Unable to complete S phase, a fission yeast MCM mutant evades the mitotic checkpoint, causing aneuploidy, chromosome fragments, and bridges. The formation of apparent yeast micronuclei that are membrane bound is shown in real time; they develop DNA damage signals and may rejoin the parent nucleus.
Abstract
DNA replication stress causes genome mutations, rearrangements, and chromosome missegregation, which are implicated in cancer. We analyze a fission yeast mutant that is unable to complete S phase due to a defective subunit of the MCM helicase. Despite underreplicated and damaged DNA, these cells evade the G2 damage checkpoint to form ultrafine bridges, fragmented centromeres, and uneven chromosome segregations that resembles micronuclei. These micronuclei retain DNA damage markers and frequently rejoin with the parent nucleus. Surviving cells show an increased rate of mutation and chromosome rearrangement. This first report of micronucleus-like segregation in a yeast replication mutant establishes underreplication as an important factor contributing to checkpoint escape, abnormal chromosome segregation, and chromosome instability.
INTRODUCTION
DNA replication stress is a well-known source of genome instability and results in increased mutations, chromosome rearrangements, and missegregation (reviewed in Naim and Rosselli, 2009; Crasta et al., 2012; Holland and Cleveland, 2012; Hatch et al., 2013). Tempering replication stress by adding extra nucleosides (Burrell et al., 2013) or inducing a checkpoint response (Casper et al., 2002) can stabilize slowly replicated regions and diminish the effect on chromosome missegregation. Of importance, genome instability is also correlated with carcinogenesis (e.g., Bagley et al., 2012; Burrell et al., 2013; Hirsch et al., 2013), particularly within fragile regions of the genome that are unable to replicate efficiently (e.g., Chan et al., 2009; Lukas et al., 2011; Naim et al., 2013). Thus cellular ability to appropriately manage replication stress prevents malignant transformation (Bartkova et al., 2005; Gorgoulis et al., 2005; Halazonetis et al., 2008).
MCM4 is an essential subunit of the minichromosome maintenance (MCM) helicase that is required for DNA replication (reviewed in Forsburg, 2004; Bochman et al., 2008). Mice with minor mcm4 mutations show evidence of replication stress, including double-strand breaks, micronuclei, and increased formation of mammary tumors (Shima et al., 2007a) or leukemia (Bagley et al., 2012). Disruptions in replication correlate with chromosome fragile sites (reviewed in Debatisse et al., 2012), and the murine mcm4 phenotype is consistent with a failure to license dormant replication origins (reviewed in Kawabata et al., 2011; McIntosh and Blow, 2012). N-terminal truncation of Mcm4 is associated with chromosome breaks and DNA repair defects in an inbred human population (Casey et al., 2012; Gineau et al., 2012; Hughes et al., 2012). MCM overexpression has been correlated with hyperproliferation and carcinogenesis in tumors (Ishimi et al., 2003b; Guida et al., 2005; Lau et al., 2010; Majid et al., 2010; Suzuki et al., 2012). Thus changes in this single MCM4 subunit have profound consequences for genome stability.
We report a novel genome-instability phenotype in a specific allele of mcm4. In fission yeast cells, most conditional mcm mutations at the restrictive temperature show significant DNA accumulation, accompanied by activation of the DNA damage checkpoint and robust cell cycle arrest (e.g., Nasmyth and Nurse, 1981; Coxon et al., 1992; Liang and Forsburg, 2001) consistent with replication fork collapse generating DNA double-strand breaks. Cells with the mcm4-degron allele show DNA underreplication with accumulation of DNA damage markers at the restrictive temperature but are unable to maintain a checkpoint arrest. After release from replication stress, these damaged cells continue to divide. The divisions are abnormal, producing ultrafine DNA bridges, multipolar spindles, and uneven chromosome segregation accompanied by the formation of small, satellite nuclei. These apparent micronuclei retain DNA damage markers and frequently rejoin with the parent nucleus. Significantly, the cells that survive this stress show substantially increased rates of mutation and chromosome rearrangement. This phenotype is distinct from mutants that block replication initiation, which fail to undergo chromosome segregation. Our data suggest that underreplication is a critical factor associated with genomic instability and establish a genetic model to investigate the links between replication stress, disruptions in chromosome segregation, and genome rearrangements.
RESULTS
Most temperature-sensitive MCM helicase mutants duplicate the majority of their genome at 36°C (Figure 1A). However, these mcm-ts mutants accumulate DNA damage and trigger cell division cycle arrest (e.g., Nasmyth and Nurse, 1981; Coxon et al., 1992; Liang and Forsburg, 2001). On the other hand, temperature-sensitive mutants in other genes that bypass replication initiation do not replicate DNA but enter mitosis. This causes a cell untimely torn (cut) phenotype in which the unreplicated DNA is cleaved by the septum (e.g., cdc18∆, rad4ts, or orp1-ts; Kelly et al., 1993b; Saka and Yanagida, 1993; Grallert and Nurse, 1996). Presumably, initiation mutants never begin DNA replication and do not generate signals to trigger the checkpoint (Kelly et al., 1993a).
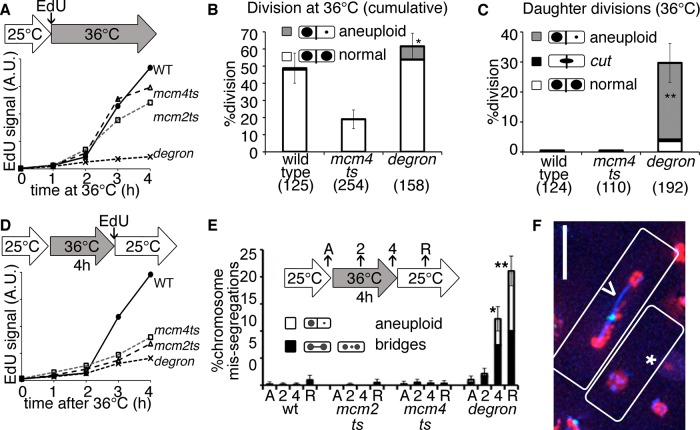
FIGURE 1:
Underreplicated mcm4-degron mutants divide during and after replication stress. (A) EdU incorporation is lowest in mcm4-degron cells during incubation at 36°C. Asynchronous cultures were shifted to 36°C during EdU exposure, and synthesis was measured by EdU-FACS fluorescence in arbitrary units (A.U.). (B) Wild-type (wt) and mcm4-degron cells divide at least once at 36°C (6-h total videomicroscopy of asynchronous cultures at 36°C). Significantly more mcm4-degron cells undergo reductional anaphase (gray) in this first division at 36°C. Proportions in B, C, and E are shown with 95% CI, Z test of significance (*p < 0.01, **p << 0.001). (C) mcm4-degron daughter cells divide their DNA unevenly at 36°C, whereas wild-type daughters delay division at 36°C. Samples as in B, daughter cell divisions only. (D) Mutants incorporate less EdU after release at 25°C, as measured by EdU FACS. The initial asynchronous population was treated for 4 h at 36°C before release at 25°C in the presence of EdU. (E) Proportion of abnormal nuclear divisions in single-time-point images acquired before heat (asynchronous [A]), at 36°C (2, 4 h), and after 2 h of recovery at 25°C (R). (F) mcm4-degron cells during 25°C release with RPA-CFP (blue), Rad52-YFP (green), and histone-RFP (red). Unequal histone division in one cell (*) and a bridge (>) with a lagging chromosome and repair focus are common. Scale bar, 10 μm.
We observe a novel phenotype in the temperature-sensitive mcm4-degron allele. This mutant has a degron cassette fused to the mcm4 temperature-sensitive (ts) allele to enhance protein turnover (Lindner et al., 2002). Unlike the well-characterized mcm4-ts allele (cdc21-M68), Mcm4degron protein is <10% of the original level during incubation at 36°C (Supplemental Figure S1A). We monitored DNA synthesis by nucleoside analogue incorporation (5-ethynyl-2′-deoxyuridine [EdU]) and detected very little accumulation in mcm4-degron at the restrictive temperature (Figure 1A and Supplemental Figure S1B). In contrast, mcm4-ts at 36°C incorporates nearly wild type EdU amounts. Only early replication origins fire in mcm4-degron, whereas mcm4-ts cells activate a combination of early and late origins more efficiently (Supplemental Figure S1C). Surprisingly, despite these global replication defects, mcm4-degron cells divide multiple times at 36°C (Figure 1, B and C, and Supplemental Video S1), causing uneven DNA segregation in daughters and granddaughters.
When cells are returned to 25°C, wild-type cells robustly continue DNA synthesis and proliferation (Figure 1D and Supplemental Figure S1D). In contrast, neither mcm4-ts nor mcm4-degron cells incorporate much EdU after release to 25°C, a period that we call “recovery.” This low incorporation suggests that either there is limited, residual synthesis across the genome or just a few cells returned to the cell cycle. The mcm4-ts cells remain cell cycle arrested after release, consistent with persistent DNA damage (Bailis et al., 2008). Surprisingly, mcm4-degron cells continue to divide (Supplemental Figure S1E and Supplemental Video S2). Spindle pole body (SPB) duplication and separation occurs with timing similar to that for wild type (Supplemental Figure S1F). However, the segregation of the DNA in mcm4-degron is highly abnormal (Figure 1E), forming lagging chromosomes, replication protein A (RPA)–labeled ultrafine bridges, and unequal DNA segregation into aneuploid and anucleate cells (Figure 1F and Supplemental Figure S1G).
Apparent micronuclei form in underreplicated mcm4-degron cells
We examined abnormal segregations in mcm4-degron more closely using live-cell video microscopy. The nuclear histone signal shows uneven segregation and fragmentation in >50% of mcm4-degron cells (Figure 2, A and B). These nuclear fragments form during mitosis and are enclosed in separate nuclear membranes, which is the definition of a micronucleus (Hatch et al., 2013; Figure 2, A–C, Supplemental Figure S2, A and B, and Supplemental Video S3). The proportion of fragmented histone masses was similar to the number of membrane-bound micronuclei, indicating that a membrane initially surrounds most wandering DNA fragments.
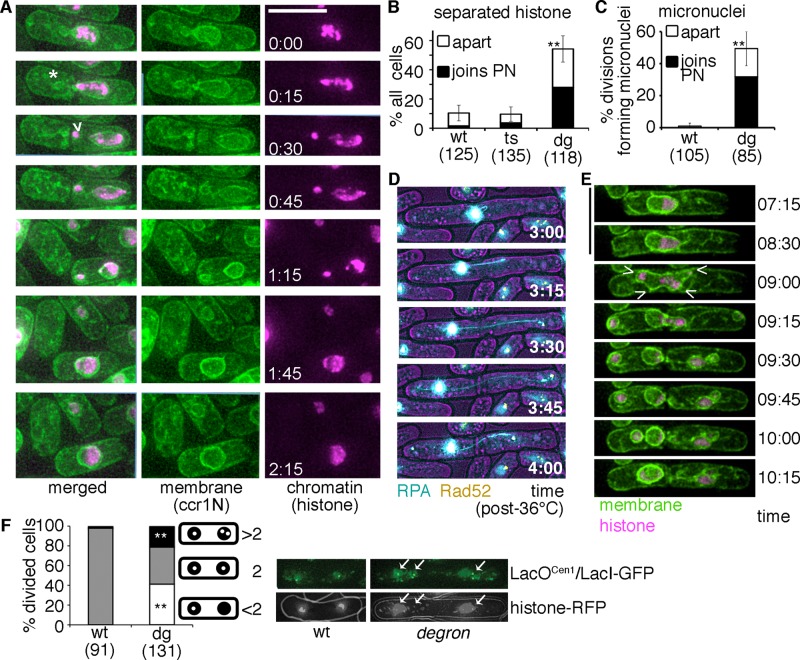
FIGURE 2:
Underreplication followed by division promotes micronuclei and genomic rearrangement in fission yeast. (A) mcm4-degron cells released to 25°C (after 4 h, 36°C) form anucleate cells (*) and apparent micronuclei (>). Histone-RFP and membrane (ccr1N-GFP) are shown; scale, 10 μm. Micronuclei are often resorbed back into the main nucleus (time 1:15–1:45 h:min) after release to 25°C. (B) Chromatin (histone-RFP) frequently separates into discrete, condensed fragments that are separate from the main nuclear mass in mcm4-degron cells. More than 50% of these fragments rejoin the parent nucleus (PN). Asynchronous cultures were treated (4 h, 36°C) and then shifted to 25°C for microscopy during recovery (12-h cumulative data, two or three biological replicates per strain). The proportion of cells that form separate histone bodies is shown relative to total number of cells monitored. (C) Membrane-enclosed chromatin masses frequently separate from the main nucleus in mcm4-degron cells but are rarely detected in wild type (wt). More than 60% of these micronuclei fuse and rejoin the parent nucleus (PN) during recovery at 25°C (12-h cumulative data, two to four biological replicates per strain). (D) mcm4-degron cells develop dynamic ssDNA (RPA-CFP, blue) bridges dotted with Rad52-YFP (yellow) during release at 25°C (time in hours:minutes). (E) Evidence for two spindles resulting in apparent micronuclei in some mcm4-degron cells during recovery (after 4 h, 36°C). Conditions as in A; scale, 10 μm. (F) A LacO array near the centromere 1 (lys1+-lacOCen1) unevenly separates in mcm4-degron divisions after 4 h at 36°C. More than two dots are frequently observed in mcm4-degron, which suggests that the array is rearranged or fragmented. LacI-GFP (green) bound to lacOCen1 is shown below relative to DNA signal (histone-RFP). Stacked histogram for pooled data from three biological replicates, with chi-squared test of significance for proportion of single dots (gray) or more than three dots (black) segregating (**p << 0.001).
There are no obvious connections between the micronucleus and the parent nucleus. Some membrane stalks remain independently attached to the septum (Supplemental Figure S2B). When these membrane-enclosed fragments remain in the same cell, they frequently rejoin the mother nucleus (∼60% of the time). Others segregate into a daughter cell during division, forming aneuploid cells. Subsequent divisions often show repeated segregation/fusion cycles (Supplemental Videos S4 and S5). Supplemental Video S4 shows delayed and failed mitosis followed by a later division, suggesting a dual spindle (e.g., Figure 2E and Supplemental Video S4). Thus these missegregations may also be linked to mitotic defects such as multipolar spindle formations.
Another mitotic abnormality observed in mammalian cells after replication stress is ultrafine DNA bridges (UFBs) between fragile DNA regions (reviewed in Chan and Hickson, 2009). UFBs are not detected using DNA stains (e.g., 4′,6-diamidino-2-phenylindole [DAPI], histone; Chan et al., 2009) but can be visualized with RPA (Chan and Hickson, 2009). We observed twisting threads of RPA spanning unequal DNA masses in 20% of mcm4-degron divisions (Figure 2D, Supplemental Figure S2, C and D, and Supplemental Video S6). The RPA signal was often separate from the histone signal, suggesting that single-strand DNA (ssDNA) has pulled apart from the bulk chromatin.
These division anomalies resemble mitosis with unreplicated genomes (MUGs), which happens in replication-arrested human cells that bypass the G2 damage checkpoint (Wise and Brinkley, 1997). One MUG characteristic is centromere fragmentation (Beeharry et al., 2013), which we detected in strains expressing a tagged centromere-associated histone Cnp1–red fluorescent protein (RFP; CENPA homologue; Supplemental Figure S2E). Fission yeast centromeres replicate early (Zhu et al., 1992) and then cluster with the SPB, except briefly during metaphase-to-anaphase transition. We observed early centromere replication in mcm4-degron (Supplemental Figure S1C) as Cnp1 foci scatter, indicating that centromeres replicate, separate, and possibly fragment. These multiple mitotic abnormalities promote DNA missegregation after replication stress in mcm4-degron.
We examined evidence for chromosome rearrangement using a lacO array near centromere I (Figure 2F). Many mcm4-degron cells failed to separate lacOCen1 foci to both daughters, causing >2 green fluorescent protein (GFP) foci/nucleus or none at all. Because lacO arrays are potential fragile sites in Schizosaccharomyces pombe (Sofueva et al., 2011), a lacOCen1 rearrangement or duplication may occur after mcm4-degron replication stress, causing cells with greater than two separating foci. This is also consistent with evidence for centromere fragmentation and rearrangement.
Increased mutations and rearrangements in surviving mcm4-degron cells
We next asked whether the 10% of surviving mcm4-degron cells show lasting signs of genome instability after transient replication stress (Figure 3A). We tested surviving cells for forward mutations that cause canavanine resistance (can1+(S) to can1−(R); Figure 3B). The baseline mutation frequency in mcm4-degron cells is higher than for wild type and mcm4-ts and significantly increases after incubation at 36°C. We also saw high rates of marker loss at other loci, including loss of an integrated marker at his7 (Supplemental Figure S3).
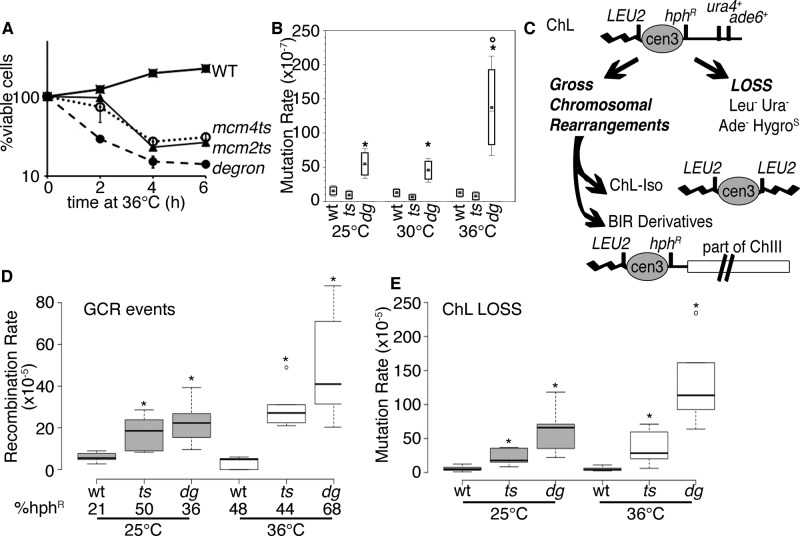
FIGURE 3:
Transient replication instability causes mutation in surviving mcm4-degron cells. (A) Relative viability of cultures at 36°C (strains FY4743, FY4857, FY5279). Cells were shifted to 36°C and plated at time points to determine viability relative to the starting culture. (B) Mutation rate (can1+) increases in mcm4-degron after 4 h at 36°C (n = 7). *p < 0.001 comparing wt or mcm4-ts with mcm4-degron; °p << 0.001 change from 25 to 36°C in mcm4-degron. Plots in B, D, and E show a center median line bounded by 25th and 75th percentiles. (C) Schematic for the minichromosome (ChL) assay, followed by markers (also see Nakamura et al., 2008; Li et al., 2013). Cells may lose ChL or undergo gross chromosomal rearrangements (GCRs). An isochromosome (ChL-iso) is formed by duplication of the left arm with the LEU2+ marker producing a smaller chromosome. Break-induced replication (BIR) products may occur between ChL and chromosome III, producing a longer product that is frequently hygromycin resistant. (D) GCR events are highest in mcm4-degron after 4 h at 36°C. Significant median differences from wild type at 25 or 36°C are reported as p < 0.001 (*) with outliers (o). (E) ChL loss is highest in mcm4-degron after 4 h at 36°C and even before replication stress at 25°C. Loss is also higher in mcm4-degron compared with mcm4-ts (p < 0.02. all conditions). Conditions and analysis as in D.
To assess potentially catastrophic chromosome rearrangements, we introduced a nonessential minichromosome into the mcm4-ts and mcm4-degron strains. This minichromosome carries multiple genetic markers to maintain its overall stability or monitor its structural integrity (Figure 3C). A low level of chromosome rearrangement is observed in wild-type cells, including break-induced replication and isochromosome formation (Figure 3, D and E; Nakamura et al., 2008). Increased rearrangements are also observed in replication fork protection complex mutants (Li et al., 2013).
Consistent with the minichromosome maintenance (mcm) phenotype, we observed an increased rate of minichromosome loss in both mcm4ts and mcm4-degron relative to wild type at 25°C (Figure 3E). Chromosome loss is modestly increased after incubation at 36°C in the mcm4-degron strain but shows no significant change in the mcm4-ts background.
Both mcm4-degron and mcm4-ts strains show an increased rate of rearrangement relative to wild type at 25°C (Figure 3D). However, after a 4-h pulse at 36°C, the mcm4-degron mutant shows a dramatic increase in chromosome rearrangements that is not observed in mcm4-ts. Thus the division abnormalities observed in mcm4-degron are accompanied by increased mutations, chromosome rearrangements, and chromosome loss.
Damage persists in mcm4-degron mutants
The ability of underreplicated mcm4-degron cells to divide repeatedly suggests either that there is little DNA damage or that the damage checkpoint is not activated. To address the first point, we examined DNA repair proteins during arrest and release by visualizing fluorescently tagged versions of the ssDNA-binding protein RPA, an early damage marker, and the Rad52 recombination protein. The mcm4-ts mutant forms many discrete RPA and Rad52 foci during arrest at 36°C, which coalesce into a bright, pannuclear signal upon release (Figure 4A and Supplemental Figure S4, A–C). This is consistent with earlier observations (Bailis et al., 2008) suggesting widespread late replication fork collapse at multiple sites, similar to the checkpoint mutant cds1∆ in HU (Sabatinos et al., 2012). In contrast, mcm4-degron mutants form one or two large, distinctive RPA “megafoci.” RPA and Rad52 colocalization in both mcm4 mutants (Supplemental Figure S4D), coupled with their low viabilities, suggests that these are dominated by stalled or damaged replication forks (e.g., Lambert et al., 2010) and not stably stalled replication forks (Irmisch et al., 2009).
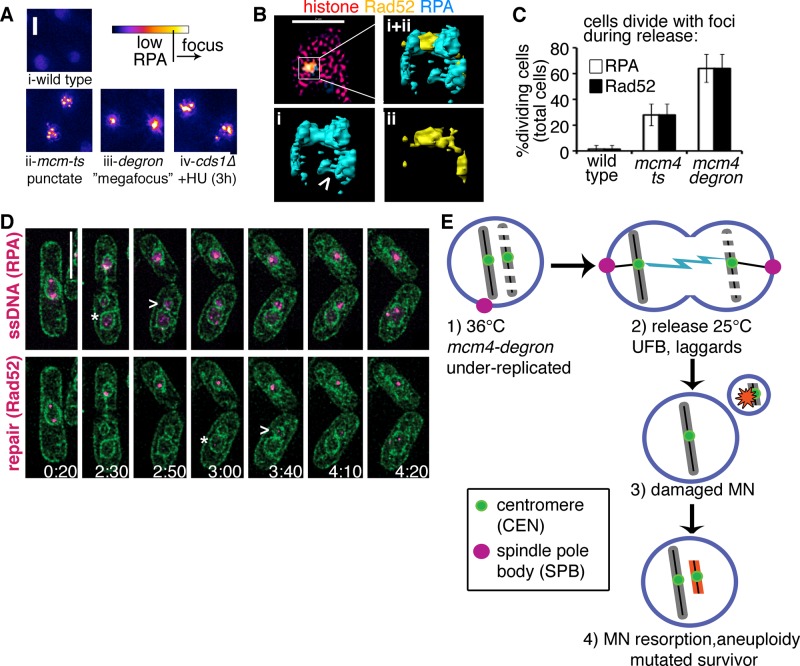
FIGURE 4:
Divisions occur in the presence of DNA damage and repair signals. (A) RPA focus patterns during replication collapse are different in each mutant but rarely develop in wild type at 36°C (i). Multiple (more than three) punctate RPA foci form in mcm-ts nuclei after 4h 36°C (ii) and later become a pannuclear RPA signal like that observed in cds1∆+HU (iv). A unique “megafocus” of bright, compact RPA forms in mcm4-degron (iii). Heat map scale (top) and 2- μm scale. (B) 3D-SIM images of mcm4-degron nucleus after 4 h 36°C (top left) in one midfocal z-section (xy); scale bar, 2 μm. (i, ii) Enlarged yz-perspectives of surface-rendered megafocus. Also see Supplemental Video S5. (C) RPA and Rad52 foci are present at division in mcm4-degron (also see Supplemental Videos S2 and S6). (D) DNA damage (RPA-CFP, top magenta) and DNA repair foci (Rad52-YFP, bottom magenta) develop in newly formed micronuclei (MN; assessed with membrane marker ccr1N-GFP; green). Cells were incubated at 36°C, 4 h before videomicroscopy during release at 25°C for 6 h. A time scale is indicated on the bottom right corner of each panel (hours:minutes). The MN form damage and repair signals after they are first detected in the parent nucleus (*, parent; >, MN), before rejoining. Scale bar, 5 μm. (E) A proposed model for transient replication-stress inducing mutations in surviving cells after mcm4-degron inactivation, shown at the level of the nucleus (nuclear membrane in blue). Cells treated 4 h at 36°C are underreplicated (step 1) but divide, causing UFBs and fragmented DNA during mitosis (step 2). Fragments are membrane-bound MN that develop DNA damage (step 3). Resorption of MN back into the parent nucleus promotes further genome instability during 25°C recovery, leading to the development of a mutated surviving population (step 4).
We used superresolution microscopy to examine the megafocus substructure. Three-dimensional (3D) structured illumination microscopy (SIM) images show that the megafocus is an RPA complex, with filaments that extend from the center forming cups and voids that contain Rad52 (Figure 4B). At this higher resolution, the RPA/Rad52 foci are not simple dots but instead highly structured patterns within a 0.2-μm diameter. The 3D-SIM images show that Rad52 and RPA fit together end to end and that RPA tendrils loop out into surrounding histone regions (Supplemental Figure S4E). Further, we observe that the megafocus occurs in histone-deficient nuclear regions (Supplemental Video S7).
The presence of Rad52 repair foci in ∼15% of untreated mcm4-degron cells suggests that the cells suffer damage even under permissive conditions. Pulsed-field gels show that untreated mcm4-degron chromosomes migrate poorly and generate a low–molecular weight smear indicating DNA breaks (Supplemental Figure S4F). This genome instability may be due to Mcm4degron protein instability compared with wild-type Mcm4 before temperature shift (Supplemental Figure S1A). These observations are consistent with our previous observation that reduced MCM levels cause genome instability before replication is noticeably affected (Liang et al., 1999).
Surprisingly, RPA and Rad52 foci persist in dividing mcm4-degron cells (Figure 4C and Supplemental Videos S2 and S8). We also see RPA and Rad52 foci in the apparent micronuclei (Figure 4D); these may be markers of ongoing DNA synthesis, stalled forks, or DNA damage. Consistently, we find that these signals appear later in the putative micronuclei than in the primary nucleus (Figure 4D, arrowhead vs. asterisk in the primary nucleus) and can be reincorporated into the parent nucleus (Figure 4E).
The phenotypes we observe with mcm4-degron are different from those seen in other replication initiation mutants. Orp1ORC1 marks replication origins (orp1-4; Grallert and Nurse, 1996), and Rad4TopBP1 is essential for replication initiation and also activation of the DNA damage checkpoint (rad4-116; Saka and Yanagida, 1993). These mutants enter a lethal mitosis with unreplicated DNA that is cleaved by the cell septum (cut). Both orp1ts and rad4ts formed some RPA and Rad52 foci, but the quantity and patterns were different from those for mcm4-degron (Supplemental Figure S5, A–C). The rad4ts mutants are much shorter at division, typical of cut mutants, with a sub-1C DNA content and increased cell death (Supplemental Figure S5, D and E). Fewer chromosome missegregations occur in either orp1ts or rad4ts than in mcm4-degron, particularly during recovery at 25°C (Supplemental Figure S5F). Thus mcm4-degron defines a new class of early replication mutant.
mcm4-degron transiently activates the damage checkpoint and then escapes
RPA contributes directly to fork stability and damage checkpoint activation (Zou et al., 2003; Toledo et al., 2013). The checkpoint kinase, Chk1, is phosphorylated in asynchronous mcm4-degron cells. Chk1 activation, detected by a band shift Western blot (Figure 5B), is required for both checkpoint initiation and maintenance (Latif et al., 2004). In mcm4-degron, activated Chk1 is present in asynchronous cells but decreases at 36°C during replication stress (Figure 5B) even as RPA and Rad52 foci form (Figure 4C). After release to 25°C, Chk1 is moderately phosphorylated in mcm4-degron, and cells continue to divide. In contrast, Chk1 is inactive in asynchronous mcm4-ts cells but becomes highly phosphorylated at 36°C and during release at 25°C. Consistent with these data, the inhibitory Cdc2 phosphorylation on threonine 15 that prevents mitosis (O’Connell et al., 1997) is reduced in mcm4-degron (Figure 5C). This checkpoint activation explains the robust cell cycle arrest of the mcm4-ts compared with mcm4-degron. Moreover, under these conditions, we Crb253BP1 levels drop sharply in mcm4-degron at 36°C, suggesting that the checkpoint signal is interrupted upstream of Chk1 (Figure 5B).
FIGURE 5:
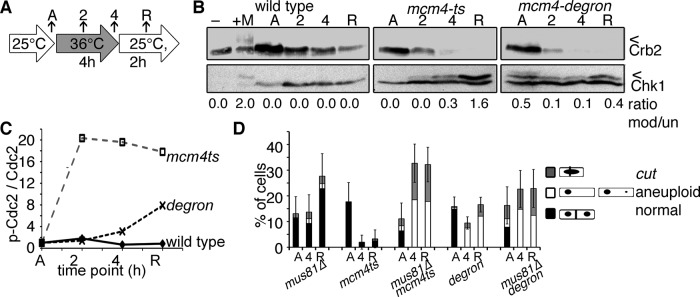
Underreplication promotes micronuclei, DNA damage, and aneuploidy in fission yeast. (A) Experimental scheme. Asynchronous cells were shifted to 36°C for 4 h total and then released to 25°C for 2 h. (B) The DNA damage checkpoint becomes activated by Chk1-HA phosphorylation (*) in methyl methanesulfonate (MMS)-treated wild-type cells (+M) and in mcm4-ts. Chk1-HA is moderately phosphorylated in asynchronous mcm4-degron and never attains activated levels of mcm4-ts, as assessed by the ratio of modified (top) to unmodified (bottom) Chk1. The 53BP1 homologue Crb2 is phosphorylated in response to MMS treatment and stable in wild type but is rapidly lost in mcm4-degron at 36°C. Arrowheads (<) indicate modified forms of proteins, and the bar (–) indicates a non–HA-tagged control lysate. (C) Cdc2 is not phosphorylated in mcm4-degron at 36°C and only minimally during recovery (25°C). In contrast, high-level, sustained Cdc2 phosphorylation occurs in mcm4-ts. Cdc2 modified and unmodified protein levels were detected on Western blots and quantified to plot the ratio at each time point. (D) Loss of Mus81 endonuclease (mus81∆) increases divisions in mcm4-ts mus81∆, forming aneuploid and cut cells.
Without Chk1, the mcm-ts chk1∆ double mutants enter premature mitosis and cut at 36°C (Supplemental Figure S6, A and B; Liang et al., 1999). In mcm4-degron chk1∆ double mutants, the fraction of cut cells is only slightly higher than in mcm4-degron alone. This suggests that the Chk1 checkpoint transiently restrains division in mcm4-degron at 36°C. Once returned to 25°C, there is no difference between division numbers and morphology in mcm4-degron and mcm4-degron chk1∆ double mutants.
Mus81 promotes checkpoint arrest during late-replication failure
What is different about the late replication fork failure in mcm4-ts and the early collapse in mcm4-degron? Whereas mcm4-ts accumulates DNA breaks and robustly activates the damage checkpoint, mcm4-degron does not. The contrast in their RPA patterns and timing suggests that the two mutants generate different replication arrest structures. Because Mus81 endonuclease reportedly cleaves stalled replication forks in late S phase to promote fork restart (Froget et al., 2008; Saugar et al., 2013), we reasoned that Mus81 might cleave mcm4-ts arrested forks to form DNA breaks and generate a robust damage signal
Consistent with this model, we found that a mcm4-ts mus81∆ double mutant showed a dramatic increase in dividing cells at 36°C compared with mcm4-ts alone (Figure 5D) and thus resembles the mcm4-degron. In contrast, mus81∆ did not change the proportion of mcm4-degron cells forming micronuclei or undergoing asymmetric divisions during release (Supplemental Figure S6, C–E). We infer that Mus81-dependent damage formed in mcm4-ts generates a signal for robust G2 checkpoint activation and cell cycle arrest. In contrast, the early-failing replication forks in mcm4-degron fail to activate fully or maintain the G2 checkpoint.
DISCUSSION
Mcm4 is an essential subunit of the MCM helicase, the primary replicative helicase of eukaryotic cells (e.g., Maiorano et al., 2000; Labib et al., 2001; Ishimi et al., 2003a; Yabuta et al., 2003). Disrupting Mcm4 function drives genome instability in many models. Mouse mcm4 mutations are associated with chromosome breaks, genome rearrangements, micronucleus formation, and breast or blood cancers (Shima et al., 2007a, b; Bagley et al., 2012). It has been proposed that this reflects a failure to license additional replication origins that allow rescue of a failed replication fork (Kawabata et al., 2011; McIntosh and Blow, 2012). In humans, MCM4 truncation mutations are associated with chromosome instability and DNA repair defects (Casey et al., 2012; Gineau et al., 2012; Hughes et al., 2012). MCM overexpression is correlated with hyperproliferation and carcinogenesis (e.g., Ishimi et al., 2003b; Guida et al., 2005). Therefore Mcm4 plays a fundamental role in maintaining genome stability.
We characterized a novel temperature-sensitive allele of mcm4 in fission yeast (mcm4-degron) that generates a distinct form of early replication stress in which early replication forks fire but undergo little DNA synthesis. This is accompanied by transient DNA damage checkpoint activation and then escape, suggesting that cells are unable to initiate or maintain a checkpoint response (Latif et al., 2004). Because there are low levels of checkpoint mediator protein Crb2 at 36°C, the checkpoint activation step in mcm4-degron may not be amplified (Lin et al., 2012); this agrees with recent work proposing that Chk1 activation is linked to the MCM complex (Han et al., 2014). Alternatively, mcm4-degron checkpoint maintenance might fail, allowing escape, as is the case at telomeres where Crb2 is absent (Carneiro et al., 2010).
This contrasts with mcm4-ts mutants, which synthesize almost a wild-type amount of DNA before undergoing robust checkpoint-dependent arrest. We observe that Mus81 endonuclease is required to maintain activation of Chk1 in mcm4-ts cells. This suggests that Mus81 recognizes and acts upon a specific structure formed during late fork collapse in mcm4-ts, and this generates the robust checkpoint signal that maintains cell cycle arrest. This may be due to inactivation of Mus81 during early S phase, as is observed in Saccharomyces cerevisiae (Saugar et al., 2013). Alternatively, there may be no Mus81-susceptible substrates formed in mcm4-degron early replication arrest, preventing a strong G2 checkpoint activation.
The mcm4-degron cells show replication stress even without a temperature shift, as indicated by their constitutive DNA repair foci (Rad52), smeared chromosomes by pulsed-field gel electrophoresis (PFGE) analysis, higher mutation rates, and constitutively activated Chk1. This is consistent with other work showing that reduced MCM protein levels contribute to genome instability (Liang et al., 1999; Gineau et al., 2012). The mcm4-degron cells also acquire a novel RPA/Rad52 structure that is not seen in other replication-initiation mutants. We propose that this “megafocus” represents early-firing replication origins that are clustered during initiation (Knott et al., 2012) and then collapse as Mcm4degron protein is lost. Our superresolution analysis of the mcm4-degron megafocus shows that the structures of ssDNA and Rad52-bound DNA are intertwined. These megafoci of colocalized RPA and Rad52 do not stably activate the DNA damage checkpoint, similar to replication stress–induced foci of brc1∆ mutants (Bass et al., 2012). In our model, Mcm4degron preassembled at early origins is protected from immediate inactivation at 36°C, and the protein becomes vulnerable during the transition to replication elongation, causing replication failure and ssDNA accumulation at an early stage. In contrast, the mcm4-ts mutants arrest with numerous dispersed RPA foci, consistent with late fork collapse detected by phosphorylated H2A(x) (Bailis et al., 2008). This may occur stochastically or at specific fragile sites.
Unexpectedly, we observe that despite underreplication, most mcm4-degron cells divide during both replication stress at 36°C and again after release (Figures 2 and 4). These abnormal mitoses produce UFBs marked with RPA and apparent centromere fragmentation. Cells undergo continued, abnormal divisions that generate small, membrane-bound bodies that contain a subset of the genome. These may segregate into separate daughter cells, generating aneuploidy, or remain in the mother cell, where they may rejoin the parent nucleus. These structures are intriguingly suggestive of micronuclei.
In mammalian cells, micronuclei form when a subset of the genome is separated into distinct membrane-bound bodies. These may form after irradiation (e.g., Kato and Sandberg, 1968) or when cells with replication defects enter mitosis (Kato and Sandberg, 1968; Shima et al., 2007a; Chan et al., 2009; Utani et al., 2010; Bagley et al., 2012). Micronuclei may contain acentric genome fragments or whole chromosomes and may be associated with dicentrics and chromosome bridges. These data indicate that they may result from different forms of genetic stress or mitotic failure (e.g., Fenech et al., 2011).
Although micronuclei are common markers in cancer cells (e.g., Crasta et al., 2012; Hatch et al., 2013), the relationship between their formation, stability, and overall genome instability is not understood. For example, micronuclei clearly form in response to whole-genome damage and replication stress, as seen in mouse Mcm4 mutants (Shima et al., 2007a; Gineau et al., 2012), and yet spindle poisons that perturb mitosis also cause whole-chromosome missegregation and micronuclei (Crasta et al., 2012; Hatch et al., 2013; Zhang et al., 2015). In the latter studies, DNA replication is delayed in micronuclei compared with the parent nucleus (Crasta et al., 2012), leading to DNA damage. Indeed, there is long-standing evidence that chromosomes within micronuclei are severely damaged or pulverized (Kato and Sandberg, 1968; Crasta et al., 2012; Zhang et al., 2015). The resulting chromosome rearrangements may be incorporated into the genome if the micronuclear DNA merges with the parent nucleus during mitosis (reviewed in Forment et al., 2012; Holland and Cleveland, 2012; Zhang et al., 2013). These observations have led to the suggestion that aberrant micronucleus segregations are associated with the catastrophic chromosome rearrangements termed chromothripsis (Crasta et al., 2012; Holland and Cleveland, 2012; Zhang et al., 2015).
The events that generate micronuclei are likely linked to other cytogenetic abnormalities, including chromosome bridging, breakage-fusion-bridge cycles, and centromere fission (e.g., Fenech et al., 2011; Martinez and van Wely, 2011; Sorzano et al., 2013). In mammalian cells, caffeine-induced checkpoint bypass produces evidence of centromere fragmentation in underreplicated cells (Burrell et al., 2013). Centromere breaks and fission have been associated with micronucleus formation and chromosome rearrangements (Guerrero et al., 2010; Martinez and van Wely, 2011). Consistent with this, we previously described the fission yeast pericentromere repeats as vulnerable to rearrangement during replication stress (Li et al., 2013). The unusual mcm4-degron mutant phenotype establishes a yeast model to examine missegregation events in which we observe evidence for centromere fission, UFBs, and abnormal/aneuploid segregation.
We predicted that these abnormal segregations and apparent micronuclei should be associated with increased evidence of genome instability, and genetic studies showed this to be the case. The mcm4-degron strain is a mutator, with increased accumulation of forward mutations after incubation at 36°C. Using a nonessential minichromosome (e.g., Nakamura et al., 2008; Li et al., 2013), we observed a striking increase in chromosome rearrangements in the mcm4-degron cells that survive replication stress compared with wild-type or mcm4-ts cells, which maintain checkpoint arrest.
We infer that the abnormal divisions of mcm4-degron establish a source of continuing genome instability (model in Figure 4D). A fraction of the underreplicated genome is separated during mitosis and shows accumulation of RPA and Rad52 foci later than in the parent nucleus. This could reflect DNA damage or asynchronous DNA replication. Apparent nuclear fusion or rejoining between the separated body and the parent nucleus reincorporates the damaged DNA into the parent nucleus after mitosis. We hypothesize that this is one cause of enhanced mutation rate after transient mcm4-degron inactivation. Intriguingly, data in mammalian systems suggest that DNA damage that occurs during mitosis can be masked until the next cell cycle (e.g., Lukas et al., 2011). Of importance, we show that transient replication instability has long-reaching effects and that genome instability (persistent RPA/Rad52 foci, bridges, and apparent yeast micronuclei) is established and transmitted over multiple divisions during growth reestablishment (Supplemental Videos S4 and S5).
Of course, nuclear membrane dynamics differs in yeast and mammals. We observe that ∼70% of fission yeast micronuclei fuse with the parent nucleus. This is similar to the frequency observed for micronuclear DNA rejoining the parent DNA during mitosis in microtubule-destabilized mammalian cells (Hatch et al., 2013). However, micronuclear membrane fusion is not reported in mammalian cells (Crasta et al., 2012). The open mammalian mitosis, with nuclear envelope breakdown, allows micronuclear DNA to rejoin the parent nucleus when the nuclear membrane is degraded during mitosis. In contrast, the fission yeast mitosis is closed, and the nuclear envelope does not degrade. Therefore the mechanism of yeast micronuclear DNA rejoining may be different and appears to be postmitotic. Of importance, the abnormal segregation we observe is clearly mitotic in origin. It is led by centromeres and spindle pole bodies (Figure 2E and Supplemental Figure S2E) and is distinct from abnormal nuclear budding, such as that observed in fission yeast mutants with disrupted nuclear membrane dynamics (Sazer et al., 2014).
This is the first report of micronucleus-like divisions in fission yeast, and it is not observed in the other early replication mutants tested (orc1ts, rad4ts). Thus these divisions are a feature of a very specific early replication defect that allows some fraction of the genome to undergo segregation, evading the checkpoint by circumventing DNA breakage through Mus81.
Micronuclei induced by a yeast mcm4 mutation are particularly intriguing, given the association of micronuclei, chromosome breaks, and cancers in mouse Mcm4 mutants (Shima et al., 2007a; Bagley et al., 2012). It is possible that disruptions in the MCM4 subunit are particularly linked to damage that evades the checkpoint and promotes abnormal mitosis. Significantly, yeast genetic tools now allow a detailed investigation of contributing factors and description of outcomes. This provides a powerful genetic model to investigate the mechanisms of aberrant segregation and micronucleus formation caused by replication instability and the potential of large-scale genetic damage.
MATERIALS AND METHODS
Cell growth and physiology
Fission yeast strains are described in Supplemental Table S1 and were grown as in Sabatinos et al. (2012). Physiology experiments for viability, DNA synthesis, Chk1 protein, PFGE, and flow cytometry were performed in supplemented Edinburgh minimal medium (EMM). Live-cell imaging cultures were grown in fully supplemented EMM with 5 μM thiamine and photographed in the same medium. Septation and nuclear counts were performed on fixed samples. Briefly, cells were fixed in 70% ethanol, rehydrated, and then stained in 1 mg/ml aniline blue (M6900; Sigma-Aldrich) for 15 min. Stained cells were mounted on glass slides with SlowFade Gold antifade mount with DAPI (S36938; Invitrogen, ThermoFisher Scientific) and photographed. More than 200 cells were counted from two biological replicates and pooled, and then proportions and 95% confidence intervals (CIs) were calculated. Differences in proportions were assessed with a two-tailed Z test.
Micronucleus measurement
An initial assessment of the micronucleus-forming potential in cultures was made by incubating cultures at 36°C for 4 h and then imaging over 12 h at 25°C. Using a process similar to that of Hatch et al. (2013; Figure 1C), we monitored histone-RFP (hht1-RFP) in cells resolving division. The presence of smaller chromatin bodies away from the primary nucleus was scored as a “free chromatin body”; these were monitored to determine whether they rejoined the primary nucleus (resorbed).
To determine whether free chromatin bodies were membrane-enclosed micronuclei, the membrane marker ccr1 N-terminal fragment (ccr1N-GFP) was monitored with histone (hht1-RFP) in live cells after 4 h at 36°C. Cells were scored as micronucleus forming if they met three criteria: 1) the micronuclear histone mass was surrounded by membrane and separated from the parent nucleus; 2) the micronucleus formed after nuclear division, excluding rare spontaneous micronuclei; and 3) if micronuclei were retained after septation, to exclude fragmented bodies that formed transiently during mitosis. Videos from more than two biological replicates were used, and numbers were pooled. The combined proportions with 95% CI are presented. Deconvolved and projected images from a time course are shown in Figure 2A. A projected image of a single cell is shown in Supplemental Figure S2 and Supplemental Video S3.
Protein methods
Protein extracts were prepared from equal numbers of cells treated with 0.3 M sodium hydroxide. Cells were lysed by boiling for 5 min in acidic SDS–PAGE buffer (4% SDS, 60 mM Tris-HCl, pH 6.8, 5% glycerol, 4% 2-mercaptoethanol, 0.01% bromophenol blue, 0.1 M dithiothreitol). Samples were run on Tris-glycine gels and transferred to polyvinylidene fluoride membrane. Primary antibodies for Chk1HA (16B12 anti-hemagglutinin [HA]; Covance), phospho–Cdc2-Y15 (Cell Signaling Technology, Danvers, MA), and S. pombe Cdc2, Mcm4, and Crb2 (polyclonal antibodies) were incubated overnight. Blots were washed in phosphate-buffered saline (PBS)–Tween buffer, exposed to horseradish peroxidase–conjugated secondary antibody for 1 h, and then washed and exposed using enhanced chemiluminescence (Pierce). Quantitation of Mcm4 and Chk1-HA (phosphorylated and unmodified forms) was performed using QuantityOne software (Bio-Rad) as in Furuya et al. (2010).
DNA synthesis detection
To monitor DNA synthesis by nucleoside analogue incorporation, cultures were treated with either 10 μM EdU or 10 μg/ml bromodeoxyuridine (BrdU) for appropriate times before harvest. EdU-treated cells were fixed with 70% ethanol and processed using the Click-iT EdU Alexa Fluor 488 Imaging Kit according to directions (C10337; Life Technologies, ThermoFisher Scientific). BrdU chromatin immunoprecipitation (IP) was performed as described in Knott et al. (2012) with the following modifications. Cells were pelleted, snap-frozen, and then stored at −80°C. After lysis in TES (100 mM Tris, pH 8.0, 50 mM EDTA, 1% SDS) with glass beads, chromatin was sheared by sonication, resulting in ∼500–base pair fragments. DNA was phenol-chloroform extracted, isopropanol precipitated, and then resuspended in TE. Samples were diluted with IP buffer (1× PBS, 0.05% Triton X-100) before overnight incubation with anti-BrdU (RPN202; GE Healthcare, Sigma-Aldrich). Antibody-BrdU-DNA complexes were precipitated on magnetic protein A–Sepharose (Dynabeads, 10002D; Invitrogen, ThermoFisher Scientific), washed three times in IP buffer and once in TE, and then incubated in TES at 65°C (15 min). DNA was then purified using a Qiagen PCR purification kit and quantitatively amplified on a PerkinElmer HT9700 using origin-specific primers (Supplemental Table S2) and iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, CA). The Pfaffl method was used to determine percentage IP for each region relative to input DNA.
Flow cytometry and microscopy
Cells were fixed in cold 70% ethanol for cell cycle analysis or microscopy. For DAPI/septa staining, cells were rehydrated in water and incubated for 10 min in 1 mg/ml aniline blue (M6900; Sigma-Aldrich). Cells in mount (50% glycerol, 1 μg/ml DAPI, and 1 μg/ml p-phenylenediamine) were photographed on a Leica DMR wide-field epifluorescence microscope using a 63× objective lens (numerical aperture [NA] 1.62 Plan Apo), 100-W Hg arc lamp for excitation, and a 12-bit Hamamatsu ORCA-100 charge-coupled device (CCD) camera. OpenLab version 3.1.7 (ImproVision, Lexington, MA) software was used at acquisition and ImageJ (National Institutes of Health, Bethesda, MD) for analysis.
Whole-cell SYTOX Green and EdU flow cytometry (fluorescence-activated cell sorting [FACS]) were performed as described in Sabatinos et al. (2012, 2013). DNA synthesis by EdU incorporation was assessed by adding 10 μM EdU to cultures before harvest. Whole-cell FACS for EdU was performed on rehydrated cells using Click-iT (Invitrogen, ThermoFisher Scientific) with Alexa Fluor 488.
Live-cell imaging
Medium for all live-cell imaging was EMM plus supplements plus thiamine. Live-cell videomicroscopy experiments at 36°C were performed on 2% agarose pads sealed with VaLaP (1/1/1 [wt/wt/wt] Vaseline/lanolin/paraffin). Long-term videomicroscopy at 25°C was performed in CellAsics microfluidics plates (Y04C series; EMD Millipore), with constant temperature and medium flow. Fluorescent-tag images of live cells were acquired using a DeltaVision microscope (with softWoRx version 4.1; GE, Issaquah, WA) using a 60× (NA 1.4 PlanApo) lens, solid-state illuminator, and 12-bit CCD camera. Sections of static time points were eighteen 0.3-μm z-sections. Long-term time-lapse videos used nine z-steps of 0.5 μm. Images were deconvolved and maximum intensity projected (softWoRx). Transmitted light images were added to projected fluorescence images. Images were contrast adjusted using an equivalent histogram stretch on all samples. A threshold of signal 2× over the average nuclear background was used for RPA-CFP and Rad52-YFP focus discrimination. Foci are presented as the proportion of nuclei per category of focus with ±95% CI. Significance was assessed with chi-squared tests and differences between proportions with two-tailed Z tests.
Mutation analysis
The forward canavanine mutation rate at can1+ was determined as described (Sabatinos et al., 2013). Briefly, cultures were diluted in yeast extract with supplements (YES) medium and plated on 15-cm canavanine plates (70 μg/ml in pombe minimal medium with glutamate [PMG] plus supplements plus phloxine B). Plates were scored after 8 d at 25°C, for the number of can1− colonies compared with total cells plated, calculated from titer plates. Grouped experiments were performed independently, and then the mutation rate was calculated using the MSS-MLE algorithm in FALCOR (www.keshavsingh.org/protocols/FALCOR.html). Results were plotted in Delta Graph and compared by two-tailed t test.
The frequency of hsv-tk+ loss and sectoring was scored in 500–2000 cells plated on YES, grown at 25°C, and then replica plated onto fluorodeoxyuridine (FUdR) plates (20 μg/ml in EMM plus supplements plus phloxine B). The number of FUdR-resistant and sectored colonies was counted per total number of colonies and the proportions assessed with Z tests (vassarstats.net/propdiff_ind.html). Grouped experiments were performed independently and pooled for analysis. A box plot of sectored data was made using BoxPlotR, showing median, 25th/75th percentile boundaries, and 1.5× interquartile whiskers (boxplot.tyerslab.com/).
The ChL minichromosome strains were grown as in Nakamura et al. (2008) and Li et al. (2013). Cultures were plated for viability and on PMG-HULA plates with 5-fluoroacetate (5-FOA; Zymo Research, Irvine, CA). Cultures were then incubated at 36°C for 4 h and then plated as at the start. All plates were grown at 25°C, and the number of Ura– colonies counted and compared with the total number of surviving cells. Ura– colonies were replica plated onto PMG-HUA and PMG-HUL with 5-FOA to assess Ura– Leu– and Ura– Ade– colonies, respectively. Ura– Leu– colonies were patched or replica plated onto PMG-HUL and YES-hygromycin to assess hygromycin and Ade status as in Li et al. (2013). FALCOR was used to calculate recombination/mutation rates, and a Mann–Whitney two-tailed U test was used to assess significance between observed sets.
Supplementary Material
Acknowledgments
We thank the University of Southern California Center for Electron Microscopy and Microanalysis for support with 3D-SIM microscopy and Stephen Kearsey, Jian Qu Wu, Xie Tang, and Zac Cande for strains. This work was supported by National Institutes of Health Grants R01 GM081418 and GM111040.
Abbreviations used:
- A.U
arbitrary units
- BIR
break-induced replication
- CFP
cyan fluorescent protein
- cut
cell untimely torn
- 3D-SIM
3-dimensional structured illumination microscopy
- EdU
5-ethynyl-2′-deoxyuridine
- FUdR
5-fluoro-2′-deoxyuridine
- GCR
gross chromosomal rearrangements
- GFP
green fluorescent protein
- HU
hydroxyurea
- iso
isochromosome
- MCM
minichromosome maintenance
- MN
micronucleus
- MUG
mitosis with unreplicated genomes
- PMG
pombe minimal medium with glutamate
- PN
parent nucleus
- RFP
red fluorescent protein
- RPA
replication protein A
- ssDNA
single-strand DNA
- UFB
ultrafine bridge
- wt
wild type
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-05-0318) on August 5, 2015.
REFERENCES
- Bagley BN, Keane TM, Maklakova VI, Marshall JG, Lester RA, Cancel MM, Paulsen AR, Bendzick LE, Been RA, Kogan SC, et al. A dominantly acting murine allele of mcm4 causes chromosomal abnormalities and promotes tumorigenesis. PLoS Genet. 2012;8:e1003034. doi: 10.1371/journal.pgen.1003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis JM, Luche DD, Hunter T, Forsburg SL. Minichromosome maintenance proteins interact with checkpoint and recombination proteins to promote s-phase genome stability. Mol Cell Biol. 2008;28:1724–1738. doi: 10.1128/MCB.01717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bass KL, Murray JM, O’Connell MJ. Brc1-dependent recovery from replication stress. J Cell Sci. 2012;125:2753–2764. doi: 10.1242/jcs.103119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeharry N, Rattner JB, Caviston JP, Yen T. Centromere fragmentation is a common mitotic defect of S and G2 checkpoint override. Cell Cycle. 2013;12:1588–1597. doi: 10.4161/cc.24740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Bell SP, Schwacha A. Subunit organization of Mcm2-7 and the unequal role of active sites in ATP hydrolysis and viability. Mol Cell Biol. 2008;28:5865–5873. doi: 10.1128/MCB.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, Domingo E, Kanu N, Dewhurst SM, Gronroos E, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro T, Khair L, Reis CC, Borges V, Moser BA, Nakamura TM, Ferreira MG. Telomeres avoid end detection by severing the checkpoint signal transduction pathway. Nature. 2010;467:228–232. doi: 10.1038/nature09353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JP, Nobbs M, McGettigan P, Lynch S, Ennis S. Recessive mutations in MCM4/PRKDC cause a novel syndrome involving a primary immunodeficiency and a disorder of DNA repair. J Med Genet. 2012;49:242–245. doi: 10.1136/jmedgenet-2012-100803. [DOI] [PubMed] [Google Scholar]
- Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- Chan KL, Hickson ID. On the origins of ultra-fine anaphase bridges. Cell Cycle. 2009;8:3065–3066. doi: 10.4161/cc.8.19.9513. [DOI] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- Coxon A, Maundrell K, Kearsey SE. Fission yeast cdc21+ belongs to a family of proteins involved in an early step of chromosome replication. Nucleic Acids Res. 1992;20:5571–5577. doi: 10.1093/nar/20.21.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O. Common fragile sites: mechanisms of instability revisited. Trends Genet. 2012;28:22–32. doi: 10.1016/j.tig.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, Norppa H, Eastmond DA, Tucker JD, Thomas P. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 2011;26:125–132. doi: 10.1093/mutage/geq052. [DOI] [PubMed] [Google Scholar]
- Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froget B, Blaisonneau J, Lambert S, Baldacci G. Cleavage of stalled forks by fission yeast Mus81/Eme1 in absence of DNA replication checkpoint. Mol Biol Cell. 2008;19:445–456. doi: 10.1091/mbc.E07-07-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K, Miyabe I, Tsutsui Y, Paderi F, Kakusho N, Masai H, Niki H, Carr AM. DDK phosphorylates checkpoint clamp component Rad9 and promotes its release from damaged chromatin. Mol Cell. 2010;40:606–618. doi: 10.1016/j.molcel.2010.10.026. [DOI] [PubMed] [Google Scholar]
- Gineau L, Cognet C, Kara N, Lach FP, Dunne J, Veturi U, Picard C, Trouillet C, Eidenschenk C, Aoufouchi S, et al. Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J Clin Invest. 2012;122:821–832. doi: 10.1172/JCI61014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Grallert B, Nurse P. The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- Guerrero AA, Gamero MC, Trachana V, Futterer A, Pacios-Bras C, Diaz-Concha NP, Cigudosa JC, Martinez AC, van Wely KH. Centromere-localized breaks indicate the generation of DNA damage by the mitotic spindle. Proc Natl Acad Sci USA. 2010;107:4159–4164. doi: 10.1073/pnas.0912143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guida T, Salvatore G, Faviana P, Giannini R, Garcia-Rostan G, Provitera L, Basolo F, Fusco A, Carlomagno F, Santoro M. Mitogenic effects of the up-regulation of minichromosome maintenance proteins in anaplastic thyroid carcinoma. J Clin Endocrinol Metab. 2005;90:4703–4709. doi: 10.1210/jc.2004-2459. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Han X, Aslanian A, Fu K, Tsuji T, Zhang Y. The interaction between checkpoint kinase 1 (Chk1) and the minichromosome maintenance (MCM) complex is required for DNA damage-induced Chk1 phosphorylation. J Biol Chem. 2014;289:24716–24723. doi: 10.1074/jbc.M114.575035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch D, Kemmerling R, Davis S, Camps J, Meltzer PS, Ried T, Gaiser T. Chromothripsis and focal copy number alterations determine poor outcome in malignant melanoma. Cancer Res. 2013;73:1454–1460. doi: 10.1158/0008-5472.CAN-12-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat Med. 2012;18:1630–1638. doi: 10.1038/nm.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CR, Guasti L, Meimaridou E, Chuang CH, Schimenti JC, King PJ, Costigan C, Clark AJ, Metherell LA. MCM4 mutation causes adrenal failure, short stature, and natural killer cell deficiency in humans. J Clin Invest. 2012;122:814–820. doi: 10.1172/JCI60224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch A, Ampatzidou E, Mizuno K, O’Connell MJ, Murray JM. Smc5/6 maintains stalled replication forks in a recombination-competent conformation. EMBO J. 2009;28:144–155. doi: 10.1038/emboj.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimi Y, Komamura-Kohno Y, Kwon HJ, Yamada K, Nakanishi M. Identification of MCM4 as a target of the DNA replication block checkpoint system. J Biol Chem. 2003a;278:24644–24650. doi: 10.1074/jbc.M213252200. [DOI] [PubMed] [Google Scholar]
- Ishimi Y, Okayasu I, Kato C, Kwon HJ, Kimura H, Yamada K, Song SY. Enhanced expression of Mcm proteins in cancer cells derived from uterine cervix. Eur J Biochem. 2003b;270:1089–1101. doi: 10.1046/j.1432-1033.2003.03440.x. [DOI] [PubMed] [Google Scholar]
- Kato H, Sandberg AA. Chromosome pulverization in human cells with micronuclei. J Natl Cancer Inst. 1968;40:165–179. [PubMed] [Google Scholar]
- Kawabata T, Luebben SW, Yamaguchi S, Ilves I, Matise I, Buske T, Botchan MR, Shima N. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell. 2011;41:543–553. doi: 10.1016/j.molcel.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993a;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Nurse P, Forsburg SL. Coupling DNA replication to the cell cycle. Cold Spring Harb Symp Quant Biol. 1993b;58:637–644. doi: 10.1101/sqb.1993.058.01.071. [DOI] [PubMed] [Google Scholar]
- Knott SR, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavare S, Aparicio OM. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell. 2012;148:99–111. doi: 10.1016/j.cell.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Kearsey SE, Diffley JF. MCM2–7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1-phase and are required to establish, but not maintain, the S-phase checkpoint. Mol Biol Cell. 2001;12:3658–3667. doi: 10.1091/mbc.12.11.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Mizuno K, Blaisonneau J, Martineau S, Chanet R, Freon K, Murray JM, Carr AM, Baldacci G. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol Cell. 2010;39:346–359. doi: 10.1016/j.molcel.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Latif C, den Elzen NR, O’Connell MJ. DNA damage checkpoint maintenance through sustained Chk1 activity. J Cell Sci. 2004;117:3489–3498. doi: 10.1242/jcs.01204. [DOI] [PubMed] [Google Scholar]
- Lau KM, Chan QK, Pang JC, Li KK, Yeung WW, Chung NY, Lui PC, Tam YS, Li HM, Zhou L, et al. Minichromosome maintenance proteins 2, 3 and 7 in medulloblastoma: overexpression and involvement in regulation of cell migration and invasion. Oncogene. 2010;29:5475–5489. doi: 10.1038/onc.2010.287. [DOI] [PubMed] [Google Scholar]
- Li PC, Petreaca RC, Jensen A, Yuan JP, Green MD, Forsburg SL. Replication fork stability is essential for the maintenance of centromere integrity in the absence of heterochromatin. Cell Rep. 2013;3:638–645. doi: 10.1016/j.celrep.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang DT, Forsburg SL. Characterization of Schizosaccharomyces pombe mcm7(+) and cdc23(+) (MCM10) and interactions with replication checkpoints. Genetics. 2001;159:471–486. doi: 10.1093/genetics/159.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang DT, Hodson JA, Forsburg SL. Reduced dosage of a single fission yeast MCM protein causes genetic instability and S phase delay. J Cell Sci. 1999;112:559–567. doi: 10.1242/jcs.112.4.559. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Wardlaw CP, Morishita T, Miyabe I, Chahwan C, Caspari T, Schmidt U, Carr AM, Garcia V. The Rad4(TopBP1) ATR-activation domain functions in G1/S phase in a chromatin-dependent manner. PLoS Genet. 2012;8:e1002801. doi: 10.1371/journal.pgen.1002801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner K, Gregan J, Montgomery S, Kearsey SE. Essential role of MCM proteins in premeiotic DNA replication. Mol Biol Cell. 2002;13:435–444. doi: 10.1091/mbc.01-11-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- Maiorano D, Lemaitre JM, Mechali M. Stepwise regulated chromatin assembly of MCM2–7 proteins. J Biol Chem. 2000;275:8426–8431. doi: 10.1074/jbc.275.12.8426. [DOI] [PubMed] [Google Scholar]
- Majid S, Dar AA, Saini S, Chen Y, Shahryari V, Liu J, Zaman MS, Hirata H, Yamamura S, Ueno K, et al. Regulation of minichromosome maintenance gene family by microRNA-1296 and genistein in prostate cancer. Cancer Res. 2010;70:2809–2818. doi: 10.1158/0008-5472.CAN-09-4176. [DOI] [PubMed] [Google Scholar]
- Martinez AC, van Wely KH. Centromere fission, not telomere erosion, triggers chromosomal instability in human carcinomas. Carcinogenesis. 2011;32:796–803. doi: 10.1093/carcin/bgr069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh D, Blow JJ. Dormant origins, the licensing checkpoint, and the response to replicative stresses. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a012955. pii: a012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol. 2009;11:761–768. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- Naim V, Wilhelm T, Debatisse M, Rosselli F. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat Cell Biol. 2013;15:1008–1015. doi: 10.1038/ncb2793. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Okamoto A, Katou Y, Yadani C, Shitanda T, Kaweeteerawat C, Takahashi TS, Itoh T, Shirahige K, Masukata H, et al. Rad51 suppresses gross chromosomal rearrangement at centromere in Schizosaccharomyces pombe. EMBO J. 2008;27:3036–3046. doi: 10.1038/emboj.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1981;182:119–124. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- O’Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatinos SA, Green MD, Forsburg SL. Continued DNA synthesis in replication checkpoint mutants leads to fork collapse. Mol Cell Biol. 2012;32:4986–4997. doi: 10.1128/MCB.01060-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatinos SA, Mastro TL, Green MD, Forsburg SL. A mammalian-like DNA damage response of fission yeast to nucleoside analogs. Genetics. 2013;193:143–157. doi: 10.1534/genetics.112.145730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Yanagida M. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+ Cell. 1993;74:383–393. doi: 10.1016/0092-8674(93)90428-s. [DOI] [PubMed] [Google Scholar]
- Saugar I, Vazquez MV, Gallo-Fernandez M, Ortiz-Bazan MA, Segurado M, Calzada A, Tercero JA. Temporal regulation of the Mus81-Mms4 endonuclease ensures cell survival under conditions of DNA damage. Nucleic Acids Res. 2013;41:8943–8958. doi: 10.1093/nar/gkt645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S, Lynch M, Needleman D. Deciphering the evolutionary history of open and closed mitosis. Curr Biol. 2014;24:R1099–R1103. doi: 10.1016/j.cub.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, Hartford SA, Tye BK, Schimenti JC. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007a;39:93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- Shima N, Buske TR, Schimenti JC. Genetic screen for chromosome instability in mice: Mcm4 and breast cancer. Cell Cycle. 2007b;6:1135–1140. doi: 10.4161/cc.6.10.4250. [DOI] [PubMed] [Google Scholar]
- Sofueva S, Osman F, Lorenz A, Steinacher R, Castagnetti S, Ledesma J, Whitby MC. Ultrafine anaphase bridges, broken DNA and illegitimate recombination induced by a replication fork barrier. Nucleic Acids Res. 2011;39:6568–6584. doi: 10.1093/nar/gkr340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorzano CO, Pascual-Montano A, Sanchez de Diego A, Martinez AC, van Wely KH. Chromothripsis: breakage-fusion-bridge over and over again. Cell Cycle. 2013;12:2016–2023. doi: 10.4161/cc.25266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Kurata M, Abe S, Miyazawa R, Murayama T, Hidaka M, Yamamoto K, Kitagawa M. Overexpression of MCM2 in myelodysplastic syndromes: association with bone marrow cell apoptosis and peripheral cytopenia. Exp Mol Pathol. 2012;92:160–166. doi: 10.1016/j.yexmp.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, Bekker-Jensen S, Mailand N, Bartek J, Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- Utani K, Kohno Y, Okamoto A, Shimizu N. Emergence of micronuclei and their effects on the fate of cells under replication stress. PLoS One. 2010;5:e10089. doi: 10.1371/journal.pone.0010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DA, Brinkley BR. Mitosis in cells with unreplicated genomes (MUGs): spindle assembly and behavior of centromere fragments. Cell Motil Cytoskeleton. 1997;36:291–302. doi: 10.1002/(SICI)1097-0169(1997)36:3<291::AID-CM9>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Yabuta N, Kajimura N, Mayanagi K, Sato M, Gotow T, Uchiyama Y, Ishimi Y, Nojima H. Mammalian Mcm2/4/6/7 complex forms a toroidal structure. Genes Cells. 2003;8:413–421. doi: 10.1046/j.1365-2443.2003.00645.x. [DOI] [PubMed] [Google Scholar]
- Zhang CZ, Leibowitz ML, Pellman D. Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 2013;27:2513–2530. doi: 10.1101/gad.229559.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Brun C, Kurooka H, Yanagida M, Huberman JA. Identification and characterization of a complex chromosomal replication origin in Schizosaccharomyces pombe. Chromosoma. 1992;102:S7–S16. doi: 10.1007/BF02451780. [DOI] [PubMed] [Google Scholar]
- Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.