Abstract
Unlike mammals with adaptive immunity, plants rely on their innate immunity based on pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) for pathogen defense. Reactive oxygen species, known to play crucial roles in PTI and ETI, can perturb cellular redox homeostasis and lead to changes of redox-sensitive proteins through modification of cysteine sulfhydryl groups. Although redox regulation of protein functions has emerged as an important mechanism in several biological processes, little is known about redox proteins and how they function in PTI and ETI. In this study, cysTMT proteomics technology was used to identify similarities and differences of protein redox modifications in tomato resistant (PtoR) and susceptible (prf3) genotypes in response to Pseudomonas syringae pv tomato (Pst) infection. In addition, the results of the redox changes were compared and corrected with the protein level changes. A total of 90 potential redox-regulated proteins were identified with functions in carbohydrate and energy metabolism, biosynthesis of cysteine, sucrose and brassinosteroid, cell wall biogenesis, polysaccharide/starch biosynthesis, cuticle development, lipid metabolism, proteolysis, tricarboxylic acid cycle, protein targeting to vacuole, and oxidation–reduction. This inventory of previously unknown protein redox switches in tomato pathogen defense lays a foundation for future research toward understanding the biological significance of protein redox modifications in plant defense responses.
Introduction
Pseudomonas syringae, a hemibiotrophic bacterial pathogen, causes bacterial speck disease in a wide range of plant species, including crops such as tomato (Solanum lycopersicum) and the reference plant Arabidopsis thaliana. The disease leads to serious decreases in crop yield and significant economic loss in agriculture and therefore threatens global food security.1 According to the Food and Agriculture Organization of the United Nations, the global demand for food is projected to rise by 50% by 2030.2 As the world’s population continues to grow, arable lands decrease, and pathogens continue to evolve resistance to plant defense, the study of plant–pathogen interaction has become an important and urgent area of biological research. Recent development in research tools and technologies has enabled scientists to gain novel mechanistic insights into the molecular processes underlying plant–pathogen interactions. The knowledge may be employed in the development of crops with enhanced defense through genetic engineering and breeding.
Plant–pathogen interactions involve sophisticated molecular interplay underlying an evolutionary arms race.3 Plants are able to recognize pathogens through pathogen-associated molecular patterns (PAMPs). This response and subsequent changes within the plant cells are often called PAMP-triggered immunity (PTI). The microbe recognition is achieved by plasma membrane pattern recognition receptors, which are leucine-rich receptor-like kinases or receptor-like proteins.4 After PAMP recognition, plant cells undergo alkalization, change in ion flux, increase in reactive oxygen species (ROS), and activation of mitogen-activated protein kinase cascade.5 Bypassing or suppressing these responses benefits the bacteria. Through a type III secretion system, the bacteria can inject effector proteins (T3Es) into the host cells to attenuate PTI. Recognition of the T3Es by plant resistance (R) proteins is important for the plant’s survival and effector-triggered immunity (ETI). ETI initiates an incompatible interaction characterized by a hypersensitive response (HR), programmed cell death, defense gene transcription, increase in salicylic acid (SA), and induction of systemic acquired resistance (SAR).6 Beyond transcriptional and protein level changes, post-translational modifications (PTMs) such as nitrosylation, disulfide bond formation, and phosphorylation known to regulate protein activities, may play a role in the plant–pathogen interactions.
PTMs of proteins may function as molecular switches to turn on or off signaling and/or metabolic processes in plant response to external stimuli.7–9 As previously reported, both PTI and ETI responses lead to increases in ROS production.4,10,11 Thiol is a nucleophile, which when exposed to oxidative stress, is capable of undergoing reversible inter- and intra-molecular disulfide bond formation, nitrosylation, glutathionylation, sulfinic acid modification, and irreversible sulfonic acid modification.12 The high pKa values of protein cysteines (8–9) make these residues responsive to small redox perturbation by forming reactive ionized thiolate groups.13 H2O2 is able to permeate the cell membrane and serves as a signaling molecule to induce redox changes and protein thiol modifications.7,14,15 In recent years, several reversible cysteine modifications (e.g., S-nitrosylation, S-sulfenation, S-thiolation, and S-glutathionylation) have been characterized and shown to play a role in modulating the functions of regulatory proteins.16–18 However, only a limited number of defense-related proteins have been shown to be redox-regulated in plants, e.g., a cytokine-induced apoptosis inhibitor 1 (AtCIAPIN 1),19 natriuretic peptide receptor 1,20 transcription factor 1, nicotinamide adenine dinucleotide phosphate-oxidase (rubidium hydroxide),22 S-nitrosoglutathione reductase 1,23 SA-binding protein,24 and BRI1-associated receptor-like kinase 1.25 Redox modifications induced in response to P. syringae pv tomato (Pst) infection have not been identified between resistant (PtoR) and susceptible (prf3) genotypes of tomato at early (4 h) and late (24 h) time points after infection. Therefore, it is hypothesized that during the infection process, Pst will cause differential redox regulation of proteins amongst different time points and between genotypes. This hypothesis is supported by the change in H2O2 production observed between the early and late time points.26 In addition, the identification of novel redox-sensitive proteins in response to pathogens will allow improved understanding of the roles of redox regulation and redox signaling in plant defense.
Among the proteomics approaches used for identification of proteins that undergo oxidative modifications, Cysteine Labeling Tandem Mass Tags (cysTMT with m/z 126-131, Thermo Scientific Inc., San Jose, CA, USA) have been utilized in the identification of disulfide bond formation and thiol nitrosylation.27,28 A reverse-labeling approach allows alkylation of free thiols in protein samples to prevent artificial oxidation, and then reduction of oxidation sensitive cysteines, which will be labeled with multiplex cysTMT tags. Using this approach, 4348 proteins have been confidently identified. A total of 3258 cysteine-containing peptides and 1580-associated proteins were observed. Among these proteins, 19, 27, and 51 potential redox-sensitive proteins were identified in prf3 at 4 h after inoculation (hai), PtoR at 4 hai, and PtoR at 24 hai, respectively. The redox-sensitive cysteines in these proteins were mapped and their sensitivity levels were quantified. In addition, the redox changes were compared to the data of total protein level changes23 using the same set of samples. Some of the interesting proteins include serine–threonine receptor kinase, WD repeat protein, vacuolar-sorting receptor, and 3-ketoacyl-CoA thilase (KAT2). This study reports an inventory of potential redox switches and protein redox modulation as an interesting mechanism in plant response to pathogen infection.
Materials and methods
Plant materials and bacteria inoculation
The plant materials tomato (S. lycopersicum) genotypes PtoR and prf3 used for this study were the same as those used for the global protein abundance study,26 except that the prf3 24 hai samples were not included due to the limitation of six-plexes of the cysTMT.28 Please refer to Parker et al.26 for plant growth and bacterial inoculation conditions.
Protein extraction, CysTMT labeling, and gel fractionation
Protein extraction was performed in three biological replicates of PtoR 4 hai, prf3 4 hai, and PtoR 24 hai, respectively. Samples were grounded with 5× the volume of 20% tricarboxylic acid (TCA). The samples were then vortexed and put on ice for 30 min. Samples were stored at −20 °C for 1 h and subsequently centrifuged at 12 000 rpm at 4 °C for 15 min. The supernatant was removed, and the pellet was washed with 80% cold acetone, centrifuging at 10 000 rpm at 4 °C for 15 min. This step was repeated three times. Samples were resuspended in a sample buffer (7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) and quantified as previously described.26
Proteins were labeled as previously described.28 For each sample, 100 μg protein was treated with iodoacetamide (IAM) for alkylating-free cysteine thiols, reduced with tris-(2-carboxyethyl)phosphine (TCEP), and then labeled with the cysTMT tags as described in Parker et al. 28 with slight modifications. Mock inoculated samples (PtoR 4 hai, prf3 4 hai, and PtoR 24 hai) were labeled with cysTMT tags 126 m/z, 128 m/z, and 130 m/z, respectively. Pst-treated samples (PtoR 4 hai, prf3, and PtoR 24 hai) were labeled with cysTMT tags 127 m/z, 129 m/z, and 131 m/z, respectively. The samples were then combined and subjected to non-reducing 1D gel fractionation. From the gel, 12 fractions were excised for trypsin digestion according to manufacturer’s instructions (Thermo Scientific Inc., San Jose, CA, USA).
Liquid chromatography–mass spectrometry (LC-MS)/MS of the cysTMT samples
LC-MS/MS was performed on a Thermo Scientific Q-Exactive system, which is a hybrid Quadrupole-Orbitrap Mass Spectometer. Samples were loaded onto a C18 EASY-Spray column (75 micron × 25 cm, with 2 micron particle size) (Thermo Scientific Inc., Bellefonte, PA, USA) heated to 35 °C. The LC flow rate was 300 nL min–1. A gradient was set up with a 60 min linear ramp from 2% acetonitrile with 0.1% formic acid to 30% acetonitrile with 0.1% formic acid, followed by a 10 min linear ramp to 98% acetonitrile with 0.1% formic acid, where it was held for 20 min. Data were acquired on the Q-Exactive hybrid mass spectrometer using the full MS/data dependent MS2, targeting the top 5 peptides. A full scan was performed at 70 000 resolution, scanning from 400 m/z to 2000 m/z at a target of 3E6 in the Orbitrap. MS/MS scanning was performed in the Orbitrap at 17 500 resolution with a quadrupole isolation width of 2 m/z and a 28% normalized collision energy, using a fixed first mass of 105 m/z. The MS/MS target was 1E6 with a 100 ms maximum injection time. The underfill ratio was set to 1.0%, and the apex trigger was turned on with a minimum and maximum triggering time of 3–12 s. Peptide match was set to ‘preferred with exclude isotopes’, and dynamic exclusion duration was set to 60 s.
Database searching and data analysis
Raw data files were submitted to Proteome Discoverer 1.4 for database searching (Thermo Scientific Inc., Bremen, Germany). The spectrum grouper node used a precursor mass tolerance of 10 ppm and a maximum retention time difference of 1.1 min. Proteome Discover 1.4 with SEQUEST algorithm was used to search the ITAG 2.3 tomato database (34 727 entries, downloaded from http://solgenomics.net) combined with a list ESTs derived from the Harvard DFCI index (52 542 entries, from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4205239). These sequences were merged with a list of contaminants from the Max Planck Institute of Biochemistry (http://maxquant.org/contaminants.zip). Dynamic modifications included methionine oxidation, cysteine carbamidomethyl, and cysTMT6plex. Percolator was set to a target false discovery rate (FDR) of 0.05. Quantification was performed using the cysTMT6plex reporter ion method.
Unique peptide peak intensities for each label of cysTMT were exported, and ratios were calculated accordingly from the median-normalized peak intensity values. Student’s t-test (two-tailed) on the log2-transformed treated/control ratios was performed. A peptide with a p > 0.05 and a fold change greater than 1.2 or less than 0.8 were considered to be statistically significant. The isobaric tags for relative and absolute quantitation (iTRAQ) data by Parker et al.26 were searched using the database mentioned above in order to generate information about total protein level changes (Supplementary Table S1). The significant peptides labeled with cysTMT were compared and contrasted with the significant proteins quantified via iTRAQ. Student’s t-test was conducted between the fold change of cysTMT-labeled peptides and the fold change of the corresponding proteins quantified via iTRAQ. A correction factor was applied to the fold change of cysTMT-labeled peptides, taking into account the fold change of the protein quantified via iTRAQ.
Quantitative real-time PCR
In order to gain indication of potential protein level changes of the proteins not identified in the proteomics study,23 relative quantification in real-time RT-PCR (qRT-PCR) was performed (Supplementary Table S2). VeriQuest Sybr with Fluorescein Kit (Affymetrix Inc., Santa Clara, CA, USA) was used with CFX90 (Bio-Rad, Hercules, CA, USA). For each reaction, two technical and three biological replicates were included. Relative expression of the target genes was calculated using the Ct method. The differences in Ct values (ΔCt) between the target genes and reference gene (ACT7, Supplementary Table S2) were calculated to normalize the differences in the complimentary DNA concentrations for each reaction. Differential gene expression between inoculated and control samples was assessed using t-test.
Results
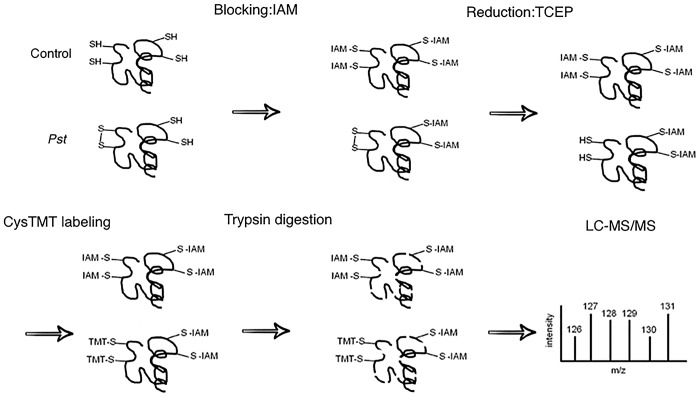
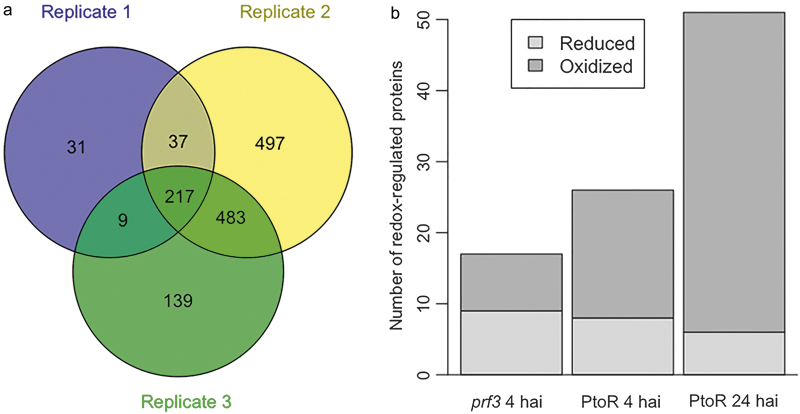
Identification of redox-responsive cysteines, peptides, and proteins Redox proteomics approaches are based on differential labeling of redox-modified cysteines in proteins and have shown utility in unraveling important biological mechanisms. Here we chose to use the TCA method to precipitate proteins because TCA not only lyses the cells, but also protonates all thiolates to prevent thiol-disulfide exchange reactions.29 In addition, a reverse-labeling procedure was performed, in which IAM was used to block free thiol groups. After alkylation of the free thiols, reduction of reversibly oxidized cysteine residues and labeling with cysTMT reagents were conducted. This reverse-labeling procedure maintains the initial redox state of the proteins and prevents artificial oxidation during sample preparation. Therefore, the increases of the cysTMT signals from specific cysteine peptides derived from treated samples compared to control samples indicate the presence of oxidation responsive/sensitive cysteines (Figure 1). The use of this powerful methodology enabled identification of proteins with cysteines that underwent redox modifications in the resistant PtoR and susceptible prf3 genotypes of tomato at early and late time points after infection. In total, 4348 proteins were confidently identified among three biological replicates with a FDR of 0.05. A total of 3258 cysteine-containing peptides and 1580-associate proteins were observed. Of the cysteine-containing peptides, 2749 peptides (84.3%) were labeled with cysTMT tags and correlated to 1413 proteins. A comparison of the three biological replicates showed that 217 proteins were present in all the replicates, and 529 were present in at least two replicates (Figure 2). It should be noted that the three replicates represent independent biological replicates using the dip-inoculation method, which is close to natural infection, but known to generate more variations than the infiltration method.30 A peptide abundance greater than 1.2 or less than 0.8 with a p < 0.05 was considered to be redox-sensitive (Supplementary Figure S1). As previously addressed, the identification of redox proteins may be complicated due to protein turnover. This issue may be solved by comparing the redox proteomics data with protein level change data31,32 (Supplementary Figure S2). In this report, the total protein level data generated by Parker et al.26 was used to correct the redox changes. It is important to highlight that the same set of samples were used in the proteomics study23 and in our redox proteomics study in order to avoid sample variation. For example, a dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex-like protein was identified as a potential redox-regulated protein in the prf3 susceptible genotype with a fold change of 1.46 at 4 hai (Table 1). However, the iTRAQ analysis revealed a significant fold change of 2.06. Therefore, the redox-fold change may be due to the protein level change rather than a cysteine redox response. Cleary, our redox proteomics method showed utility in the identification of redox-responsive cysteines, peptides, and proteins.
Figure 1.
Reverse cysTMT-labeling workflow. Control and Pst-treated samples were prepared at the same time. Blocking reduced thiols with IAM followed by reduction of the oxidized thiols with TCEP allows the previously oxidized thiols due to Pst treatment to be labeled with the isobaric cysTMT reagents.
Figure 2.
Comparison of redox proteomes and identified redox proteins in different tomato genotypes. (a) Venn diagram showing the number of shared proteins among three different biological replicates. (b) The number of proteins underwent reduction (gray) or oxidation (dark) in the susceptible prf3 and resistant PtoR genotypes at early and late stage of Pst infection. PtoR clearly showed more oxidized proteins at both 4 hai and 24 hai, compared to prf3.
Table 1. Potential redox-responsive proteins identified in prf3 at 4 h after Pst inoculation.
| Peptide | Protein ID | Protein description | p-value | cysTMT FCa | iTRAQ FCb | mRNA FCc | DiANNAd |
|---|---|---|---|---|---|---|---|
| AVSAIVScLLGPER | Solyc03g111610.2.1 | Protein CbbY-like | 0.014 | 0.66 | 1.21NS | ✓ | |
| AGScSScAGK | Solyc10g044520.1.1 | Ferredoxin-1, chloroplastic | 0.031 | 0.68 | 0.96NS | ||
| SDFHVAAQNcWVK | Solyc04g011510.2.1 | Triosephosphate isomerase | 0.011 | 0.73 | 0.90NS | ||
| AcLKVPSNEGVLVR | Solyc06g084480.2.1 | Protease Do-like 2, chloroplastic-like | 0.015 | 0.73 | n/a | 0.76NS | |
| AcAFSEDTNLLLTGGFEK | Solyc03g120010.2.1 | Serine–threonine kinase receptor-associated protein like | 0.003 | 0.74 | 0.89NS | ✓ | |
| FANIAcK | Solyc03g078290.2.1 | 40S ribosomal protein S18-like | 0.018 | 0.75 | 1.42NS | ||
| VcEcPTVQGVK | Solyc10g074980.1.1 | Vacuolar-sorting receptor 1-like | 0.014 | 0.75 | 2.12S | ✓ | |
| IPFVcGCR | Solyc06g081980.1.1 | Probable pyridoxal biosynthesis protein PDX1-like | 0.045 | 0.77 | 1.02NS | ✓ | |
| IQDEcGQVVScSDIVAIAAR | Solyc04g071890.2.1 | PREDICTED: peroxidase 12-like | 0 | 0.79 | 1.20NS | ✓ | |
| EAFVEMcADEYVQK | Solyc03g115980.1.1 | Geranylgeranyl diphosphate reductase, chloroplastic-like | 0.028 | 0.8 | 1.03NS | ||
| VcQVIGAVVDVR | Solyc05g008460.2.1 | ATP synthase subunit beta, mitochondrial-like | 0.038 | 1.25 | 1.09NS | ||
| DSVGQPINVAcEVQQLLGNNR | Solyc01g007320.2.1 | ATP synthase CF1 beta subunit, chloroplastic | 0.005 | 1.28 | 0.95NS | ||
| QGFTNLPIcMAK | Solyc01g006280.2.1 | Formate-tetrahydrofolate ligase-like | 0.049 | 1.28 | 0.86NS | ✓ | |
| GQEGAGIVcVNDNVLK | Solyc01g110520.2.1 | Amidophosphoribosyltransferase 2, chloroplastic-like | 0.029 | 1.29 | 1.40NS | ||
| GNNILVMcDAYTPAGEPIPTNKR | Solyc11g011380.1.1 | Glutamine synthetase | 0.003 | 1.33 | 1.17NS | ||
| VAAKLESMEPcSSVK | Solyc09g082060.2.1 | Cysteine synthase-like | 0.035 | 1.36 | 1.42NS | ||
| ATALALVQHPVVNSScR | Solyc11g017250.1.1 | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex-like | 0.013 | 1.46 | 2.06S | ||
| FYTEcFGMK | Solyc02g080630.2.1 | Lactoylglutathione lyase-like | 0.03 | 1.52 | 0.91NS | ||
| GVDNSQIKVVcAVAAPPALTK | Solyc09g011300.2.1 | Uracil phosphoribosyltransferase-like isoform 1 | 0.013 | 1.55 | 1.13NS |
Fold change between inoculated and control samples obtained from cycTMT.
Fold change between inoculated and control samples obtained from iTRAQ,23 NS, not significant (p ≧ 0.05), S, significant (p < 0.05).
Fold change between inoculated and control samples obtained from qRT-PCR, NS, not significant (p ≧ 0.05), S, significant (p < 0.05).
Prediction of intra-molecular disulfide bond formation.
Redox-responsive proteins at early stage of Pst infection of prf3
In the susceptible prf3 genotype, 19 cysteine-containing peptides from 19 different proteins were found to be redox-regulated upon pathogen infection (Table 1). Ten of the peptides were found to be in more reduced state at 4 hai with Pst, while the other nine were in more oxidized state compared to the mock control samples (Figure 2). The identification of proteins in a more reduced state under Pst inoculation suggests differential compartmentalization of subcellular redox environment and the dynamic nature of protein redox state. When the total protein-level change and transcriptional change were taken into account, two out of the 19 peptides containing redox-sensitive cysteines also showed significant protein amount changes in the same direction (Table 1). Vacuolar-sorting receptor 1-like and dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex-like were initially assigned as potentially redox-regulated based on the cysTMT data. However, when the protein level correction (based on the iTRAQ data) was applied, these proteins also showed total protein level change. Thus, the observed redox-fold change of these proteins may be due to the protein level change rather than a cysteine redox response. The sequences of the identified redox-regulated proteins were analyzed for intra-molecular disulfide bond formation using DiANNA software (http://clavius.bc.edu/~clotelab/DiANNA/). Five of the 17 redox-sensitive peptides were predicted to form intra-molecular disulfide bonds (Table 1). It should be noted that disulfide bonds represent only one possibility of thiol modifications, and other modifications, such as sulfenic acid, S-nitrosylation and glutathionylation33 are also chemically reversible. The cysTMT labeling method will allow the identification of these cysteine modifications if modification specific reductants are used, e.g., ascorbate and copper for S-nitrosylation.34,35 As shown in Figure 3, analysis of GO terms of relevant biological processes revealed that the potential redox-regulated proteins were involved cellular carbohydrate metabolism, energy processes (plasma membrane adenosine triphosphate (ATPase), ATP synthesis, photosynthesis, and pentose-phosphate shunt), as well as gluconeogenesis and cysteine biosynthetic process. The oxidation–reduction process was represented by five proteins (ferredoxin, peroxidase 12-like, glutamine synthetase (GS), cysteine synthase, and lactoylglutathione lyase-like) in the prf3 genotype inoculated with Pst for 4 h.
Figure 3.
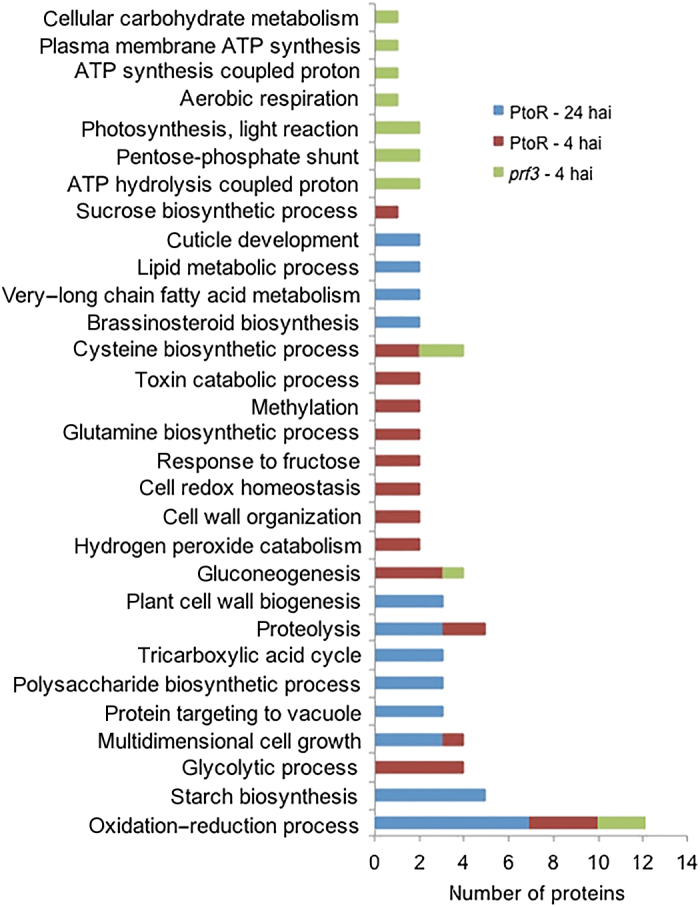
Comparative ontology analysis of the redox proteins after Pst infection between susceptible and resistant genotypes at early and late stages of infection. Relevant biological process GO terms are shown on the y-axis. Numbers of proteins significantly redox-regulated in each genotype and time point after infection are shown on the x-axis.
Redox proteins at early stage of Pst infection of PtoR
A total of 27 cysteine-containing peptides from 27 different proteins were identified being redox-regulated in the PtoR samples at 4 hai (Table 2). Among these redox-regulated peptides eight were in more reduced state under Pst inoculation; while 19 were in more oxidized state compared to the control samples (Figure 2). Analysis of total protein level and transcriptional change revealed that two peptides identified as being redox-regulated showed significant protein/transcript level change (Table 2). Carbohydrate esterase exhibited a significant fold change of 1.31 in redox modification and a significant fold change of 2.10 in protein level. In addition, proteinase inhibitor type-2 CEVI57-like isoform 1 showed a significant fold change of 1.30 in redox modification and a significant fold change of 1.69 in transcriptional level. Thirteen out of the 26 redox-regulated proteins were predicted to form intra-molecular disulfide bonds (Table 2). As noted in the previous section, other thiol modifications could be responsible for the rest of cysteines identified here. Biological processes of the potential redox-regulated proteins of PtoR plants at 4 hai include sucrose biosynthetic process, cysteine biosynthetic process, gluconeogenesis, and other processes such as proteolysis, methylation, glycolytic process, toxin catabolic process, cell wall organization, and cell growth. Most of all, proteins in the following three biological processes, i.e., cell redox homeostasis, hydrogen peroxide catabolic process, and oxidation-reduction process may play an important role in regulating the dynamic protein redox state during Pst infection (Figure 3). Although cysteine biosynthesis, gluconeogenesis, and oxidation–reduction process were shared with prf3, PtoR at 4 hai has many differences (e.g., in sucrose biosynthesis, glutatmine biosynthesis, and glycolytic process).
Table 2. Potential redox-responsive proteins identified in PtoR at 4 h after Pst innoculation.
| Peptide | Protein ID | Protein description | p-value | cysTMT FC | iTRAQ FC | mRNA FC | DiANNA |
|---|---|---|---|---|---|---|---|
| AVMEAVGScLTNK | Solyc08g065490.2.1 | Serine hydroxymethyltransferase, mithocondrial-like | 0.014 | 0.55 | 1.38NS | ||
| YVEScKFPTDGSSPK | Solyc12g056530.1.1 | Fructose-1 6-bisphosphatase - cytosolic-like | 0.002 | 0.7 | 1.08NS | ✓ | |
| FWEVIcDEHGVDPTGR | Solyc04g081490.2.1 | Tubulin beta-1 chain | 0.031 | 0.72 | 0.96NS | ✓ | |
| TGcEIVVAEKEK | Solyc01g091580.2.1 | Cell division protein FtsY homolog, chloroplastic-like | 0.009 | 0.73 | 0.90NS | ✓ | |
| cALVYGQMNEPPGAR | Solyc05g008460.2.1 | ATP synthase subunit beta, mitochondrial-like | 0.028 | 0.75 | 1.09NS | ||
| RKDTVVIALADETcDEPK | Solyc11g069720.1.1 | Cell division cycle protein 48 homolog | 0.016 | 0.76 | 1.06NS | ||
| SGPFYFISGNKDNcDK | Solyc12g013900.1.1 | Early nodulin-like protein 2-like | 0.009 | 0.78 | n/a | 0.86NS | |
| YAADVcAGVILGR | Solyc12g010040.1.1 | Neutral leucine aminopeptidase preprotein | 0.035 | 0.8 | 1.07NS | ||
| LPGFHVcVGSGGER | Solyc09g009390.2.1 | Monodehydroascorbate reductase-like | 0.013 | 1.2 | 0.93NS | ||
| SAPQcGFSQR | Solyc09g005620.2.1 | Glutaredoxin-1 protein | 0.022 | 1.21 | 1.24NS | ||
| ATEGPIVADKNcEK | Solyc05g013820.2.1 | Proteasome subunit beta type-7-A-like | 0.008 | 1.23 | 1.11NS | ||
| IcEEcK | Solyc06g060790.1.1 | 3-isopropylmalate dehydratase small subunit-like isoform 1 | 0.017 | 1.24 | 0.93NS | ||
| GGNNILVIcDAYTPAGEPIPTNK | Solyc01g080280.2.1 | Glutamine synthetase, chloroplastic | 0.022 | 1.24 | 1.06NS | ✓ | |
| GNNILVMcDAYTPAGEPIPTNKR | Solyc11g011380.1.1 | Glutamine synthetase | 0.039 | 1.24 | 0.92NS | ||
| LAcVIAECGEM | Solyc01g096520.2.1 | Peptidyl-prolyl cis-trans isomerase H-like isoform 1 | 0.015 | 1.25 | n/a | 0.88NS | |
| HDPcVVPR | Solyc04g049350.2.1 | Chorismate synthase 1, chloroplastic precursor | 0.004 | 1.27 | 0.95NS | ✓ | |
| DTWVPAYDDDDFcLR | Solyc03g006140.2.1 | Neutral ceramidase-like isoform 1 | 0.038 | 1.27 | 0.75NS | ✓ | |
| SHSEIQDSNGQNIGEITSGGFSPcLK | Solyc02g080810.2.1 | Aminomethyltransferase, mitochondrial-like | 0.024 | 1.29 | 1.08NS | ||
| RAEAFGcPISYYTR | Solyc03g120670.2.1 | Glyoxylate/hydroxypyruvate reductase A HPR2-like | 0.037 | 1.29 | 0.97NS | ✓ | |
| VAYMTcPPESTK | Solyc03g020030.2.1 | Proteinase inhibitor type-2 CEVI57-like isoform 1 | 0.02 | 1.3 | n/a | 1.69S | |
| EcESNPSILR | Solyc01g102330.2.1 | Probable carbohydrate esterase At4g34215-like | 0.04 | 1.31 | 2.10S | ✓ | |
| EEcENMKPEL | Solyc08g014340.2.1 | Cysteine synthase | 0.031 | 1.32 | 1.08NS | ✓ | |
| NPVPPPEcR | Solyc02g091920.2.1 | Xyloglucan endotransglucosylase-hydrolase XTH7 | 0.008 | 1.4 | n/a | 0.65NS | ✓ |
| ADDcIGPEVDK | Solyc07g066610.2.1 | Phosphoglycerate kinase, chloroplastic-like | 0.043 | 1.43 | 1.05NS | ✓ | |
| SLAVIGPNANLAYTMVGSYEGSPcK | Solyc01g104950.2.1 | Beta-xylosidase/alpha-L-arabinofuranosidase 2 | 0.042 | 1.59 | 1.41NS | ✓ | |
| VLTGEETGNNAFPScSPDGK | Solyc06g008620.1.1 | Uncharacterized protein LOC101247825 | 0.005 | 1.75 | 1.09NS | ✓ | |
| DDQVSGQcAK | Solyc11g039830.1.1 | Glycine-tRNA ligase 1, mitochondrial-like | 0.034 | 2.13 | 1.03NS | ✓ |
Redox-responsive proteins at late stage of Pst infection of PtoR
During the late stage of infection in the tomato resistant genotype PtoR, 51 cysteine-containing peptides from 50 different proteins were found to be potentially redox-regulated (Table 3). Six out of the 51 redox-regulated peptides were in more reduced state at this late stage of Pst infection, whereas the others 45 were in more oxidized state compared to the control samples (Figure 2). When the total protein level and transcriptional level changes were analyzed, eight of the peptides containing redox-sensitive cysteines exhibited significant protein/transcript amount changes (Table 3). DNA damage repair/tolerance protein DRT100-like and calreticulin-like isoform 2 exhibited a significant fold change of 0.73 and 2.88, respectively in redox modification, and a significant fold change of 0.015 and 2.87, respectively at the transcriptional level. Therefore, the observed redox-fold change of these proteins may be due to protein level change rather than a cysteine redox response. Additionally, glutamate dehydrogenase and oxidoreductase UcpA-like showed a significant fold change of 5.34 and 7.4, respectively in redox modification, and a significant fold change of 3.03 and 3.50, respectively at the transcriptional level. Since the redox modification fold change was substantially greater than the transcriptional level fold change, it is possible that both protein level and redox regulation occurred upon infection. Furthermore, 3-oxoacyl-(acyl-carrier-protein) synthase II, chloroplastic alpha-1,4 glucan phosphorylase L-2 isozyme, glucoronokinase-1-like, and elongation factor 1-aplha presented a significant positive fold change (1.24, 1.52, 1.87, 2.5, respectively) in redox modification, and a significant negative fold change (0.28, 0.74, 0.60, 0.57, respectively) at the transcriptional level. In these specific cases, it is likely that the redox modification is even greater than the observed, due to negative regulation of transcriptional/protein level. Interestingly, the protein KAT2 was found to undergo oxidation, with the highest 22-fold change in oxidation and no significant change in protein abundance (Table 3). The identified Cys138 in peptide GDAVDDLcNAR is localized in active site of the enzyme and it was shown to form a disulfide bond with Cys192 during redox regulation. An analysis of tomato and Arabidopsis KAT2 amino acid sequences revealed that the cysteine identified in our study undergoing oxidation in response to Pst is the cysteine localized at the active site of the enzyme (Figure 4). Thus, it can be concluded that KAT2 is in its inactive form at late stage of infection. Of the 51 peptides containing redox-sensitive cysteines, 19 were predicted to form intra-molecular disulfide bonds (Table 3). Biological processes of the potential 51 redox-regulated proteins were sucrose biosynthetic process, brassinosteroid biosynthesis, cell wall biogenesis, polysaccharide biosynthetic process, and starch biosynthetic process, cuticle development, some lipid metabolic process, proteolysis, TCA cycle, protein targeted to vacuole, and oxidation–reduction process (Figure 3). Except sharing proteolysis, cell growth and oxidation–reduction processes with PtoR 4 hai, the PtoR 24 hai showed many different functional processes in line with ETI responses.
Table 3. Potential redox-responsive proteins identified in PtoR at 24 h after Pst inoculation.
| Peptide | Protein ID | Protein description | p-value | cysTMT FC | iTRAQ FC | mRNA FC | DiANNA |
|---|---|---|---|---|---|---|---|
| AVSAIVScLLGPER | Solyc03g111610.2.1 | Protein CbbY-like | 0.003 | 0.31 | 0.90NS | ✓ | |
| TLFGFHENGcVSK | Solyc03g121180.2.1 | GDSL esterase/lipase APG-like | 0.015 | 0.41 | n/a | 0.66 NS | |
| DcTSVQQcYLELSK | Solyc04g005340.2.1 | Alpha-1,4-glucan-protein synthase | 0.04 | 0.73 | 0.98NS | ✓ | |
| GITGSIPScITSLPFLR | Solyc11g056680.1.1 | DNA-damage-repair/toleration protein DRT100-like | 0.036 | 0.73 | n/a | 0.015S | ✓ |
| YGTVPIcASTGGLVDTVK | Solyc08g083320.2.1 | Granule-bound starch synthase 1, chloroplastic | 0.045 | 0.75 | 1.25NS | ||
| QFDcEILIR | Solyc05g056290.2.1 | Biotin carboxylase carrier protein | 0.039 | 0.8 | 0.98NS | ||
| GLIScVTGK | Solyc07g005390.2.1 | NADP glyceraldehyde-3-phosphate dehydrogenase-like | 0.046 | 1.22 | 1.13NS | ✓ | |
| TMGVc | Solyc11g011920.1.1 | Glutamate decarboxylase isoform2 | 0.004 | 1.24 | 1.02NS | ||
| GTGVILcVEK | TC242228_4 | 3-oxoacyl-[acyl-carrier-protein] synthase II, chloroplastic-like | 0.042 | 1.24 | n/a | 0.28 S | |
| ScSEEGDEEAIYR | Solyc02g066950.2.1 | Uncharacterized protein LOC101249042 | 0.013 | 1.24 | n/a | 0.78 NS | ✓ |
| GVAGLYDYGPPGcAVK | Solyc03g118170.2.1 | Glycine-tRNA ligase 1, mitochondrial-like | 0.038 | 1.28 | 0.89NS | ||
| NEYcMEVTPK | Solyc11g011960.1.1 | UTP-glucose-1-phosphate uridylyltransferase-like isoform 1 | 0.033 | 1.3 | 0.89NS | ||
| ATALALVQHPVVNSScR | Solyc11g017250.1.1 | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex-like | 0.012 | 1.31 | 0.96NS | ||
| SKFDNLYGcR | Solyc09g092380.2.1 | Adenosylhomocysteinase-like isoform X1 | 0.032 | 1.35 | 1.09NS | ✓ | |
| cYESPQASLR | Solyc04g047770.2.1 | VHS domain-containing protein At3g16270-like | 0.045 | 1.38 | 0.33NS | ✓ | |
| cSKPVVIADcGQL | Solyc01g111170.2.1 | Peptidyl-prolyl cis-trans isomerase | 0.01 | 1.39 | 0.87NS | ||
| FWEVIcDEHGVDPTGR | Solyc04g081490.2.1 | Beta-tubulin | 0.043 | 1.39 | 0.88NS | ||
| TGGLSVYPFTSSVHGKVEPcSK | Solyc01g108600.2.1 | Presequence protease 1, chloroplastic/mitochondrial-like | 0.02 | 1.41 | 0.98NS | ||
| YTQGFSGADITEIcQR | Solyc10g084050.1.1 | Cell division cycle protein 48 homolog | 0.009 | 1.43 | 1.66NS | ✓ | |
| DMFSTYGTITScK | Solyc01g107870.2.1 | Polyadenylate-binding protein 2-like | 0.032 | 1.44 | 1.07NS | ||
| IFNSDGSEPEMcGNGIR | Solyc09g005700.2.1 | Diaminopimelate epimerase, chloroplastic-like | 0.006 | 1.49 | 0.97NS | ||
| IcYVLYPGDESLEGK | Solyc05g012510.2.1 | Alpha-1,4 glucan phosphorylase L-2 isozyme, chloroplastic | 0.034 | 1.5 | n/a | 0.53S | |
| SPATDGcEYLFTGSR | TC223221_1 | WD repeat-containing protein 48 homolog | 0.002 | 1.52 | n/a | 0.74NS | |
| GALVcIDGTFATPLNQK | Solyc02g067180.2.1 | Cystathionine gamma synthase | 0.015 | 1.54 | 1.04NS | ✓ | |
| SFEASHcSK | Solyc03g117040.2.1 | Uncharacterized protein LOC101244470 | 0.005 | 1.55 | n/a | 1.1NS | |
| VLSANPEAPLNIEcLMDEKDVR | Solyc12g043120.1.1 | Heat shock 70 kDa protein 15-like | 0.008 | 1.59 | 0.89NS | ||
| EAGVTVcESPAK | Solyc01g007910.2.1 | Succinyl-CoA ligase [ADP-forming] subunit alpha-1, mitochondrial-like | 0.012 | 1.59 | 1.08NS | ✓ | |
| EVScLVK | Solyc04g049990.1.1 | Protein EIN4-like | 0.024 | 1.59 | n/a | 0.64NS | |
| GGQQVQAcR | Solyc01g073690.2.1 | Protein EIN4-like | 0.043 | 1.69 | n/a | 1.58NS | |
| cDVIASGIVNAAK | Solyc06g083790.2.1 | Succinyl CoA ligase beta subunit-like protein | 0.013 | 1.73 | 1.13NS | ||
| DGVGIcFPDLK | Solyc07g065120.2.1 | Uncharacterized protein LOC101262834 | 0.046 | 1.74 | n/a | 1.15NS | |
| TcSYLTSSAK | Solyc07g053650.2.1 | 26S proteasome non-ATPase regulatory subunit 2 1A-like | 0.049 | 1.85 | n/a | 1.12NS | |
| FTGSGGAVVVYcPDGTSQVK | Solyc02g093440.2.1 | Glucuronokinase 1-like | 0.037 | 1.87 | n/a | 0.60S | ✓ |
| NGTcSWLRPDGK | Solyc12g099000.1.1 | S-adenosylmethionine synthase 2-like isoform 1 | 0.022 | 2.07 | 0.81NS | ✓ | |
| SVVEEcGENISK | Solyc01g073740.2.1 | Citrate synthase, mitochondrial-like | 0.028 | 2.16 | 0.81NS | ||
| TVIDcFEK | Solyc07g054640.1.1 | NAD(P)H dehydrogenase B1, mitochondrial-like | 0.015 | 2.23 | n/a | 1.3NS | |
| VPNVQYIcTSPK | Solyc04g040180.2.1 | Putative methyltransferase DDB_G0268948-like | 0.002 | 2.28 | 1.25NS | ||
| TYSAcIDDHTK | Solyc10g074980.1.1 | Vacuolar-sorting receptor 1-like | 0.017 | 2.42 | 1.37NS | ✓ | |
| TVcGGAGINAcSNPAK | Solyc12g017460.1.1 | GDSL esterase/lipase At1g28590-like | 0.012 | 2.46 | n/a | 1.08NS | ✓ |
| QMIcccNK | TC222139_2 | Elongation factor 1-alpha | 0.046 | 2.5 | n/a | 0.57S | ✓ |
| AQQIcAVLYPGDATENGK | Solyc09g031970.2.1 | Alpha-glucan phosphorylase, H isozyme-like isoform 1 | 0.037 | 2.58 | 1.06NS | ||
| IFTSSSALYcTSTVEGDNEVKR | Solyc06g008620.1.1 | Uncharacterized protein LOC101247825 | 0.018 | 2.6 | 1.05NS | ✓ | |
| AANRLPGFGVITNIINGGLEcGR | Solyc10g055800.1.1 | Endochitinase 3-like | 0.008 | 2.63 | 1.18NS | ✓ | |
| YcAPDPEQDFSSGYEGKDVVVENLR | Solyc01g107130.2.1 | Vacuolar-sorting receptor 3-like isoform 2 | 0.005 | 2.73 | n/a | 1.18NS | ✓ |
| FGGDTPYSIMFGPDIcGYSTK | Solyc01g100380.2.1 | Calreticulin-like isoform 2 | 0.042 | 2.88 | n/a | 2.87S | |
| VcEcPTVQGVK | Solyc10g074980.1.1 | Vacuolar-sorting receptor 1-like | 0.018 | 2.99 | 1.37NS | ||
| GGIGcSPSDLSISELER | TC226243_3 | Glutamate dehydrogenase | 0.013 | 5.34 | n/a | 3.03S | |
| QSIIDEcFSK | Solyc12g011160.1.1 | 3-hydroxyisobutyryl-CoA hydrolase-like protein 5-like | 0.017 | 5.42 | 0.90NS | ||
| LLAAISSRPGQcGR | Solyc07g065170.2.1 | 40S ribosomal protein S8-like | 0.018 | 7.11 | n/a | 2.26NS | |
| SLcDQINSNSEETPR | Solyc12g044940.1.1 | Oxidoreductase UcpA-like | 0.024 | 7.43 | n/a | 3.50S | |
| GDAVDDLcNAR | Solyc09g061840.2.1 | 3-ketoacyl-CoA thiolase 2, peroxisomal | 0.033 | 21.98 | 0.67NS | ✓ |
Figure 4.
Alignment of tomato KAT2 sequence with its Arabidopsis ortholog. The shaded cysteine residue with a star underneath was identified as being redox-regulated upon Pst infection in tomato. This cysteine is localized in the active site of the enzyme and known to form a disulfide bond with the other cysteine at position 192 in Arabidopsis (highlighted in gray).
Discussion
Identification of redox proteins using cysTMT reverse labeling with correction of protein level change
During plant–pathogen interaction, both PTI and ETI responses lead to increases in ROS production,4,10,11 and the ROS can lead to oxidation of cysteine thiol groups.36 To map and quantify cysteine redox modifications at the proteome scale, cysTMT was developed to specifically label protein cysteine residues with free thiol groups from six different samples.28 This methodology has been used in the identification of disulfide bond formation as well as thiol nitrosylation.27,28 Here a reverse-labeling approach was used, where the reduced cysteines were first blocked using IAM, followed by the reduction of the oxidized cysteines and further labeling of the free thiols. Thus, the increases of the cysTMT signal from treated samples compared to control samples indicate oxidation of sensitive cysteines (Figure 1). On the other hand, since the reduced cysteines are blocked prior to the reduction of reversible-oxidized cysteines residues and labeling, decreases of the cysTMT signal from treated samples compared to control samples indicate the reduction of sensitive-cysteines. Therefore, this methodology is able to identify proteins that undergo oxidation as well as reduction in addition to mapping the redox-sensitive cysteines.
Although cysTMT methodology is able to identify and quantify potential redox-regulated proteins, it does not address the issue of protein turnover in the course of the experiments (Supplementary Figure S2). Many researches have overlooked this issue, which could have led to misleading results.37–40 Here, the data of protein level changes generated by Parker et al.,26 which used the same set of samples as in this study, were used to correct the redox-proteomics results. Although comparing potential redox changes with results for protein level changes obtained from parallel or different studies have been done41–43 and helped researches gain confidence in redox protein identification, it has been a challenge when proteins quantified in redox experiments are either absent or not quantified with high confidence in the total proteomics experiment. In the results presented here, 20% of the potential redox-regulated proteins were not identified in the total proteomics experiments. To overcome this issue, transcriptional level information was obtained through qRT-PCR for those proteins not identified in the total proteomics experiment.23 Please note that transcript levels are not always consistent with the abundance of cognate proteins.44 For any interesting potential redox proteins, further validation experiments and functional studies need to be done.
Protein redox regulation in susceptible and resistant genotypes at early stage of Pst infection
In S. lycopersicum, it is known that the R proteins Pto and Prf are required for resistance against Pst strains expressing the effector AvrPto. In this plant–pathogen interaction, the R protein Pto, is a protein kinase that recognizes AvrPto and initiates specific resistance to Pst.45 However, the resistance mediated by Pto–AvrPto interaction and further activation of HR is dependent on the nucleotide-binding site-leucine rich repeat protein Prf.46 It is known that the Pto–AvrPto interaction triggers the production of ROS leading to an oxidative burst in the cells, which results in a HR. Tai47 demonstrated that the oxidative burst is Prf dependent because NO and H2O2 production as well as HR were not detected in tomato prf3 infiltrated with the constitutively active mutant ptoL205D. In contrast, Parker et al.26 showed H2O2 production in prf3 plants when inoculated with Pst. The authors suggest that the increase in H2O2 is a result of rapid pathogen infiltration and tissue collapse. Although in this work the susceptible genotype showed a smaller number of redox-regulated proteins, the results revealed that Pst infection caused redox changes and PTMs of proteins in both genotypes. This can be explained by the fact that cell death, where ROS plays a crucial role, is associated with both immunity and disease susceptibility.48
Some studies have shown the association between defense responses and primary metabolism in processes involved in energy production, such as glycolysis and the pentose phosphate pathway, TCA cycle, mitochondrial electron transport, ATP biosynthesis, and biosynthesis of some amino acids.49,50 In PtoR plants, Pst infection caused redox regulation of serine hydromethyltranferase, ATP synthase, aminomethyltransferase, phosphoglycerate kinase, and fructose 1,6-biphosphatase that were identified as being more reduced upon Pst infection (Table 2). Among the proteins, ATP synthase and phosphoglycerate kinase were found to be redox-regulated in previous studies.19,42 With the exception of aminomethyltransferase, all the other proteins are known to be thioredoxin targets.51 In the prf3 plants, only three proteins related to primary metabolism were found to be redox-regulated after Pst infection. Triosephosphate isomerase was reduced, while ATP synthase CF1 beta subunit (chloroplastic) and ATP synthase subunit beta (mitochondrial) were oxidized in response to pathogen infection (Table 1). Triosephosphate isomerase protein was already previously reported to be S-nitrosylated during HR.52 All the three proteins are known to be thioredoxin targets.51 It is accepted that the complexity of plant defense responses requires abundant amount of energy.49 In addition, the primary metabolic pathways play a role as a source of signaling molecules to directly or indirectly trigger defense responses.53 Based on the above results, it is plausible that redox regulation of these proteins serves as an activity switch to turn on or off the different connections between carbohydrate metabolism and defense responses (Figure 5a).
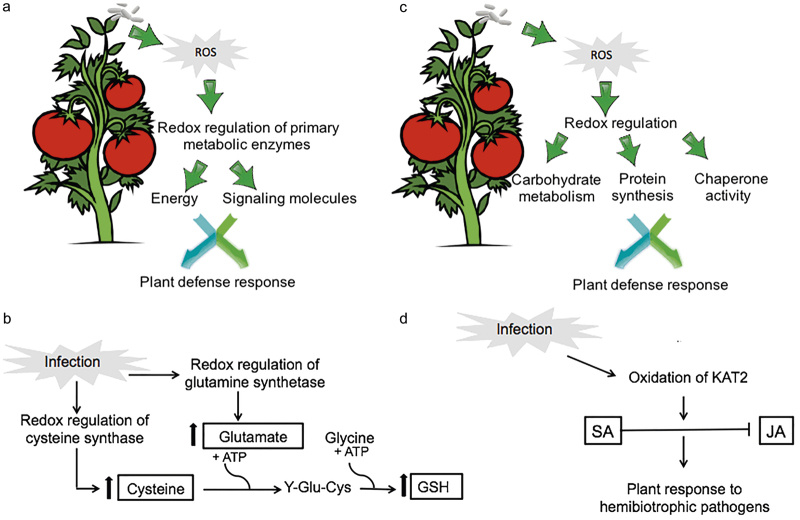
Figure 5.
The role of redox regulation in plant defense response. (a) Role of redox regulation of primary metabolic enzymes in plant defense response at early stage of infection. Redox regulation may serve as an activity switch to turn on or off the different connections between primary metabolism and defense response, playing a role in energy as well as signaling to directly or indirectly trigger defense responses. (b) Role of cysteine synthase oxidation and GS in plant defense response. Cysteine is the sulfur amino acid precursor of GSH, which plays a crucial role in maintaining cellular redox homeostasis. Oxidation of cysteine synthase may cause an increase in enzyme activity, leading to synthesis of more cysteines and further increase in the amount of GSH. Glutamate is a precursor of GSH and glutamine. Oxidation of GS decreases its activity, leading to less glutamine. This may cause an increase in the availability of glutamate for GSH production. (c) Role of redox regulation of enzymes belonging to carbohydrate metabolism, protein synthesis, and chaperone activity in plant defense response. It is likely that a synchronized interaction between sugar and hormonal signaling pathways leads to effective immune responses. Redox regulation of proteins of carbohydrate metabolism might affect apoplastic sugar levels, through which SAR is regulated. Redox regulation of proteins involved in protein synthesis may regulate enzyme activity in protein synthesis, which is an energy-consuming process, and therefore it is considered an important regulation step in stress responses. Oxidative stress is known to enhance misfolded proteins in the endoplasmic reticulum (ER), causing ER stress or unfolded protein response where proteins with chaperone activity play a crucial role. (d) Role of oxidation of KAT2 in the antagonistic interaction between SA and JA. Oxidation of KAT2, which is one of the three core enzymes that catalyze β-oxidation of JA synthesis, may lead to inactivation of KAT2.
Glutathione (GSH) is a non-protein thiol metabolite that is known to play a crucial role in many cellular processes as development, growth, and responses to biotic and abiotic stresses.54 Due to its high abundance and antioxidant activity, GSH plays an important role in maintaining cellular redox homeostasis. Biosynthesis of GSH occurs in the cytosol in two ATP-dependent enzymatic steps: formation of γ-glutamylcysteine from glutamate and cysteine and formation of GSH from γ-glutamylcysteine and glycine. One of the key determinants of GSH synthesis is the availability of the sulfur amino acid precursor cysteine.55 In our data, cysteine synthase and cysteine synthase-like were oxidized in response to Pst PtoR and prf3 plants, respectively (Tables 1 and 2). Currently, there is no report showing how oxidation affects cysteine synthase activity, it is possible that oxidized form of the enzyme is more active, leading to synthesis of more cysteines and increased amount of GSH in the defense against Pst (Figure 5b). As mentioned above, glutamate is another precursor of GSH. It is also a precursor of glutamine. GS catalyzes the ATP-required formation of glutamine from glutamate and ammonia. Here it was shown that both cytoplasmic and chloroplastic forms of GS, which are thioredoxin targets51 in PtoR plants, and the cytoplasmic form in prf3 plants were oxidized in response to Pst infection (Tables 1 and 2). A study in Canavalia lineata showed a redox-responsive site in the chloroplastic form GS and the reduction requirement of the sulfhydryl group for its high activity.56 Consequently, it is plausible that oxidation of GS decreases its activity. Therefore, less glutamine is synthesized, which can increase the availability of the precursor glutamate for the production GSH.
Proteins related to the antioxidant systems were also found to be redox-regulated in response to Pst infection. Some are known to be involved in ROS scavenging and reduction of other proteins. It is expected that they become oxidized while acting to reduce proteins. In the PtoR plants, glutaredoxin-1 and monodehydroascorbate reductase known to be thioredoxin targets51 were oxidized upon Pst infection. Peroxidase 12-like was found to be reduced in response to Pst in prf3 susceptible plants (Table 1). Increases in peroxidase activities in several crops such as rice, wheat, barley, cotton, and sugar cane appeared to correlate with their resistance to pathogens.57 A study with Arabidopsis ascorbate peroxidase (cAPX) showed that incubating cAPX with thioredoxin-h5 drastically inhibits its activity in vitro, suggesting that the active form of cAPX requires cysteine oxidization.58 In summary, the results presented here demonstrated that ROS produced in both PtoR and prf3 plants lead to redox regulation of proteins with similar functions in plant defense at early stage of infection. Targeted functional characterization of these potential redox proteins will reveal detailed molecular mechanisms as to how redox-regulation plays a role in plant immune responses.
Protein redox regulation in the resistant genotype at early and late stages of Pst infection
In incompatible plant–pathogen interaction, the SAR is induced to confer long-lasting protection against the pathogen. It is known that ETI is able to trigger SAR through local and systemic synthesis of SA. SAR requires the accumulation of SA, and this is associated with an increased expression of a large number of pathogenesis-related genes (PR genes) in both local and systemic tissues. Over the course of incompatible interaction, different proteins may be redox-regulated at early and late stages of infection. The results presented here showed that during late stage of infection a group of proteins related to carbohydrate metabolism were redox-regulated, which were not observed at early stage of infection. Among them, a biotin carboxylase carrier protein was found in a more reduced state after Pst infection. On the other hand, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, alpha-glucan phosphorylase, UTP-glucose-1-phosphate uridylyltransferase-like isoform 1, alpha-1,4 glucan phosphorylase L-2 isozyme (chloroplastic), and alpha-glucan H isozyme-like isoform 1 were all found in a more oxidized state (Table 3). With the exception of biotin carboxylase carrier protein and alpha-glucan H isozyme-like isoform 1, all the other redox proteins identified at the late stage of infection were known thioredoxin targets. Recent studies have shown the importance of sugar signaling in plant defense response under biotic stress.59 It is likely that a synchronized interaction between sugar and hormonal signaling pathways leads to effective immune responses. Thus, it can be speculated that redox-regulation of proteins of carbohydrate metabolism might affect apoplastic sugar levels, through which SAR is regulated (Figure 5c). Such a hypothesis is certainly worth testing in the future.
Protein synthesis is an energy-consuming process in the cells, and therefore it is considered an important regulation step in stress responses. Previous studies have shown oxidative modifications of some proteins involved in translational process when the cells are subjected to oxidative stress.19,38 Elongation factor 1-alpha (eEF1α) was found to undergo oxidation at the late stage of Pst infection (Table 3). In mammals and yeast, oxidation stress causes inhibition of protein synthesis by phosphorylation of the elongation initiation factor eIF2α.60,61 The results presented here show another putative level of PTM regulation of protein synthesis through regulation of translational elongation process by oxidation of eEF1α. eEF1α was identified as a thioredoxin target and redox-regulated under oxidative stress in previous studies.42,51 In addition, 40S ribosomal protein S8-like was found at the late stage of infection, and glycine-tRNA ligase 1 underwent oxidation in both early and late stages of Pst infection. Tubulin beta-1, which is a component of the cytoskeleton and target of thioredoxin,51 was found to be more reduced at early stage of infection (Table 2) and more oxidized later in the infection (Table 3). Components of cytoskeleton have previously been demonstrated to be sensitive to oxidation in both animals62,63 and Arabidopsis,64 but how they play a role in response to the infection is not clear.
Two other classes of proteins related to oxidative stress and β-oxidation were found to be redox-regulated at the late stage but not at the early stage of infection. Oxidative stress is known to enhance misfolded proteins in the endoplasmic reticulum (ER), causing ER stress or unfolded protein response. Two proteins with chaperone activity were identified as redox-regulated under pathogen infection. Heat shock 70 kDa protein (Hsp70) was found to undergo oxidation at late stage of infection (Table 3). DnaK, major Hsp70 in Escherichia coli was shown to form a dimer through disulfide bond in the ATP-bound state, and this dimer formation is important for the chaperone activity.65 The other protein was calreticulin 2, a calcium-binding chaperone that was identified to be oxidized based on cysTMT information. However, transcriptional level information revealed that the observed redox regulation fold change might be due to protein level change rather than a cysteine redox response (Table 3). Finally, two proteins involved in β-oxidation, 3-hydroxyisobutyryl-CoA hydrolase-like66 and KAT2 (see Role of 3-ketoacyl-CoA thiolase 2 oxidation in the antagonism between SA and JA section), were found to be highly oxidized in response to Pst at the late stage of infection (Table 3). 3-hydroxyisobutyryl-CoA hydrolase was already reported as redox-sensitive protein that undergoes oxidation in fungal pathogen infection.67 These interesting potential redox-regulated proteins in the early and late stages of Pst infection in PtoR plants are targeted for future studies on the functions of redox regulation in PtoR defense.
Role of KAT2 oxidation in the antagonism between SA and JA
It has been broadly accepted that the JA, SA, and ET defense signaling pathways are part of a complex signaling network, in which they influence each other in positive or negative manners.68 SA is considered as one of the key hormones that regulate the pathogen-induced immune responses in both ETI and SAR signaling. The SA response is characterized by transcriptional activation of PR genes and a local hypersensitive reaction to restrict proliferation of biotrophic pathogens.68 JA, a lipid-derived signaling molecule that is synthetized from the β-oxidation of the precursor molecule 3-oxo-2-(2′-pentenyl)-cyclopentane-1-octanoic acid (OPC:8). JA is involved in several aspects of plant biology including responses to biotic stress. Although JA is known to modulate defense against necrotrophic pathogens, there is a growing body of literature indicating the role of JA in other aspects of plant–pathogen interactions such as SAR.69 The interaction between SA and JA signaling is a complex process, and the primary mode of interaction between them appears to be mutually antagonistic. In tomato several studies have shown the inhibitory effect of SA on JA signaling.70–72 In addition, genetic studies in Arabidopsis showed that mutants impaired in SA accumulation exhibit enhanced responses to inducers of JA-dependent gene expression.73 It has been shown in Arabidopsis that mutant plants impaired in JA perception (e.g., coronatine insensitive1, coi1) exhibit enhanced SA-dependent defenses and resistance to P. syringae.74,75
Here it was shown that KAT2 had the highest level of oxidation at the late stage of Pst infection in PtoR (Table 3). KAT2 is one of the three core enzymes that catalyze β-oxidation, which is an important pathway for JA synthesis.76 Pye et al. (2010)77 showed that A. thaliana KAT2 is a sensitive redox switch protein, which regulates β-oxidation at minor perturbations of the peroximal redox environment. They also showed that oxidized form of the enzyme is inactive, while the reduced form is active. In addition, Cys138 that is localized in active site of the enzyme was shown to form a disulfide bond with Cys192 during redox regulation. Oxidation of Cys138 indicated that KAT2 was in its inactive for at late stage of infection. At 24 h after pathogen inoculation, SA is expected to play a big role in plant defense. Therefore, oxidation of KAT2 may be the functional switch in the antagonistic interaction between SA and JA. However, some further studies need to be conducted to confirm this hypothesis and the role of KAT2 oxidation in plant immunity response (Figure 5d).
Acknowledgments
We thank Prof. Greg Martin and Ms. Diane Dunham from Boyce Thompson Institute for Plant Research for providing the experimental materials and invaluable advice. The redox-proteomics work was partly supported by awards from the National Science Foundation (MCB 0818051 and MCB 1412547) to S. Chen.
The authors declare no conflict of interest.
References
- Engl C, Waite CJ, McKenna JF et al. Chp8, a diguanylate cyclase from Pseudomonas syringae pv. Tomato DC3000, suppresses the pathogen-associated molecular pattern flagellin, increases extracellular polysaccharides, and promotes plant immune evasion. mBio 2014; 5: e01168–e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agricultural development economics: long-term perspectives. Available at http://www.fao.org/economic/esa/esag/en/(accessed 13 April 2015).
- Hamdoun S, Gill M, Yao N, Lu H. Dynamics of defense responses and cell fate change during Arabidopsis-Pseudomonas syringae interactions. PLoS One 2013; 8: e83219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre V, Spallek T, Häweker H et al. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol 2008; 18: 1824–1832. [DOI] [PubMed] [Google Scholar]
- Nicaise V, Roux M, Zipfel C. Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol 2009; 150: 1638–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE. Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 2015; 66: 487–511. [DOI] [PubMed] [Google Scholar]
- Di Simplicio P, Franconi F, Frosalí S, Di Giuseppe D. Thiolation and nitrosation of cysteines in biological fluids and cells. Amino Acids 2003; 25: 323–339. [DOI] [PubMed] [Google Scholar]
- Laloi C, Mestres-Ortega D, Marco Y, Meyer Y, Reichheld J-P. The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol 2004; 134: 1006–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran A, Cotter TG. Redox regulation of protein kinases. FEBS J 2013; 280: 1944–1965. [DOI] [PubMed] [Google Scholar]
- Jeworutzki E, Roelfsema MRG, Anschütz U et al. Early signaling through the Arabidopsis pattern recognition receptors FLS2 and EFR involves Ca-associated opening of plasma membrane anion channels. Plant J Cell Mol Biol 2010; 62: 367–378. [DOI] [PubMed] [Google Scholar]
- Xin X-F, He SY. Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 2013; 51: 473–498. [DOI] [PubMed] [Google Scholar]
- Charles R, Jayawardhana T, Eaton P. Gel-based methods in redox proteomics. Biochimica et Biophysica Acta 2014; 1840: 830–837. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Loake GJ. Redox-based protein modifications: the missing link in plant immune signalling. Curr Opin Plant Biol 2011; 14: 358–364. [DOI] [PubMed] [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hurst RD, Hancock JT. Hydrogen peroxide and nitric oxide as signalling molecules in plants. J Exp Bot 2002; 53: 1237–1247. [PubMed] [Google Scholar]
- Stone JR, Yang S. Hydrogen peroxide: a signaling messenger. Antioxid Redox Signal 2006; 8: 243–270. [DOI] [PubMed] [Google Scholar]
- Spadaro D, Yun B-W, Spoel SH et al. The redox switch: dynamic regulation of protein function by cysteine modifications. Physiol Plant 2010; 138: 360–371. [DOI] [PubMed] [Google Scholar]
- Leitner M, Vandelle E, Gaupels F, Bellin D, Delledonne M. NO signals in the haze: nitric oxide signalling in plant defence. Curr Opin Plant Biol 2009; 12: 451–458. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Tada Y, Loake GJ. Post-translational protein modification as a tool for transcription reprogramming. New Phytol 2010; 186: 333–339. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang S, Lu Y et al. Proteomic analysis of early-responsive redox-sensitive proteins in arabidopsis. J Proteome Res 2012; 11: 412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003; 113: 935–944. [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Sell S, Müller B, Leister D, Durner J. Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 2010; 22: 2894–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B-W, Feechan A, Yin M et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 2011; 478: 264–268. [DOI] [PubMed] [Google Scholar]
- Frungillo L, Skelly MJ, Loake GJ, Spoel SH, Salgado I. S-nitrosothiols regulate nitric oxide production and storage in plants through the nitrogen assimilation pathway. Nat Commun 2014; 5: 5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-Q, Feechan A, Yun B-W et al. S-nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J Biol Chem 2009; 284: 2131–2137. [DOI] [PubMed] [Google Scholar]
- Bender K, Wang X, Cheng G et al. Glutaredoxin AtGRXC2 catalyzes inhibitory glutathlonylation of Arabidopsis BRI1-associated receptor-like kinase 1 (BAK1) in vitro. Biochem J 2015; 467: 399–413. [DOI] [PubMed] [Google Scholar]
- Parker J, Koh J, Yoo M-J et al. Quantitative proteomics of tomato defense against Pseudomonas syringae infection. Proteomics 2013; 13: 1934–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CI, Uhrigshardt H, O’Meally RN et al. Identification and quantification of S-nitrosylation by cysteine reactive tandem mass tag switch assay. Mol Cell Proteomics 2012; 11: M111.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J, Zhu N, Zhu M, Chen S. Profiling thiol redox proteome using isotope tagging mass spectrometry. J Vis Exp 2012; 24: pii: 3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichert LI, Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol 2004; 2: e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng WL, Rehm AH, Charkowski AO et al. Pseudomonas syringae exchangeable effector loci: sequence diversity in representative pathovars and virulence function in P. syringae pv. syringae B728a. J Bacteriol 2003; 185: 2592–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez S, Zhu M, Chen S. Proteomics of Arabidopsis redox proteins in response to methyl jasmonate. J Proteomics 2009; 73: 30–40. [DOI] [PubMed] [Google Scholar]
- Fu C, Wu C, Liu T et al. Elucidation of thioredoxin target protein networks in mouse. Mol Cell Proteomics 2009; 8: 1674–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt M, Messens J, Collet J-F. How proteins form disulfide bonds. Antioxid Redox Signal 2011; 15: 49–66. [DOI] [PubMed] [Google Scholar]
- Kovacs I, Lindermayr C. Nitric oxide-based protein modification: formation and site-specificity of protein S-nitrosylation. Plant Proteomics 2013; 4: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JM (ed.). The identification of protein S-nitrosocysteine New York: Springer, Humana Press, 2009. Available at http://link.springer.com/protocol/10.1007%2F978-1-59745-198-7_155 (accessed 13 April 2015). [Google Scholar]
- Go Y-M, Jones DP. The redox proteome. J Biol Chem 2013; 288: 26512–26520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y-M, Roede JR, Orr M, Liang Y, Jones DP. Integrated redox proteomics and metabolomics of mitochondria to identify mechanisms of Cd toxicity. Toxicol Sci 2014; 139: 59–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Zhang H, Wang H, Xia Y. Identification of redox-sensitive cysteines in the Arabidopsis proteome using OxiTRAQ, a quantitative redox proteomics method. Proteomics 2014; 14: 750–762. [DOI] [PubMed] [Google Scholar]
- Su D, Gaffrey MJ, Guo J et al. Proteomic identification and quantification of S-glutathionylation in mouse macrophages using resin-assisted enrichment and isobaric labeling. Free Radic Biol Med 2014; 67: 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuramalingam M, Matros A, Scheibe R, Mock H-P, Dietz K-J. The hydrogen peroxide-sensitive proteome of the chloroplast in vitro and in vivo. Front Plant Sci 2013; 4: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Hu J, Liu T et al. Quantitative analysis of redox-sensitive proteome with DIGE and ICAT. J Proteome Res 2008; 7: 3789–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Zhu N, Song W et al. Thiol-based redox proteins in abscisic acid and methyl jasmonate signaling in Brassica napus guard cells. Plant J 2014; 78: 491–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo-Álvarez A, Ramos E, Villanueva J et al. Differential redox proteomics allows identification of proteins reversibly oxidized at cysteine residues in endothelial cells in response to acute hypoxia. J Proteomics 2012; 75: 5449–5462. [DOI] [PubMed] [Google Scholar]
- Chen S, Harmon AC. Advances in plant proteomics. Proteomics 2006; 6: 5504–5516. [DOI] [PubMed] [Google Scholar]
- Salmeron JM, Barker SJ, Carland FM et al. Tomato mutants altered in bacterial disease resistance provide evidence for a new locus controlling pathogen recognition. Plant Cell 1994; 6: 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Oldroyd GE, Rommens CM et al. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 1996; 86: 123–133. [DOI] [PubMed] [Google Scholar]
- Tai Y-S. Detection of reactive oxygen species can be used to distinguish ToxA-induced cell death from the hypersensitive response. Res J Bot 2007; 2: 1–12. [Google Scholar]
- Abramovitch RB, Anderson JC, Martin GB. Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol 2006; 7: 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton MD. Primary metabolism and plant defense – fuel for the fire. Mol Plant Microbe Interact 2009; 22: 487–497. [DOI] [PubMed] [Google Scholar]
- Less H, Angelovici R, Tzin V, Galili G. Coordinated gene networks regulating Arabidopsis plant metabolism in response to various stresses and nutritional cues. Plant Cell 2011; 23: 1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montrichard F, Alkhalfioui F, Yano H et al. Thioredoxin targets in plants: the first 30 years. J Proteomics 2009; 72: 452–474. [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Campostrini N, Mattè A et al. Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics 2008; 8: 1459–1469. [DOI] [PubMed] [Google Scholar]
- Rojas CM, Senthil-Kumar M, Tzin V, Mysore KS. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front Plant Sci 2014; 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil-Maurizi C, Poinssot B. Role of glutathione in plant signaling under biotic stress. Plant Signal Behav 2012; 7: 210–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SC. Glutathione synthesis. Biochim Biophys Acta 2013; 1830: 3143–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YA, Kim SG, Kwon YM. The plastidic glutamine synthetase activity is directly modulated by means of redox change at two unique cysteine residues. Plant Sci 1999; 149: 175–182. [Google Scholar]
- Datta SK, Muthukrishnan S. Pathogenesis-related proteins in plants Boca Raton, FL: CRC Press, 1999. [Google Scholar]
- Correa-Aragunde N, Foresi N, Lamattina L. Nitric oxide is a ubiquitous signal for maintaining redox balance in plant cells: Regulation of ascorbate peroxidase as a case study. J Exp Bot 2015; 66: 2913–2921. [DOI] [PubMed] [Google Scholar]
- Moghaddam MRB, Van den Ende W. Sugars and plant innate immunity. J Exp Bot 2012; 63: 3989–3998. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 2003; 11: 619–633. [DOI] [PubMed] [Google Scholar]
- Dunand-Sauthier I, Walker CA, Narasimhan J et al. Stress-activated protein kinase pathway functions to support protein synthesis and translational adaptation in response to environmental stress in fission yeast. Eukaryot Cell 2005; 4: 1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle-Donne I, Rossi R, Milzani A et al. The actin cytoskeleton response to oxidants: from small heat shock protein phosphorylation to changes in the redox state of actin itself. Free Radic Biol Med 2001; 31: 1624–1632. [DOI] [PubMed] [Google Scholar]
- Lassing I, Schmitzberger F, Björnstedt M et al. Molecular and structural basis for redox regulation of beta-actin. J Mol Biol 2007; 370: 331–348. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Skipsey M, Grundy NM, Edwards R. Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol 2005; 138: 2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbeng EB, Liu Q, Tian X et al. A functional DnaK dimer is essential for the efficient interaction with Hsp40 heat shock protein. J Biol Chem 2015; 290: 8849–8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Thompson B et al. chy1, an Arabidopsis mutant with impaired β-oxidation, is defective in a peroxisomal β-hydroxyisobutyryl-CoA hydrolase. J Biol Chem 2001; 276: 31037–31046. [DOI] [PubMed] [Google Scholar]
- Ilyas S, Rehman A, Varela AC, Sheehan D. Redox proteomics changes in the fungal pathogen Trichosporon asahii on arsenic exposure: identification of protein responses to metal-induced oxidative stress in an environmentally-sampled isolate. PLoS One 2014; 9: e102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 2002; 5: 325–331. [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JDG. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 2011; 49: 317–343. [DOI] [PubMed] [Google Scholar]
- Doherty HM, Selvendran RR, Bowles DJ. The wound response of tomato plants can be inhibited by aspirin and related hydroxy-benzoic acids. Physiol Mol Plant Pathol 1988; 33: 377–384. [Google Scholar]
- Pena-Cortés H, Albrecht T, Prat S et al. Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta 1993; 191: 123–128. [Google Scholar]
- Doares SH, Narvaez-Vasquez J, Conconi A, Ryan CA. Salicylic acid inhibits synthesis of proteinase inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiol 1995; 108: 1741–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Willits MG, Glazebrook J. Arabidopsis thaliana EDS4 contributes to salicylic acid (SA)-dependent expression of defense responses: evidence for inhibition of jasmonic acid signaling by SA. Mol Plant-Microbe Interact 2000; 13: 503–511. [DOI] [PubMed] [Google Scholar]
- Kloek AP, Verbsky ML, Sharma SB et al. Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J Cell Mol Biol 2001; 26: 509–522. [DOI] [PubMed] [Google Scholar]
- Feys B, Benedetti CE, Penfold CN, Turner JG. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 1994; 6: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Antonenkov VD, Glumoff T, Hiltunen JK. Peroxisomal β-oxidation – a metabolic pathway with multiple functions. Biochimica et Biophysica Acta 2006; 1763: 1413–1426. [DOI] [PubMed] [Google Scholar]
- Pye VE, Christensen CE, Dyer JH et al. Peroxisomal plant 3-ketoacyl-CoA thiolase structure and activity are regulated by a sensitive redox switch. J Biol Chem 2010; 285: 24078–24088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.