Abstract
The timing of the onset and release of dormancy impacts the survival, productivity and spatial distribution of temperate horticultural and forestry perennials and is mediated by at least three main regulatory programs involving signal perception and processing by phytochromes (PHYs) and PHY-interacting transcription factors (PIFs). PIF4 functions as a key regulator of plant growth in response to both external and internal signals. In poplar, the expression of PIF4 and PIF3-LIKE1 is upregulated in response to short days, while PHYA and PHYB are not regulated at the transcriptional level. Integration of light and environmental signals is achieved by gating the expression and transcriptional activity of PIF4. During this annual cycle, auxin promotes the degradation of Aux/IAA transcriptional repressors through the SKP–Cullin-F–boxTIR1 complex, relieving the repression of auxin-responsive genes by allowing auxin response factors (ARFs) to activate the transcription of auxin-responsive genes involved in growth responses. Analyses of transcriptome changes during dormancy transitions have identified MADS-box transcription factors associated with endodormancy induction. Previous studies show that poplar dormancy-associated MADS-box (DAM) genes PtMADS7 and PtMADS21 are differentially regulated during the growth-dormancy cycle. Endodormancy may be regulated by internal factors, which are specifically localized in buds. PtMADS7/PtMADS21 may function as an internal regulator in poplar. The control of flowering time shares certain regulatory hierarchies with control of the dormancy/growth cycle. However, the particularities of different stages of the dormancy/growth cycle warrant comprehensive approaches to identify the causative genes for the entire cycle. A growing body of knowledge also indicates epigenetic regulation plays a role in these processes in perennial horticultural and forestry plants. The increased knowledge contributes to better understanding of the dormancy process and consequently to precise manipulation of dormancy-related horticultural traits, such as flowering time.
Introduction
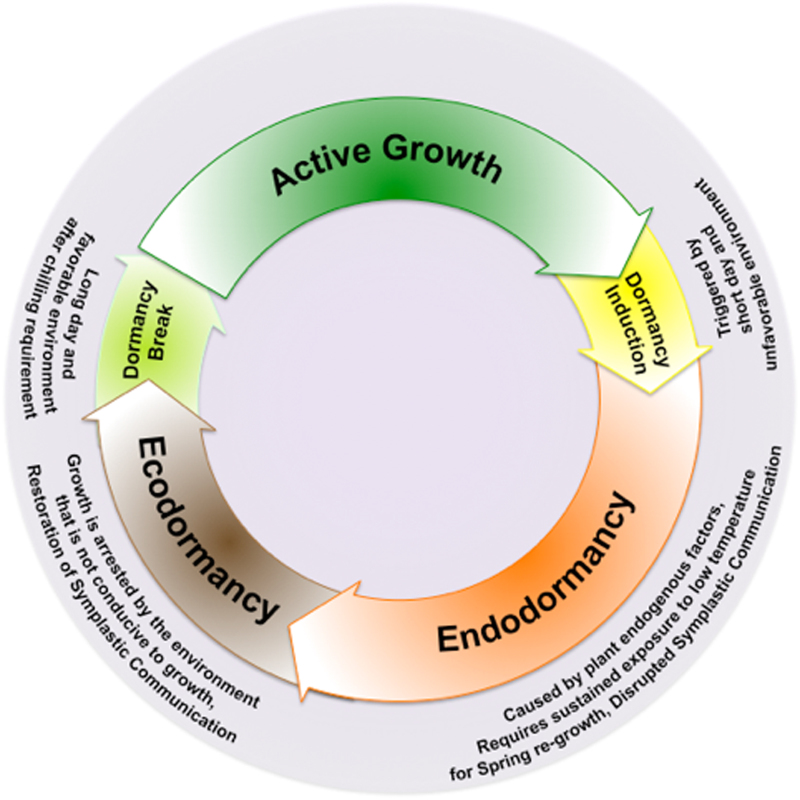
Because of their multiyear life spans, temperate perennial horticultural and forestry plants have an adaptive mechanism that alters active growth and vegetative dormancy in accord with seasonal climate changes. These plants use cyclically changing environmental signals, such as day-length and temperature, to coordinate their growth and development with seasonal changes in climate (Figure 1).1–3 Thus, the proper timing of the onset and release of dormancy impacts the survival, productivity and spatial distribution of temperate perennials. Dormancy is frequently defined as the ‘absence of visible growth in any plant structure containing a meristem’.4,5 In that context, Lang4 further described three types of vegetative dormancy: (i) paradormancy, also known as apical dominance, which is the suppression of lateral bud growth by the actively growing portion, such as the apical meristem; (ii) ecodormancy, in which growth is arrested by environmental conditions that are not conducive to growth but resumes when conditions again become favorable; and (iii) endodormancy, which is caused by plant endogenous factors and requires a sustained exposure to low temperatures for spring regrowth to occur. Plants in the endodormancy state are not capable of growth even if external physiological signals are removed and returned to growth-promoting conditions.6 However, this widely accepted definition of dormancy relies on visible physiological changes in the plant, such as changes in bud phenology, and, while evidencing the result of the developmental processes within the plants, does not actually account for the cellular and molecular changes occurring within the meristems during transitions into and out of dormancy. Recently, Rohde and Bhalerao7 introduced a new definition of dormancy as ‘the inability to initiate growth from meristems (and other organs and cells with the capacity to resume growth) under favorable conditions’.
Figure 1.
Transitions in seasonal growth-dormancy cycling in temperate woody perennials. Temperate perennials synchronize the onset of vegetative dormancy in accord with the changes in the environment. Decreasing day-length is sensed by phytochromes and triggers growth cessation and dormancy, coupled with low temperature. Prolonged exposure to chilling temperatures is required for dormancy release. Growth resumes once the environment becomes conducive to growth (i.e., day-length and temperature above critical thresholds).
The overarching question of the molecular mechanisms that govern the dormancy and growth changes in perennial plants in response to seasonal climatic variation remains largely unanswered. While temporal coincidence between the seasonal changes and the internal biological clock is thought to be prerequisite for these cyclical processes, the underlying genetic control must be more complex than simply a response to shifts in these two environmental stimuli given the highly complex and physiologically different responses to these stimuli in spring and fall. Indeed, in many localities, comparable temperatures and day lengths are present during spring and fall, yet in one instance, dormancy is induced and in the other the initiation of annual growth occurs. Fulfilling a chilling requirement prior to breaking dormancy has been suggested as the reason for this differential response,5 but the genes or regulation of gene expression associated with such a mechanism have not been identified. By what mechanisms then do perennial plants, such as fruit trees, use the roughly equivalent photoperiods and temperatures of spring and fall, as well as the intervening seasons of warmth and cold, to regulate these two dramatically different physiological responses?
Better understanding of the molecular mechanisms controlling the annual growth and dormancy cycle has the potential to help mitigate the impact of climate change on plant productivity and survival by providing vital information about how temperate perennials utilize the environmental cues to trigger adaptive mechanisms. Increases in winter temperature would be expected to cause delayed and erratic flower development in tree species with large chilling requirements and both early spring bud break and increased risk of frost damage in species with low chilling requirements. Dormancy also determines winter survival of both native and invasive perennials and thus affects agricultural productivity and biodiversity in the ecosystem. Identification of expression-based molecular markers for dormancy regulation may facilitate cultivar selection and breeding for development of regionally suited crops in accord with changes in the global climate.
Recent efforts to elucidate the transcriptional regulatory mechanisms by which woody plants use environmental stimuli to coordinate their periods of growth with the annual cycle of environmental conditions have contributed much to our understanding of these processes. The growth-dormancy cycle in temperate perennials appears to be tightly controlled by complex genetic regulatory networks in response to endogenous and environmental signals.3,8–14 Examination of available literature on dormancy regulation suggests that growth cessation and dormancy onset is mediated by at least three main regulatory programs: (i) short-day (SD) signal perception and processing by phytochromes (PHYs) and PHY-interacting transcription factors (PIFs);7,15–17 (ii) dormancy cycle stage-specific alterations in the auxin responsiveness of the transcriptome, possibly through the action of auxin receptor F-box protein TIR1;8 and (iii) internal factors, such as dormancy-associated MADS-box (DAM) transcription factors, controlling endodormancy.18 Recently, we carried out a large-scale RNA-Seq-based Digital Gene Expression profiling using the stems of field-grown poplars and have identified 305 dormancy-associated transcription factors (unpublished data). These studies show that dormancy regulation in poplar is achieved at both transcriptional (e.g., transcription factors PIFs and DAMs) and post-transcriptional (e.g., TIR1) levels.
Perception of short-day photoperiods induces growth cessation and transition to dormancy
Due to their lack of variation from year to year, seasonal changes in photoperiod provide a consistent source of information for the timing of developmental events in perennial plants. Plants monitor photoperiod changes by means of light-sensitive pigments, including PHY, which play central roles in modulating key physiological and developmental processes.19,20 PHYs, red and far-red light photoreceptors, are synthesized in the cytosol and translocated to the nucleus as the active Pfr form in response to red light.21–23 The light-activated PHY then triggers signaling events that alter the expression of target genes, modulating key physiological and developmental processes of plants.24–26 Plants have multiple PHYs (e.g., PHYA, PHYB, etc.) that differentially respond to the variation in the light spectrum found in common terrestrial environments20,27 and promote an array of growth responses in seeds, seedlings and mature plants. However, the regulatory gene networks targeted by all classes of plant PHYs significantly overlap and share common regulatory factors.28,29
PIF4 functions as a key regulator of plant growth in response to external signals, such as temperature and light, as well as internal signals, including gibberellin and the circadian clock. Light-activated photoreceptor PHYs (Pfr) directly interact with the basic helix–loop–helix transcription factors named phytochrome interacting factors, PIFs.28,30–32 PIF4/PIF5 acts as a negative regulator of PHYB signaling.33 They accumulate to a high level in the dark but are degraded upon interaction with the Pfr form of PHYB.32,34 The transcription of PIF4/PIF5 is regulated by circadian clock-mediated coincidence mechanisms. PIF4/PIF5 transcripts are accumulated in the nighttime of SDs due to coincidence between the internal (circadian clock) and external (photoperiod) time cues, while the night-time expression disappears in long days.35 It appears that both transcriptional (i.e., the clock-mediated upregulation of PIF4/PIF5) and post-transcriptional (i.e., the degradation of PIF4/PIF5 by light-activated PHYB) regulation govern the photoperiodic control of hypocotyl elongation in SDs.36 In poplar, the expression of PIF4 and PIF3-LIKE1 is upregulated in response to SDs, while PHYA and PHYB are not regulated at the transcriptional level.18 Along with PIF4/PIF5, PIF3 has also been shown to be involved in photoperiodic control of plant growth.37 PIF3 interacts with the Pfr form of both PHYA and PHYB.28 However, in sharp contrast to PIF4/PiF5, PIF3 is not subject to transcriptional regulation by the circadian clock, but its protein abundance oscillates under diurnal conditions as a result of declining protein degradation governed by PHYB.37
Both light and temperature signaling serve as critical environmental cues for growth cessation and dormancy onset. Perception of SD photoperiods induces growth cessation and transition to dormancy.38 Poplar responds to SDs with a PHYA-mediated cessation of elongation growth, terminal bud set and dormancy.7,15–17 Temperature signals function as an additional environmental factor to modify the sensitivity to photoperiod signals at growth cessation.39 Integration of light and environmental signals is achieved by gating the expression and transcriptional activity of PIF4. Considering that other environmental conditions (e.g., temperature and nutrient conditions) also regulate photomorphogenesis,40,41 it is important to also understand whether PIF4/PIF5 plays a role in plant responses to environmental cues other than light. In addition to their crucial roles in the diurnal and SD-specific control of hypocotyl elongation, PIF4 and its close homolog PIF5 are involved in auxin signaling in Arabidopsis thaliana.42 PIF4 increases auxin levels to the point necessary to promote elongation growth at high temperature,43 further confirming the earlier discovery that PIF4 integrates both light and temperature signals to regulate plant growth and development.28,34,40,44 PIF4/PIF5 also acts as modulators of auxin signaling in photoperiodic control of plant growth.36,37,42 Transcriptional activity of PIF4 is blocked by DELLA proteins, which function as key repressors of GA-responsive growth through inhibition of GA-regulated gene expression, by binding the DNA-recognition domain of PIF4.45 GA induces ubiquitin-mediated degradation of DELLA proteins, thereby preventing DELLA from binding to PIF4, which then results in concomitant accumulation of free PIF4 in the nucleus.45 Brassinosteroids play a crucial role in light- and temperature-mediated growth regulation through their promotion of cell elongation, cell expansion and vascular differentiation. PIF4 interacts directly with a brassinosteroid-activated transcription factor BZR1 and then together bind to thousands of common target genes to interdependently regulate the transcription of the targets.46 Based on these findings, it appears that PIFs function as cellular ‘hubs’ where environmental signals are integrated to regulate plant responses to the changing environmental conditions.28,44 However, the mechanisms by which PIFs control dormancy cycle remain unknown.
Cellular response to plant growth regulators is a part of seasonal dormancy regulation
The plant hormone auxin regulates virtually every aspect of plant growth and development. Therefore, how the auxin signal is perceived and interpreted by plant cells is also a central question in the biology of dormancy. In poplar, SD-induced growth cessation and dormancy onset involves dormancy cycle stage-specific alterations in auxin responsiveness of the transcriptome in the stem tissues.8 The cellular response to auxin is quite rapid, with transcription regulation of a large and diverse set of genes detectable within minutes of exposure.47 Proteins involved in auxin distribution and signaling are induced by auxin, including auxin efflux machinery components (e.g., PIN proteins), influx (e.g., LAX proteins) carriers and Aux/IAA proteins that are unstable transcriptional repressors of auxin responses.48 The expression of Aux/IAAs, primary auxin response genes, is rapidly upregulated by auxin. The Aux/IAA proteins interact with auxin response factors (ARFs), a family of transcription factors that bind auxin-responsive promoters and regulate transcription.49 This interaction negatively regulates auxin-responsive gene expression by keeping ARFs from forming functional dimers.50 Auxin promotes the degradation of Aux/IAA transcriptional repressors through the ubiquitin protein ligase SKP–Cullin-F–boxTIR1 complex (SCFTIR1),51,52 thereby relieving the active repression of the auxin-responsive genes by allowing ARFs to activate the transcription of auxin-responsive genes.53
The F-box protein subunit of the SCF complex (TIR1 in the case of Aux/IAA) confers target specificity. Direct binding of auxin to TIR1 allows the TIR1-Aux/IAA binding. Crystal structures show that auxin binds to a single surface pocket of TIR1 and the Aux/IAA substrate peptides, docked on top of auxin, occupy the rest of the TIR1 pocket.54,55 TIR1 is required for auxin response in Arabidopsis.56 Quadruple mutants of TIR1 and its three homologs AFB (tir1 afb1 afb2 afb3) are auxin-insensitive,57 further supporting the hypothesis that auxin stimulates binding of SCFTIR1 to the AUX/IAA repressor, and its degradation, resulting in the activation of auxin-responsive genes. The expression of PttTIR1, a poplar homolog of TIR1, was reduced by 50% after 42 SDs preceding endodormancy, suggesting its role in SD-induced growth cessation and dormancy onset in poplar.8 In order to gain insight into how auxin signaling induces growth cessation and dormancy onset, it is imperative to identify the molecular regulators that mediate SD modulation of auxin responsiveness in perennial plants.
In Japanese apricot (Prunus persica), various hormone response genes, such as INDOLE-3-ACETIC ACID-INDUCIBLE 33 (IAA33), the gibberellin-regulated gene family, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5 (ERF5), the auxin-responsive gene family and a member of the F-box/RNI-LIKE gene family, were differentially regulated at four different seasonal dormancy stages, suggesting their involvement in dormancy regulation.58 Exogenous application of GA4 resulted in earlier budbreak in Japanese apricot.59 The proteomic and transcriptomic analysis after the GA4 treatment showed that proteins and genes associated with energy metabolism appear to be involved in dormancy release.
Dam genes may function as a positive regulator of endodormancy induction
Analyses of transcriptome changes during dormancy transitions in several perennial plant species, including poplars, have identified several MADS-box transcription factors associated with endodormancy induction. The first definitive evidence that DAM genes directly impact endodormancy was obtained from the analysis of evergrowing (evg) mutant of peach [Prunus persica (L.) Batsch], which fails to cease growth and enter dormancy under dormancy-inducing conditions.60,61 The DAM genes are expressed in terminal buds of the wild type, whereas their expression is lost in the evg mutant.62 Expression of DAM3, DAM5 and DAM6 during winter indicated involvement of the transcription factors in endodormancy were expressed during winter, with expression of DAM5 and DAM6 higher in high chill cultivars and at a minimum coincident to bud break, indicating a potential role in repressing bud break.63 Deletion of six tandem-arrayed DAM genes (PpMADS1-6) in the EVG locus resulted in loss of endodormancy induction.64,65 DAM homologs in leafy spurge (Euphorbia esula), EeDAM1 and EeDAM2, were associated with endodormancy induction.13
DAM genes have a high sequence similarity to two different transcription factors, SHORT VEGETATIVE PHASE (SVP) and AGAMOUS-LIKE24 (AGL24), which were previously associated with flowering regulation in the winter annual Arabidopsis. Yamane et al.66 identified a MADS-box gene with endodormancy-associated expression in Japanese apricot (Prunus mume). This gene has a high sequence similarity with SVP and AGL24 and is upregulated during endodormancy induction and downregulated during endodormancy release.63 SVP inhibits flowering through negative regulation of a floral integrator FLOWERING LOCUS T (FT),67,68 while AGL24 acts as a promoter of flowering.69 SVP is upregulated under short day conditions in Arabidopsis.13 In poplar, overexpression of FT inhibits SD-induced growth cessation.70,71 Transgenic poplars overexpressing PHYA have elevated expression levels of FT and FT-like genes, such as CENL1.69 Expression of FT or CENL1 is downregulated in poplar18,72 and spruce during induction of endodormancy.73 FT expression in leafy spurge buds was coordinately down regulated as DAM gene expression was induced,13 indicating coordinated regulation of genes associated with flowering and with dormancy onset and release in accord with cyclical changes in the growth environment.
The analysis of the phylogenetic relationship between Arabidopsis, peach and poplar MIKCC-type MADS-box genes shows that the peach DAM genes belong to a clade with two Arabidopsis (AGL22/SVP and AGL24) and eight poplar (PtMADS7, PtMADS21, PtMADS26, PtMADS27, PtMADS28, PtMADS29, PtMADS47 and PtMADS48) MADS-box genes.74 In the clade, two poplar homologs, PtMADS7 and PtMADS21, formed a monophyletic group with the six peach homologs. Transgenic poplar plants constitutively overexpressing an apricot DAM (PmDAM6) showed growth cessation and terminal bud set under environmental conditions that are conducive to continued growth.75 Our preliminary data show that the two poplar DAM genes (PtMADS7 and PtMADS21) are differentially regulated during the growth-dormancy cycle. Based on the findings reported in the literature and our preliminary study, it is possible that endodormancy is regulated by internal factors (e.g., internal growth inhibitors), which are specifically localized in buds during endodormancy and prevent the buds from resuming growth. Thus, it is prudent to consider that PtMADS7/PtMADS21 may function as such an internal regulator in poplar.
Transcriptome changes occurring during the growth-dormancy cycle
Studies on phenological traits in Populus suggest that the timing of bud flush may be controlled by a modest number of major genes with low environmental variation.76,77 However, no such genes have yet been identified. The changes in gene expression during both the onset and breaking of dormancy in poplar have been described.6,7,18 Schrader et al.78,79 and Druart et al.10 reported a comprehensive analysis of cambial dormancy utilizing cryosection-isolated cambial cells from P. tremula during active growth and dormancy. These studies used cDNA microarray experiments (POP1)80 to investigate transcriptional changes underlying cambial dormancy. However, the POP1 cDNA microarray consists of only 13 490 clones with high redundancy, which is not enough to cover the entire transcriptome of poplar in which currently at least 45 778 protein-coding loci have been identified (Phytozome, http://www.phytozome.net/poplar.php). Nonetheless, very significant insights into dormancy/growth cycle regulation were gained when Bhalerao and his colleagues carried out global transcript profiling (POP2 cDNA Arrays) during the activity-dormancy transition in the cambial meristem in a hybrid aspen.8 Their findings suggested that dormancy stage-specific changes of auxin responsiveness of the transcriptome might be involved in SD-induced growth cessation. Recently, Ko et al.14 obtained whole genome transcriptional profiles (Affymetrix Poplar GeneChip Arrays) covering the winter and summer stems of field-grown P. trichocarpa trees. The study showed high levels of transcriptional activity and differential regulation of a broad array of genes beyond those associated with environmental stress during the dormancy period. In grape (Vitis vinifera), global transcriptome analysis during bud development showed that a MADS-box gene (VvFLC2) and other transcripts with similar expression patterns appear to be involved in the regulation of endodormancy induction.81 In kiwifruit, upregulation of the glutathione S-transferase class of genes, which are associated with stress responses, was observed in response to application of hydrogen cyanamide used to synchronize budbreak.82 In raspberry, microarray analysis of gene expression during dormancy transition enabled identification of key dormancy-related genes, with a high percentage of stress-response genes present in the expression profiles.83 Hedley et al.84 identified 842 differentially expressed ESTs (expressed sequence tags) during dormancy release in blackcurrant (Ribes nigra) with high representation of genes encoding transcription factors such as zinc finger family members (B-box, CCCH, C2H2 and C3HC4/RING types), which are known to play important roles in signal transduction pathways. The EST analysis also showed that expression of a Short Vegetative Phase (SVP)-type MADS box transcription factor gene peaked in early winter and gradually declined into spring-time. Homologues of SVP in apricot also showed similar expression pattern to that of blackcurrant under dormancy release inducing condition.66,83
Recent discoveries have shown that the control of flowering time shares certain regulatory hierarchies with control of the seasonal dormancy/growth cycle.85,86 Overexpression of a poplar homolog of FLOWERING LOCUS T2 (FT2) suppressed SD-induced growth cessation and dormancy onset and its downregulation by RNAi led to less vegetative growth.71 In addition, overexpression of a poplar FLC-like MADS box gene delayed SD-induced growth cessation and leaf senescence in poplar.87 In leafy spurge, analysis of 1000 genes differentially expressed through dormancy transitions led to the identification of a MADS-box transcription factor related to the DAM genes from peach. The authors hypothesize that it may play a direct role in dormancy induction and maintenance through regulation of FLOWERING LOCUS T.13 Rinne et al.88 investigated the molecular and cellular-level changes occurring during the dormancy cycle at the shoot apical meristem (SAM) of poplar. That study showed that chilling-induced expression of 1,3-β-glucanases at the SAM reopens signal conduits (i.e., plasmodesmata) and subsequently enables the symplastic movement of FT to its target, resulting in dormancy release.89 While these commonalities between flowering and dormancy regulation offer new avenues to critically assess the transcriptional network for seasonal regulation of the dormancy/growth cycle, the particularities of different stages of the dormancy/growth cycle, especially in terms of gene expression in the meristems, warrant various comprehensive approaches to identify the causative genes involved in the entire dormancy cycle.
Epigenetic regulation of dormancy
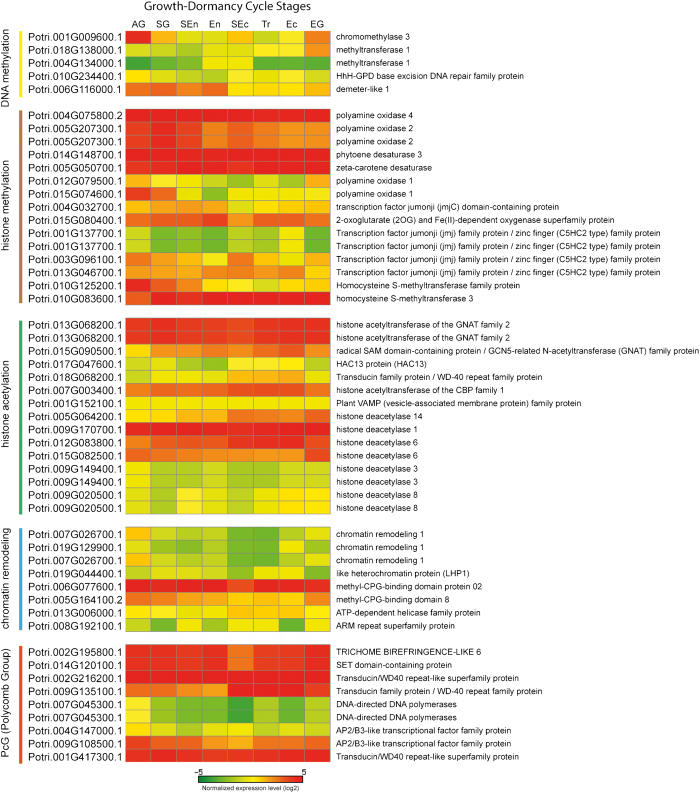
There is growing evidence that genome-wide epigenetic regulation of gene expression is involved in dormancy regulation (for reviews) (Table 1).90,91 Whole-genome DNA methylation and acetylated histone 4 (H4) analyses in chestnut (Castanea sativa) showed higher DNA methylation ratios and lower H4 acetylation levels in dormant buds compared to actively growing tissues,92,93 suggesting gene silencing concomitant with bud dormancy. In hybrid aspen, putative histone deacetylases (HDA14 and HDA08), histone lysine methyltranferase (SUVR3) and histone ubiquitination involved gene (HUB2) were upregulated during growth-dormancy transition, while several Trithorax family genes counteracting the repressive effect of Polycomb complex and putative DEMETER-like DNA glycosylases are downregulated.89 These results suggest that some unknown active growth regulating genes might be repressed during dormancy induction through chromatin compaction mechanisms, such as histone deacetylation and methylation, histone ubiquitination, DNA methylation and Polycomb activity. Ko et al.14 also reported that many genes involved in chromatin remodeling/modification in poplar (e.g., including BRAT1, SWI2/SNF2, NFC2 and PAF1) showed upregulation in winter/dormancy stems, suggesting the dynamic nature of winter/dormancy-responsive gene regulation at chromatin level (Figure 2).
Table 1. List of the genes that are associated with epigenetic regulation during dormancy.
| Subgroup | P. trichocarpa v3.0 ID | Description | AGI | Reference |
|---|---|---|---|---|
| DNA methylation | Potri.001G009600.1 | Chromomethylase 3 | At1g69770 | 100 |
| Potri.018G138000.1 | Methyltransferase 1 | At4g08990 | 100 | |
| Potri.004G134000.1 | Methyltransferase 1 | At4g13610 | 100 | |
| Potri.010G234400.1 | HhH-GPD base excision DNA repair family protein | At5g04580 | 101 | |
| Potri.006G116000.1 | Demeter-like 1 | At3g10010 | 101 | |
| Histone methylation | Potri.004G075800.2 | Polyamine oxidase 4 | At1g65840 | 102 |
| Potri.005G207300.1 | Polyamine oxidase 2 | At2g43020 | 102 | |
| Potri.005G207300.1 | Polyamine oxidase 2 | At2g43020 | 102 | |
| Potri.014G148700.1 | Phytoene desaturase 3 | At4g14210 | 102 | |
| Potri.005G050700.1 | Zeta-carotene desaturase | At3g04870 | 102 | |
| Potri.012G079500.1 | Polyamine oxidase 1 | At5g13700 | 102 | |
| Potri.015G074600.1 | Polyamine oxidase 1 | At5g13700 | 102 | |
| Potri.004G032700.1 | transcription factor jumonji domain-containing protein | At1g62310 | 102 | |
| Potri.015G080400.1 | 2-oxoglutarate and Fe(II)-dependent oxygenase superfamily protein | At5g63080 | 102 | |
| Potri.001G137700.1 | Transcription factor jumonji family protein/zinc finger (C5HC2 type) family protein | At5g46910 | 102 | |
| Potri.001G137700.1 | Transcription factor jumonji family protein/zinc finger (C5HC2 type) family protein | At5g46910 | 102 | |
| Potri.003G096100.1 | Transcription factor jumonji family protein/zinc finger (C5HC2 type) family protein | At5g46910 | 102 | |
| Potri.013G046700.1 | Transcription factor jumonji family protein/zinc finger (C5HC2 type) family protein | At1g08620 | 102 | |
| Potri.010G125200.1 | Homocysteine S-methyltransferase family protein | At3g25900 | ||
| Potri.010G083600.1 | Homocysteine S-methyltransferase 3 | At3g22740 | ||
| Histone acetylation | Potri.013G068200.1 | Histone acetyltransferase of the GNAT family 2 | At5g56740 | 103 |
| Potri.013G068200.1 | Histone acetyltransferase of the GNAT family 2 | At5g56740 | 103 | |
| Potri.015G090500.1 | Radical SAM domain-containing protein/GCN5-related N-acetyltransferase (GNAT) family protein | At5g50320 | 103 | |
| Potri.017G047600.1 | HAC13 protein (HAC13) | At3g19040 | 103 | |
| Potri.018G068200.1 | Transducin family protein/WD-40 repeat family protein | At1g55970 | 103 | |
| Potri.007G003400.1 | Histone acetyltransferase of the CBP family 1 | At1g55970 | 103 | |
| Potri.001G152100.1 | Plant VAMP (vesicle-associated membrane protein) protein | At1g55970 | 103 | |
| Potri.005G064200.1 | Histone deacetylase 14 | At4g33470 | 103 | |
| Potri.009G170700.1 | Histone deacetylase 1 | At4g38130 | 103 | |
| Potri.012G083800.1 | Histone deacetylase 6 | At5g63110 | 103, 104 | |
| Potri.015G082500.1 | Histone deacetylase 6 | At5g63110 | 103, 104 | |
| Potri.009G149400.1 | Histone deacetylase 3 | At3g44750 | 103 | |
| Potri.009G149400.1 | Histone deacetylase 3 | At3g44750 | 103 | |
| Potri.009G020500.1 | Histone deacetylase 8 | At1g08460 | 103 | |
| Potri.009G020500.1 | Histone deacetylase 8 | At1g08460 | 103 | |
| Chromatin remodeling | Potri.007G026700.1 | Chromatin remodeling 1 | At5g66750 | |
| Potri.019G129900.1 | Chromatin remodeling 1 | At5g66750 | 105 | |
| Potri.007G026700.1 | Chromatin remodeling 1 | At5g66750 | 105 | |
| Potri.019G044400.1 | Like heterochromatin protein (LHP1) | At5g17690 | 104 | |
| Potri.006G077600.1 | Methyl-CPG-binding domain protein 02 | At5g35330 | ||
| Potri.005G164100.2 | Methyl-CPG-binding domain 8 | At1g22310 | ||
| Potri.013G006000.1 | ATP-dependent helicase family protein | At1g08060 | 104 | |
| Potri.008G192100.1 | ARM repeat superfamily protein | At3g06400 | ||
| PcG (Polycomb Group) | Potri.002G195800.1 | TRICHOME BIREFRINGENCE-LIKE 6 | At4g02020 | 104, 106 |
| Potri.014G120100.1 | SET domain-containing protein | At4g02020 | 104, 106 | |
| Potri.002G216200.1 | Transducin/WD40 repeat-like superfamily protein | At5g58230 | 104, 106 | |
| Potri.009G135100.1 | Transducin family protein/WD-40 repeat family protein | At2g16780 | 104, 106 | |
| Potri.007G045300.1 | DNA-directed DNA polymerases | At5g67100 | 106 | |
| Potri.007G045300.1 | DNA-directed DNA polymerases | At5g67100 | 106 | |
| Potri.004G147000.1 | AP2/B3-like transcriptional factor family protein | At1g49480 | 106 | |
| Potri.009G108500.1 | AP2/B3-like transcriptional factor family protein | At3g18990 | 106 | |
| Potri.001G417300.1 | Transducin/WD40 repeat-like superfamily protein | At3g20740 | 104, 106 |
Figure 2.
Heat map of 52 genes that are associated with epigenetic regulation during the dormancy cycle. To show their normalized expression levels according to eight different growth-dormancy cycle stages, we used our unpublished RNA-Seq data to create a heat map for the genes involved in DNA methylation, histone modification, chromatin remodeling and polycomb group (PcG). The gene expression values were normalized by RPKM method. Red and green in the heat map mean upregulated and downregulated genes, respectively. Growth-dormancy cycle stages are active growth (AG), stop growth (SG), start of endodormancy (SEn), endodormancy (En), start of ecodormancy (SEc), transition of ecodormancy (Tr), ecodormancy (Ec) and growth resumption (EG).
Two genes similar to FERTILIZATION INDEPENDENT ENDOSPERM (FIE) and PICKLE (PKL), which are involved in epigenetic regulation, were strongly up-regulated in poplar immediately after transfer to SD conditions.18 FIE is known as a component of the Polycomb repressive complex 2 involved in the trimethylation of H3 lysine 27 (H3K27me3), while PKL is an ATP-dependent chromatin remodeler that regulates embryo identity traits.94,95 In fact, transgenic hybrid aspen lines with RNAi-mediated suppression of FIE were not able to establish dormancy, even though growth cessation and bud formation processes did not seem to be affected.96
A time period of quantitative and cumulative chilling is required for both dormancy release and vernalization for flowering in Arabidopsis. In this process, transcriptional regulation of MADS box genes, FLOWERING LOCUS C (FLC) and DAM, appears to include common epigenetic mechanisms. Horvath et al.86 reported that the chromatin in the promoter region of leafy spurge DAM1 had a decrease in the level of trimethylation of H3 lysine 4 (H3K4me3) (i.e., active histone modification) and an increase of H3K27me3 (i.e., repressive histone modification) after a prolonged cold exposure. Likewise, the two histone modifications were also found in the vernalization-dependent repression of FLC in Arabidopsis.97,98 In addition, downregulation of PdDAM6 after dormancy release was associated with a decrease of H3 acetylation in the chromatin around the ATG region.99
Taken together, epigenetic mechanisms appear to be involved in the seasonal growth-dormancy regulation of perennial plants. However, the details of these mechanisms are so far unknown. Further research, such as annual growth-dormancy stage-specific genome-wide DNA methylation and histone acetylation analysis in perennial trees, is required to identify the main components.
Future perspective
Many avenues of research remain to be explored before the genetic regulation of the complex physiological processes associated with the onset and breaking of dormancy and the integration of these processes with the regulation of flowering are understood. It is apparent that this involves a broad array of genes beyond those just associated with environmental stress. That auxin plays a major role has long been evident, but changes in the auxin responsiveness of the transcriptome need to be more fully addressed as PIF involvement is better understood. Continued identification and classification of DAM genes from additional species is needed to identify the common functional features of clades of these sequence-specific proteins. As microarray technologies continue to evolve and be widely used in gene expression studies, next-generation sequencing technologies such as Illumina, 454 Life Sciences (Roche) and Applied Biosystems (ABI) offer unparalleled depth, specificity and sensitivity for confident discovery of novel and rare transcripts. The growing availability of high quality annotation in many horticultural species now provides the opportunity for large-scale RNA-Seq-based Digital Gene Expression to examine the critical changes in transcriptome during growth-dormancy cycle. Such future work should yield deeper insights into the transcriptional regulation of the annual growth cycle. Clearly also, genome-wide epigenetic regulation of gene expression is involved in dormancy regulation with possible regulatory gene repression through chromatin compaction. Future research to elucidate the role of gene silencing in dormancy onset is needed.
Economically, dormancy regulation represents a very significant physiological problem that is critical to fruit and other horticultural sectors, and also to attaining increased, sustainably produced energy and fuel from intensive, short rotation plantation production of biomass. In agriculture, inappropriate dormancy responses in fruit tree crops will result in reduced productivity due to winter damage in flower buds. In fruit trees with a low chilling requirement for spring dormancy release, increased or erratic winter temperature will result in early spring bud-break and an increased risk of frost damage. On the other hand, species with a greater chilling requirement may not undergo proper flower development with higher winter temperatures. Increased understanding of the genetic regulation of the timing of dormancy induction and release, as well as the depth of endodormancy and thus cold hardiness, could profoundly impact horticultural production systems. In many regions, local climate is a key factor in selecting sites for the production of cherries, blueberries, peaches, apricots, wine grapes and other fruits and vegetables. For these crops, the lower limit of the sustained cold temperatures during winter and other microclimatic factors involving erratic spring temperatures or early frosts are often as critical as soil type and water availability in defining, and thus limiting, production zones. In these areas, producers are challenged annually by seasonal climate variation. For instance, a warm spring followed by a cold snap resulted in about 90% crop loss in tart cherries and apples in Michigan in 2012. In many fruits including apples and cherries, developing cultivars with even a one-week delay in the breaking of dormancy would greatly reduce the annual risk of crop loss to early frost and thus increase farm profitability in regions where early frosts regularly cause severe damage. It would also enable the development of new cultivars with growing seasons and hardiness levels tailored to specific planting zones and sites where these crops cannot currently be grown due to erratic frosts or cold temperatures, thereby increasing the geographic range over which some fruit crops could be produced and expand the production of locally grown fruits and vegetables.
More efficient woody biomass production for biofuels could also be accomplished by tailoring the growth and dormancy cycle to the environmental conditions where the plants are grown and could lead to increased primary productivity of temperate perennials grown in short rotation biomass plantations. For instance, carefully shortening the dormancy period, with the understanding that excessive prolonging of the growth period carries a tradeoff between winter survival and biomass production, may result in significant increase in growing period. This would be particularly effective for increasing biomass accumulation by indeterminant growth habit species like Populus, where current growth rates for biofuels are often marginally profitable. Tailoring the onset of dormancy to maximize utilization of the full growing season would result in increased late season growth and thus, accumulation of additional feedstock annually. Although this would not be of as great of economic significance as potentially expanding fruit and vegetable production regions, it could well lift these short rotation biofuel systems above the cutoff for profitability and enable successful deployment of short rotation woody biofuel crops in areas where the climate and marginal profitability currently limit production of these crops on retired farm lands.
The authors declare no conflict of interest.
References
- Horvath DP, Anderson JV, Chao WS et al. Knowing when to grow: signals regulating bud dormancy. Trends Plant Sci 2003; 8: 534–540. [DOI] [PubMed] [Google Scholar]
- Dennis FG. Dormancy—what we know (and don’t know). HortScience 1994; 29: 1249–1255. [Google Scholar]
- Park S, Keathley DE, Han K-H. Transcriptional profiles of the annual growth cycle in Populus deltoids. Tree Physiol 2008; 28: 321–329. [DOI] [PubMed] [Google Scholar]
- Lang GA. Dormancy: a new universal terminology. HortScience 1987; 22: 817–820. [Google Scholar]
- Arora R, Rowland LJ, Tanino K. Induction and release of bud dormancy in woody perennials: a science comes of age. HortScience 2003; 38: 911–921. [Google Scholar]
- Anderson JV, Horvath DP, Chao WS et al. Bud dormancy in perennial plants: a mechanism for survival. Top Curr Genet 2010; 21: 69–90. [Google Scholar]
- Rohde A, Bhalerao RP. Plant dormancy in the perennial context. Trends Plant Sci 2007; 12: 217–223. [DOI] [PubMed] [Google Scholar]
- Baba K, Karlberg A, Schmidt J et al. Activity-dormancy transition in the cambial meristem involves stage-specific modulation of auxin response in hybrid aspen. Proc Natl Acad Sci USA 2011; 108: 3418–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao R, Keskitalo J, Sterky F et al. Gene expression in autumn leaves. Plant Physiol 2003; 131: 430–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druart N, Johansson A, Baba K et al. Environmental and hormonal regulation of the activity-dormancy cycle in the cambial meristem involves stage-specific modulation of transcriptional and metabolic networks. Plant J 2007; 50: 557–573. [DOI] [PubMed] [Google Scholar]
- Frewen BE, Chen TH, Howe GT et al. Quantitative trait loci and candidate gene mapping of bud set and bud flush in Populus. Genetics 2000; 154: 837–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DE, Jonsson P, Bylesjö M et al. Changes in diurnal patterns within the Populus transcriptome and metabolome in response to photoperiod variation. Plant Cell Environ 2010; 33: 1298–1313. [DOI] [PubMed] [Google Scholar]
- Horvath DP, Chao WS, Suttle JC et al. Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genomics 2008; 9: 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Prassinos C, Keathley D et al. Novel aspects of transcriptional regulation in the winter survival and maintenance mechanism of poplar. Tree physiol 2011; 31: 208–225. [DOI] [PubMed] [Google Scholar]
- Howe G, Hackett W, Furnier G et al. Photoperiodic responses of a northern and southern ecotype of black cottonwood. Physiol Plant 1995; 93: 695–708. [Google Scholar]
- Olsen J, Junttila O, Nilson J et al. Ectopic expression of oat phytochrome A in hybrid aspen changes critical daylength for growth and prevents cold acclimatization. Plant J 1997; 12: 1339–1350. [Google Scholar]
- Welling A, Moritz T, Palva ET et al. Independent activation of cold acclimation by low temperature and short photoperiod in hybrid aspen. Plant Physiol 2002; 129: 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttink T, Arend M, Morreel K et al. A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 2007; 19: 2370–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 2011; 21: 664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot 2010; 61: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A. Nuclear localization activity of phytochrome B. Plant J 1996; 10: 859–868. [DOI] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L et al. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 1999; 11: 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Gil P, Kozma-Bognár L et al. Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 2002; 14: 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet 2007; 8: 217–230. [DOI] [PubMed] [Google Scholar]
- Quail PH. Photosensory perception and signalling in plant cells: new paradigms? Curr Opin Cell Biol 2002; 14: 180–188. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 2006; 57: 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews SS. Seeing the light. Nat Genet 2006; 38: 606–608. [DOI] [PubMed] [Google Scholar]
- Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 2011; 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Park J, Kim J et al. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J 2012: 72: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 2007; 12: 514–521. [DOI] [PubMed] [Google Scholar]
- Duek PDP, Fankhauser CC. bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci 2005; 10: 51–54. [DOI] [PubMed] [Google Scholar]
- Khanna RR, Huq EE, Kikis EAE et al. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 2004; 16: 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq EE, Quail PHP. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 2002; 21: 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD et al. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 2008; 53: 312–323. [DOI] [PubMed] [Google Scholar]
- Niwa Y, Yamashino T, Mizuno T. The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol 2009; 50: 838–854. [DOI] [PubMed] [Google Scholar]
- Kunihiro A, Yamashino T, Nakamichi N et al. PHYTOCHROME-INTERACTING FACTOR 4 and 5 (PIF4 and PIF5) Activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol 2011; 52: 1315–1329. [DOI] [PubMed] [Google Scholar]
- Soy J, Leivar P, González-Schain N et al. Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J 2012; 71: 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resman L, Howe G, Jonsen D et al. Components acting downstream of short day perception regulate differential cessation of cambial activity and associated responses in early and late clones of hybrid poplar. Plant Physiol 2010; 154: 1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Bastien C, Boerjan W. Temperature signals contribute to the timing of photoperiodic growth cessation and bud set in poplar. Tree Physiol 2011; 31: 472–482. [DOI] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T et al. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 2009; 19: 408–413. [DOI] [PubMed] [Google Scholar]
- Stewart JL, Maloof JN, Nemhauser JL. PIF genes mediate the effect of sucrose on seedling growth dynamics. PLoS ONE 2011; 6: e19894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto Y, Kubozono S, Yamashino T et al. Circadian clock- and PIF4-controlled plant growth: a coincidence mechanism directly integrates a hormone signaling network into the photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol 2012; 53: 1950–1964. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 2011; 108: 20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD et al. Rhythmic growth explained by coincidence between internal and external cues. Nature 2007; 448: 358–361. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M et al. A molecular framework for light and gibberellin control of cell elongation. Nature 2008; 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 2012; 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 2008; 24: 55–80. [DOI] [PubMed] [Google Scholar]
- Chapman E, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet 2009; 43: 265–285. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen GG. Auxin response factors. Curr Opin Plant Biol 2007; 10: 453–460. [DOI] [PubMed] [Google Scholar]
- Benjamins R, Scheres B. Auxin: the looping star in plant development. Ann Rev Plant Biol 2008; 59: 443–465. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature 2005a435: 441–445. [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005; 435: 446–451. [DOI] [PubMed] [Google Scholar]
- Maraschin, Fdos S, Memelink J, and Offringa R. Auxin-induced, SCFTIR1-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J 2009; 59: 100–109. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderón Villalobos LIA, Sharon M et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007; 446: 640–645. [DOI] [PubMed] [Google Scholar]
- Lee D, Park J, Lee H et al. Genome-wide analysis of the auxin-responsive transcriptome downstream of iaa1 and its expression analysis reveal the diversity and complexity of auxin-regulated gene expression. J Exp Bot 2009; 60: 3935–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM et al. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev 1998; 12: 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 2005b; 9: 101–119. [DOI] [PubMed] [Google Scholar]
- Zhong W, Gao Z, Zhuang W et al. Genome-wide expression profiles of seasonal bud dormancy at four critical stages in Japanese apricot. Plant Mol Biol 2013; 83: 247–264. [DOI] [PubMed] [Google Scholar]
- Zhuang W, Gao Z, Wang L et al. Comparative proteomic and transcriptomic approaches to address the active role of GA4 in Japanese apricot flower bud dormancy release. J Exp Bot 2013; 64: 4953–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-A J, Sherman WB, Scorza R et al. ‘Evergreen’ peach, its inheritance and dormant behavior. J Am Soc Hort Sci 1994; 119: 789–792. [Google Scholar]
- Wang Y, Georgi LL, Reighard GL et al. Genetic mapping of the evergrowing gene in peach [Prunus persica (L.) Batsch]. J Hered 2002; 93: 352–358. [DOI] [PubMed] [Google Scholar]
- Bielenberg DG, Wang Y, Fan S et al. A deletion affecting several gene candidates is present in the evergrowing peach mutant. J Hered 2004; 95: 436–444. [DOI] [PubMed] [Google Scholar]
- Jiménez S, Reighard GL, Bielenberg DG. Gene expression of DAM5 and DAM6 is suppressed by chilling temperatures and inversely correlated with bud break rate. Plant Mol Biol 2010; 73: 157–167. [DOI] [PubMed] [Google Scholar]
- Bielenberg DG, Wang YE, Li Z et al. Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Gen Genom 2008; 4: 495–507. [Google Scholar]
- Li Z, Reighard GL, Abbott AG et al. Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J Exp Bot 2009; 60: 3521–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H, Kashiwa Y, Ooka T et al. Suppression subtractive hybridization and differential screening reveals endodormancy-associated expression of an SVP/AGL24-type MADS-box gene in lateral vegetative buds of Japanese apricot. J Am Soc Hort Sci 2008; 133: 708–716. [Google Scholar]
- Hartmann U, Höhmann S, Nettesheim K et al. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 2000; 21: 351–360. [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH et al. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 2007; 21: 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C et al. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J 2003; 33: 867–874. [DOI] [PubMed] [Google Scholar]
- Böhlenius H, Huang T, Charbonnel-Campaa L et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 2006; 312: 1040–1043. [DOI] [PubMed] [Google Scholar]
- Hsu CY, Adams JP, Kim H et al. FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc Natl Acad Sci USA 2011; 108: 10756–10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruonala R, Rinne PL, Kangasjärvi J et al. CENL1 expression in the rib meristem affects stem elongation and the transition to dormancy in Populus. Plant Cell 2008; 20: 59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllenstrand N, Clapham D, Källman T et al. A Norway spruce FLOWERING LOCUS T homolog is implicated in control of growth rhythm in conifers. Plant Physiol 2007; 144: 248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenéz S, Lawton-Rauh AL, Reighard GL et al. Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach. BMC Plant Biol 2009; 9: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki R, Yamane H, Ooka T et al. Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol 2011; 157: 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw HD, Stettler RF. Molecular genetics of growth and development in populus. IV. Mapping QTLs with large effects on growth, form, and phenology traits in a forest tree. Genetics 1995; 139: 963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Howe G, Bradshaw H. Molecular genetic analysis of dormancy-related traits in poplars. Weed Sci 2002; 50: 232–240. [Google Scholar]
- Schrader J, Nilsson J, Mellerowicz E et al. A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 2004; 16: 2278–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader J, Moyle R, Bhalerao R et al. Cambial meristem dormancy in trees involves extensive remodeling of the transcriptome. Plant J 2004; 40: 173–187. [DOI] [PubMed] [Google Scholar]
- Andersson A, Keskitalo J, Sjödin A et al. A transcriptional timetable of autumn senescence. Genome Biol 2004; 5: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Riquelme J, Grimplet J, Martínez-Zapater JM et al. Transcriptome variation along bud development in grapevine (Vitis vinifera L.). BMC Plant Biol 2012; 12: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton EF, Wu RM, Richardson AC et al. A rapid transcriptional activation is induced by the dormancy-breaking chemical hydrogen cyanamide in kiwifruit (Actinidia deliciosa) buds. J Exp Bot 2009; 60: 3835–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzitelli L, Hancock RD, Haupt S et al. Co-ordinated gene expression during phases of dormancy release in raspberry (Rubus idaeus L.) buds. J Exp Bot 2007; 58: 1035–1045. [DOI] [PubMed] [Google Scholar]
- Hedley PE, Russell JR, Jorgensen L et al. Candidate genes associated with bud dormancy release in blackcurrant (Ribes nigrum L.) BMC Plant Biol 2010; 10: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath D. Common mechanisms regulate flowering and dormancy. Plant Sci 2009; 177: 523–531. [Google Scholar]
- Horvath DP, Sung S, Kim D et al. Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol Biol 2010; 73: 169–179. [DOI] [PubMed] [Google Scholar]
- Coleman GD, Chen KY, Parmentier-Line C. Over-expression of a poplar FLC-like MADS box gene impinges on low-temperature responses during vegetative bud dormancy. In: Proceedings of Plant and Animal Genome Conference; 14–18 January 2012; San Diego, CA, USA. [Google Scholar]
- Rinne PL, Welling A, Vahala J et al. Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-beta-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 2011; 23: 130–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni G. Dormancy cycling in Populus: the symplasmic connection. Plant Cell 2011; 23: 3. [Google Scholar]
- Cooke JE, Eriksson ME, Junttila O. The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant Cell Environ 2012; 35: 1707–1728. [DOI] [PubMed] [Google Scholar]
- Rios G, Leida C, Conejero A et al. Epigenetic regulation of bud dormancy events in perennial plants. Front Plant Sci 2014; 5: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría ME, Hasbún R, Valera MJ et al. Acetylated H4 histone and genomic DNA methylation patterns during bud set and bud burst in Castanea sativa. J Plant Physiol 2009; 166: 1360–1369. [DOI] [PubMed] [Google Scholar]
- Karlberg A, Englund M, Petterle A et al. Analysis of global changes in gene expression during activity-dormancy cycle in hybrid aspen apex. Plant Biotechnol 2010; 27: 1–16. [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 1999; 96: 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bishop B, Ringenberg W et al. The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol 2012; 159: 418–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petterle A. ABA and chromatin remodeling regulate the activity-dormancy cycle in hybrid aspen. PhD thesis, Swedish University of Agricultural Sciences, Uppsala, Sweden, 2011. [Google Scholar]
- Bastow R, Mylne JS, Lister C et al. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 2004; 427: 164–167. [DOI] [PubMed] [Google Scholar]
- Kim SY, He Y, Jacob Y et al. Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 2005; 17: 3301–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leida C, Conesa A, Llácer G et al. Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytol 2012; 193: 67–80. [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES. DNA methylation, a key regulator of plant development and other processes. Curr Opinion Genet Devel 2000; 10: 217–223. [DOI] [PubMed] [Google Scholar]
- Penterman J, Uzawa R, Fischer RL. Genetic interactions between DNA demethylation and methylation in Arabidopsis. Plant Physiol 2007; 145: 1549–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Ma H. Evolutionary history of histone demethylase families: distinct evolutionary patterns suggest functional divergence. BMC Evol Biol 2008; 8: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Müller A, Napoli CA et al. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucl Acids Res 2002; 30: 5036–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A and Kovalchuk I. Epigenetic control of plant stress response. Environ Mol Mutagen 2008; 49: 61–72. [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet 1999; 22: 94–97. [DOI] [PubMed] [Google Scholar]
- Schubert D, Clarenz O, Goodrich J. Epigenetic control of plant development by Polycomb-group proteins. Curr Opin Plant Biol 2005; 8: 553–561. [DOI] [PubMed] [Google Scholar]