Protein disulfide isomerase acts as a reductase to reduce a mutant proinsulin called Akita, priming it for retrotranslocation across the endoplasmic reticulum (ER) membrane by using the Sel1L-Hrd1-p97 ER-associated degradation machinery.
Abstract
In mutant INS gene–induced diabetes of youth (MIDY), characterized by insulin deficiency, MIDY proinsulin mutants misfold and fail to exit the endoplasmic reticulum (ER). Moreover, these mutants bind and block ER exit of wild-type (WT) proinsulin, inhibiting insulin production. The ultimate fate of ER-entrapped MIDY mutants is unclear, but previous studies implicated ER-associated degradation (ERAD), a pathway that retrotranslocates misfolded ER proteins to the cytosol for proteasomal degradation. Here we establish key ERAD machinery components used to triage the Akita proinsulin mutant, including the Hrd1-Sel1L membrane complex, which conducts Akita proinsulin from the ER lumen to the cytosol, and the p97 ATPase, which couples the cytosolic arrival of proinsulin with its proteasomal degradation. Surprisingly, we find that protein disulfide isomerase (PDI), the major protein oxidase of the ER lumen, engages Akita proinsulin in a novel way, reducing proinsulin disulfide bonds and priming the Akita protein for ERAD. Efficient PDI engagement of Akita proinsulin appears linked to the availability of Hrd1, suggesting that retrotranslocation is coordinated on the lumenal side of the ER membrane. We believe that, in principle, this form of diabetes could be alleviated by enhancing the targeting of MIDY mutants for ERAD to restore WT insulin production.
INTRODUCTION
Insulin biosynthesis is initiated when preproinsulin is translocated into the endoplasmic reticulum (ER) (Steiner et al., 1967) and its signal sequence is cleaved to generate proinsulin. Oxidative folding of proinsulin ensues (Liu et al., 2012), characterized by formation of three highly conserved disulfide bonds (denoted as B7-A7, B19-A20, and A6-A11). Properly folded proinsulin then exits the ER, is transported to the Golgi, and is delivered to secretory granules, where pro–hormone convertases excise the C-peptide, with B and A chains of mature insulin remaining attached via B7-A7 and B19-A20 interchain disulfide bonds.
Recently a subset of human insulin gene mutations has been discovered to cause an autosomal-dominant syndrome that we call mutant INS gene–induced diabetes of youth (MIDY; Stoy et al., 2007; Liu et al., 2010b; Weiss, 2013): insulin-deficient diabetes without islet autoantibodies. MIDY is a protein-misfolding disease, and most MIDY proinsulin mutants misfold within the ER, causing permanent neonatal or later-onset diabetes, depending upon the particular mutation. Among these, the most-studied MIDY mutant is Akita proinsulin: substitution at the A7 cysteine causes severe early-onset diabetes in both humans and Akita mice.
MIDY mutants fail to be transported from the ER to the Golgi complex. Moreover, MIDY mutants impair wild-type (WT) proinsulin exit from the ER, thereby limiting WT insulin production, provoking increased blood glucose excursions, and stimulating expression of even more proinsulin (both mutant and WT), which remains entrapped within the ER (Liu et al., 2007; Weiss, 2013). This triggers ER stress that may ultimately result in β-cell death.
Eliminating buildup of mutant proinsulin in the ER might restore β-cell function and survival. This possibility motivates us to investigate cellular mechanisms of degradation of misfolded proinsulin. One such mechanism is the ER-associated degradation (ERAD) pathway (Tsai et al., 2002; Smith et al., 2011). In ERAD, a misfolded ER substrate must first be identified by ER lumenal components and then delivered across (retrotranslocated) to the cytosolic side of the ER membrane for ubiquitination, with the assistance of ERAD membrane components (Carvalho et al., 2010; Stein et al., 2014). Finally, the ubiquitinated substrate is extracted from the ER membrane into the cytosol for delivery to proteasomes for degradation (Ye et al., 2001).
There is reason to suspect that MIDY mutants may use the ERAD pathway (Allen et al., 2004; Hartley et al., 2010; Tiwari et al., 2013), although how misfolded proinsulin in the ER lumen is recognized and primed for retrotranslocation remains unknown. Here we establish that canonical ERAD membrane components, including the E3 ubiquitin ligase Hrd1, its membrane-binding partner Sel1L, and cytosolic p97, help to conduct Akita proinsulin from the ER lumen to the cytosol. Strikingly, we find that protein disulfide isomerase (PDI), a profolding enzyme known to drive oxidation of disulfide bonds (Hatahet and Ruddock, 2009; Benham, 2012), instead exhibits a novel interaction with misfolded proinsulin to exert a prodegradative role that includes reducing proinsulin disulfide bonds. Our findings provide a framework for rational therapeutic approaches to preventing and treating diabetes provoked by proinsulin misfolding.
RESULTS
The Hrd1-Sel1L complex promotes ERAD of Akita proinsulin
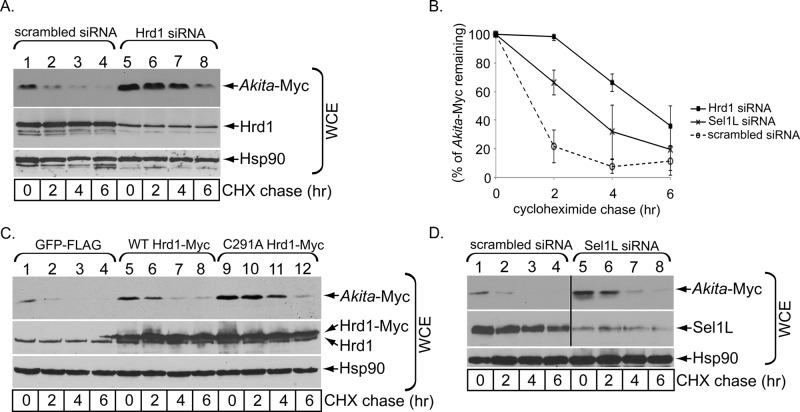
To determine whether Akita proinsulin is degraded by ERAD, we first evaluated whether Hrd1, the multitransmembrane E3 ubiquitin ligase that is also proposed to function as the retrotranslocon (Carvalho et al., 2010; Stein et al., 2014), controls the fate of transfected (Myc-tagged) Akita proinsulin. Using cycloheximide chase in 293T cells (and in β-cells; see later discussion), we found that Hrd1 knockdown markedly impaired degradation of Akita proinsulin (Figure 1A; quantified in Figure 1B). Of importance, depleting Hrd1 (or any other protein described in this study) did not trigger massive ER stress, as XBP1 remained largely unspliced under these conditions (Supplemental Figure S1A). Our analyses also demonstrated that overexpressing enzymatically inactive, dominant-negative Hrd1-C291A (Bernardi et al., 2010) potently stabilized Akita proinsulin (Figure 1C, top). A smaller degree of inhibition of degradation of Akita proinsulin was also observed upon WT Hrd1-Myc overexpression (this is likely due to perturbation of the stoichiometric ratio of Hrd1 to its natural binding partners, in this way decreasing the efficiency of functional Hrd1-associated complexes). Overexpression of WT, mutant Hrd1, or any other construct in this study also did not provoke enhanced XBP1 splicing (Supplemental Figure S1B). Together the results in Figure 1, A–C, indicate that Hrd1 participates in the degradation of Akita proinsulin. Next we asked whether Sel1L also promotes ERAD of Akita proinsulin. Sel1L is a membrane protein that serves as a scaffold to engage ERAD lumenal components (Hosokawa et al., 2008; Mueller et al., 2008; Williams et al., 2013), interacts with Hrd1 (Mueller et al., 2006), and recruits client substrates to Hrd1 (Bernasconi et al., 2010; Stanley et al., 2011). Using a similar cycloheximide-chase protocol, we found that Sel1L knockdown also impaired Akita ERAD (Figure 1D; quantified in Figure 1B), strongly implicating the Hrd1-Sel1L membrane complex in the degradation of Akita proinsulin.
FIGURE 1:
The Hrd1-Sel1L complex promotes ERAD of Akita. (A) 293T cells expressing Akita-Myc were transfected with a scrambled or Hrd1-specific siRNA. Cells were then treated with cycloheximide for the indicated time and harvested, and the resulting WCE was analyzed by using the indicated antibodies. (B) The Akita band intensity in A and D was quantified with ImageJ (National Institutes of Health, Bethesda, MD). Data represent mean ± SD of at least three independent experiments. (C) As in A, except that cells were transfected with GFP-FLAG, WT Hrd1-Myc, or C291A Hrd1-Myc construct. (D) As in A, except cells were transfected with a siRNA against Sel1L. The black vertical line indicates that an intervening lane from the same immunoblot has been spliced out.
Because Akita appears to use the classic ERAD pathway in which the Hrd1-Sel1L complex serves as the core membrane component, we asked whether expressing WT proinsulin might affect ERAD of Akita, as patients with MIDY harbor one WT copy of the proinsulin gene. Indeed, coexpressing superfolder green fluorescent protein–tagged WT proinsulin (SfGFP-WT proinsulin, which largely behaves as WT proinsulin; Haataja et al., 2013) significantly impaired degradation of Myc-tagged Akita (Supplemental Figure S1C, top). This result suggests that the established interaction between WT proinsulin and Akita (Liu et al., 2007) can functionally disrupt degradation of Akita, potentially exacerbating ER stress, which contributes to MIDY.
p97 facilitates Akita degradation
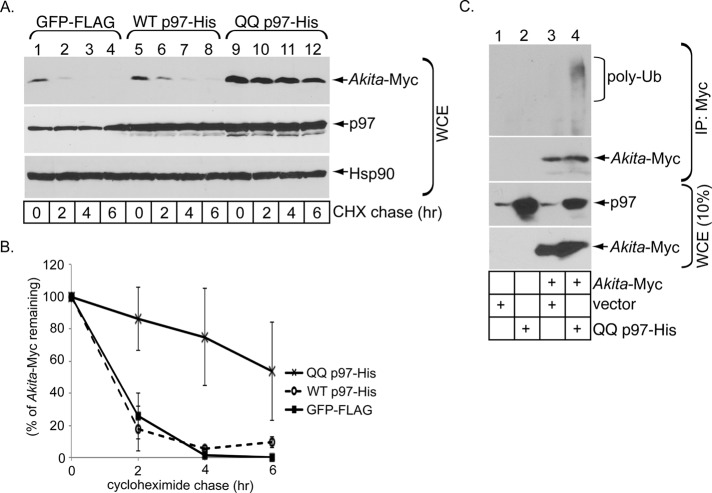
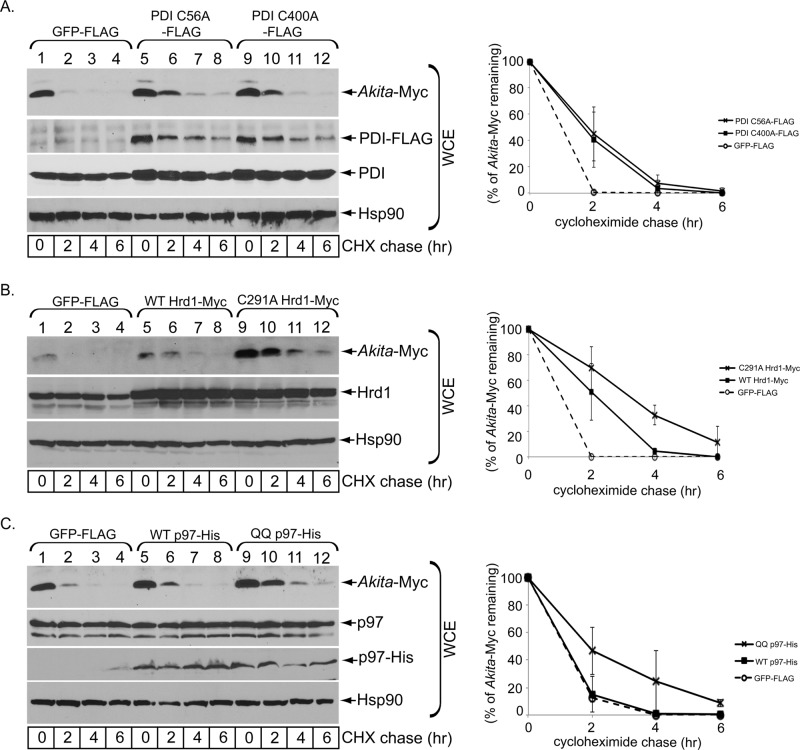
Because Hrd1 orients its catalytic E3 ligase domain on the cytosolic surface of the ER membrane, retrotranslocated substrates become available for ubiquitination, and the ubiquitinated substrates are then extracted into the cytosol—in most cases by the cytosolic p97 ATPase (Ye et al., 2001)—before being degraded by cytosolic proteasomes. To assess whether p97 participates in ERAD of Akita proinsulin, we overexpressed a histidine (His)-tagged WT or ATPase-defective dominant-negative version of p97 (QQ-p97). Whereas WT p97 was largely without effect, overexpression of QQ p97-His markedly stabilized Akita proinsulin (Figure 2A; quantified in Figure 2B), implicating p97 in the extraction of Akita proinsulin from the ER membrane to the cytosol.
FIGURE 2:
p97 facilitates Akita degradation. (A) As in Figure 1C, except that 293T cells were transfected with WT p97-His or QQ p97-His. (B) As in Figure 1B, except that the Akita band intensity in A is quantified. (C) 293T cells expressing Akita-Myc or not were transfected with an empty vector or QQ p97-His as indicated. Cells were lysed in a RIPA buffer and Akita immunoprecipitated from the resulting WCE using a Myc antibody. The precipitated material was subjected to SDS–PAGE and immunoblotted with the indicated antibodies. The WCE was also immunoblotted.
Because p97 binds to ubiquitinated substrates, blocking Akita proinsulin extraction into the cytosol should cause Akita proinsulin to accumulate as an ubiquitinated species. To test this, we cotransfected cells to express Akita proinsulin and QQ p97-His. When Myc-tagged Akita proinsulin was immunoprecipitated (Figure 2C, second from top), the polyubiquitinated species accumulated only in cells expressing QQ p97-His (Figure 2C, top). This finding is consistent with the idea that p97 extracts ubiquitinated Akita proinsulin from the ER membrane and supports that the classic Hrd1/Sel1L-p97 pathway promotes ERAD of Akita proinsulin.
PDI promotes Akita degradation
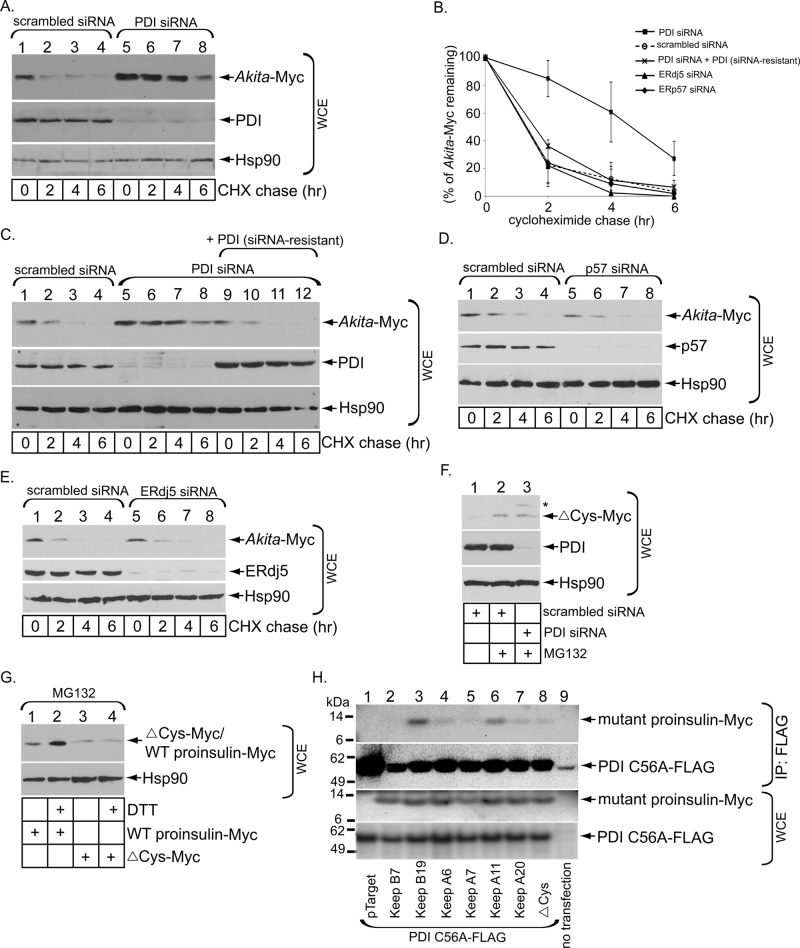
One major impediment to ERAD is the presence of disulfide bonds that hold the substrate in a plicated conformation believed to be incompatible with retrotranslocation. Because Akita proinsulin still harbors five cysteine residues in an aberrant disulfide configuration (Izumi et al., 2003; Liu et al., 2007, 2010a) that is unknown, enzymatic reduction of disulfide bonds is likely important for its retrotranslocation. The major ER-resident oxidoreductase is PDI, which is believed to promote oxidative folding of many newly synthesized secretory proteins (Hatahet and Ruddock, 2009; Benham, 2012). However, PDI does not appear to facilitate oxidative folding of proinsulin in the ER of pancreatic β-cells (Rajpal et al., 2012) and in fact has been implicated in retrotranslocation of certain ERAD substrates and a bacterial toxin (Gillece et al., 1999; Forster et al., 2006). Indeed, we found that in cells in which PDI was knocked down for 3 d, Akita proinsulin was robustly stabilized (Figure 3A; quantified in Figure 3B). A similar effect on Akita degradation was observed when PDI was knocked down for only 2 d (Supplemental Figure S1D). When a small interfering RNA (siRNA)–resistant PDI construct was expressed in PDI-knockdown cells, degradation of Akita proinsulin was largely restored (Figure 3C; quantified in Figure 3B). However, knockdown of ERp57 (Figure 3D) or ERdj5 (Figure 3E), ER-resident oxidoreductases (Apperizeller-Herzog and Ellgaard, 2008; Ushioda et al., 2008), did not significantly affect ERAD of Akita proinsulin (quantified in Figure 3B). These data strongly suggest that ERAD of Akita proinsulin is specifically targeted by PDI.
FIGURE 3:
PDI exerts a key role in regulating Akita degradation. (A) As in Figure 1A, except that 293T cells were transfected with a PDI-specific siRNA for 3 d. (B) As in Figure 1C, except that the Akita band intensity in A and C–E are quantified. (C) As in A, except where indicated cells transfected with a PDI-specific siRNA for 3 d were cotransfected with a PDI construct resistant to the PDI-specific siRNA. (D) As in A, except hat cells were transfected with an ERp57-specific siRNA. (E) As in A, except that cells were transfected with an ERdj5-specific siRNA. (F) 293T cells expressing either a control or PDI-specific siRNA were cotransfected with ΔCys-Myc and treated or not with MG132 as indicated. (G) Cells expressing WT proinsulin-Myc or ΔCys-Myc were treated with DTT (10 μM) in the presence of MG132 and the WCE analyzed as in F. (H) 293T cells expressing the indicated Myc-tagged mutant proinsulins were cotransfected with PDI C56A-FLAG. The cells were lysed and immunoprecipitated with M2 FLAG antibody-conjugated beads, analyzed by reducing SDS–PAGE, and immunoblotted using the appropriate antibodies. The WCE was also directly immunoblotted with anti-Myc or anti-FLAG antibodies. pTarget was the plasmid used as a control for the mutant proinsulin constructs.
PDI can function as both an ER molecular chaperone and an ER oxidoreductase (Cai et al., 1994). To understand the predominant role of PDI in the ERAD of Akita proinsulin, we tested whether knockdown of PDI affected degradation of a proinsulin construct lacking all Cys residues (ΔCys-Myc; Liu et al., 2010a), which is detected intracellularly in the presence, but not the absence, of MG132 treatment (Figure 3F). Of importance, silencing of PDI did not augment the steady-state level of ΔCys-Myc (Figure 3F), suggesting that the mechanism by which PDI facilitates retrotranslocation of Akita proinsulin likely involves interaction with one or more of its cysteine residues. A low level of a slower-migrating species (Figure 3F, asterisk) that appeared in the PDI-depleted cells may represent a different folded conformation of this artificial mutant proinsulin that persists even under SDS–PAGE, consistent with PDI being a chaperone that unfolds ERAD substrates during retrotranslocation (Gillece et al., 1999). Of interest, treating cells with the reducing agent dithiothreitol (DTT) did not alter ΔCys-Myc’s steady-state level but did for WT proinsulin-Myc (Figure 3G), suggesting that degradation of the cysteine-less proinsulin mutant is unaffected by changing the cellular redox state.
PDI interacts with, and acts as a reductase for, Akita proinsulin
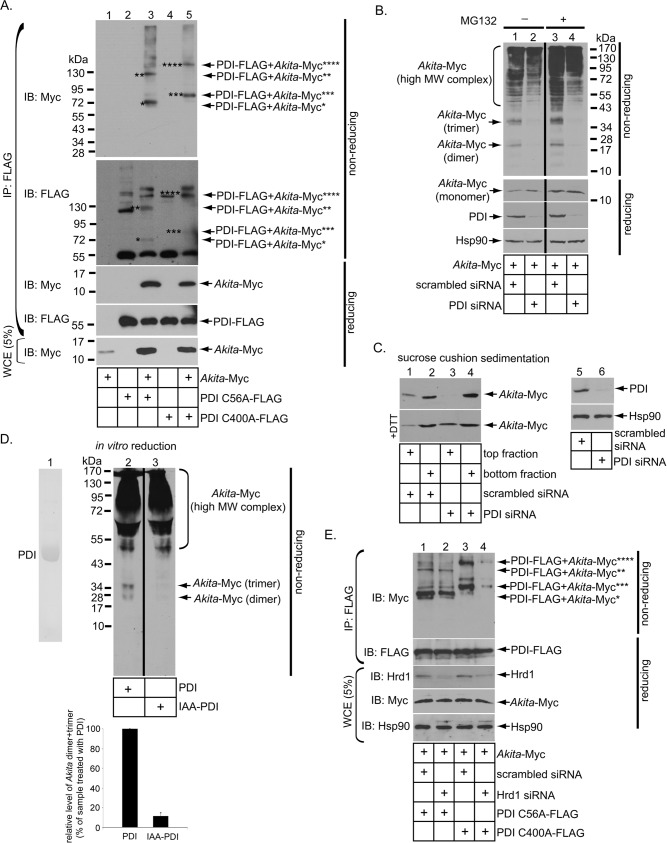
To determine whether initiation of ERAD of Akita proinsulin involves a direct interaction with PDI, we exploited so-called PDI trap mutants, in which a CXXC thioredoxin motif (where C = cysteine and X = any amino acid) is mutated to CXXA. When functioning as a reductase, the first cysteine thiol of the CXXC motif attacks a disulfide bond in the substrate to form a mixed-disulfide intermediate that is rapidly broken by intradomain disulfide pairing with the second CXXC cysteine thiol. These steps reduce the substrate disulfide bond while internally oxidizing the thioredoxin motif. However, CXXA trap mutants can only cross-link with the substrate and cannot form the intradomain disulfide pair, thereby “trapping” mixed disulfide-bonded adducts between PDI and its substrates. Because PDI possesses two thioredoxin domains, we used two (FLAG-tagged) trap mutants: PDI C56A, to create the first CXXA, and PDI C400A, to create the second CXXA. When coexpressed with Akita proinsulin, we found that precipitation of either PDI trap mutant recovered disulfide-bonded adducts engaging both PDI and Akita under nonreducing conditions (Figure 4A, top). As a control, precipitating WT PDI did not recover any disulfide-bonded adduct with Akita (Supplemental Figure S2A, compare lane 2 to lane 4). One of the main adducts recovered by precipitating PDI C56A had a molecular mass of ∼69 kDa (Figure 4A, asterisk) which corresponds precisely to the cumulative molecular masses of one PDI (59 kDa) plus one proinsulin (∼10 kDa), strongly suggesting a direct disulfide bond rather than binding via intermediary proteins. Of interest, precipitation of PDI C400A recovered an adduct of ∼79 kDa (Figure 4A, triple asterisks), likely representing one PDI molecule plus an Akita dimer. Other proinsulin-containing PDI adducts (Figure 4A, double and quadruple asterisks) were also recovered. By contrast, neither of two different ERp57 trap mutants nor three distinct ERp72 trap mutants captured Akita proinsulin (Supplemental Figure S2B), strongly suggesting that PDI specifically engages Akita proinsulin.
FIGURE 4:
PDI acts as a reductase against Akita. (A) 293T cells expressing Akita were transfected with or without PDI C56A-FLAG or PDI C400A-FLAG. Tagged PDI proteins were immunoprecipitated from the resulting WCE using a FLAG antibody. The precipitated material was subjected to nonreducing or reducing SDS–PAGE as indicated and immunoblotted using the appropriate antibodies. The WCE was also immunoblotted with a Myc antibody. (B) 293T cells expressing Akita and transfected with either a scrambled or PDI-specific siRNA were treated with or without MG132. The resulting WCE was subjected to nonreducing or reducing SDS–PAGE as indicated and immunoblotted using the indicated antibodies. The black vertical line indicates that an intervening lane from the same immunoblot has been spliced out. (C) Lanes 1–4, WCE in B was treated with or without DTT, layered over a 20% sucrose cushion, and centrifuged. The top and bottom fractions were separated and subjected to SDS–PAGE, followed by immunoblotting using a Myc antibody. Lanes 5 and 6, WCE was subjected to SDS–PAGE, followed by immunoblotting using the indicated antibodies. (D) His-tagged PDI purified from bacteria is shown in the Coomassie gel. Immunoprecipitated Akita-Myc from PDI-knockdown cells was incubated with PDI or IAA-modified PDI in the presence of 1 mM GSH and 1 mM GSSG, and the samples were subjected to nonreducing SDS–PAGE, followed by immunoblotting with a Myc antibody. Bottom graph, the intensity level of Akita dimer and trimer was quantified by ImageJ. Data represent the mean ± SD of three independent experiments. The black vertical line indicates that an intervening lane from the same immunoblot has been spliced out. (E) Cells expressing Akita transfected with a scrambled or Hrd1-specific siRNA were cotransfected with either PDI C56A-FLAG or PDI C400A-FLAG. The ability of tagged PDI protein to interact with Akita was analyzed as in A.
We also found that the steady-state level of Akita proinsulin was higher in cells expressing PDI C56A than with other trap mutants (Supplemental Figure S2B, fourth panel), suggesting that such trapping inhibits ERAD of Akita proinsulin. Indeed, cycloheximide-chase experiments confirmed that expression of PDI-C56A and PDI-C400A stabilized Akita proinsulin in comparison to expressing ERp57-C60A or P5 C58A (Supplemental Figure S2C, top), where P5 represents yet another PDI family member. These analyses provide independent evidence that PDI specifically plays a functional role in targeting Akita proinsulin for ERAD.
Because Akita proinsulin lacks a cysteine at position A7, the Cys-B7 lacks its natural partner for disulfide bonding. Although this might render Cys-B7 especially available for cross-linking with PDI, proinsulin misfolding might involve a variety of disulfide mispairings, making other Cys residues more available for adduct formation with PDI. To simplify our analysis, we constructed a family of proinsulin mutants each bearing only a single Cys residue. To our surprise, when coexpressed with the PDI-C56A trap mutant, two of the six single-Cys proinsulin mutants consistently coprecipitated to a greater degree with PDI: a proinsulin construct that harbors only the B19 Cys residue (Keep B19), and Keep A11 (Figure 3H; quantified in Supplemental Figure S3A). However, even when Cys-B7 was the only available unpaired cysteine, the Keep B7 mutant did not preferentially interact with overexpressed PDI trap mutant (Figure 3H; quantified in Supplemental Figure S3A). Similarly, when the various proinsulin mutants were precipitated (with anti-myc), Keep B7 consistently coprecipitated less PDI C56A-FLAG than did Keep B19 or Keep A11 (Supplemental Figure S3B). (Curiously, Keep A20 was also able to coprecipitate PDI trap mutant more efficiently than others, suggesting that A20 is also a reactive cysteine, although association of Keep A20 with PDI trap might affect epitope exposure during immunoprecipitation with anti-FLAG.) Together these coprecipitation results support that even when freely available, the proinsulin B7 cysteine is disfavored from forming a disulfide bond with PDI; thus it is more likely that a complex pattern of intramolecular disulfide mispairing within Akita proinsulin accounts for the robust formation of Akita proinsulin adducts with PDI (Figure 4A).
We then examined the status of Akita in cells with PDI knockdown. When evaluated by nonreducing SDS–PAGE, Akita proinsulin in control cells formed high–molecular weight (MW) protein complexes and additionally migrated as a disulfide-bonded dimer and trimer (Figure 4B, top, lane 1). In PDI-knockdown cells (lane 2), the dimer and trimer disappeared and likely entered the high-MW complex. The Akita monomer was revealed when the samples were subjected to reducing SDS–PAGE, with the Akita level being higher in PDI-depleted cells (Figure 4B, second from top, lanes 1 and 2). Addition of the proteasome inhibitor MG132 did not restore loss of the Akita dimer or trimer observed by nonreducing SDS–PAGE in PDI-depleted cells (Figure 4B, top, compare lanes 4 and 3), although the steady-state level of low-MW Akita did increase in the MG132-treated sample as expected (Figure 4B, top, compare lane 3 with lane 1). Whereas incubation with the autophagy inhibitor LY294002 (Blommaart et al., 1997) did enhance the steady-state level of Akita in control cells (Supplemental Figure S4, compare lane 3 to lane 1), this inhibitor clearly did not restore the loss of Akita dimer or trimer in PDI-knockdown cells (Supplemental Figure S4, top, compare lanes 4 and 3). These results suggest that autophagy is unlikely to be responsible for the disappearance of Akita dimer or trimer when PDI is depleted, although this process may contribute to quality control of Akita in control cells. Thus the data are consistent with the idea that disappearance of dimeric and trimeric Akita in PDI-depleted cells reflects a loss of proinsulin reductase activity rather than ERAD or ER-dependent autophagy of dimeric and trimeric Akita proinsulin, suggesting that enzymatically active PDI helps to limit Akita proinsulin from becoming engaged in high-MW disulfide-linked complexes.
As yet another independent approach, we layered cell extracts expressing Akita proinsulin over a 20% sucrose cushion and centrifuged, generating bottom and top fractions. The top fraction harbors lower-MW species, likely dimeric and monomeric species of Akita proinsulin (Figure 4C, top, lane 1), whereas the bottom fraction (Figure 4C, top, lane 2) contains Akita proinsulin predominantly in high-MW complexes whose sizes are heterogeneous and appear to be larger than ∼55 kDa. In control cells, a small portion of Akita proinsulin was recovered in the top fraction, with most detected at bottom (Figure 4C, top, lanes 1 and 2) whereas in PDI-knockdown cells, Akita proinsulin in the top fraction was eliminated (lanes 3 and 4). Of importance, when DTT was added to cell extracts before fractionation, even the PDI-knockdown cells yielded recovery of Akita proinsulin in the top fraction (Figure 4C, second from top), mimicking the effect of PDI. In PDI-depleted cells, the combined level of Akita in top plus bottom fractions of DTT-treated samples is similar to the Akita level found in the bottom fraction only in non–DTT-treated samples (Figure 4C, compare second from top and top, lanes 3 and 4). As expected, DTT reduced Akita in cell extracts derived from PDI-depleted cells, generating Akita monomer and dimer (Supplemental Figure S5). However, a significant fraction of Akita proinsulin remained in high-MW complexes even after reduction by DTT (Figure 4C, bottom, lane 4), presumably due to noncovalent associations with each other or with other cellular proteins. This is consistent with a report suggesting that Akita engages in both covalent and noncovalent interactions to remain in the high-MW complexes (Yoshinaga et al., 2005). Our data thus strongly suggest that the inability to partition to the top fraction after PDI knockdown is due to a lack of reducing power for Akita proinsulin in the ER, supporting the notion that PDI acts as a reductase for Akita proinsulin. To assess whether PDI can reduce Akita in vitro, we used a modified protocol from the cell-based assay. Akita was first isolated from PDI-depleted cells by immunoprecipitation, followed by elution of the bound material using SDS and then dialysis to remove the detergent. Isolated Akita was subsequently incubated with bacterially purified PDI (Figure 4D, lane 1) or, as a negative control, PDI alkylated with iodoacetamide (IAA; IAA-PDI), which should not reduce substrates, because its active cysteines are blocked. When this experiment was performed under a condition that mimics the ER redox environment (1 mM reduced glutathione [GSH] and 1 mM oxidized glutathione [GSSG]; Hwang et al., 1992), PDI but not IAA-PDI induced formation of Akita dimer and trimer (Figure 4D, lanes 2 and 3; the dimer and trimer levels are quantified in the bottom graph), indicating that PDI uses its catalytic cysteines to reduce Akita. Note that Akita migrated slightly differently in this experiment (Figure 4D) compared with the cell-based assay (Figure 4B), possibly due to the harsh conditions used to isolate Akita in the in vitro assay. Nonetheless, these findings support the idea that PDI directly reduces Akita present in high-MW disulfide-bonded complexes to generate lower-MW species.
We previously reported that a subset of PDI localizes to the lumenal side of the Hrd1 membrane complex (Bernardi et al., 2010), which may allow this subset of PDI molecules to reduce ERAD substrates more efficiently. To test this, we examined the extent to which PDI trap mutants can capture Akita proinsulin when Hrd1 expression is silenced. Indeed, upon Hrd1 knockdown, the trapping of Akita proinsulin by PDI-C56A and PDI-C400A declined (Figure 4E, top). Thus Hrd1 availability enhances the efficiency of PDI in engaging Akita to reduce proinsulin disulfide bonds before retrotranslocation.
PDI, Hrd1, and p97 promote ERAD of Akita in pancreatic β-cells
We next explored the roles of PDI, Hrd1, and p97 in the ERAD of Akita proinsulin in the INS-1 832/13 pancreatic β-cell line. Similar to results observed in 293T cells (Supplemental Figure S2C), expressing the PDI trap mutants in β-cells slowed degradation of Akita proinsulin compared with controls (GFP-FLAG, Figure 5A, top; the Akita band intensity is quantified in the graph at right). In addition, in INS-1 832/13 cells, Akita proinsulin was robustly stabilized by dominant-negative Hrd1-C291A but more modestly by WT Hrd1 (Figure 5B, top; the Akita band intensity is quantified in the graph at right). Finally, expressing QQ p97 potently inhibited Akita degradation (Figure 5C, top; the Akita band intensity is quantified in the graph at right). Together these findings indicate that, as in 293T cells, the role of the PDI-Hrd1-p97 axis in the ERAD of Akita proinsulin is indeed maintained within the context of pancreatic β-cells.
FIGURE 5:
PDI, Hrd1, and p97 promote ERAD of Akita in pancreatic β-cells. (A) As in Supplemental Figure S2C, except that rat pancreatic INS-1 832/13 β-cells were used and GFP-FLAG was expressed instead of ERp57 C60A-FLAG. The Akita band intensity is quantified as in Figure 1B. (B) As in Figure 1C, except that the experiment was performed in INS-1 832/13 β-cells and GFP-FLAG was transfected instead of vector. The Akita band intensity is quantified as in Figure 1B. (C) As in Figure 2A, except that the experiment was performed in INS-1 832/13 β-cells and GFP-FLAG was transfected instead of vector. The Akita band intensity is quantified as in Figure 1B.
DISCUSSION
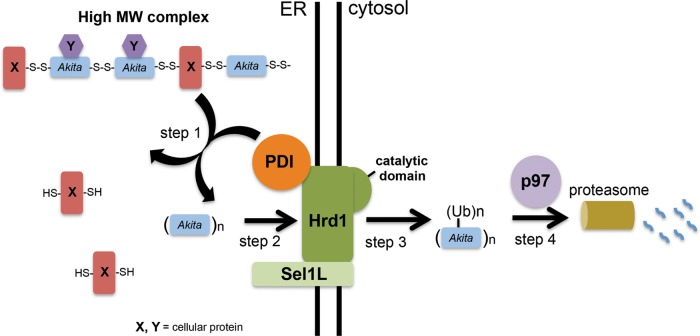
We propose the following model for cellular disposal of Akita mutant proinsulin (Figure 6): disulfide bonds engaged by Akita proinsulin, including intermolecular disulfide bonds, are reduced by PDI (step 1), whereupon Akita proinsulin retrotranslocates across the ER membrane via the Hrd1/Sel1L membrane complex (indeed, Hrd1 availability enhances PDI engagement of Akita proinsulin; step 2). Consistent with the known enzymatic activity of Hrd1, Akita proinsulin becomes polyubiquitinated upon arrival at the cytosolic side of the ER membrane (step 3) before extraction into the cytosol by p97, delivering Akita proinsulin to cytosolic proteasomes (step 4).
FIGURE 6:
Model depicting ERAD of Akita. To dispose of Akita via the ERAD pathway, PDI reduces the disulfide bonds in this mutant proinsulin to initiate its retrotranslocation into the cytosol (step 1). The precise pairing of disulfide bonds in Akita proinsulin is not known. In the second step, Akita retrotranslocates across the ER membrane by using the Hrd1/Sel1L membrane complex (step 2). Once it is presented to the cytosol, Akita becomes polyubiquitinated using Hrd1’s catalytic activity (step 3). In the final step, p97 propels polyubiquitinated Akita into the cytosol, targeting it to the proteasome for degradation (step 4).
Degradation of Akita proinsulin engages key elements of the classic ERAD pathway
Classic ERAD involves the Hrd1-Sel1L membrane complex and the cytosolic p97 ATPase (Tsai et al., 2002; Smith et al., 2011). The core component is the six-transmembrane-spanning Hrd1 (Kikkert et al., 2004), which associates with numerous adaptor proteins. Hrd1 is believed to serve, first, as the physical conduit for transfer of client substrates from the ER lumen to the cytosol (Carvalho et al., 2010; Stein et al., 2014) and, second, as the major E3 ligase that ubiquitinates ERAD substrates upon arrival at the cytosolic face of the ER membrane (Claessen et al., 2012). Using cycloheximide chase in conjunction with selective siRNA-mediated knockdown of Hrd1, we find that inhibiting Hrd1 function robustly blocks the degradation of Akita proinsulin, increasing its steady-state level (Figure 1, A and B). We also find that overexpression of enzymatically inactive Hrd1 (or, to a much lesser extent, WT Hrd1) similarly produces dominant-negative inhibition of Akita proinsulin degradation (Figure 1C). The dominant effects of recombinant Hrd1 are believed to arise by competing for natural Hrd1 binding partners, thereby impairing the proper stoichiometry and function of Hrd1-associated protein complexes. Although the results of our WT Hrd1 overexpression experiments differ slightly from those reported previously (Allen et al., 2004), our conclusions are entirely concordant with the idea that the Hrd1 E3 ligase is involved in ERAD of Akita proinsulin, ubiquitinating the substrate when it emerges on the cytosolic side of the ER membrane.
Moreover, our findings demonstrate that Sel1L, a membrane binding partner of Hrd1, also participates in ERAD of Akita proinsulin (Figure 1, B and D). Yet another Hrd1 interactor, Herp, was also reported to modestly increase the steady-state level of Akita proinsulin, suggesting that Herp may also be involved in ERAD of this substrate (Hartley et al., 2010). The findings with both Herp and Sel1L support our contention that Hrd1 and its interacting partners guide ER-to-cytosol retrotranslocation of misfolded proinsulin.
On presentation on the cytosolic side of the ER membrane, polyubiquitinated ERAD substrates encounter the cytosolic p97 ATPase, which (in conjunction with its binding partners, including Ufd1 and Npl4; Woodman, 2003) uses the energy of ATP hydrolysis to extract the substrate into the cytosol (Zhang and Ye, 2014). We found that overexpressing a catalytically inactive p97 dramatically blocked the degradation of Akita proinsulin (Figure 2, A and B), supported by a recent report that chemical inhibition of p97 modestly inhibits Akita proinsulin degradation in a manner generally similar to that achieved by chemical inhibition of proteasomes (Zhang et al., 2014). The notion that p97 at the cytosolic face of the ER membrane facilitates the degradation of ubiquitinated Akita proinsulin is further demonstrated by the accumulation of polyubiquitinated Akita proinsulin in the presence of the dominant-negative p97 mutant (Figure 2C). Taken together, these findings strongly suggest that p97 mobilizes mutant proinsulin into the cytosol for delivery to proteasomes.
PDI’s reductase activity primes Akita proinsulin for ERAD
A key and surprising finding of our study involves the role of PDI in initiating retrotranslocation of Akita proinsulin for ERAD. Such a conclusion is not unreasonable: whereas PDI does not serve as an oxidase for native proinsulin disulfide bond formation (Rajpal et al., 2012), disulfide isomerase activity may be used to help break nonnative disulfide bonds. Moreover, the development of nonremediable misfolding may “tag” Akita proinsulin as a potential ERAD substrate. PDI can operate as a chaperone, facilitating the unfolding of substrates targeted for ERAD before their retrotranslocation (Tsai et al., 2001; Schelhaas et al., 2007), but it is likely that, once selected for ERAD, nonnative (and probably even remaining native) proinsulin disulfide bonds must be reduced so that the unfolded substrate can thread through the retrotranslocon.
In the context of genetic mutation of one of the six evolutionarily conserved proinsulin cysteine residues, the development of nonremediable proinsulin misfolding is guaranteed. An unpaired proinsulin cysteine residue may have the opportunity to attack other cysteines within the same proinsulin molecule or other proteins present in the ER (including other copies of proinsulin, ER oxidoreductases, or others). PDI appears to preferentially interact with the Cys-B19, Cys-A11, and even Cys-A20 of proinsulin (Figure 3H and Supplemental Figure S3), but Cys-B7 (the normal disulfide bonding partner of Cys-A7) is not a preferential site of adduct formation with PDI. Thus Akita proinsulin is likely to be involved in a more complex pattern of disulfide mispairing that leads to the generation of PDI adducts. Consistent with this, intermolecular disulfide bonds are observed for Akita proinsulin, including both nonnative disulfide-linked proinsulin dimers and trimers and disulfide bonds with other proteins in high- MW complexes (Izumi et al., 2003; Liu et al., 2007, 2010b; Figure 4), and knockdown of PDI causes loss of the dimers and trimers (Figure 4B) while specifically blocking ERAD of Akita proinsulin (Figure 3).
Several new and novel observations support the idea that the role of PDI in the ERAD of Akita proinsulin is as a reductase. First, trap mutants that can mimic initial PDI reductase activity capture Akita proinsulin in disulfide-linked adducts, including adducts that correspond to a direct 1:1 interaction between PDI and Akita proinsulin (Figure 4A). Second, expression of these same PDI trap mutants inhibits degradation of Akita proinsulin (Supplemental Figure S2C). Third, PDI knockdown causes simple disulfide-bonded Akita dimers to be lost (Figure 4B) and to fractionate with high-MW complexes, whereas addition of a chemical reductant mimics the effect of PDI (Figure 4C), strongly suggesting that lack of PDI results in a lack of reducing power for Akita proinsulin in the ER. Fourth, purified PDI is capable of reducing Akita proinsulin in the high-MW complexes to generate Akita dimers and trimers (Figure 4D). Thus our collective findings argue strongly that PDI acts as a reductase to initiate retrotranslocation of Akita proinsulin. Because many MIDY mutations involve a loss or gain of a cysteine that promotes formation of nonnative disulfide bonds (Liu et al., 2010b), the reductase activity of PDI is also likely to promote ERAD of other MIDY mutants. Indeed, the role of PDI in ERAD is far more widespread than for proinsulin alone. We note that the classic components of ERAD, including Sel1L, Hrd1, and p97, are conserved down to yeast; indeed, it is well established that the yeast Sel1L homologue (Hrd3p) recruits lumenal factors for ERAD of substrates that also engage yeast Hrd1 (Denic et al., 2006). With this in mind, we note that even in this simple eukaryote, the redox activity of PDI has been shown to be important for ERAD of the mutant CPY* (Grubb et al., 2012). Thus it is not surprising that our results for ERAD of Akita proinsulin in heterologous HEK 293T cells are also obtained in pancreatic β-cells (Figure 5), as ERAD uses common, ancient, essential pathways.
The pathogenesis of MIDY is attributed to the inability of WT proinsulin to exit the ER, which is a critical step for normal insulin production (Liu et al., 2010b; Weiss, 2013). Because MIDY proinsulin entraps WT proinsulin in the ER, the less abundant the misfolded proinsulin, the less is the risk of diabetes (Hodish et al., 2011). In principle, restoration of WT proinsulin exit from the ER, resulting in more WT insulin production, should be possible if efficient disposal of misfolded proinsulin can be achieved. Although there is still much to be learned about the degradation of misfolded proinsulin in the ER, exploiting ERAD may serve as a crucial platform for the development of future therapeutic approaches to combat diabetes caused by ER stress–induced β-cell failure linked to misfolded proinsulin within the ER.
MATERIALS AND METHODS
Reagents
Antibodies used were as follows: rabbit anti-Myc (Immunology Consultants Laboratories, Portland, OR); mouse anti-VCP (p97), anti-PDI (RL90), and anti-ERp57 (Abcam, Cambridge, MA); rabbit anti-Hrd1 (Proteintech, Chicago, IL); rabbit anti-Hsp90 (H-114; Santa Cruz Biotechnology, Santa Cruz, CA); mouse anti-ubiquitin (Invitrogen, Carlsbad, CA); rabbit anti-FLAG, anti-Sel1L, and mouse and rabbit horseradish peroxidase (HRP) secondary antibodies (Sigma-Aldrich, St. Louis, MO); rabbit anti-Myc (9E10) for immunoprecipitation (IP; K. Verhey, University of Michigan, Ann Arbor, MI). Chemicals were from Sigma-Aldrich except when noted otherwise. Phenylmethylsulfonyl fluoride (PMSF) was from Acros Organics; DMEM, RPM1 1640 medium, and other cell culture reagents were from Life Technologies (Carlsbad, CA). An expression plasmid encoding Myc-epitope–tagged human C(A7)Y Akita mutant proinsulin (Akita-Myc) was previously described (Liu et al., 2010b). A second expression plasmid encoding Myc-tagged human C(A7)Y proinsulin was constructed in which all five remaining Cys residues were mutated to Ser residues; this kind of construct, previously referred to as proinsulin-DelCys (Liu et al., 2010a), is also known here as ΔCys-Myc. Six related expression plasmids encoding Myc-tagged human proinsulin were constructed in which five Cys residues were mutated to Ser; each construct retains only one Cys residue, and they are referred to as Keep B7, Keep B19, Keep A6, Keep A7, Keep A11, and Keep A20, respectively. Purification of His-tagged PDI from bacteria was as previously described (Forster et al., 2009). Human PDI trap mutants were gifts from T. Rapoport (Harvard University, Cambridge, MA). The INS-1 832/13 β-cell line was a generous gift from Christopher Newgard (University of Texas Southwestern, Dallas, TX).
Cell culture, plasmid transfection, cycloheximide chase, immunoprecipitation, and immunoblotting
Rat INS-1 832/13 or human HEK 293T cells were cultured at 37°C in a humid incubator (5% CO2). Cells were seeded on six-well plates 1 d before transfection of 1–3 μg of plasmid DNA using home-made polyethylenimine (PEI) or Lipofectamine 2000 (Invitrogen). At 4 h posttransfection, cells were treated with cycloheximide (100 μg/ml) and chased for 0, 2, 4, and 6 h. In experiments using ΔCys-Myc or single-Cys Keep mutants, cells were treated with MG132 (10 μM) for 3 h before cell lysis. Cells were harvested in phosphate-buffered saline (PBS) containing 10 mM N-ethylmaleimide (NEM). For IP, cells were lysed in 400 μl of RIPA buffer (Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 1% sodium dexoycholate, 0.1% SDS) supplemented with 10 mM NEM and 1 mM PMSF; in Figure 3H, the modified RIPA buffer contained 0.25% SDS. Cells were incubated on ice for 10 min and centrifuged, and the resulting whole-cell extract (WCE) was incubated with antibody (1:200) at 4°C for 24 h, followed by 2 h of incubation with precleared protein A/G-agarose beads at 4°C. Beads were washed three times and boiled in SDS sample buffer with or without 100 mM DTT. For immunoblot analyses, cells were lysed in 100 μl of RIPA buffer to generate WCE, which was boiled in SDS sample buffer with 100 mM DTT. Then samples were resolved on 8–15% SDS–PAGE gels, transferred to nitrocellulose, and blotted with specific primary and HRP-conjugated secondary (1:2000) antibodies according to manufacturer’s protocols (Sigma-Aldrich).
siRNA knockdown of Hrd1, Sel1L, and PDI
siRNA was transfected into cells using RNAiMAX (Invitrogen), and cells were chased and harvested 48–72 h posttreatment. The sequences of the siRNAs are as follows: Dharmacon human Hrd1, 5′-GGAGACUGCCACUACAGUUGUUU-3′, at 10 nM for 48-h knockdown; Dharmacon human Sel1L, 5′-GAAACAAACAUUCGAGAUAUU-3′, at 25 nM for 48-h knockdown; and human PDI, 5′-CAACUUUGAAGGGGAGGUCTT-3′, and rat PDI, 5′-GCGCAUACUUGAGUUCUUUTT-3′, at 75 nM for 72-h knockdown.
siRNA-resistant PDI construct
siRNA-resistant WT PDI was generated with KOD Hot Start DNA polymerase (EMD Chemicals, Billerica, MA) from mouse WT PDI according to the manufacturer’s protocol. The sequences of forward and reverse primers are as follows: forward, 5′-GCAGCTAGCCCCCATTTGGGACAAGCTCGGCGAAACCTACAAGGATCATGAGAATATC-3′; and reverse, 5′-GATATTCTCATGATCCTTGTAGGTTTCGCCGAGCTTG TCCCAAATGGGGGCTAGCTGC-3′. The mutated siRNA-resistant PDI construct was confirmed by sequencing. A 2-μg amount of siRNA-resistant PDI plasmid was transfected into HEK 293T cells with PEI 24 h posttransfection of the PDI siRNA.
Sucrose cushion sedimentation
Cells transfected with scrambled or PDI siRNA were lysed in RIPA buffer to generate a WCE as described. WCE was centrifuged at 50,000 rpm at 4°C for 20 min using a TLA 100 rotor (Beckman, Brea, CA). Half of the resulting supernatant (50 μl) was treated or not with DTT (100 mM) for 30 min on ice, layered over an equal volume of a 20% sucrose solution, and centrifuged at 75,000 rpm at 4°C for 30 min using the same rotor. Top (50 μl) and bottom (50 μl) fractions were separated and subjected to immunoblot analyses.
In vitro PDI reduction assay
PDI was expressed in Escherichia coli strain BL21-Pro and purified as previously described (Forster et al., 2009). Purified PDI was aliquoted and stored at −80°C. Akita-Myc was expressed in PDI-depleted HEK 293T cells for 24 h, followed by cell lysis (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], 150 mM NaCl, 1% Triton X-100, 10 mM NEM, and 1 mM PMSF) and overnight immunoprecipitation as described. After three washes, Akita-Myc was eluted in 20 μl of a 0.5% SDS buffer at room temperature for 30 min, dialyzed, and diluted to 200 μl in a buffer containing 50 mM HEPES and 150 mM NaCl. Isolated Akita-Myc was incubated in the presence of 1 mM GSH and 1 mM GSSG with either PDI or PDI alkylated with 25 mM IAA, and the reaction was continued at 37°C for 30 min. Samples were separated on a nonreducing SDS–PAGE and subjected to immunoblot analysis.
Supplementary Material
Acknowledgments
We thank Leena Haataja and Huan Guo (University of Michigan, Ann Arbor, MI) for assistance. B.T. is funded by the National Institutes of Health (RO1 083252-05). The contributions of P.A. and M.L. were funded by National Institutes of Health Grants DK48280 and DK088856, respectively. This work was also partially supported by the Protein Folding Disease Initiative of the University of Michigan Medical School. C.C. is supported by Cellular and Molecular Biology Program National Institutes of Health Training Grant 5T32GM00731538.
Abbreviations used:
- CHX
cycloheximide
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- GFP
green fluorescent protein
- GSH
glutathione
- GSSG
oxidized glutathione
- IAA
iodoacetamide
- IAA-PDI
iodoacetamide-treated protein disulfide isomerase
- IB
immunoblot
- IP
immunoprecipitation
- MIDY
mutant INS gene–induced diabetes of youth
- MW
molecular weight
- PDI
protein disulfide isomerase
- PEI
polyethylenimine
- SfGFP
superfolder green fluorescent protein
- siRNA
small interfering RNA
- Ub
ubiquitin
- WCE
whole cell extract
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-01-0034) on August 12, 2015.
REFERENCES
- Allen JR, Nguyen LX, Sargent KEG, Lipson KL, Hackett A, Urano F. High ER stress in beta-cells stimulates intracellular degradation of misfolded insulin. Biochem Biophys Res Commun. 2004;324:166–170. doi: 10.1016/j.bbrc.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Apperizeller-Herzog C, Ellgaard L. The human PDI family: versatility packed into a single fold. Biochim Biophys Acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Benham AM. The protein disulfide isomerase family: key players in health and disease. Antioxid Redox Signal. 2012;16:781–789. doi: 10.1089/ars.2011.4439. [DOI] [PubMed] [Google Scholar]
- Bernardi KM, Williams JM, Kikkert M, van Voorden S, Wiertz EJ, Ye YH, Tsai B. The E3 ubiquitin ligases Hrd1 and gp78 bind to and promote cholera toxin retro-translocation. Mol Biol Cell. 2010;21:140–151. doi: 10.1091/mbc.E09-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-L-S substrates. J Cell Biol. 2010;188:223–235. doi: 10.1083/jcb.200910042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- Cai H, Wang CC, Tsou CL. Chaperone-like activity of protein disulfide isomerase in the refolding of a protein with no disulfide bonds. J Biol Chem. 1994;269:24550–24552. [PubMed] [Google Scholar]
- Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen JHL, Kundrat L, Ploegh HL. Protein quality control in the ER: balancing the ubiquitin checkbook. Trends Cell Biol. 2012;22:22–32. doi: 10.1016/j.tcb.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Forster ML, Mahn JJ, Tsai B. Generating an unfoldase from thioredoxin-like domains. J Biol Chem. 2009;284:13045–13056. doi: 10.1074/jbc.M808352200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster ML, Sivick K, Park YN, Arvan P, Lencer WI, Tsai B. Protein disulfide isomerase-like proteins play opposing roles during retrotranslocation. J Cell Biol. 2006;173:853–859. doi: 10.1083/jcb.200602046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillece P, Luz JM, Lennarz WJ, de La Cruz FJ, Romisch K. Export of a cysteine-free misfolded secretory protein from the endoplasmic reticulum for degradation requires interaction with protein disulfide isomerase. J Cell Biol. 1999;147:1443–1456. doi: 10.1083/jcb.147.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb S, Guo L, Fisher EA, Brodsky JL. Protein disulfide isomerases contribute differentially to the endoplasmic reticulum-associated degradation of apolipoprotein B and other substrates. Mol Biol Cell. 2012;23:520–532. doi: 10.1091/mbc.E11-08-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haataja L, Snapp E, Wright J, Liu M, Hardy AB, Wheeler MB, Markwardt ML, Rizzo M, Arvan P. Proinsulin intermolecular interactions during secretory trafficking in pancreatic beta cells. J Biol Chem. 2013;288:1896–1906. doi: 10.1074/jbc.M112.420018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley T, Siva M, Lai E, Teodoro T, Zhang LL, Volchuk A. Endoplasmic reticulum stress response in an INS-1 pancreatic beta-cell line with inducible expression of a folding-deficient proinsulin. BMC Cell Biol. 2010;11:59. doi: 10.1186/1471-2121-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatahet F, Ruddock LW. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- Hodish I, Absood A, Liu L, Liu M, Haataja L, Larkin D, Al-Khafaji A, Zaki A, Arvan P. In vivo misfolding of proinsulin below the threshold of frank diabetes. Diabetes. 2011;60:2092–2101. doi: 10.2337/db10-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Nagasawa K, Moriyama T, Okawa K, Nagata K. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J Biol Chem. 2008;283:20914–20924. doi: 10.1074/jbc.M709336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- Izumi T, Yokota-Hashimoto H, Zhao SL, Wang A, Halban PA, Takeuchi T. Dominant negative pathogenesis by mutant proinsulin in the Akita diabetic mouse. Diabetes. 2003;52:409–416. doi: 10.2337/diabetes.52.2.409. [DOI] [PubMed] [Google Scholar]
- Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, Thanedar S, Roitelman J, Chau V, Wiertz E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem. 2004;279:3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- Liu M, Haataja L, Wright J, Wickramasinghe NP, Hua QX, Phillips NF, Barbetti F, Weiss MA, Arvan P. Mutant INS-gene induced diabetes of youth: proinsulin cysteine residues impose dominant-negative inhibition on wild-type proinsulin transport. PLoS One. 2010a;5:e13333. doi: 10.1371/journal.pone.0013333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Hodish I, Haataja L, Lara-Lemus R, Rajpal G, Wright J, Arvan P. Proinsulin misfolding and diabetes: mutant INS gene-induced diabetes of youth. Trends Endocrinol Metab. 2010b;21:652–659. doi: 10.1016/j.tem.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Hodish I, Rhodes CJ, Arvan P. Proinsulin maturation, misfolding, and proteotoxicity. Proc Natl Acad Sci USA. 2007;104:15841–15846. doi: 10.1073/pnas.0702697104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Lara-Lemus R, Shan SO, Wright J, Haataja L, Barbetti F, Guo H, Larkin D, Arvan P. Impaired cleavage of preproinsulin signal peptide linked to autosomal-dominant diabetes. Diabetes. 2012;61:828–837. doi: 10.2337/db11-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci USA. 2008;105:12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B, Lilley BN, Ploegh HL. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J Cell Biol. 2006;175:261–270. doi: 10.1083/jcb.200605196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpal G, Schuiki I, Liu M, Volchuk A, Arvan P. Action of protein disulfide isomerase on proinsulin exit from endoplasmic reticulum of pancreatic beta-cells. J Biol Chem. 2012;287:43–47. doi: 10.1074/jbc.C111.279927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelhaas M, Malmstrom J, Pelkmans L, Haugstetter J, Ellgaard L, Grunewald K, Helenius A. Simian virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell. 2007;131:516–529. doi: 10.1016/j.cell.2007.09.038. [DOI] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley AM, Carvalho P, Rapoport T. Recognition of an ERAD-L substrate analyzed by site-specific in vivo photocrosslinking. FEBS Lett. 2011;585:1281–1286. doi: 10.1016/j.febslet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A, Ruggiano A, Carvalho P, Rapoport TA. Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell. 2014;158:1375–1388. doi: 10.1016/j.cell.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner DF, Cunningham D, Spigelman L, Aten B. Insulin biosynthesis—evidence for a precursor. Science. 1967;157:697–700. doi: 10.1126/science.157.3789.697. [DOI] [PubMed] [Google Scholar]
- Stoy J, Edghill EL, Flanagan SE, Ye HG, Paz VP, Pluzhnikov A, Below JE, Hayes MG, Cox NJ, Lipkind GM, et al. Insulin gene mutations as a cause of permanent neonatal diabetes. Proc Natl Acad Sci USA. 2007;104:15040–15044. doi: 10.1073/pnas.0707291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari A, Schuiki I, Zhang L, Allister EM, Wheeler MB, Volchuk A. SDF2L1 interacts with the ER-associated degradation machinery and retards the degradation of mutant proinsulin in pancreatic beta-cells. J Cell Sci. 2013;126:1962–1968. doi: 10.1242/jcs.117374. [DOI] [PubMed] [Google Scholar]
- Tsai B, Rodighiero C, Lencer WI, Rapoport TA. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 2001;104:937–948. doi: 10.1016/s0092-8674(01)00289-6. [DOI] [PubMed] [Google Scholar]
- Tsai B, Ye YH, Rapoport TA. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev Mol Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- Weiss MA. Diabetes mellitus due to the toxic misfolding of proinsulin variants. FEBS Lett. 2013;587:1942–1950. doi: 10.1016/j.febslet.2013.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JM, Inoue T, Banks L, Tsai B. The ERdj5-Sel1L complex facilitates cholera toxin retrotranslocation. Mol Biol Cell. 2013;24:785–795. doi: 10.1091/mbc.E12-07-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman PG. p97, a protein coping with multiple identities. J Cell Sci. 2003;116:4283–4290. doi: 10.1242/jcs.00817. [DOI] [PubMed] [Google Scholar]
- Ye YH, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Yoshinaga T, Nakatome K, Nozaki J, Naitoh M, Hoseki J, Kubota H, Nagata K, Koizumi A. Proinsulin lacking the A7-B7 disulfide bond, Ins2Akita, tends to aggregate due to the exposed hydrophobic surface. Biol Chem. 2005;386:1077–1085. doi: 10.1515/BC.2005.124. [DOI] [PubMed] [Google Scholar]
- Zhang L, Nosak C, Sollazzo P, Odisho T, Volchuk A. IRE1 inhibition perturbs the unfolded protein response in a pancreatic β-cell line expressing mutant proinsulin, but does not sensitize the cells to apoptosis. BMC Cell Biol. 2014;15 doi: 10.1186/1471-2121-15-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Ye YH. The final moments of misfolded proteins en route to the proteasome. DNA Cell Biol. 2014;33:477–483. doi: 10.1089/dna.2014.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.