and NOR, was used as the criterion for prediction of grisandienes as the products of many uncharacterized group V1 clusters (Table 1).
Comparison of the trypacidin, pestheic acid, and geodin clusters to clusters from other fungi allows us to speculate on products from undefined fungal clusters. A PKS (AEO66245) encoded by Thielavia terrestris is closely related to the geodin and trypacidin PKSs, and the cluster to which it belongs has homologs to eight of the thirteen genes in the trypacidin cluster (Figure 5). Importantly, these include genes encoding the enzymes required for the anthraquinone to benzophenone transition, a BVO and an NOR, as well as the benzophenone to grisandiene transition, which is catalyzed by a MCO in this subgroup of clusters. The presence of genes encoding these key enzymes suggests that this cluster might ultimately produce a grisandiene (trypacidin or geodin-like molecule). This fungus is a little-known species that can cause human infections [85]. Interestingly, trypacidin is a toxic spore metabolite produced by the opportunistic pathogen A. fumigatus [86], which may suggest that the metabolite produced by the T. terrestris cluster could play a role in pathogenicity.
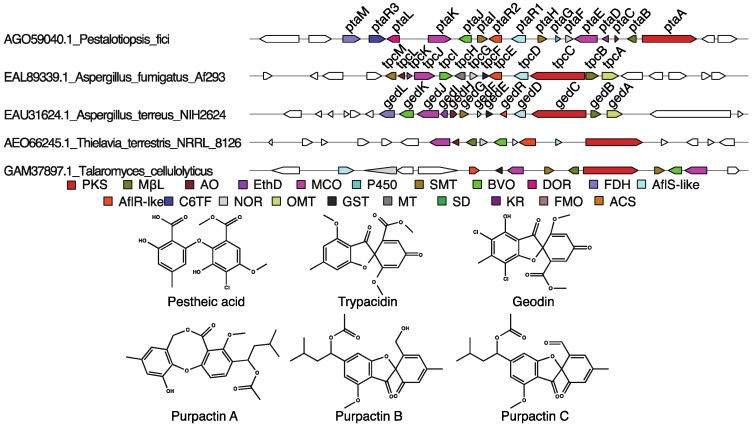
Figure 5.
Examples of tpc-like clusters of interest. At top, the gene cluster diagrams are shown for the characterized pta, tpc, and ged clusters as well as two uncharacterized gene clusters from the group V1 chosen from the larger group V phylogenetic tree, Figure S2. Genes are represented as arrows with a color corresponding to the proteins they encode which are detailed in the color key below the cluster diagrams. Genes with no color were not identified as homologous to any group V1 cluster gene. The structures of the characterized products from this clade, pestheic acid, trypacidin, and geodin, are shown below the color key. Below these are three metabolites reported to be produced by T. cellulolyticus (now considered synonymous with T. pinophilus), purpactins A–C [87]. PKS = Polyketide synthase, MβL = Metallo-β-lactamase-type thioesterase, AO = Anthrone oxidase, EthD = EthD domain-containing protein, a putative decarboxylase [51,53], MCO = multicopper oxidase, P450 = cytochrome P450, SMT = S-adenosylmethionine-dependent methyltransferase, BVO = Baeyer-Villiger oxidase, DOR = Pyridine nucleotide-disulfide oxidoreductase, FDH = Flavin-dependent halogenase, AflS = Transcriptional co-regulator of the aflatoxin biosynthetic gene cluster [76], AflR = Transcriptional regulator of the aflatoxin biosynthetic gene cluster, C6TF = GAL4-like Zn(II)2Cys6-domain and fungal-specific transcription factor domain-containing protein, NOR = NADH-dependent oxidoreductase, OMT = O-methyltransferase, GST = Glutathione S-transferase, MT = Methyltransferase, SD = Scytalone dehydratase, KR = Ver-1-like ketoreductase [77,78], FMO = Flavin-dependent monooxygenase, ACS = Acyl-CoA synthase.
Inspection of another of the PKSs closely related to the trypacidin, geodin, and pestheic acid PKSs, GAM37897.1 from Talaromyces cellulolyticus (now recognized as synonymous with T. pinophilus) (Figure 5), showed that the gene encoding this protein is part of a thirteen-gene cluster with homologs of nine of the thirteen genes in the trypacidin-producing gene cluster. This cluster also has genes encoding the key BVO and NOR enzymes and so likely produces a grisandiene, whether as an intermediate or an end-product. This species is known to produce many secondary metabolites including austin, mitorubrins, penicillides/purpactins/vermixocins, rubropunctatin, vermicellin, vermiculin, vermistatin and (3-O-methyl-, 3-O-methyl-5,6-epoxy-) funicones, MC-141, pestalacin A, stromemycin, dinapinone A1 and A2, and monoapinone A–E [87]. The structures of vermixocins and purpactins suggest they are products of this cluster. These compounds have grisandiene- and depsidone-like scaffolds, which are known or speculated to derive from the biosynthetic pathways of group V clusters such as geodin and pestheic acid [66,88]. Notably, these compounds appear to be prenylated despite the lack of a prenyltransferase identified in the cluster; however, the modification of SMs by prenyltransferases encoded outside of the gene cluster has been observed in monodictyphenone derivatives [79].
3.2. Group V2
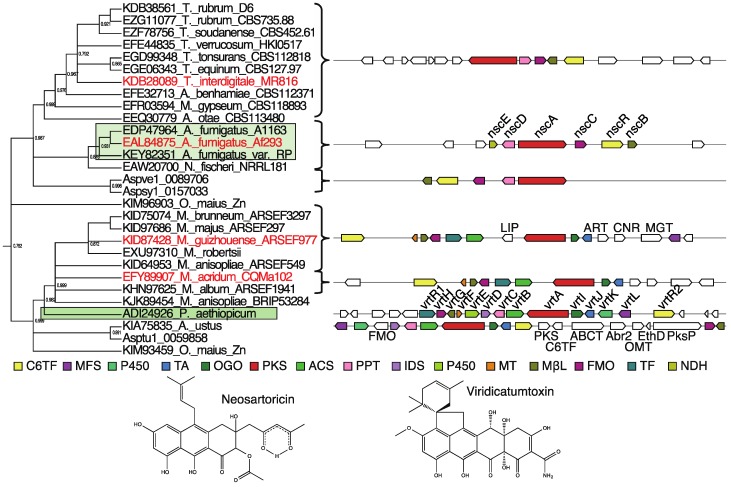
Another subgroup of group V is group V2, which includes nona- and decaketide synthases with C6–C11 cyclization. Characterized examples include asperthecin [46], viridicatumtoxin [48], TAN-1612 [64], and neosartoricin [49]. Enzymatic activities unique to this group include a fourth-ring cyclization facilitated by a flavin-monooxygenase (FMO) and MβL-TE combination. In contrast to the tricyclic (anthracene) backbones produced by most group V PKSs, two characterized examples in group V2 have the ability to generate tetracyclic (naphthacenedione) backbones, TAN-1612, and viridicatumtoxin. This ability depends on several factors including the ability to synthesize a long, i.e., nona- or decaketide, backbone and the presence of both an MβL-TE with Claisen-cyclase activity and a unique FMO [64]. Though VrtA is only a nonaketide synthase, it accepts the very unusual malonamoyl-CoA starter unit produced by VrtB and VrtJ, and thus has a long enough chain for a fourth cyclization [48]. Phylogenetic analysis of polycyclic prenyltransferases (PPTs) associated with these clusters was previously used to identify a group of clusters with this triad of a unique PKS, MβL-TE, and FMO in dermatophytic fungi [89] (Figure 6).
Figure 6.
A clade of nsc-like clusters in Trichophyton, a clade of vrt-like clusters in Metarhizium, and other closely related clusters. An excerpt of the group V phylogenetic tree made with FastTree [60], Figure S2, containing the PKSs from the neosartoricin-producing cluster, NscA (EAL84875) [49], and the viridicatumtoxin-producing cluster, VrtA (ADI24926) [48], and groups of related uncharacterized PKSs, primarily in Trichophyton and Metarhizium, respectively, is shown at top left. The bootstrap values are presented next to their corresponding nodes. The green boxes indicate PKSs from the same species in which the characterized clusters were originally described. Next to the tree are the gene clusters corresponding to the PKSs that were identifiable through MultiGeneBLAST analysis. Gene cluster diagrams next to brackets depict the cluster corresponding to the PKS with its accession number highlighted in red, but all of the bracketed PKSs belong to clusters which are identical in terms of the presence and synteny of their nsc- or vrt-cluster homologs. Genes are represented as arrows with a color corresponding to the proteins they encode which are detailed in the color key below the tree and cluster diagrams. Genes with no color were not identified as homologous to any group V2 cluster gene. The products of the characterized examples from this clade, neosartoricin and viridicatumtoxin, are shown at bottom. C6TF = GAL4-like Zn(II)2Cys6-domain and fungal-specific transcription factor domain-containing protein, VrtR1-like, MFS = Major Facilitator Superfamily transporter, P450 = Cytochrome P450, TA = Threonine aldolase, OGO = 2-oxoglutarate-Fe(II)-type oxidoreductase, PKS = Polyketide synthase, ACS = Acetoacetyl-CoA synthase, PPT = Polycyclic prenyltransferase, IDS = Isoprenyl diphosphate synthase, MT = Methyltransferase, MβL = Metallo-β-lactamase-type thioesterase, FMO = Flavin-dependent monooxygenase, TF = GAL4-like Zn(II)2Cys6-domain and fungal-specific transcription factor domain-containing protein, VrtR2-like, NDH = NAD-dependent dehydratase, LIP = Secretory lipase, ART = Arrestin, CNR = Copper-containing nitrite reductase, MGT = Magnesium transporter, ABCT = ABC transporter, Abr2 = Conidial pigment laccase, OMT = O-methyltransferase, EthD = EthD domain-containing protein, putative decarboxylase, PksP = Conidial pigment polyketide synthase.
3.2.1. Asperthecin-Like Clusters
Asperthecin, associated with sexual spore color in A. nidulans (Palmer and Keller unpublished data), is produced from a three-gene cluster encoding the NR-PKS, the MβL-TE, and a FAD-dependent oxidoreductase [46]. As seen in Figure S2, the three close relatives A. versicolor, A. sydowii, and A. ustus contain the same cluster, which we hypothesize is also likely to be associated with ascospore color in these fungi. The presence of this minimal triad of genes encoding a PKS, MβL-TE, and FMO, as well as the close relationship of these PKSs to the characterized asperthecin-producing PKS, were the criteria used to predict asperthecin as a product of several uncharacterized clusters (Table 1).
3.2.2. TAN-1612-Like Clusters
TAN-1612, identified in A. niger [64] and also present in A. kawachii, is produced by a five-gene cluster, and three of these genes are homologous to the apt cluster genes. In addition to the three proteins described above, this cluster is differentiated from the apt cluster by the presence of genes encoding a methyltransferase (MT) and a GAL4-like Zn(II)2Cys6-domain and fungal-specific transcription factor domain-containing protein (C6TF). It is tempting to speculate that TAN-1612 might be associated with ascospore pigmentation however the sexual stage of neither A. niger nor A. kawachii has been described yet for assessment of such a hypothesis. The presence of this set of five genes, as well as the close relationship of these PKSs to the characterized TAN-1612-producing PKS, were used as the criteria for predicting TAN-1612 as the product of several uncharacterized clusters (Table 1).
3.2.3. Neosartoricin-Like Clusters
Neosartoricin is produced by a six-gene cluster in A. fumigatus and N. fischeri [49]. This cluster was also identified as producing the related fumicyclines [63]. The cluster’s PKS, NscA/FccA, produces a decaketide chain, the longest known of all PKSs so far described along with the TAN-1612-producing PKS, AdaA [90]. Five of the six genes in the nsc/fcc cluster are conserved in the dermatophytic genera, Trichophyton, Arthroderma, and Microsporum (Figure 6) some of which have previously been noted [89]. Notably, the gene for which there is no conserved homolog in these species, the NAD-dependent dehydratase (NDH)-encoding nscE/fccE, has no proposed role in neosartoricin or fumicycline biosynthesis in A. fumigatus [49,63]. Compared to the nsc cluster, these conserved clusters in the Arthrodermataceae appear to have two two-gene inversions, but are otherwise syntenically conserved. Four of these five genes are homologous to genes in the TAN-1612-producing cluster of A. niger. The neosartoricin-producing cluster is differentiated from TAN-1612-producing clusters by the presence of genes encoding a PPT and an NDH and the absence of a homolog of the MT-encoding gene from the TAN-1612-producing cluster. The presence of the unique set of five genes of the nsc cluster, excluding the NDH-encoding nscE, as well as the close relationship of these PKSs to the characterized neosartoricin-producing PKS, were used as the criteria for predicting neosartoricin and fumicycline as the product of a group of uncharacterized clusters (Table 1). Fumicyclines are induced in the presence of Streptomyces rapamycinicus and neosartoricin has demonstrated immunosuppressive activity [49,63]. This potentially suggests an important role for this cluster in virulence of dermatophytes.
3.2.4. Viridicatumtoxin-Like Clusters
Viridicatumtoxin is a tetracyclic mycotoxin produced by Penicillium species. The 14-gene vrt cluster contains homologs of the 5 conserved nsc genes mentioned above. We have identified a group of Metarhizium species that have gene clusters with homologs of ten of the vrt-cluster genes (Figure 6). Metarhizium species are entomopathogenic fungi in the Clavicipitaceae family. These Metarhizium clusters contain two regions of conserved synteny with four and five gene regions of the vrt cluster and are predicted to yield a tetracyclic polyketide. Interestingly, in A. ustus a vrt-like cluster contains homologs of abr2 and pksP of the conidial pigment biosynthetic gene cluster, suggesting that this may be one large cluster or two interwoven clusters perhaps similar to that of the intermingled fumagillin/pseurotin supercluster in A. fumigatus [91]. Several of these species have been noted to have vrt-like clusters in recent studies [83,92]. The presence of the minimal triad of genes encoding a PKS, MβL-TE, and FMO, multiple other homologs of genes unique to the vrt cluster amongst group V cluster genes, as well as the close relationship of these PKSs to the characterized viridicatumtoxin-producing PKS, were used as the criteria for prediction of “viridicatumtoxin-like naphthacenedione” as the product of a group of uncharacterized clusters (Table 1).
3.3. Group V3: Griseofulvin and Various Alternariol-Like Pathways
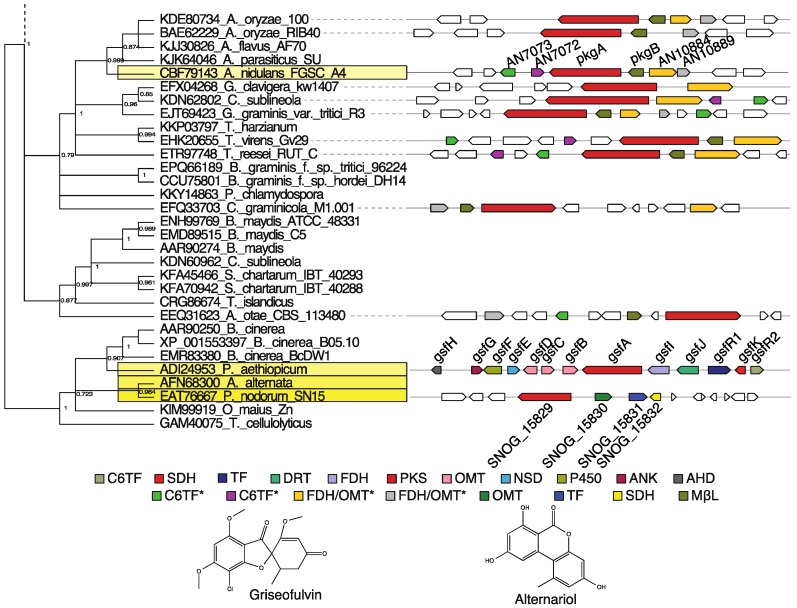
This group V subgroup includes heptaketide synthases catalyzing an unusual C1–C6 or C2–C7 first cyclization followed by a C8–C13 second cyclization. Characterized examples include alternariol and griseofulvin. Alternariol is an important mycotoxin produced by members of Alternaria, Aspergillus, and Phaeosphaeria [54,61,62]. This metabolite is a fairly common crop contaminant with carcinogenic, phytotoxic, and antifungal activity. Despite its importance, genetic studies characterizing the biosynthesis of alternariol were only recently undertaken. To date, three gene clusters have been implicated in the synthesis of alternariol, one in Alternaria alternata, one in A. nidulans, and one in Phaeosphaeria nodorum (syn. Parastagonospora nodorum) (Figure 7). Initially, PksJ (AFN68301) was identified in A. alternata using siRNA and gene deletion approaches as the PKS primarily responsible for alternariol production. However, two other PKSs, PksH (AFN68299) and PksI (AFN68300), were shown to be affected by the knockdown of pksJ expression. Notably, no MβL-TE was identified adjacent to PksJ in this study [61]. In A. nidulans, promoter replacement experiments were used to show that PkgA (CBF79143) and PkgB produce alternariol and coumarins [54]. Most recently, in P. nodorum, SNOG_15829 (EAT76667) was also found to produce alternariol. The cluster associated with this NR-PKS includes a gene encoding an MβL-TE (SNOG_15826), but it bears little similarity to the other MBL-TEs of group V clusters [62], which could be due to poor sequence quality. Interestingly, the NR-PKS EAT76667 is most similar to PksI from A. alternata, suggesting that PksI, and not PksJ or PksH, is the alternariol-producing PKS in A. alternata. These clusters were not characterized further than the identification of a PKS and an MβL-TE; the additional genes analyzed by MGB as part of the PkgA (CBF79143) and SNOG_15829 (EAT76667) clusters were included based only on their reported co-regulation with the PKS- and MβL-TE-encoding genes [62,93]. Because of these limitations in the studies characterizing alternariol-producing gene clusters, our product predictions for group V3 PKSs are based only on the presence or absence of genes encoding a PKS and MβL-TE and the closeness of the relationships of these PKSs with the characterized group V3 PKSs (Table 1).
Figure 7.
A clade of gsf-, pkg-, PskI-, and SNOG_15820-like clusters. An excerpt of the group V phylogenetic tree made with FastTree [60], Figure S2, containing the PKSs from the griseofulvin-producing cluster, GsfA (ADI24953) [48], three alternariol-producing clusters [54,61,62], and a group of related uncharacterized PKSs, is shown at top left. The bootstrap values are presented next to their corresponding nodes. The yellow boxes indicate PKSs from the same species in which the characterized clusters were originally described. Next to the tree are the gene clusters corresponding to the PKSs that were identifiable through MultiGeneBLAST analysis. Genes are represented as arrows with a color corresponding to the proteins they encode which are detailed in the color key below the tree and cluster diagrams. Asterisks signify potential gene truncation due to misannotation. Genes with no color were not identified as homologous to any group V3 cluster gene. The products of the characterized examples from this clade, griseofulvin and alternariol, are shown at bottom. C6TF = GAL4-like Zn(II)2Cys6-domain and fungal-specific transcription factor domain-containing protein, GsfR2-like, SDH = Short chain dehydrogenase, TF = GAL4-like Zn(II)2Cys6-domain and fungal-specific transcription factor domain-containing protein, GsfR1-like, DRT = Drug resistance transporter, EmrB subfamily, FDH = Flavin-dependent halogenase, PKS = Polyketide synthase, OMT = O-methyltransferase, NSD = Nucleoside-diphosphate-sugar dehydratase, P450 = Cytochrome P450, ANK = Ankyrin repeat-containing protein, AHD = YcaC-related amidohydrolase, FDH/OMT = Flavin-dependent halogenase and O-methyltransferase bifunctional protein, MβL = Metallo-β-lactamase-type thioesterase. * Asterisks signify potential gene truncation due to misannotation.
Griseofulvin, produced by Penicillium species, is an antifungal drug widely used against dermatophytic infections [94,95,96]. Despite its resemblance to other grisandienes like trypacidin and geodin, the griseofulvin biosynthetic pathway [48] is quite unique and constitutes an interesting example of convergent evolution at the biochemical level. The NR-PKS responsible for griseofulvin production, GsfA (ADI24953), generates a benzophenone directly and by a different cyclization register than that of the group V1 PKSs MdpG, TpcC, GedC, and PtaA, i.e., C1–C6 as opposed to C6–C11 for group V1 PKSs or even C4–C9 for the aflatoxin-producing group IV PKS, AflC [84]. Thus, whereas the biosynthetic pathways of group V1 clusters proceed through an anthraquinone intermediate to a benzophenone intermediate by action of a BVO, GsfA synthesizes a benzophenone as its initially released product. This is made possible by the unusual C1–C6 and C8–C13 connections it catalyzes. This latter connection is also observed in the biosynthesis of alternariol, but the two differ in their initial cyclization, i.e., C1–C6 for griseofulvin and C2–C7 for alternariol [62]. Further, the benzophenone to grisandiene transition in griseofulvin biosynthesis is catalyzed by a cytochrome P450 as opposed to a MCO in the cases of trypacidin, geodin, and pestheic acid [97]. Curiously, the release mechanism of GsfA has yet to be elucidated, as it lacks the MβL-TE that is characteristic of group V. We speculate that the unique cyclization catalyzed by these PKSs might obviate the MβL-TE activity and explain the lack of an MβL-TE in the gsf cluster and the dissimilarity of the MβL-TE of the P. nodorum alternariol-producing cluster. It has been noted that the close relationship of the griseofulvin- and alternariol-producing PKSs is likely not coincidental and that unusual modes of cyclization may be unique to this clade [57].
4. Uncharacterized Clusters in Symbionts and Pathogens
As expected, considering the number of sequenced Aspergillus species, many of the NR-PKSs identified from this study are from Aspergillus species. However, of the non-Aspergillus genera and species, many are symbiotic, including pathogenic fungi associated with specific hosts. These findings support reviews of pathogenic fungi which highlight the potential role of secondary metabolites in virulence [1]. Below we touch on this emerging theme from our analysis.
4.1. Plant Pathogens
Several plant pathogenic fungi are present in two main groupings in Figure S2. Notably, the subclade to which the griseofulvin and alternariol PKSs belong contains many plant pathogenic species (Figure 7), including the pine pathogen Grosmannia clavigera, the sorghum pathogen Colletotrichum sublineola, the wheat pathogen Gaeumannomyces graminis, the maize pathogen Bipolaris maydis, the grape pathogen Botrytis cinerea, and two grass powdery mildew species from the genus Blumeria. This latter finding is especially intriguing as the genomes of obligate biotrophs such as powdery mildews contain few secondary metabolite genes [98,99]. As alternariol produced by related clusters from Alternaria, Aspergillus, and Phaeosphaeria spp. is known to be an important phytotoxin, it is possible these NR-PKSs produce a metabolite important in the fungal/plant host interaction.
Several plant pathogenic fungi also are present in the subclades producing trypacidin, geodin, pestheic acid, and monodictyphenone. The genus Bipolaris and allied genera Setophaeria and Pyrenophora—all grass pathogens—are particularly well represented in this clade. Another grass pathogen, Claviceps purpurea, is also present in this subclade, although it is taxonomically distant from the Bipolaris species and is best known for its suite of alkaloid-producing clusters [100]. It would be interesting to see if these clusters produce a metabolite specialized to interactions with grass hosts.
4.2. Mycorrhizal Fungi
Oidiodendron maius belongs to the rare group of fungi establishing mycorrhizal relationships with the plant family Ericaceae (heather) [101,102]. Most unusually, this species contains four NR-PKSs associated with every group V subgroup. Poor annotation of the genome prevented full assessment of the associated clusters; hopefully future sequencing efforts will provide data for accurate cluster predictions.
4.3. Fungal Pathogens
The mycoparasitic genus Trichoderma is represented twice in our analysis, once where they contain a gene cluster quite similar to the pkg cluster (PKS = CBF79143) and one similar to the pestheic acid cluster of group V1. Considering the mycoparasitic lifestyle of these fungi, the presence of these clusters raises the question whether their respective products could play a role in mycoparasitism.
4.4. Animal Pathogens
Several animal pathogens, from insect to human, contain NR-PKSs also found throughout group V. Entomopathogenic fungi. The genus Metarhizium, along with Beauveria bassiana not represented in Group V, is well known for its potential in biological control of various insects [103]. A putative group V1 cluster was identified in B. bassiana including EJP67854, but it did not include an annotated PKS and so was excluded from further analysis. It is unclear whether this is related to any of the several NR-PKSs that have previously been noted in B. bassiana [104]. As mentioned earlier, several species in Metarhizium contain clusters with significant similarity to the Penicillium viridicatumtoxin cluster (Figure 6). The Metarhizium genus is also well represented in group V1, suggesting the ability to produce a metabolite similar to these compounds. Toxicity is associated with all of these metabolites and may afford virulence properties to these insect pathogens.
Dermatophytes. Several dermatophytic genera, Trichophyton, Arthoderma, and Microsporum contain a neosartoricin-like gene cluster. This compound exhibits antiproliferative activity which may be suggestive of an immunosuppressive role in human infection by dermatophytic fungi [49]. The causal agent of a devastating bat disease in North America known as white nose syndrome, Pseudogymnoascus destructans, is a dermatophyte of bats [105]. This species and other members of the genus are found in two sections of group V, both grouping to the trypacidin, geodin, pestheic acid, and monodictyphenone clades. Metabolites produced by these clusters could possibly play a role in virulence of P. destructans.
5. Experimental Section
To retrieve the amino acid sequences of NR-PKSs in our initial search, the monodictyphenone PKS, MdpG [51], was analyzed with the NCBI’s BLASTP [106] against fungi (taxid: 4751) with max target sequences increased to 1000 and other parameters set to defaults. A similar search was executed on AspGD (aspgd.org) and 25 non-duplicate PKSs were identified from the top 50 hits and added to the list retrieved from the NCBI. The human fatty acid synthase, FASN, was added as a marker for the outgroup, which also included 70 HR-PKSs and hybrid PKS/NRPSs. The KS domains for these 908 PKSs were retrieved using the NCBI’s Conserved Domain Database (CDD) utility [107,108,109] and aligned using MAFFT [110] with default parameters. Alignment columns with greater than 40% gaps were removed using TrimAl [111], and the sequences were realigned. A maximum likelihood phylogenetic tree was constructed using FastTree [60] in Geneious 8.1.5 [112] with 1000 bootstrap replicates and otherwise default parameters. Nodes with bootstrap support values of less than 70% were collapsed to polytomy in Figure 6, Figure 7, and Figure S2. This tree was modified for presentation using FigTree [113] (Figure 2, Figure S1). To extract the sequences for the group V KS domains, a neighbor-joining tree was constructed in Geneious with default parameters, and the sequences for the smallest monophyletic group containing the characterized group V PKSs were selected and exported as a sub-alignment. These 188 sequences were realigned with MAFFT and used to construct a maximum likelihood phylogenetic tree using FastTree as above. This tree was modified for presentation using FigTree [113] (Figure 3, Figure S2). Excerpts of this tree were used to create Figure 4, Figure 5, Figure 6 and Figure 7.
MultiGeneBLAST [74] (MGB) was used to facilitate the analysis of uncharacterized clusters associated with the PKSs identified by the methods described above. MGB architecture searches were executed with the synteny conservation weight set to 0, the percent identity threshold set to 25%, the maximum intergenic distance threshold set to 25 kb, and otherwise default parameters. MGB searches were carried out with multifasta files containing one representative of each group of orthologous genes from a given subclade of group V, e.g., the group V1 search used a file containing one PKS, one MβL-TE, etc. (Tables S1–S3). If multiple genes in a single cluster encoded similar types of proteins, e.g., C6TFs, TFs, P450s, or MTs, they were considered as separate and included in the multifasta file for that subgroup (i.e., V1, V2, or V3) despite their potential redundancy. This was done to identify potential patterns in the distribution of orthologs across other clusters, i.e., if a given ortholog was more similar to one or the other of the potentially redundant genes. Alternatively, if the potentially redundant genes were not differentiable with our MGB parameters they and all their orthologs were considered to encode the same type of protein and represented as such in the cluster diagrams and corresponding color keys. The database available online (http://multigeneblast.sourceforge.net/index.html) containing all GenBank entries, updated 1/2015, was used to obtain the majority of the cluster diagrams (Figure S2, Figure 4, Figure 5, Figure 6 and Figure 7). Others were obtained from a custom fungal database generated by downloading 578 annotated fungal genomes from NCBI using a custom python script (https://github.com/nextgenusfs/NR-PKS_ms/get_ncbi_genomes.py). These genomes were then incorporated into a MGB database using the `makedb` program from the command line distribution of MGB. Still others, such as the T. cellulolyticus (GAM37897) and A. terreus (EAU31624) clusters (Figure 5, Figure S2), were manually created or modified (to reflect re-annotation of ATEG_08457 [66]), respectively. The protein descriptors in the keys or adjacent to the cluster diagrams (Figure 4, Figure 5, Figure 6 and Figure 7, Figure S2) were derived from the function or conserved domain of the protein according to published studies describing group V SM gene clusters [46,47,48,49,51,52,53,54,61,62,63,64,66,78] or individual BLAST searches.
6. Conclusions
The potential to synthesize polyketides is widespread in the fungal taxa ascomycetes and basidiomycetes, but examination of biochemical PKS classes indicates taxonomic specificities [57]. Illustrating this point, the TE-less NR-PKSs described in this work and others [54,57], group V, are notably absent in certain mycotoxigenic genera (e.g., Fusarium) and basidiomycetes but, surprisingly, found in genera not noted for secondary metabolism (e.g., Oidiodendron and Blumeria) (Figure 5, Figure 6 and Figure 7, Figure S2). Many of the described products derived from these NR-PKSs exhibit toxic activities. For example, questin, trypacidin, and endocrocin have been assessed for their impact on virulence as they are produced by the human pathogen A. fumigatus. Trypacidin and endocrocin in particular have been shown to exhibit toxic and neutrophil inhibitory properties, respectively, in pathogenicity studies [75,86]. The other A. fumigatus group V metabolite, neosartoricin, exhibits T-cell antiproliferative activity, which may be suggestive of an immunosuppressive role in human infection [49]. Furthermore, griseofulvin is a potent antifungal [94], viridicatumtoxin is a mycotoxin [114,115], and alternariol is known for its phytotoxic properties [116,117]. Thus, it is not improbable that the products of the NR-PKS clusters identified in the pathogenic and symbiotic fungi in this study (Figure S2) could impact fungal/host interactions, as virulence factors or signaling molecules. However, it is also possible that such roles could be coincidental or in addition to other functions in fungal biology.
As predicted by their structure, it is likely all are UV absorbing pigmented molecules observed in the visible yellow-orange spectrum. Possibly one significant role of group V PKSs may lie in protection or development of spores. Asexual spores are common, air-dispersed spores essential for propagation in the kingdom Fungi and must be equipped with defenses against abiotic stresses, such as UV radiation, oxidative stress, and desiccation. Several studies have shown that loss of pigmentation of spores leads to reduced viability and/or virulence in pathogenic fungi [118,119,120,121]. Sexual spores are the product of meiosis and essential for genetic recombination and species diversity and, in several species, are also airborne spores that would be exposed to UV radiation similarly to asexual spores [122,123]. Both trypacidin and endocrocin are produced in the asexual spore [75,86] and our studies (Palmer and Keller, unpublished data)—supported by an earlier investigation [124]—suggest that asperthecin is the red pigment characterizing the color of A. nidulans sexual ascospores. Assessment of spore viability of NR-PKS fungal mutants under UV conditions might shed light on a conserved role of these molecules to protect from specific abiotic stresses.
In this study we have examined all of the group V NR-PKSs available from NCBI and all of the corresponding gene clusters that were readily available using MultiGeneBLAST. The relatively high number of studies characterizing SM gene clusters from group V [46,47,48,49,51,52,53,54,61,62,63,64,66,78], has enabled us to predict the products of uncharacterized clusters in this group (Table 1). For group V1 this is largely based on the presence or absence of genes encoding key enzymes known to catalyze particular reactions, e.g., anthraquinone ring-opening by BVOs and NORs or benzophenone ring-closure by MCOs. For groups V2 and V3 these predictions are based only on the presence of homologs of a majority of the genes in the most closely related characterized group V cluster, e.g., neosartoricin is predicted to be produced from clusters containing homologs of five of the six nsc cluster genes. This study demonstrates that, by comparison to characterized examples in a given group of NR-PKSs, predictions can be made from phylogenetic analysis and used to help choose SM gene clusters to characterize. For example, future research efforts might be better spent studying SM gene clusters of interest highlighted by this study and not on those that are highly similar to characterized clusters and therefore likely to produce a known compound. Application of these methods to other groups of NR-PKSs might provide a similarly beneficial perspective.
Acknowledgments
The authors would like to thank Dr. Jonathan M. Palmer for assistance with the computational analysis associated with the construction of phylogenetic trees and MultiGeneBLAST databases. We would also like to thank our funding sources, grants from the National Science Foundation (EFRI-1136903), the United States Fish and Wildlife Service (F14AP00618), and the National Institutes of Health training grant to The University of Wisconsin-Madison Genetics Department (5T32GM07133).
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/2072-6651/7/9/3572/s1.
Author Contributions
The text was written by KT, PW, and NPK. Sequences were obtained and processed by PW and KT. Phylogenetic trees were constructed by KT. MultiGeneBLAST analyses were carried out by KT and PW. Figures were constructed by KT and PW. General oversight and guidance was provided by NPK.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Perez-Nadales E., Nogueira M.F., Baldin C., Castanheira S., el Ghalid M., Grund E., Lengeler K., Marchegiani E., Mehrotra P.V., Moretti M., et al. Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genet. Biol. 2014;70:42–67. doi: 10.1016/j.fgb.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rohlfs M., Churchill A.C. Fungal secondary metabolites as modulators of interactions with insects and other arthropods. Fungal Genet. Biol. 2011;48:23–34. doi: 10.1016/j.fgb.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Scherlach K., Graupner K., Hertweck C. Molecular bacteria-fungi interactions: Effects on environment, food, and medicine. Annu. Rev. Microbiol. 2013;67:375–397. doi: 10.1146/annurev-micro-092412-155702. [DOI] [PubMed] [Google Scholar]
- 4.Allam N.G., Abd el-Zaher E.H.F. Protective role of Aspergillus fumigatus melanin against ultraviolet (UV) irradiation and Bjerkandera adusta melanin as a candidate vaccine against systemic candidiasis. Afr. J. Biotech. 2012;11:6566–6577. [Google Scholar]
- 5.Chiang Y.M., Oakley B.R., Keller N.P., Wang C.C. Unraveling polyketide synthesis in members of the genus Aspergillus. Appl. Microbiol. Biotechnol. 2010;86:1719–1736. doi: 10.1007/s00253-010-2525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushley K.E., Turgeon B.G. Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol. Biol. 2010;10:26. doi: 10.1186/1471-2148-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wawrzyn G.T., Bloch S.E., Schmidt-Dannert C. Discovery and characterization of terpenoid biosynthetic pathways of fungi. Methods Enzymol. 2012;515:83–105. doi: 10.1016/B978-0-12-394290-6.00005-7. [DOI] [PubMed] [Google Scholar]
- 8.Umemura M., Nagano N., Koike H., Kawano J., Ishii T., Miyamura Y., Kikuchi M., Tamano K., Yu J., Shin-ya K., et al. Characterization of the biosynthetic gene cluster for the ribosomally synthesized cyclic peptide ustiloxin B in Aspergillus flavus. Fungal Genet. Biol. 2014;68:23–30. doi: 10.1016/j.fgb.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Fedorova N.D., Moktali V., Medema M.H. Bioinformatics approaches and software for detection of secondary metabolic gene clusters. Methods Mol. Biol. 2012;944:23–45. doi: 10.1007/978-1-62703-122-6_2. [DOI] [PubMed] [Google Scholar]
- 10.Gallo A., Ferrara M., Perrone G. Phylogenetic study of polyketide synthases and nonribosomal peptide synthetases involved in the biosynthesis of mycotoxins. Toxins. 2013;5:717–742. doi: 10.3390/toxins5040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finn R.D., Clements J., Arndt W., Miller B.L., Wheeler T.J., Schreiber F., Bateman A., Eddy S.R. Hmmer web server: 2015 update. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang I.K., Reeves C., Gaucher G.M. Isolation and sequencing of a genomic DNA clone containing the 3’ terminus of the 6-methylsalicylic acid polyketide synthetase gene of Penicillium urticae. Can. J. Microbiol. 1991;37:86–95. doi: 10.1139/m91-013. [DOI] [PubMed] [Google Scholar]
- 13.Fujii I., Ono Y., Tada H., Gomi K., Ebizuka Y., Sankawa U. Cloning of the polyketide synthase gene atX from Aspergillus terreus and its identification as the 6-methylsalicylic acid synthase gene by heterologous expression. Mol. Gen. Genet. 1996;253:1–10. doi: 10.1007/s004380050289. [DOI] [PubMed] [Google Scholar]
- 14.Beck J., Ripka S., Siegner A., Schiltz E., Schweizer E. The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum. Its gene structure relative to that of other polyketide synthases. Eur. J. Biochem. 1990;192:487–498. doi: 10.1111/j.1432-1033.1990.tb19252.x. [DOI] [PubMed] [Google Scholar]
- 15.Mayorga M.E., Timberlake W.E. The developmentally regulated Aspergillus nidulans wA gene encodes a polypeptide homologous to polyketide and fatty acid synthases. Mol. Gen. Genet. 1992;235:205–212. doi: 10.1007/BF00279362. [DOI] [PubMed] [Google Scholar]
- 16.Yu J., Chang P.K., Cary J.W., Wright M., Bhatnagar D., Cleveland T.E., Payne G.A., Linz J.E. Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl. Environ. Microbiol. 1995;61:2365–2371. doi: 10.1128/aem.61.6.2365-2371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trail F., Mahanti N., Rarick M., Mehigh R., Liang S.H., Zhou R., Linz J.E. Physical and transcriptional map of an aflatoxin gene cluster in Aspergillus parasiticus and functional disruption of a gene involved early in the aflatoxin pathway. Appl. Environ. Microbiol. 1995;61:2665–2673. doi: 10.1128/aem.61.7.2665-2673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J.H., Leonard T.J. Sterigmatocystin biosynthesis in Aspergillus nidulans requires a novel type I polyketide synthase. J. Bacteriol. 1995;177:4792–4800. doi: 10.1128/jb.177.16.4792-4800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang G., Rose M.S., Turgeon B.G., Yoder O.C. A polyketide synthase is required for fungal virulence and production of the polyketide T-toxin. Plant Cell. 1996;8:2139–2150. doi: 10.1105/tpc.8.11.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker S.E., Kroken S., Inderbitzin P., Asvarak T., Li B.Y., Shi L., Yoder O.C., Turgeon B.G. Two polyketide synthase-encoding genes are required for biosynthesis of the polyketide virulence factor, T-toxin, by Cochliobolus heterostrophus. Mol. Plant Microbe Interact. 2006;19:139–149. doi: 10.1094/MPMI-19-0139. [DOI] [PubMed] [Google Scholar]
- 21.Díaz-Sánchez V., Avalos J., Limón M.C. Identification and regulation of fusA, the polyketide synthase gene responsible for fusarin production in Fusarium fujikuroi. Appl. Environ. Microbiol. 2012;78:7258–7266. doi: 10.1128/AEM.01552-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song Z., Cox R.J., Lazarus C.M., Simpson T.J. Fusarin C biosynthesis in Fusarium moniliforme and Fusarium venenatum. ChemBioChem. 2004;5:1196–1203. doi: 10.1002/cbic.200400138. [DOI] [PubMed] [Google Scholar]
- 23.Gaffoor I., Trail F. Characterization of two polyketide synthase genes involved in zearalenone biosynthesis in Gibberella zeae. Appl. Environ. Microbiol. 2006;72:1793–1799. doi: 10.1128/AEM.72.3.1793-1799.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y.T., Lee Y.R., Jin J., Han K.H., Kim H., Kim J.C., Lee T., Yun S.H., Lee Y.W. Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae. Mol. Microbiol. 2005;58:1102–1113. doi: 10.1111/j.1365-2958.2005.04884.x. [DOI] [PubMed] [Google Scholar]
- 25.Proctor R.H., Desjardins A.E., Plattner R.D., Hohn T.M. A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet. Biol. 1999;27:100–112. doi: 10.1006/fgbi.1999.1141. [DOI] [PubMed] [Google Scholar]
- 26.Yu F., Zhu X., Du L. Developing a genetic system for functional manipulations of FUM1, a polyketide synthase gene for the biosynthesis of fumonisins in Fusarium verticillioides. FEMS Microbiol. Lett. 2005;248:257–264. doi: 10.1016/j.femsle.2005.05.053. [DOI] [PubMed] [Google Scholar]
- 27.Shimizu T., Kinoshita H., Ishihara S., Sakai K., Nagai S., Nihira T. Polyketide synthase gene responsible for citrinin biosynthesis in Monascus purpureus. Appl. Environ. Microbiol. 2005;71:3453–3457. doi: 10.1128/AEM.71.7.3453-3457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Awakawa T., Kaji T., Wakimoto T., Abe I. A heptaketide naphthaldehyde produced by a polyketide synthase from Nectria haematococca. Bioorg. Med. Chem. Lett. 2012;22:4338–4340. doi: 10.1016/j.bmcl.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Cary J.W., Harris-Coward P.Y., Ehrlich K.C., Di Mavungu J.D., Malysheva S.V., De Saeger S., Dowd P.F., Shantappa S., Martens S.L., Calvo A.M. Functional characterization of a veA-dependent polyketide synthase gene in Aspergillus flavus necessary for the synthesis of asparasone, a sclerotium-specific pigment. Fungal Genet. Biol. 2014;64:25–35. doi: 10.1016/j.fgb.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Cary J.W., Uka V., Han Z., Buyst D., Harris-Coward P.Y., Ehrlich K.C., Wei Q., Bhatnagar D., Dowd P.F., Martens S.L., et al. An Aspergillus flavus secondary metabolic gene cluster containing a hybrid PKS-NRPS is necessary for synthesis of the 2-pyridones, leporins. Fungal Genet. Biol. 2015;81:88–97. doi: 10.1016/j.fgb.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 31.Schindler D., Nowrousian M. The polyketide synthase gene pks4 is essential for sexual development and regulates fruiting body morphology in Sordaria macrospora. Fungal Genet. Biol. 2014;68:48–59. doi: 10.1016/j.fgb.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Hashimoto M., Nonaka T., Fujii I. Fungal type III polyketide synthases. Nat. Prod. Rep. 2014;31:1306–1317. doi: 10.1039/C4NP00096J. [DOI] [PubMed] [Google Scholar]
- 33.Chiang Y.M., Szewczyk E., Davidson A.D., Keller N., Oakley B.R., Wang C.C. A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J. Am. Chem. Soc. 2009;131:2965–2970. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown D.W., Butchko R.A., Baker S.E., Proctor R.H. Phylogenomic and functional domain analysis of polyketide synthases in Fusarium. Fungal Biol. 2012;116:318–331. doi: 10.1016/j.funbio.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Crawford J.M., Dancy B.C., Hill E.A., Udwary D.W., Townsend C.A. Identification of a starter unit acyl-carrier protein transacylase domain in an iterative type I polyketide synthase. Proc. Natl. Acad. Sci. USA. 2006;103:16728–16733. doi: 10.1073/pnas.0604112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe C.M., Townsend C.A. Initial characterization of a type I fatty acid synthase and polyketide synthase multienzyme complex NorS in the biosynthesis of aflatoxin B1. Chem. Biol. 2002;9:981–988. doi: 10.1016/S1074-5521(02)00213-2. [DOI] [PubMed] [Google Scholar]
- 37.Zhou H., Qiao K., Gao Z., Meehan M.J., Li J.W., Zhao X., Dorrestein P.C., Vederas J.C., Tang Y. Enzymatic synthesis of resorcylic acid lactones by cooperation of fungal iterative polyketide synthases involved in hypothemycin biosynthesis. J. Am. Chem. Soc. 2010;132:4530–4531. doi: 10.1021/ja100060k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu T., Sanchez J.F., Chiang Y.M., Oakley B.R., Wang C.C. Rational domain swaps reveal insights about chain length control by ketosynthase domains in fungal nonreducing polyketide synthases. Org. Lett. 2014;16:1676–1679. doi: 10.1021/ol5003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crawford J.M., Thomas P.M., Scheerer J.R., Vagstad A.L., Kelleher N.L., Townsend C.A. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320:243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerber R., Lou L., Du L. A PLP-dependent polyketide chain releasing mechanism in the biosynthesis of mycotoxin fumonisins in Fusarium verticillioides. J. Am. Chem. Soc. 2009;131:3148–3149. doi: 10.1021/ja8091054. [DOI] [PubMed] [Google Scholar]
- 41.Bok J.W., Chiang Y.M., Szewczyk E., Reyes-Dominguez Y., Davidson A.D., Sanchez J.F., Lo H.C., Watanabe K., Strauss J., Oakley B.R., et al. Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schroeckh V., Scherlach K., Nützmann H.W., Shelest E., Schmidt-Heck W., Schuemann J., Martin K., Hertweck C., Brakhage A.A. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez J.F., Chiang Y.M., Szewczyk E., Davidson A.D., Ahuja M., Elizabeth Oakley C., Woo Bok J., Keller N., Oakley B.R., Wang C.C. Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol. Biosyst. 2010;6:587–593. doi: 10.1039/B904541D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujii I., Watanabe A., Sankawa U., Ebizuka Y. Identification of claisen cyclase domain in fungal polyketide synthase wA, a naphthopyrone synthase of Aspergillus nidulans. Chem. Biol. 2001;8:189–197. doi: 10.1016/S1074-5521(00)90068-1. [DOI] [PubMed] [Google Scholar]
- 45.Korman T.P., Crawford J.M., Labonte J.W., Newman A.G., Wong J., Townsend C.A., Tsai S.C. Structure and function of an iterative polyketide synthase thioesterase domain catalyzing claisen cyclization in aflatoxin biosynthesis. Proc. Natl. Acad. Sci. USA. 2010;107:6246–6251. doi: 10.1073/pnas.0913531107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szewczyk E., Chiang Y.M., Oakley C.E., Davidson A.D., Wang C.C., Oakley B.R. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl. Environ. Microbiol. 2008;74:7607–7612. doi: 10.1128/AEM.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim F.Y., Hou Y., Chen Y., Oh J.H., Lee I., Bugni T.S., Keller N.P. Genome-based cluster deletion reveals an endocrocin biosynthetic pathway in Aspergillus fumigatus. Appl. Environ. Microbiol. 2012;78:4117–4125. doi: 10.1128/AEM.07710-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chooi Y.H., Cacho R., Tang Y. Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum. Chem. Biol. 2010;17:483–494. doi: 10.1016/j.chembiol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chooi Y.H., Fang J., Liu H., Filler S.G., Wang P., Tang Y. Genome mining of a prenylated and immunosuppressive polyketide from pathogenic fungi. Org. Lett. 2013;15:780–783. doi: 10.1021/ol303435y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Awakawa T., Yokota K., Funa N., Doi F., Mori N., Watanabe H., Horinouchi S. Physically discrete beta-lactamase-type thioesterase catalyzes product release in atrochrysone synthesis by iterative type I polyketide synthase. Chem. Biol. 2009;16:613–623. doi: 10.1016/j.chembiol.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 51.Chiang Y.M., Szewczyk E., Davidson A.D., Entwistle R., Keller N.P., Wang C.C., Oakley B.R. Characterization of the Aspergillus nidulans monodictyphenone gene cluster. Appl. Environ. Microbiol. 2010;76:2067–2074. doi: 10.1128/AEM.02187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu X., Liu L., Zhang F., Wang W., Li J., Guo L., Che Y., Liu G. Identification of the first diphenyl ether gene cluster for pestheic acid biosynthesis in plant endophyte Pestalotiopsis fici. ChemBioChem. 2014;15:284–292. doi: 10.1002/cbic.201300626. [DOI] [PubMed] [Google Scholar]
- 53.Throckmorton K., Lim F.Y., Kontoyiannis D., Zheng W., Keller N.P. Redundant synthesis of a conidial polyketide by two distinct secondary metabolite clusters in Aspergillus fumigatus. Environ. Microbiol. 2015 doi: 10.1111/1462-2920.13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahuja M., Chiang Y.M., Chang S.L., Praseuth M.B., Entwistle R., Sanchez J.F., Lo H.C., Yeh H.H., Oakley B.R., Wang C.C. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J. Am. Chem. Soc. 2012;134:8212–8221. doi: 10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kroken S., Glass N.L., Taylor J.W., Yoder O.C., Turgeon B.G. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. USA. 2003;100:15670–15675. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y., Image I.I., Xu W., Image I., Tang Y. Classification, prediction, and verification of the regioselectivity of fungal polyketide synthase product template domains. J. Biol. Chem. 2010;285:22764–22773. doi: 10.1074/jbc.M110.128504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu L., Zhang Z., Shao C.L., Wang J.L., Bai H., Wang C.Y. Bioinformatical analysis of the sequences, structures and functions of fungal polyketide synthase product template domains. Sci. Rep. 2015;5 doi: 10.1038/srep10463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lackner G., Misiek M., Braesel J., Hoffmeister D. Genome mining reveals the evolutionary origin and biosynthetic potential of basidiomycete polyketide synthases. Fungal Genet. Biol. 2012;49:996–1003. doi: 10.1016/j.fgb.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 59.Hansen F.T., Gardiner D.M., Lysøe E., Fuertes P.R., Tudzynski B., Wiemann P., Sondergaard T.E., Giese H., Brodersen D.E., Sørensen J.L. An update to polyketide synthase and non-ribosomal synthetase genes and nomenclature in Fusarium. Fungal Genet. Biol. 2015;75:20–29. doi: 10.1016/j.fgb.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 60.Price M.N., Dehal P.S., Arkin A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saha D., Fetzner R., Burkhardt B., Podlech J., Metzler M., Dang H., Lawrence C., Fischer R. Identification of a polyketide synthase required for alternariol (AOH) and alternariol-9-methyl ether (AME) formation in Alternaria alternata. PLoS ONE. 2012;7:e40564. doi: 10.1371/journal.pone.0040564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chooi Y.H., Muria-Gonzalez J.M., Mead O.L., Solomon P.S. Snpks19 encodes the polyketide synthase for alternariol mycotoxin biosynthesis in the wheat pathogen Parastagonospora nodorum. Appl. Environ. Microbiol. 2015 doi: 10.1128/AEM.00278-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.König C.C., Scherlach K., Schroeckh V., Horn F., Nietzsche S., Brakhage A.A., Hertweck C. Bacterium induces cryptic meroterpenoid pathway in the pathogenic fungus Aspergillus fumigatus. ChemBioChem. 2013;14:938–942. doi: 10.1002/cbic.201300070. [DOI] [PubMed] [Google Scholar]
- 64.Li Y., Chooi Y.H., Sheng Y., Valentine J.S., Tang Y. Comparative characterization of fungal anthracenone and naphthacenedione biosynthetic pathways reveals an α-hydroxylation-dependent claisen-like cyclization catalyzed by a dimanganese thioesterase. J. Am. Chem. Soc. 2011;133:15773–15785. doi: 10.1021/ja206906d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Couch R.D., Gaucher G.M. Rational elimination of Aspergillus terreus sulochrin production. J. Biotechnol. 2004;108:171–178. doi: 10.1016/j.jbiotec.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 66.Nielsen M.T., Nielsen J.B., Anyaogu D.C., Holm D.K., Nielsen K.F., Larsen T.O., Mortensen U.H. Heterologous reconstitution of the intact geodin gene cluster in Aspergillus nidulans through a simple and versatile pcr based approach. PLoS ONE. 2013;8:e72871. doi: 10.1371/journal.pone.0072871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Henry K.M., Townsend C.A. Ordering the reductive and cytochrome p450 oxidative steps in demethylsterigmatocystin formation yields general insights into the biosynthesis of aflatoxin and related fungal metabolites. J. Am. Chem. Soc. 2005;127:3724–3733. doi: 10.1021/ja0455188. [DOI] [PubMed] [Google Scholar]
- 68.Henry K.M., Townsend C.A. Synthesis and fate of o-carboxybenzophenones in the biosynthesis of aflatoxin. J. Am. Chem. Soc. 2005;127:3300–3309. doi: 10.1021/ja045520z. [DOI] [PubMed] [Google Scholar]
- 69.Ehrlich K.C., Li P., Scharfenstein L., Chang P.K. HypC, the anthrone oxidase involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2010;76:3374–3377. doi: 10.1128/AEM.02495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cary J.W., Ehrlich K.C., Bland J.M., Montalbano B.G. The aflatoxin biosynthesis cluster gene, aflX, encodes an oxidoreductase involved in conversion of versicolorin A to demethylsterigmatocystin. Appl. Environ. Microbiol. 2006;72:1096–1101. doi: 10.1128/AEM.72.2.1096-1101.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ehrlich K.C., Montalbano B., Boué S.M., Bhatnagar D. An aflatoxin biosynthesis cluster gene encodes a novel oxidase required for conversion of versicolorin A to sterigmatocystin. Appl. Environ. Microbiol. 2005;71:8963–8965. doi: 10.1128/AEM.71.12.8963-8965.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang K.X., Fujii I., Ebizuka Y., Gomi K., Sankawa U. Molecular cloning and heterologous expression of the gene encoding dihydrogeodin oxidase, a multicopper blue enzyme from Aspergillus terreus. J. Biol. Chem. 1995;270:21495–21502. doi: 10.1074/jbc.270.37.21495. [DOI] [PubMed] [Google Scholar]
- 73.Carbone I., Ramirez-Prado J.H., Jakobek J.L., Horn B.W. Gene duplication, modularity and adaptation in the evolution of the aflatoxin gene cluster. BMC Evol. Biol. 2007;7:111. doi: 10.1186/1471-2148-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Medema M.H., Takano E., Breitling R. Detecting sequence homology at the gene cluster level with multigeneblast. Mol. Biol. Evol. 2013;30:1218–1223. doi: 10.1093/molbev/mst025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berthier E., Lim F.Y., Deng Q., Guo C.J., Kontoyiannis D.P., Wang C.C., Rindy J., Beebe D.J., Huttenlocher A., Keller N.P. Low-volume toolbox for the discovery of immunosuppressive fungal secondary metabolites. PLoS Pathog. 2013;9:e1003289. doi: 10.1371/journal.ppat.1003289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ehrlich K.C., Mack B.M., Wei Q., Li P., Roze L.V., Dazzo F., Cary J.W., Bhatnagar D., Linz J.E. Association with AflR in endosomes reveals new functions for AflJ in aflatoxin biosynthesis. Toxins. 2012;4:1582–1600. doi: 10.3390/toxins4121582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skory C.D., Chang P.K., Cary J., Linz J.E. Isolation and characterization of a gene from aspergillus parasiticus associated with the conversion of versicolorin a to sterigmatocystin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 1992;58:3527–3537. doi: 10.1128/aem.58.11.3527-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Simpson T.J. Genetic and biosynthetic studies of the fungal prenylated xanthone shamixanthone and related metabolites in Aspergillus spp. Revisited. ChemBioChem. 2012;13:1680–1688. doi: 10.1002/cbic.201200014. [DOI] [PubMed] [Google Scholar]
- 79.Sanchez J.F., Entwistle R., Hung J.H., Yaegashi J., Jain S., Chiang Y.M., Wang C.C., Oakley B.R. Genome-based deletion analysis reveals the prenyl xanthone biosynthesis pathway in Aspergillus nidulans. J. Am. Chem. Soc. 2011;133:4010–4017. doi: 10.1021/ja1096682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Keller N.P., Kantz N.J., Adams T.H. Aspergillus nidulans verA is required for production of the mycotoxin sterigmatocystin. Appl. Environ. Microbiol. 1994;60:1444–1450. doi: 10.1128/aem.60.5.1444-1450.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Studt L., Wiemann P., Kleigrewe K., Humpf H.U., Tudzynski B. Biosynthesis of fusarubins accounts for pigmentation of Fusarium fujikuroi perithecia. Appl. Environ. Microbiol. 2012;78:4468–4480. doi: 10.1128/AEM.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wiemann P., Willmann A., Straeten M., Kleigrewe K., Beyer M., Humpf H.U., Tudzynski B. Biosynthesis of the red pigment bikaverin in Fusarium fujikuroi: Genes, their function and regulation. Mol. Microbiol. 2009;72:931–946. doi: 10.1111/j.1365-2958.2009.06695.x. [DOI] [PubMed] [Google Scholar]
- 83.Pi B., Yu D., Dai F., Song X., Zhu C., Li H., Yu Y. A genomics based discovery of secondary metabolite biosynthetic gene clusters in Aspergillus ustus. PLoS ONE. 2015;10:e0116089. doi: 10.1371/journal.pone.0116089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cacho R.A., Chooi Y.H., Zhou H., Tang Y. Complexity generation in fungal polyketide biosynthesis: A spirocycle-forming P450 in the concise pathway to the antifungal drug griseofulvin. ACS Chem. Biol. 2013;8:2322–2330. doi: 10.1021/cb400541z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu D. Myceliophthora and Thielavia. In: Liu D., editor. Molecular Detection of Human Fungal Pathogens. CRC Press; Boca Raton, FL, USA: Taylor & Francis Group; Boca Raton, FL, USA: 2011. pp. 445–448. [Google Scholar]
- 86.Gauthier T., Wang X., Sifuentes Dos Santos J., Fysikopoulos A., Tadrist S., Canlet C., Artigot M.P., Loiseau N., Oswald I.P., Puel O. Trypacidin, a spore-borne toxin from Aspergillus fumigatus, is cytotoxic to lung cells. PLoS ONE. 2012;7:e29906. doi: 10.1371/journal.pone.0029906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yilmaz N., Visagie C.M., Houbraken J., Frisvad J.C., Samson R.A. Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 2014;78:175–341. doi: 10.1016/j.simyco.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Adeboya M.O., Edwards R.L., Lassoe T., Maitland D.J., Shields L., Whalley A.J.S. Metabolites of the higher fungi. Part 29. Maldoxin, maldoxone, dihydromaldoxin, isodihydromaldoxin and dechlorodihydromaldoxin. A spirocyclohexadienone, a depsidone and three diphenyl ethers: Keys in the depsidone biosynthetic pathway from a member of the fungus genus Xylaria. J. Chem. Soc. 1996;12:1419–1425. [Google Scholar]
- 89.Chooi Y.H., Wang P., Fang J., Li Y., Wu K., Tang Y. Discovery and characterization of a group of fungal polycyclic polyketide prenyltransferases. J. Am. Chem. Soc. 2012;134:9428–9437. doi: 10.1021/ja3028636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chooi Y.H., Tang Y. Navigating the fungal polyketide chemical space: From genes to molecules. J. Org. Chem. 2012;77:9933–9953. doi: 10.1021/jo301592k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wiemann P., Guo C.J., Palmer J.M., Sekonyela R., Wang C.C., Keller N.P. Prototype of an intertwined secondary-metabolite supercluster. Proc. Natl. Acad. Sci. USA. 2013;110:17065–17070. doi: 10.1073/pnas.1313258110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gibson D.M., Donzelli B.G., Krasnoff S.B., Keyhani N.O. Discovering the secondary metabolite potential encoded within entomopathogenic fungi. Nat. Prod. Rep. 2014;31:1287–1305. doi: 10.1039/C4NP00054D. [DOI] [PubMed] [Google Scholar]
- 93.Andersen M.R., Nielsen J.B., Klitgaard A., Petersen L.M., Zachariasen M., Hansen T.J., Blicher L.H., Gotfredsen C.H., Larsen T.O., Nielsen K.F., et al. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc. Natl. Acad. Sci. USA. 2013;110:E99–E107. doi: 10.1073/pnas.1205532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Finkelstein E., Amichai B., Grunwald M.H. Griseofulvin and its uses. Int. J. Antimicrob. Agents. 1996;6:189–194. doi: 10.1016/0924-8579(95)00037-2. [DOI] [PubMed] [Google Scholar]
- 95.Aggarwal N., Goindi S. Preparation and in vivo evaluation of solid lipid nanoparticles of griseofulvin for dermal use. J. Biomed. Nanotechnol. 2013;9:564–576. doi: 10.1166/jbn.2013.1569. [DOI] [PubMed] [Google Scholar]
- 96.Margarido L.A.C. Oral treatments for fungal infections of the skin of the foot. Sao Paulo Med. J. 2014;132:CD003584. doi: 10.1590/1516-3180.20141322T1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cox R. Oxidative rearrangements during fungal biosynthesis. Nat. Prod. Rep. 2014;31:1405–1424. doi: 10.1039/C4NP00059E. [DOI] [PubMed] [Google Scholar]
- 98.Wicker T., Oberhaensli S., Parlange F., Buchmann J.P., Shatalina M., Roffler S., Ben-David R., Doležel J., Šimková H., Schulze-Lefert P., et al. The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat. Genet. 2013;45:1092–1096. doi: 10.1038/ng.2704. [DOI] [PubMed] [Google Scholar]
- 99.Duplessis S., Cuomo C.A., Lin Y.C., Aerts A., Tisserant E., Veneault-Fourrey C., Joly D.L., Hacquard S., Amselem J., Cantarel B.L., et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA. 2011;108:9166–9171. doi: 10.1073/pnas.1019315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schardl C.L., Young C.A., Hesse U., Amyotte S.G., Andreeva K., Calie P.J., Fleetwood D.J., Haws D.C., Moore N., Oeser B., et al. Plant-symbiotic fungi as chemical engineers: Multi-genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 2013;9:e1003323. doi: 10.1371/journal.pgen.1003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Khouja H.R., Daghino S., Abbà S., Boutaraa F., Chalot M., Blaudez D., Martino E., Perotto S. OmGOGAT-disruption in the ericoid mycorrhizal fungus Oidiodendron maius induces reorganization of the n pathway and reduces tolerance to heavy-metals. Fungal Genet. Biol. 2014;71:1–8. doi: 10.1016/j.fgb.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 102.Elmore M.H., McGary K.L., Wisecaver J.H., Slot J.C., Geiser D.M., Sink S., O’Donnell K., Rokas A. Clustering of two genes putatively involved in cyanate detoxification evolved recently and independently in multiple fungal lineages. Genome Biol. Evol. 2015;7:789–800. doi: 10.1093/gbe/evv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu Q., Ying S.H., Li J.G., Tian C.G., Feng M.G. Insight into the transcriptional regulation of Msn2 required for conidiation, multi-stress responses and virulence of two entomopathogenic fungi. Fungal Genet. Biol. 2013;54:42–51. doi: 10.1016/j.fgb.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 104.Punya J., Swangmaneecharern P., Pinsupa S., Nitistaporn P., Phonghanpot S., Kunathigan V., Cheevadhanarak S., Tanticharoen M., Amnuaykanjanasin A. Phylogeny of type I polyketide synthases (PKSs) in fungal entomopathogens and expression analysis of PKS genes in Beauveria bassiana BCC 2660. Fungal Biol. 2015;119:538–550. doi: 10.1016/j.funbio.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 105.Chibucos M.C., Crabtree J., Nagaraj S., Chaturvedi S., Chaturvedi V. Draft genome sequences of human pathogenic fungus Geomyces pannorum sensu lato and bat white nose syndrome pathogen Geomyces (Pseudogymnoascus) destructans. Genome Announc. 2013;1 doi: 10.1128/genomeA.01045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 107.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yadav G., Gokhale R.S., Mohanty D. Towards prediction of metabolic products of polyketide synthases: An in silico analysis. PLoS Comput. Biol. 2009;5:e1000351. doi: 10.1371/journal.pcbi.1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I., et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. Trimal: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rambaut A. Figtree. [(accessed on 6 April 2015)]. Available online: http://tree.bio.ed.ac.uk/software/figtree/
- 114.Bendele A.M., Carlton W.W., Nelson G.E., Peterson R.E., Grove M.D. Viridicatumtoxin mycotoxicosis in mice and rats. Toxicol. Lett. 1984;22:287–291. doi: 10.1016/0378-4274(84)90103-6. [DOI] [PubMed] [Google Scholar]
- 115.Hutchison R.D., Steyn P.S., Van Rensburg S.J. Viridicatumtoxin, a new mycotoxin from Penicillium viridicatum westling. Toxicol. Appl. Pharmacol. 1973;24:507–509. doi: 10.1016/0041-008X(73)90057-4. [DOI] [PubMed] [Google Scholar]
- 116.Pero R.W., Posner H., Blois M., Harvan D., Spalding J.W. Toxicity of metabolites produced by the “Alternaria”. Environ. Health Perspect. 1973;4:87–94. doi: 10.1289/ehp.730487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Demuner A.J., Almeida Barbosa L.C., Mendes Miranda A.C., Geraldo G.C., da Silva C.M., Giberti S., Bertazzini M., Forlani G. The fungal phytotoxin alternariol 9-methyl ether and some of its synthetic analogues inhibit the photosynthetic electron transport chain. J. Nat. Prod. 2013;76:2234–2245. doi: 10.1021/np4005882. [DOI] [PubMed] [Google Scholar]
- 118.Esbelin J., Mallea S., Ram A.F., Carlin F. Role of pigmentation in protecting Aspergillus niger conidiospores against pulsed light radiation. Photochem. Photobiol. 2013;89:758–761. doi: 10.1111/php.12037. [DOI] [PubMed] [Google Scholar]
- 119.Romero-Martinez R., Wheeler M., Guerrero-Plata A., Rico G., Torres-Guerrero H. Biosynthesis and functions of melanin in Sporothrix schenckii. Infect. Immun. 2000;68:3696–3703. doi: 10.1128/IAI.68.6.3696-3703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang Y., Casadevall A. Decreased susceptibility of melanized Cryptococcus neoformans to UV light. Appl. Environ. Microbiol. 1994;60:3864–3866. doi: 10.1128/aem.60.10.3864-3866.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tsai H.F., Chang Y.C., Washburn R.G., Wheeler M.H., Kwon-Chung K.J. The developmentally regulated alb1 gene of Aspergillus fumigatus: Its role in modulation of conidial morphology and virulence. J. Bacteriol. 1998;180:3031–3038. doi: 10.1128/jb.180.12.3031-3038.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fourie P., Schutte T., Serfontein S., Swart F. Modeling the effect of temperature and wetness on Guignardia pseudothecium maturation and ascospore release in citrus orchards. Phytopathology. 2013;103:281–292. doi: 10.1094/PHYTO-07-11-0194. [DOI] [PubMed] [Google Scholar]
- 123.Guenther J.C., Trail F. The development and differentiation of Gibberella zeae (anamorph: Fusarium graminearum) during colonization of wheat. Mycologia. 2005;97:229–237. doi: 10.3852/mycologia.97.1.229. [DOI] [PubMed] [Google Scholar]
- 124.Brown D.W., Salvo J.J. Isolation and characterization of sexual spore pigments from Aspergillus nidulans. Appl. Environ. Microbiol. 1994;60:979–983. doi: 10.1128/aem.60.3.979-983.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.