Abstract
Scope
The increased prevalence of cardiovascular diseases has been hypothesized to be the result of an increased exposure to a host of atherogenic environmental factors, paramount among them being unhealthy dietary habits. Long-chain n-3 polyunsaturated fatty acids (PUFAs) have been shown to have cardio protective effects, partially due to their ability to regulate gene expression. In this regard, increasing attention has been devoted to the role of miRNAs as regulators of multiple metabolic pathways whose deregulation has been associated with CVD risk. In this work we investigated whether miRNA expression was regulated by docosahexanoic acid (DHA), conjugated linoleic acid (CLA) and cholesterol in Caco-2 cells.
Results
Among the modulated miRNAs, miR-107 was differentially expressed by all treatments and this modulation was independent of its hosting gene, PANK1, possibly through its own promoter, which contains binding sites for metabolically relevant transcription factors. Among the putative target genes of miR-107, we found some genes with key roles in circadian rhythm. Specifically, we demonstrated that binding of miR-107 to the CLOCK gene results in the deregulation of the circadian rhythm of the cells.
Conclusions
Since chronodisruption has been linked to metabolic disorders such as Type 2 Diabetes (T2D), atherosclerosis, obesity and Cardiovascular Disease (CVD), our findings suggests that miR-107 could represent a new approach for pharmacological treatment of these diseases.
Keywords: miR-107, DHA, dietary lipids, circadian rhythm
INTRODUCTION
Cardiovascular Disease (CVD) is the first cause of morbidity and mortality in industrialized countries. Thus, there is an urgency to develop strategies to prevent this disease and its devastating consequences. Dietary habits have been identified as major drivers of both its rise and potentially its decline, and a number of foods and dietary components have been attributed with preventive and therapeutic effects.
Dietary lipids play a key role in human health and disease and this role is controversial and depends on the type and the amount of ingested fat. Specifically, n-3 long chain polyunsaturated fatty acids (PUFAs), have been suggested to promote or maintain cardiovascular health [1, 2], mainly due to their anti-lipidemic [3, 4], anti-inflammatory [5, 6], anti-platelet [7, 8], and anti-arrhythmic [9, 10] effects. Unlike the commonly accepted cardio protective role of n-3 PUFAs, other dietary lipids are more controversial. This is the case of the conjugated linoleic acids (CLA), a group of PUFAs with two conjugated double bonds, found in red meat and dairy products. Its consumption has been associated with a reduction in total body fat and an increase in lean body mass. Nevertheless, other studies suggest that CLA isomers may induce insulin resistance [11] and an increase in total and LDL-cholesterol, but without consistency between them [12]. Different molecular mechanisms underlie the cardiovascular effects of CLA, including activation of PPARs (Peroxisome Proliferator-Activated Receptors) [13], stimulation of hepatic expression of APOB100/E receptors, [14], cholesterol-efflux mediation [15] and suppression of the production of inflammatory mediators [16, 17]. Finally, dietary cholesterol has been associated with increased CVD risk. Nevertheless, cholesterol is an essential component of the cell membrane [18], a precursor of essential steroid hormones and bile acids [19] and is involved in the control of the cell cycle's progression [20-22].
The effect of dietary lipids at the epigenetic level is poorly defined. An example of epigenetic regulation is the gene expression modulation mediated by miRNAs. The accepted role of miRNAs in cellular physiology is the “fine tuning” of gene expression, but under stress conditions, this apparently modest role becomes fundamental. The role of miRNAs in metabolic diseases including atherosclerosis, CVD, Metabolic Syndrome (MetS), obesity and T2D has been extensively reviewed recently [23, 24]. Several miRNAs have been described that modulate important metabolic processes like cholesterol efflux [25-29], hepatic metabolism [30, 31], cardiovascular development and function [32, 33], function of endothelial cells [34] and insulin signaling [35, 36]. Recent studies have suggested an effect of different dietary bioactive compounds on the expression of miRNAs. Most of these studies have focused on the chemopreventive effects of some dietary compounds through the modulation of cancer-related miRNAs [37, 38]. However, the nutritional impact on the expression of metabolically relevant miRNAs needs to be further addressed. Concerning the effect of n-3 PUFAs on metabolic miRNAs, Parra et al showed that CLA treatment influenced the expression of miRNAs in adipose tissue [39], and our previous results suggest that DHA modulates the expression of miR-30c and miR-192 in the enterocytes [40]. Additionally, we have recently shown an interaction between lipoprotein lipase (LPL) variant rs13702, which disrupts a miRNA-410 seed site, and the intake of n-3 PUFAs related to the plasma lipid profile [41]. These examples suggest that dietary n 3 PUFAs, may play an important role on the regulation of metabolically-relevant miRNAs that can be associated to CVD. However, further research is needed to assess the specific miRNAs modulated by dietary n-3 PUFAs and the physiological consequences of such modulation.
In this study we aimed to investigate the modulation of miRNAs by dietary lipids and to analyze whether this modulation was associated with CVD traits. Such modulation can contribute to clarify the effect of some controversial dietary lipids on human cardiovascular health.
MATERIAL AND METHODS
Refer to the Supporting Information of the paper for further detailed description of the methods
Cells and animals
Caco-2 and COS-7 cells were obtained from the American Type Tissue Collection (Barcelona, Spain). Cells were maintained in DMEM with 10% of Fetal Bovine Serum (FBS) supplemented with glutamine and antibiotics (Supporting Information). Caco-2 cells were differentiated on polycarbonate Transwell® filters inserts of 0.4 μm pore size (Costar, Madrid, Spain) in 6-wells plates [42] (Supporting Information).
8 week old C57BL6/J mice were fed a normal chow diet or a high fat diet containing 1.25% of cholesterol (Chol-HFD) for 16 weeks. After a 12-hour fasting, mice were sacrificed, and tissues samples were collected and immediately immersed in liquid nitrogen (Supporting Information).
Lipid micelle preparation
Cholesterol (Chol), CLA and DHA (Sigma, Madrid, Spain) were delivered to cells as micelles with Lyso-phosphatidilcholine (Lyso-PC; Sigma, Madrid, Spain) and sodium taurocholate (Tau; Sigma, Madrid, Spain). Lipid micelles were prepared as described by Field et al [43] with modifications (Supporting Information).
MiRNA-enriched RNA isolation and microarray performance
MiRNA-enriched total RNA was isolated with miRNeasy mini kit (Qiagen, Madrid, Spain) following manufacturer's instructions. The expression of miRNAs contained in the miRBase version 17 was analyzed by microarray using the μParaflo®Microfluidic Biochip Technology of LCsciences (Huston, TX, USA). Raw data were normalized, processed and analyzed for statistical significance using t-test and applying multiple testing correction (Supporting Information). Microarray data has been submitted to the GEO database complying with the Minimal Information About a Microarray Experiment (MIAME) guidelines (GSE59153). Predicted and validated target genes of significant miRNAs were examined using miRWalk, miRanda, Diana-mT, miRDB, PITA and TargetScan prediction algorithms (Supporting Information). Analysis of the overrepresentation of functional GeneOntology (GO) terms and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was made using Babelomics analysis suite (www.babelomics.bioinfo.cipf.es, Valencia, Spain).
Reverse transcription and Real-Time quantitative PCR
For mRNA quantification, total RNA was reverse transcribed with PrimeScript Reverse Transcription kit (Takara, California, USA) and the resulting cDNA was amplified by Real-Time quantitative PCR (RT-qPCR) with FastStart Universal SYBR Green Master (Roche, Barcelona, Spain). Specific Forward and Reverse primers were used (Supporting Information Table S1; Isogen Lifesciences, Barcelona, Spain). mRNA levels were calculated using the relative quantification method normalizing with RN18S.
For miRNA quantification, miRNA was reverse transcribed with miScript Reverse Transcription II Kit (Qiagen, Madrid, Spain) and amplified with miScript SYBR Green PCR Kit (Qiagen, Madrid, Spain) using a specific forward primer (Supporting Information Table S1; Isogen Lifescience, Barcelona, Spain) and a Universal reverse primer provided by the kit. miRNA levels were calculated with the relative quantification method using RNU6 as reference gene.
Promoter analysis and response to Atorvastatin and Fenofibrate
2 Kb of the miRNA-107 putative promoter sequence reported by Corcoran et al [44] was analyzed for predicted transcription factor binding sites using Transcription Element Search System (TESS, University of Pennsylvania, PA, USA) and Transcription Factor Search (TFSearch, Parallel Application TRC Laboratory, RWCP, Japan).
The response to Atorvastatin (Ator) and Fenofibrate (Feno) was studied in differentiated Caco-2 cells treated with 1, 5 or 10 μM of Ator or with 10, 25 or 50 μM of Feno for 24 and 48 hours.
miRNA inhibitors and mimics transfection
miRNA-107 inhibitor (anti-miRNA-107) and mimic (syn-miRNA-107) were obtained from Qiagen (Madrid, Spain). Caco-2 cells were transfected with 50 nM of anti-miRNA-107 or 5 nM of syn-miRNA-107 using Oligofectamine (Invitrogen, Madrid, Spain) following manufacturer's instructions. All experimental control samples were transfected with a negative inhibitor control (anti-miRNA-neg) or with syn-miRNA-1 as positive control (Qiagen, Madrid, Spain). miRNA-107 inhibition or overexpression was confirmed in all experiments by RT-qPCR as described above.
Ribonucleoprotein Ago-2 inmunoprecipitation (RIP) of miRNA-107 and its target mRNAs
Caco-2 cells were transfected with syn-miRNA-107 or control as described above. 24 hours after transfection the miRNA-mRNA binding was cross-linked and complexes were precipitated with anti-AGO2 antibodies with the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, © Merck KGaA, Darmstadt, Germany). The amount of precipitated miR-107 and CLOCK was measured by RT-qPCR (Supporting Information).
Western-Blot
Cells were lysed in ice-cold lysis buffer and washed. Total protein was quantified using the PierceTM BCA Protein assay kit (ThermoScientific, Rockford, IL, USA). Equal amounts of protein were separated by SDS/PAGE, transferred onto nitrocellulose membranes and probed with the indicated primary antibodies and the appropriate HRP-conjugated secondary antibodies. Protein bands were visualized and measured with densitometry analyses (Supporting Information).
3’ Untranslated region luciferase reporter assays
We sub-cloned a CLOCK 3’-untranslated region (3’-UTR) containing the miR-107 binding site region from the pMT01 vector (Genecopeia Inc., MD, USA) into a psiChek2 vector (Promega) (Supporting Information). Point mutations in the seed region of predicted miR-107 sites within the 3’UTR of CLOCK was generated using QuikChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer's protocol. COS-7 cells were co-transfected for 48h with 1 μg of the CLOCK 3’UTR containing vector or an empty vector and 50 nM of the syn-miR-107 (Qiagen, Madrid, Spain). Luciferase activity was measured with the Dual-Glo Luciferase Assay System (Promega). Relative luciferase activity (RLA) was calculated through the normalization with Renilla activity (Supporting Information).
Analyses of circadian rhythm in culture cells
Caco-2 cells were synchronized with a serum shock and transfected with syn-miRNA-107 or control (Supporting Information). After 24 hours of transfection, cells were harvested every 4 hours during a 24-hour period. miRNA-enriched RNA was extracted and miRNA-107 and mRNA levels of circadian genes and CLOCK-related genes were analyzed by RT-qPCR as described above. We used the cosinor test to calculate circadian period (Supporting Information).
Statistical analyses
Data are presented as mean ± SEM (n is annotated in the Figure legends). The statistical significance of differences was evaluated with either the Student's t test or the one-way analysis of variance (ANOVA) when appropriate. Significance was accepted at the level of p < 0.05. All analyses were performed with SPSS 19.0 (SPSS Inc.). All experiments were repeated at least three times and biological triplicates and technical duplicates were included in each experiment.
RESULTS
DHA modifies the miRNA expression profile in Caco-2 cells
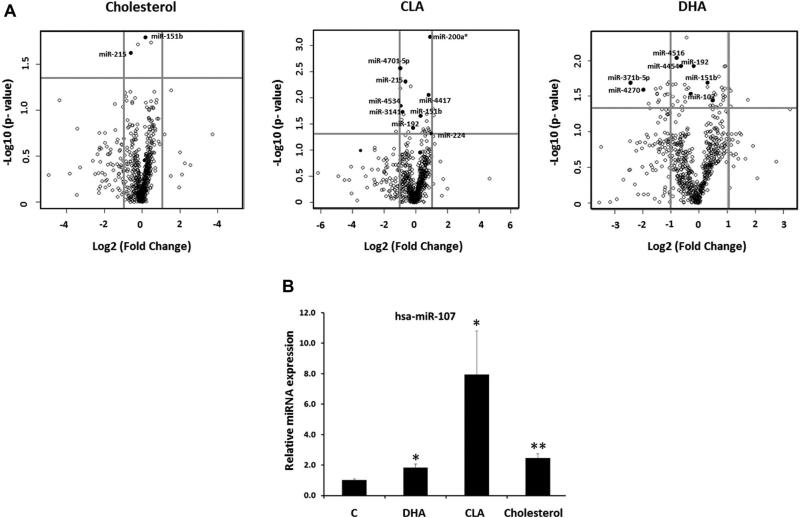
We aimed to study the effect of some dietary lipids on human enterocytes miRNA profile using an in vitro model. For this purpose, differentiated Caco-2 cells were treated with micelles containing cholesterol, DHA, CLA or empty micelles and the expression levels of the human known miRNAs (contained in the miRBase database version 17) were assessed by microarray (Figure 1A, Supporting Information Table S2).
Figure 1.
Modulation of miRNA levels by Cholesterol, DHA and CLA. Differentiated Caco-2 cells were treated with micelles containing cholesterol (250 μM), DHA (200 μM), CLA (200 μM) or nothing (empty micelles) for 24 (DHA and CLA) and 48 hours (cholesterol). miRNA levels were analysed by microarray. (A) Vulcano Plots representing the Fold Change (Log2 FC) and the p-value (−Log10 pvalue) of the miRNAs modified by cholesterol, DHA and CLA. Differentially regulated miRNAs are labelled. (B) Confirmation of the expression of miR-107 by RT-qPCR using RNU6 as housekeeping. Data are the mean of three independent experiments, each one performed in triplicate ± SEM. * p<0.05; ** p<0.001.
We identified a subset of 29 miRNAs differentially regulated by cholesterol, DHA or CLA (Figure 1A, Supporting Information Table S2). 21 miRNAs were significantly modified by DHA (12 downregulated and 9 upregulated), 11 by CLA (5 downregulated and 6 upregulated) and, surprisingly, only two miRNAs were modified by cholesterol (1 upregulated and 1 downregulated) (Figure 1A and Supporting Information Table S2). Among the miRNAs modified, we identified several whose predicted targets are involved in PPAR signaling, insulin signaling, Mitogen-Activated Protein Kinases (MAPKs) signaling, Vascular Endothelial Growth Factor (VEGF) signaling, T2D and biosynthesis of unsaturated fatty acids (data not shown). Among the differentially expressed miRNAs, we found that miR-107 was upregulated by all the three treatments, although only the DHA-mediated increase was statistically significant (Supporting Information Table S2). The increase in the miRNA-107 levels by cholesterol, DHA and CLA was further confirmed, and became significant by RT-qPCR (Figure 1B). miRNA-107 is located within the intron 5 of PANK1, a gene that encodes a key enzyme in the biosynthesis of Coenzyme A (Supporting Information Figure S1). Furthermore, the pre-miRNA-107 is highly conserved among mammals as well as its location within PANK1 (Supporting Information Figure S1). We selected miRNA-107 to further characterize its regulation by dietary lipids and its role in energetic metabolism.
Atorvastatin, Fenofibrate and a High-Cholesterol High-Fat diet modify miRNA-107 levels independently of its host gene, PANK1
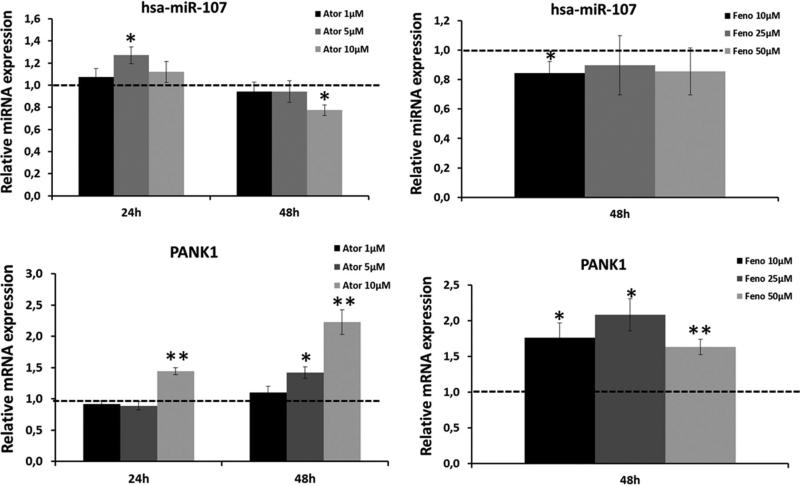
The location of miR-107 in the intron 5 of PANK1 suggests that it could be transcribed together with PANK1, which contains PPARα and SREBP responsive elements in its promoter (57). Hence, we studied the effect of a well-known PPARα agonist – fenofibrate – and a lipid-lowering drug affecting SREBP regulatory pathway – atorvastatin – on the expression levels of miRNA-107 and PANK1 (Figure 2). In order to ensure the cellular exposure to the treatments, we measured HMGR (target of Ator), CD36 and LPL (targets of Feno) at 24 and 48 hours (Supporting Information figure S2). Ator effectively activated HMGCR expression after 24 hours of treatment with any dosage. After 48 hours, the effect of the lowest dosage was lost. On the other hand, Feno increased CD36 and LPL levels only after 48 hours of treatment with 25 μM and 50 μM. For that reason, we only analyzed miR-107 and PANK1 levels after 48 hours of incubation with Feno (Figure 2). As shown in figure 2, Ator did not significantly increase the expression of miRNA-107 at 24 h, except for a 1.27-fold increase with 5 μM. After 48 hours of treatment, Ator downregulated miR-107 in a dose-dependent manner. However, the incubation of cells with 10 μM of Ator for 24 hours resulted in an increase in PANK1 levels. This effect was higher after 48 hours of incubation and became significant also with 5 μM of Ator. On the other hand, Feno downregulated miRNA-107 but upregulated PANK1 at all analyzed dosages. These results suggest that miRNA-107 levels are regulated by both Ator and Feno and that the transcription factors SREBP and PPARα may be involved in the regulation of the miRNA-107 transcription. However, the observed discordance between the response of miRNA-107 and PANK1 also suggests that the miRNA-107 regulation by both transcription factors may be independent of PANK1 regulation.
Figure 2.
Response of miRNA-107 (A) and its hosting gene, PANK1 (B) to Atorvastatin (Ator) or Fenofibrate (Feno). Differentiated Caco-2 cells were treated with the represented doses of Ator and Feno for 24 (Ator) and 48 (Ator and Feno) hours and the expression levels of miRNA-107 and PANK1 were analyzed by RT-qPCR. Data are the mean of three independent experiments, each one performed in triplicate ± SEM. * p<0.05; ** p<0.001.
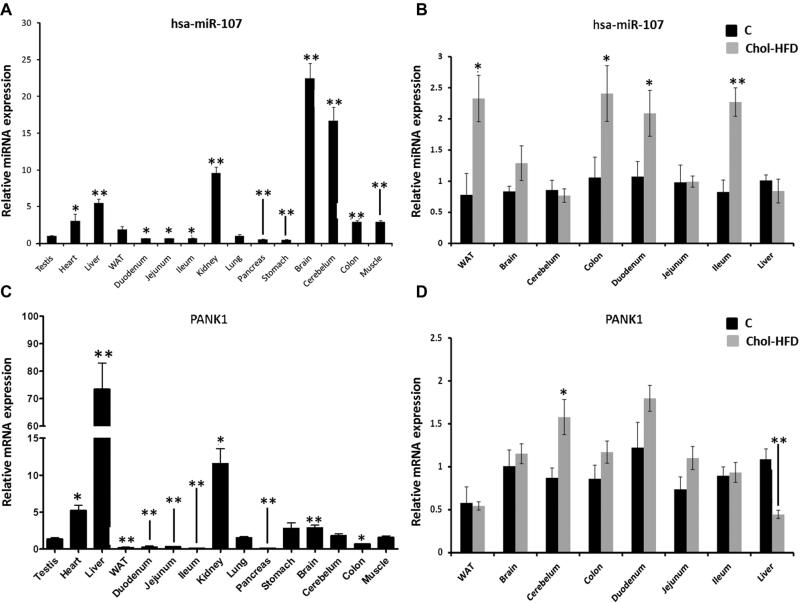
miRNA-107 was widely expressed in mouse tissues, and was particularly abundant in brain, cerebellum and kidney (Figure 3A). Unlike miR-107, PANK1 was particularly relevant in liver and its expression in brain and cerebellum was lower (Figure 3C). These results highlight the discordant expression of miRNA-107 and its hosting gene. Surprisingly, although miRNA-107 level was low in metabolic tissues such as White Adipose Tissue (WAT), intestine (duodenum, jejunum and ileum) or colon, it increased in theses tissues in mice fed a Chol-HFD, whereas remained unchanged in tissues with the highest expression levels like brain, cerebellum and liver (Figure 3B). Conversely, PANK1 was not significantly upregulated in the WAT, colon and duodenum, whereas it was in the cerebellum (Figure 3D).
Figure 3.
Analyses of miRNA-107 and PANK1 levels in mice tissues and in response to a Chol-HFD. miRNA-enriched RNA was extracted from tissues of 8-weeks old mice fed a chow or a Chol-HFD diet (n=5 per group) for 16 weeks and levels of miRNA-107 and PANK1 were measured by RT-qPCR. (A) RT-qPCR of miRNA-107 in tissues of control mice. (B) Comparison of miRNA-107 levels in tissues of mice fed a chow (black bars) or a Chol-HFD (grey bars). (C) RT-qPCR of PANK in tissues of control mice. (D) Comparison of PANK1 levels in tissues of mice fed a chow (black bars) or a Chol-HFD (grey bars). Data are the mean of 5 samples per tissue and group and two technical replicates ± SEM. * p<0.05; ** p<0.001.
The different expression pattern of miR-107 and PANK1 in mice tissues and the different response to Ator, Feno and the Chol-HFD support the previously suggested hypothesis of the regulation of miR-107 independently of PANK1 through its own promoter. Corcoran et al reported a putative miR-107 promoter located in the large intron 1 of the PANK1 gene [44] (Supporting Information Figure S1) 30.26 Kb upstream of the pre-miRNA-107 sequence (Figure 4). We aimed to check if the response of miR-107 to Ator and Feno could be mediated by the binding of SREBP and PPARs transcription factors to the miR-107 promoter. For this purpose, we carried out an in silico analysis of a region of 2Kb upstream to the predicted Transcription Start Site (TSS) to search for predicted binding sites for these transcription factors and others involved in metabolic homeostasis (Figure 4). This analysis predicted the presence of binding sites for transcription factors such as Myogenin, C/EBP, c-Fos/c-Jun, SP1, YY1, CREBP, PPARA and SREBP. These transcription factors have well-known functions in lipid metabolism and adipose tissue function, among others. Thus, the presence of theses binding sites in the putative miR-107 promoter contributes to support the hypothesis that miRNA-107 is a fundamental miRNA regulated by key nuclear receptors for energetic homeostasis.
Figure 4.
TESS and TFSearch analyses of a 2 Kb region of the predicted promoter of miRNA-107 according to Corcoran et al. The figure shows the predicted Transcription Start Site (TSS, *). The pre-miRNA-107 is underlined and the predicted length of the pri-miRNA-107 is 30260 bp. The metabolically relevant Transcription Factors Binding Sites (TFBS) are underlined. Continuous line represents TBFS in the (+) strand, whereas dashed line represents TFBS in the (−) strand.
miRNA-107 targets CLOCK gene
To gain insight into the function of miRNA-107, we analyzed its potential gene targets using different miRNA target prediction algorithms (see Materials and Methods) with stringent prediction and selection criterion (Supporting Information Table S3). We found 2805 targets. Next, we performed a functional analysis of the confidently predicted targets to identify the KEGG pathways mainly represented by these targets (Supporting Information Table S4). We found 111 KEGG pathways significantly enriched at a FDR <0.05. Among them, we found some involved in energy homeostasis and CVD: Insulin signaling, Adipocytokine signaling, MAPK signaling, T2D, Phosphatidylinositol signaling system, Biosynthesis of unsaturated fatty acids, VEGF signaling pathway (Supporting Information Tables S3 and S4). We also found significantly enriched several genes involved in Circadian rhythm, such as CLOCK, PER3 and CRY2 (Supporting Information Tables S2 and S4).
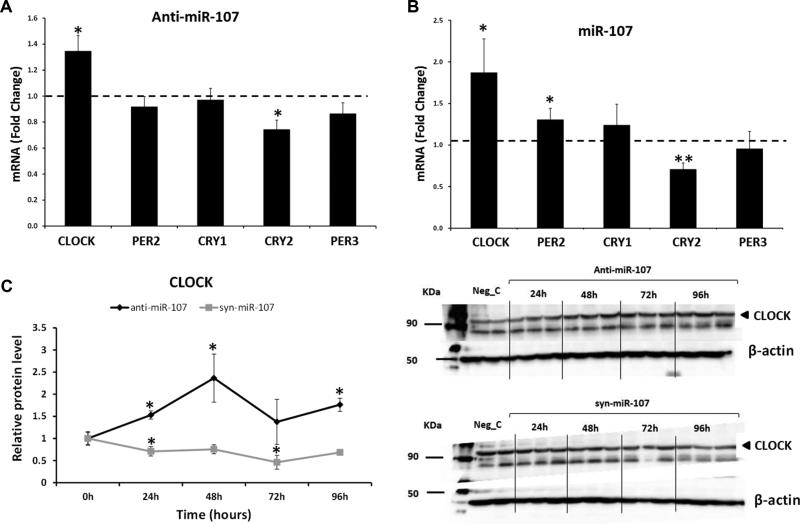
Since circadian system has been implicated in the maintenance of the metabolic homeostasis [45, 46], we focused on the effect of miRNA-107 overexpression or inhibition on the CLOCK-related genes expression (Figure 5). For this purpose, we inhibited or overexpressed miRNA-107 in Caco-2 cells and analyzed the expression of circadian CLOCK genes: CLOCK, PER2, PER3, CRY1, and CRY2. First, we assessed the effectiveness of the inhibition or overexpression of miRNA-107. (Supporting Information Figure S3). The inhibition of miRNA-107 resulted in a significant increase in the expression of CLOCK and a significant decrease in the expression of CRY2 (Figure 5A). Surprisingly, the transfection of the mimic syn-miRNA-107 also increased CLOCK levels (Figure 5B), as well as PER2 levels; whereas decreased CRY2 levels. As a positive control, we analyzed the effect of miRNA-107 inhibition and overexpression on DICER1, a validated miRNA-107 target, and we found the same effect observed with CLOCK (Supporting Information Figure S4). Thus,both, overexpression and inhibition of miR-107 upregulated CLOCK mRNA levels. This could be the result of the cellular response to the CLOCK protein deficiency provoked by miR-107 overexpression (we observed a decrease in CLOCK protein levels with the miR-107 mimic). Providing that miR-107 impinge the translation of the CLOCK mRNA but it does not lead to its degradation, the resulting deficiency in CLOCK protein could trigger a cellular response of increasing CLOCK gene transcription. This response should result in an accumulation of CLOCK mRNA molecules. On the other hand, it has been shown that the inhibition machinery associated to miRNAs could result in the accumulation of the targets in P bodies[47].
Figure 5.
Effect of miRNA-107 on CLOCK mRNA and protein levels. Caco-2 cells were transfected with anti-miRNA-107 (50 nM), syn-miRNA-107 (5 nM), anti-miRNA-C (50 nM) or MRA-1001_miRHuman_17.xl syn-miRNA-C (5 nM) for 48 hours. (A) RT-qPCR of CLOCK, CRY1, CRY2, PER2 and PER3 after miRNA-107 inhibition. (B) RT-qPCR of CLOCK, CRY1, CRY2, PER2 and PER3 after miRNA-107 overexpression. Data are the mean of three independent experiments, each one performed in triplicate ± SEM (C) Analyses of CLOCK protein levels by Western Blot. Data are the mean ± SEM of ≥ 2 experiments performed in triplicate. * p<0.05; ** p<0.001.
Next, we analyzed the effect of miRNA-107 on CLOCK protein levels (Figure 5C). As expected, CLOCK levels were increased by the miRNA-107 inhibition, peaking at 48 hours; whereas miRNA-107 overexpression decreased CLOCK protein levels in a time-dependent manner. These results suggest that the circadian system, especially the master regulator CLOCK, is actively targeted by miRNA-107.
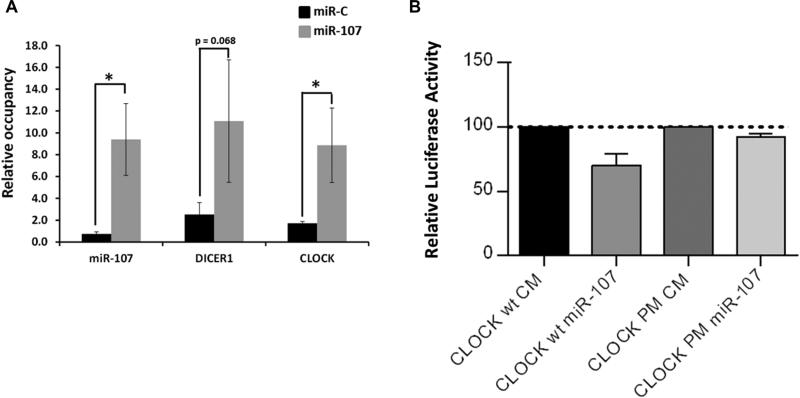
To further confirm the ability of miR-107 to bind the CLOCK gene, we carried out RIP assays with anti-AGO2 antibodies as described in Materials and Methods (Figure 6A) and luciferase reporter assays of the CLOCK 3’UTR region (Figure 6B). As expected, the levels of miRNA-107 bound to AGO2 in cells transfected with the mimic were higher than those of mock-transfected cells (12.64-fold; p<0.05). CLOCK mRNA was most concentrated in AGO2-miRNA-107 complexes in cells overexpressing miRNA-107 than in cells transfected with the scramble (5.2-fold; p<0.05). We observed a similar result with DICER1, a validated miRNA-107 target (Figure 6A). Moreover, results from luciferase reporter assay showed that miR-107 decreased the luciferase activity in the vector containing the CLOCK 3’UTR. Mutation of the miR-107 target sites relieved miR-107 repression of CLOCK (Figure 6B). These results suggest that miRNA-107 directly bind to and regulates CLOCK.
Figure 6.
Direct binding of miRNA-107 to CLOCK 3’UTR. A. Ribonucleoprotein Ago-2 inmunoprecipitation analysis. Caco-2 cells were transfected with syn-miRNA-107 or syn-miRNA-C (5 nM) for 24 hours. After a cross-linking procedure, miRNA-107-RISC complexes were inmunoprecipitated with anti-AGO2 antibody and the levels of miRNA-107, DICER1 and CLOCK were measured by RT-qPCR. B. Luciferase reporter assay. COS-7 were co-transfected with a psiCheck 2 plasmid containing the CLOCK 3’UTR or an empty vector and the syn-miR-107. The luciferase activity was measured after 48 hours and normalized to the Renilla activity. Data are the mean of three independent experiments, each one performed in triplicate ± SEM * p<0.05; ** p<0.001. CM means “Control Mimic” and PM means “Point Mutation” and refers to the vector containing the CLOCK 3’UTR with the mutated binding site.
miRNA-107 targeting of CLOCK disrupts the circadian system of cultured cells
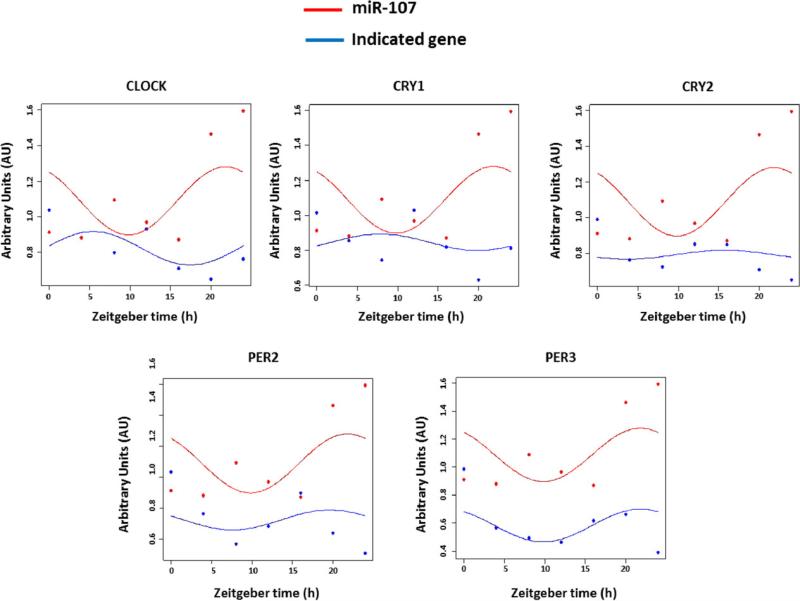
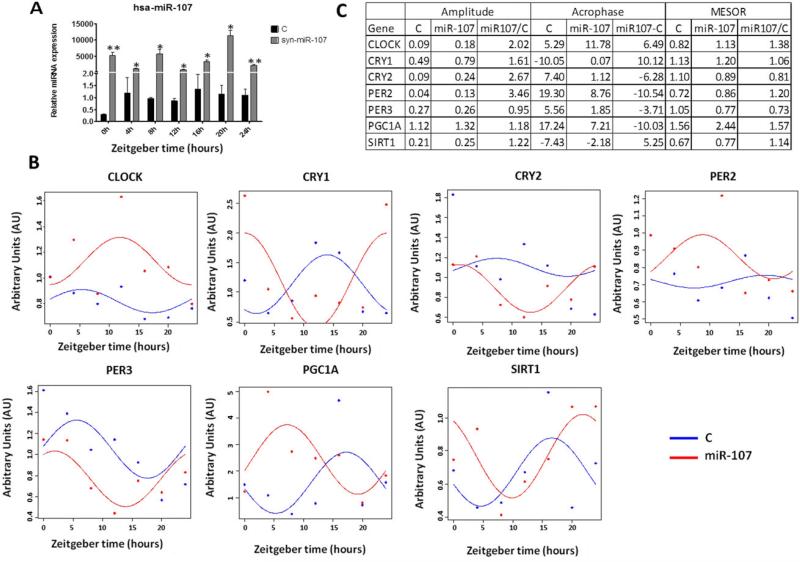
To further analyze the physiological consequences of the miRNA-107 targeting of CLOCK, we studied the circadian expression pattern of miRNA-107 and of CLOCK and CLOCK-related genes in synchronized Caco-2 cells (Figure 7). Firstly, we observed that, in synchronized untreated cells, miR-107 levels oscillate following a circadian rhythm with amplitude of 12 hours. Secondly, the circadian expression pattern of miR-107 and CLOCK is opposite with an approximate difference of 10 ± 2 hours in their acrophases. We also observed an opposite oscillation with cryptochromes, but a parallel oscillation with period genes (Figure 7). We further analyzed the effect of the miR-107 overexpression on the circadian expression of those genes. Additionally, we analyzed the expression of two metabolic genes known to be under circadian control: PGC1A and SIRT1 (Figure 8). The overexpression of miRNA-107 was confirmed in all samples (Figure 8A). As shown in figure 8B, miRNA-107 modulated the circadian rhythms of CLOCK and CLOCK-related genes, as well as other metabolic genes. The most significant finding was the modification of the acrophase that shifted among 3 and 10 hours. The amplitude was also increased in all circadian genes. The mesor of CRY2 and PER3 was lower in miR-107 treated cells, consistent with the decrease in their mRNA levels (Figure 8B and C). On the other hand, CLOCK mesor increased by miR-107 overexpression, consistent with the observed increase in its mRNA levels (Figure 8B and C). Additionally, the circadian expression of PGC1A and SIRT1 was found to be shifted by 10 and 5 hours, respectively (Figure 8C). Together, these results suggest that miR-107 regulate circadian rhythm in culture cells by targeting CLOCK genes.
Figure 7.
Circadian oscillation of miR-107 and the core CLOCK genes. Caco-2 cells were synchronized by a serum shock. Then, samples were harvested every 4 hours from a period of 24 hours. The expression levels of miR-107 and the core-CLOCK genes were analysed by RT-PCR. Data are represented as Arbitrary Units (AU) in every zeitgeber time. The represented graphs correspond to a representative experiment of a total of three, each one performed in triplicate.
Figure 8.
miRNA-107 modulates the circadian system. Caco-2 cells were synchronized by a serum shock and transfected with syn-miRNA-107 or scramble for 24 h. After that, the circadian expression of CLOCK and CLOCK-related genes were measured every 4 h by RT-qPCR. Results were analysed using the cosinor method and the parameters compared were mesor, amplitude and acrophase. (A) miR-107 levels (B) Circadian rhythms of CLOCK, PER2, PER3, CRY1, CRY2, PGC1A and SIRT1 as measured by RT-qPCR. Data are represented as Arbitrary Units (AU) in every zeitgeber time. Data are the mean ± SEM and are representative of ≥ 3 experiments performed in triplicate. * p<0.05; ** p<0.001. (C) Table showing amplitude, achropase and mesor of cells treated with miR-107 or control.
DISCUSSION
Here, we describe the effect of dietary lipids on the miRNA profile of an in vitro model of human enterocytes. We found that CLA and DHA were more effective than cholesterol modifying miRNA profile in Caco-2 cells. In fact, only two miRNAs were significantly modified by cholesterol. Another study identified a subset of 21 miRNAs differentially expressed by cholesterol [26], but they compared two opposite conditions: cholesterol loading vs cholesterol depletion in a different cellular model (THP-1 macrophages) and used LDL to load cells with cholesterol.
The potential of CLA and DHA to function as miRNA modulators have been previously studied [38, 39, 48, 49]. However, specific target miRNAs and the mechanisms involved in such modulation remain largely unknown. Faragó et al analysed the expression of 112 miRNAs in response to n-3 PUFAs in glioblastoma cells. Among other results, they found a DHA-mediated increase of miR-183 and miR-107 in GBM2 and GBM5 cells, respectively [49]. We found a similar response in these miRNAs in our model of Caco-2 cells exposed to DHA. Moreover, Davidson et al observed an increase in miR-107 levels in colonic tumours of rats fed a fish oil-enriched diet [38]. Concerning CLA, we found 12 miRNAs that were differentially expressed in our model after CLA treatment. Among them, we found an increase in miR-107. This effect of CLA on miR-107 was also found by Parra et al in the adipose tissue of mice fed a diet supplemented with different doses of CLA [39]. Together, these results highlight the potential of PUFAs to function as miRNA modulators and suggest that miRNAs modulation could be a novel mechanism that partially explains the beneficial effects of PUFAs. Among studied lipids, DHA has been proved to be the most effective one, suggesting that DHA could be instrumental for miRNA modulation related to metabolic diseases. Besides, our results, and that of the others [38, 39, 49], point to miR-107 as a novel PUFAs molecular target.
miR-107 is highly conserved among vertebrates and strongly influences relevant metabolic processes [50, 51]. miR-107 has been reported to regulate insulin sensitivity [36, 52] and to be involved in Non-alcoholic Fatty Liver Disease (NAFLD) and the development of fatty liver under ER stress [53]. It has also been reported that intestinal microbiota may be involved in the regulation of intestinal miR-107 levels [54]. However, the precise mechanisms directly regulating miR-107 levels under stress conditions and the gene targets involved in the biological effects of miR-107 need to be fully elucidated. Here, we report that bioactive lipids also contribute to the intestinal modulation of miR-107. miR-107 belongs to a cluster of miRNAs that also contains miR-103, miR-15a/b, miR-16, miR-195, miR-497, miR-503, miR-424 and miR-646 [55]. All of them share the same seed sequence and, therefore, share most targets. However, they are in different genetic context and, apart from miR-107, any other of these miRNAs was modified by dietary lipids in our conditions as assessed by microarray and by RT-qPCR (data not shown).
Although miR-107 levels in metabolic tissues such as liver, intestine or WAT were lower, we found that a Chol-HFD significantly increased the miR-107 expression in the intestine and WAT and a non-significant decrease in the liver. It has been previously reported an upregulation of miR-107 during differentiation of the adipose tissue [56], as well as in the liver of ob/ob mice [52]. Taken together, our results confirm the dietary regulation of miR-107 in metabolic tissues such as intestine, liver and WAT. On the contrary, the expression levels of miR-107 in the brain and cerebellum were not modified by the chol-HFD. The response of miR-107 to Chol-HFD was different between metabolic and neuronal tissues. This suggests a tissue-specific modulation of miR-107 involving its dietary-dependent regulation in metabolic tissues and its dietary-independent regulation in neuronal tissues.
miR-107 is located within intron 5 of PANK1 gene and this location is highly conserved among mammals. PANK1 is the rate-limiting enzyme in the biosynthesis of Coenzyme A, an essential cofactor for energetic metabolic pathways included sterol, bile and fatty acid biosynthesis as well as fatty acid oxidation [57]. PANK1 is regulated by PPARα [58] and is required to support the metabolic transition from the fed to the fasted state [59]. Although our previous results and that of the others point to an independent regulation of the miR-107 and PANK1 levels, we aimed to test if, likewise PANK1, miR-107 expression was modulated by PPARα by treating cells with fenofibrate, a synthetic PPARα ligand with hypolipidemic effect. In addition, we tested if miR-107 levels were modulated by SREBP transcription factors by treating cells with atorvastatin, a lipid-lowering drug known to activate the SREBP pathway. We found an increase in miR-107 levels after 24 hours treatment with 5 μM of Ator and with all assayed doses of Feno. However, the response of miR-107 was not correlated with that of PANK1 in times and doses. This discordant expression of miR-107 and its host gene was also observed in mice tissues. This discordant expression is supported by data showed by Poster et al [60]. Our results showing the discordant expression of PANK1 and miRNA-107 in mice tissues and the differential response to Ator, Feno or a Chol-HFD support the hypothesis that miRNA-107 expression occurs independently of its host gene transcription and is regulated by its own promoter. Together, these results point to miR-107 as a novel target of hypolipidemics pharmacological agents and suggest that both, dietary and pharmacological, modulation of miR-107 occurs through its own promoter and independently of its host gene.
We must bear in mind that miR-107 was upregulated by cholesterol. Thus, we expected than Ator, which induces a decrease in intracellular levels of cholesterol, would reduce its expression. Although we observed a weak upregulation of miR-107 after 24 hours of treatment with Ator, the same treatments resulted in a decreased in miR-107 after 48h. The cholesterol-mediated upregulation of miR-107 was observed after 48 hours of treatment.
Two putative miR-107 promoters have been reported. Monteys et al identified a putative intronic promoter for miR-107 with a TSS located 1058 bp upstream the pre-miRNA sequence [61]. On the other hand, Corcoran et al, identified a TSS located 40030 bp upstream the pre-miRNA sequence. This putative promoter is located in the large intron 1 of the PANK1 gene and results in a pri-miR-107 sequence of 40 kb. Both authors used different approaches. Whereas Monteys et al restricted their search to the 5’flanking region of the pre-miRNA sequence and searched for common promoter features using informatics algorithms, Corcoran et al carried out a Pol II inmunoprecipitation following by a microarray including 50 Kb upstream the pre-miRNA sequence. We searched for transcription factor binding sites in the promoter sequence reported by Corcoran et al who used an experimental approach to detect this putative miR-107 promoter. We found up to 9 putative binding sites for C/EBPα and one putative binding site to PPARα. These sites may explain the increase in the miR-107 levels observed during adipocyte differentiation [56]. Additionally, the binding site for PPARα may be responsible for the response we observed to Feno treatment. A SREBP putative binding site was predicted near the TSS. This binding site may be responsible for the miR-107 upregulation after 24 hours of treatment with Ator. The functionality of this SREBP binding site needs to be experimentally validated. However, the proximity of this binding site to the TSS, as well as the presence of close SP-1 and NF-Y binding sites support the hypothesis of the SREBP modulation by miR-107 levels, since this structure is characteristic of SREBP modulated promoters [62]. Additionally, we found binding sites for other metabolically important transcription factors such as CREBP, myogenin, c-Fos/c-Jun and GATA3.
Circadian rhythm was a pathway significantly overrepresented among miR-107 putative targets. There is increasing evidence suggesting that the synchrony between circadian rhythm and metabolic processes is crucial for energy balance and body weight control [63-65]. It has been reported that circadian rhythmicity is more robust in conditions of food restriction, what leads to a reduction of inflammatory and metabolic diseases markers [66]. Conversely, HFD alters the cycling expression of circadian CLOCK genes and oscillating nuclear receptors resulting in an increased susceptibility to metabolic disorders [67, 68]. Additionally, the timing of food intake is a determinant factor for weight gain [69] and can predict weight loss effectiveness in weight-loss programs [70]. Circadian rhythmicity is supported by transcriptional feedback loops that perpetuate oscillating gene expression. However, it has been recently shown that miRNAs also participate in timekeeping, providing further complexity to the CLOCK machinery [71, 72]. In this work, we describe CLOCK as a miR-107 target. We observed a modification in CLOCK protein levels that reflected the intracellular levels of miR-107 and demonstrated the miR-107 directly binds to the 3’UTR of CLOCK.
We observed that miR-107 levels oscillate following a circadian rhythm in synchronized Caco-2 cells and that the circadian expression pattern of miR-107 and CLOCK is opposite. We also observed an opposite oscillation with cryptochrome genes, but a parallel oscillation with period genes. Different groups have studied the circadian modulation of miRNAs and the circadian control mediated by miRNAs [71, 73]. The paper of Na et al described miRNA-mRNA target pairs with opposite circadian expression using microarray. They additionally showed that oscillating miRNAs were more commonly located in introns. Our results show that miR-107-CLOCK constitutes a pair of circadian modulators. Moreover, we showed that, when miR-107 was overexpressed in synchronized Caco-2 cells, circadian expression of core CLOCK genes and metabolic oscillating genes was altered. Together, these results suggest that the modulation of the circadian system may be a molecular mechanism by which miR-107 alters metabolic homeostasis.
In summary, we have shown the potential of DHA as modulator of metabolic-related miRNAs. In particular, miR-107 is a lipid-modulated miRNA that may play a role in metabolic diseases. The effect of miR-107 on lipid metabolism could be partially mediated by the modification of the circadian system. Circadian system has emerged recently as an important player in metabolic diseases. Together, the regulation of the circadian system by lipid-modified miRNAs is postulated as an important molecular mechanism underlying the effect of dietary lipids, specifically DHA, on the risk of metabolic diseases. However, our findings must be further extended to investigate the role of other miRNAs in the control of the circadian system and the effect of miRNA-mediated circadian disruption on metabolic diseases in animal models.
Supplementary Material
ACKOWLEDGEMENTS
We thank to Jesús Herranz for his assistance with the cosinor analysis of the circadian rhythm. We also thank to Roberto Martín for helping with the graphical representation of the microarray data. This work was supported by the direct finding of IMDEA-Food Institute through Comunidad de Madrid and European Union Structural Funds. Additional funding was provided by the Instituto de Salud Carlos III [FIS, PI11/00315] and the Sociedad Española de Arteriosclerosis to AD; the National Institutes of Health [R01HL107953] to CF-H; and the US Department of Agriculture Research Service [53-K06-5-10, 58e1950-9-001] and Fundación Salud 2000 – Premio Merck-Serono de Investigación 2012 to JMO.
List of abbreviations (in order of appearance)
- DHA
Docosahexanoic acid
- CLA
Conjugated Linoleic Acid
- T2D
Type 2 diabetes
- CVD
Cardiovascular Disease
- MetS
Metabolic Syndrome
- FBS
Fetal Bovine Serum
- Chol
Cholesterol
- Lyso-PC
Lyso-phosphatidilcholine
- Tau
Sodium Taurocholate
- MIAME
Minimal Information About a Microarray Experiment Guidelines
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- RT-qPCR
Real-Time quantitative PCR
- TESS
Transcription Element Search System
- TFSearch
Transcription Factor Search
- Ator
Atorvastatin
- Feno
Fenofibrate
- RIP
Ribonucleoprotein Ago-2 inmunoprecipitation
- 3′-UTR
3’-Untranslated Region
- RLA
Relative luciferase activity
- ANOVA
Analysis Of Variance
- WAT
White Adipose Tissue
- NAFLD
Non-alcoholic Fatty Liver Disease
- FC
Fold Change
- WB
Western Blott
- RT-qPCR
Room Temperature
Footnotes
AUTHOR'S CONTRIBUTIONS
LD-R, AD and JMO conceived the idea and designed the experiments. LD-R, MK, VM, JFA and BG performed the experiments. LD-R carried out the data analysis. LD-R and JMO participated in manuscript writing. CF-H, JM-B, VK and AD assisted in manuscript revision. All authors authorized the final manuscript.
CONFLICT OF INTEREST
The authors have declared no conflict of interest
REFERENCES
- 1.Mozaffarian D, Wu JH. Omega-3 fatty acids and cardiovascular disease: effects on risk factors, molecular pathways, and clinical events. J Am Coll Cardiol. 2011;58:2047–2067. doi: 10.1016/j.jacc.2011.06.063. [DOI] [PubMed] [Google Scholar]
- 2.Delgado-Lista J, Perez-Martinez P, Lopez-Miranda J, Perez-Jimenez F. Long chain omega-3 fatty acids and cardiovascular disease: a systematic review. The British journal of nutrition. 2012;107(Suppl 2):S201–213. doi: 10.1017/S0007114512001596. [DOI] [PubMed] [Google Scholar]
- 3.Harris WS, Bulchandani D. Why do omega-3 fatty acids lower serum triglycerides? Current opinion in lipidology. 2006;17:387–393. doi: 10.1097/01.mol.0000236363.63840.16. [DOI] [PubMed] [Google Scholar]
- 4.Jump DB. Fatty acid regulation of hepatic lipid metabolism. Current opinion in clinical nutrition and metabolic care. 2011;14:115–120. doi: 10.1097/MCO.0b013e328342991c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Micallef MA, Garg ML. Anti-inflammatory and cardioprotective effects of n-3 polyunsaturated fatty acids and plant sterols in hyperlipidemic individuals. Atherosclerosis. 2009;204:476–482. doi: 10.1016/j.atherosclerosis.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 6.Serhan CN. Novel lipid mediators and resolution mechanisms in acute inflammation: to resolve or not? The American journal of pathology. 2010;177:1576–1591. doi: 10.2353/ajpath.2010.100322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dangardt F, Osika W, Chen Y, Nilsson U, et al. Omega-3 fatty acid supplementation improves vascular function and reduces inflammation in obese adolescents. Atherosclerosis. 2010;212:580–585. doi: 10.1016/j.atherosclerosis.2010.06.046. [DOI] [PubMed] [Google Scholar]
- 8.Robinson JG, Stone NJ. Antiatherosclerotic and antithrombotic effects of omega-3 fatty acids. The American journal of cardiology. 2006;98:39i–49i. doi: 10.1016/j.amjcard.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 9.Brouwer IA, Raitt MH, Dullemeijer C, Kraemer DF, et al. Effect of fish oil on ventricular tachyarrhythmia in three studies in patients with implantable cardioverter defibrillators. European heart journal. 2009;30:820–826. doi: 10.1093/eurheartj/ehp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S, Sutherland F, Rosso R, Teh AW, et al. Effects of chronic omega-3 polyunsaturated fatty acid supplementation on human atrial electrophysiology. Heart rhythm : the official journal of the Heart Rhythm Society. 2011;8:562–568. doi: 10.1016/j.hrthm.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 11.Oleszczuk J, Oleszczuk L, Siwicki AK, Skopinska-Skopinska E. Biological effects of conjugated linoleic acids supplementation. Pol J Vet Sci. 2012;15:403–408. doi: 10.2478/v10181-012-0063-x. [DOI] [PubMed] [Google Scholar]
- 12.Gebauer SK, Chardigny JM, Jakobsen MU, Lamarche B, et al. Effects of ruminant trans fatty acids on cardiovascular disease and cancer: a comprehensive review of epidemiological, clinical, and mechanistic studies. Adv Nutr. 2011;2:332–354. doi: 10.3945/an.111.000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Y, Correll PH, Vanden Heuvel JP. Conjugated linoleic acid decreases production of pro-inflammatory products in macrophages: evidence for a PPAR gamma-dependent mechanism. Biochimica et biophysica acta. 2002;1581:89–99. doi: 10.1016/s1388-1981(02)00126-9. [DOI] [PubMed] [Google Scholar]
- 14.Valeille K, Gripois D, Blouquit MF, Souidi M, et al. Lipid atherogenic risk markers can be more favourably influenced by the cis-9,trans-11-octadecadienoate isomer than a conjugated linoleic acid mixture or fish oil in hamsters. The British journal of nutrition. 2004;91:191–199. doi: 10.1079/BJN20031057. [DOI] [PubMed] [Google Scholar]
- 15.Ringseis R, Wen G, Saal D, Eder K. Conjugated linoleic acid isomers reduce cholesterol accumulation in acetylated LDL-induced mouse RAW264.7 macrophage-derived foam cells. Lipids. 2008;43:913–923. doi: 10.1007/s11745-008-3226-x. [DOI] [PubMed] [Google Scholar]
- 16.Iwakiri Y, Sampson DA, Allen KG. Suppression of cyclooxygenase-2 and inducible nitric oxide synthase expression by conjugated linoleic acid in murine macrophages. Prostaglandins, leukotrienes, and essential fatty acids. 2002;67:435–443. doi: 10.1054/plef.2002.0454. [DOI] [PubMed] [Google Scholar]
- 17.Stachowska E, Dolegowska B, Dziedziejko V, Rybicka M, et al. Prostaglandin E2 (PGE2) and thromboxane A2 (TXA2) synthesis is regulated by conjugated linoleic acids (CLA) in human macrophages. Journal of physiology and pharmacology : an official journal of the Polish Physiological Society. 2009;60:77–85. [PubMed] [Google Scholar]
- 18.Sonnino S, Prinetti A. Membrane domains and the “lipid raft” concept. Curr Med Chem. 2013;20:4–21. [PubMed] [Google Scholar]
- 19.Lasunción MA. El colesterol: biosíntesis, acciones y alteraciones. Alim. Nutri. 2006;13:97–120. [Google Scholar]
- 20.Fernandez C, Lobo Md Mdel V, Gomez-Coronado D, Lasuncion MA. Cholesterol is essential for mitosis progression and its deficiency induces polyploid cell formation. Exp Cell Res. 2004;300:109–120. doi: 10.1016/j.yexcr.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez C, Martin M, Gomez-Coronado D, Lasuncion MA. Effects of distal cholesterol biosynthesis inhibitors on cell proliferation and cell cycle progression. J Lipid Res. 2005;46:920–929. doi: 10.1194/jlr.M400407-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Suarez Y, Fernandez C, Ledo B, Martin M, et al. Sterol stringency of proliferation and cell cycle progression in human cells. Biochim Biophys Acta. 2005;1734:203–213. doi: 10.1016/j.bbalip.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Thum T, Mayr M. Review focus on the role of microRNA in cardiovascular biology and disease. Cardiovasc Res. 2012;93:543–544. doi: 10.1093/cvr/cvs085. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez-Hernando C, Ramirez CM, Goedeke L, Suarez Y. MicroRNAs in metabolic disease. Arteriosclerosis, thrombosis, and vascular biology. 2013;33:178–185. doi: 10.1161/ATVBAHA.112.300144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rayner KJ, Suarez Y, Davalos A, Parathath S, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Yoon H, Ramirez CM, Lee SM, et al. MiR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Experimental neurology. 2012;235:476–483. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramirez CM, Davalos A, Goedeke L, Salerno AG, et al. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arteriosclerosis, thrombosis, and vascular biology. 2011;31:2707–2714. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez CM, Rotllan N, Vlassov AV, Davalos A, et al. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circulation research. 2013;112:1592–1601. doi: 10.1161/CIRCRESAHA.112.300626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esau C, Davis S, Murray SF, Yu XX, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonauer A, Carmona G, Iwasaki M, Mione M, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 33.Danielson LS, Park DS, Rotllan N, Chamorro-Jorganes A, et al. Cardiovascular dysregulation of miR-17-92 causes a lethal hypertrophic cardiomyopathy and arrhythmogenesis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:1460–1467. doi: 10.1096/fj.12-221994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cordes KR, Sheehy NT, White MP, Berry EC, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrera BM, Lockstone HE, Taylor JM, Ria M, et al. Global microRNA expression profiles in insulin target tissues in a spontaneous rat model of type 2 diabetes. Diabetologia. 2010;53:1099–1109. doi: 10.1007/s00125-010-1667-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trajkovski M, Hausser J, Soutschek J, Bhat B, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 37.Davis CD, Ross SA. Evidence for dietary regulation of microRNA expression in cancer cells. Nutr Rev. 2008;66:477–482. doi: 10.1111/j.1753-4887.2008.00080.x. [DOI] [PubMed] [Google Scholar]
- 38.Davidson LA, Wang N, Shah MS, Lupton JR, et al. n-3 Polyunsaturated fatty acids modulate carcinogen-directed non-coding microRNA signatures in rat colon. Carcinogenesis. 2009;30:2077–2084. doi: 10.1093/carcin/bgp245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parra P, Serra F, Palou A. Expression of adipose microRNAs is sensitive to dietary conjugated linoleic acid treatment in mice. PLoS One. 2010;5:e13005. doi: 10.1371/journal.pone.0013005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gil-Zamorano J, Martin R, Daimiel L, Richardson K, et al. Docosahexaenoic acid modulates the enterocyte Caco-2 cell expression of microRNAs involved in lipid metabolism. J Nutr. 2014;144:575–585. doi: 10.3945/jn.113.189050. [DOI] [PubMed] [Google Scholar]
- 41.Richardson K, Nettleton JA, Rotllan N, Tanaka T, et al. Gain-of-function lipoprotein lipase variant rs13702 modulates lipid traits through disruption of a microRNA-410 seed site. American journal of human genetics. 2013;92:5–14. doi: 10.1016/j.ajhg.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levy E, Mehran M, Seidman E. Caco-2 cells as a model for intestinal lipoprotein synthesis and secretion. Faseb J. 1995;9:626–635. doi: 10.1096/fasebj.9.8.7768354. [DOI] [PubMed] [Google Scholar]
- 43.Field FJ, Albright E, Mathur SN. Regulation of cholesterol esterification by micellar cholesterol in CaCo-2 cells. J Lipid Res. 1987;28:1057–1066. [PubMed] [Google Scholar]
- 44.Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, et al. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One. 2009;4:e5279. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oike H, Oishi K, Kobori M. Nutrients, Clock Genes, and Chrononutrition. Current nutrition reports. 2014;3:204–212. doi: 10.1007/s13668-014-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sancar G, Brunner M. Circadian clocks and energy metabolism. Cell Mol Life Sci. 2014;71:2667–2680. doi: 10.1007/s00018-014-1574-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 48.Mandal CC, Ghosh-Choudhury T, Dey N, Choudhury GG, Ghosh-Choudhury N. miR-21 is targeted by omega-3 polyunsaturated fatty acid to regulate breast tumor CSF-1 expression. Carcinogenesis. 2012;33:1897–1908. doi: 10.1093/carcin/bgs198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farago N, Feher LZ, Kitajka K, Das UN, Puskas LG. MicroRNA profile of polyunsaturated fatty acid treated glioma cells reveal apoptosis-specific expression changes. Lipids Health Dis. 2011;10:173. doi: 10.1186/1476-511X-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab. 2007;91:209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foley NH, O'Neill LA. miR-107: a toll-like receptor-regulated miRNA dysregulated in obesity and type II diabetes. J Leukoc Biol. 2012;92:521–527. doi: 10.1189/jlb.0312160. [DOI] [PubMed] [Google Scholar]
- 52.Li S, Chen X, Zhang H, Liang X, et al. Differential expression of microRNAs in mouse liver under aberrant energy metabolic status. J Lipid Res. 2009;50:1756–1765. doi: 10.1194/jlr.M800509-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhatia H, Verma G, Datta M. miR-107 orchestrates ER stress induction and lipid accumulation by post-transcriptional regulation of fatty acid synthase in hepatocytes. Biochimica et biophysica acta. 2014;1839:334–343. doi: 10.1016/j.bbagrm.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 54.Xue X, Cao AT, Cao X, Yao S, et al. Downregulation of microRNA-107 in intestinal CD11c(+) myeloid cells in response to microbiota and proinflammatory cytokines increases IL-23p19 expression. European journal of immunology. 2014;44:673–682. doi: 10.1002/eji.201343717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finnerty JR, Wang WX, Hebert SS, Wilfred BR, et al. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402:491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leonardi R, Zhang YM, Rock CO, Jackowski S. Coenzyme A: back in action. Prog Lipid Res. 2005;44:125–153. doi: 10.1016/j.plipres.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 58.Ramaswamy G, Karim MA, Murti KG, Jackowski S. PPARalpha controls the intracellular coenzyme A concentration via regulation of PANK1alpha gene expression. J Lipid Res. 2004;45:17–31. doi: 10.1194/jlr.M300279-JLR200. [DOI] [PubMed] [Google Scholar]
- 59.Leonardi R, Rehg JE, Rock CO, Jackowski S. Pantothenate kinase 1 is required to support the metabolic transition from the fed to the fasted state. PLoS One. 2010;5:e11107. doi: 10.1371/journal.pone.0011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Polster BJ, Westaway SK, Nguyen TM, Yoon MY, Hayflick SJ. Discordant expression of miR-103/7 and pantothenate kinase host genes in mouse. Mol Genet Metab. 2010;101:292–295. doi: 10.1016/j.ymgme.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monteys AM, Spengler RM, Wan J, Tecedor L, et al. Structure and activity of putative intronic miRNA promoters. Rna. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daimiel LA, Fernandez-Suarez ME, Rodriguez-Acebes S, Crespo L, et al. Promoter analysis of the 3beta-hydroxysterol Delta24-reductase (DHCR24) gene: characterization of sterol regulatory element-binding protein (SREBP)-mediated activation. Bioscience reports. 2012;33:57–69. doi: 10.1042/BSR20120095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turek FW, Joshu C, Kohsaka A, Lin E, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 66.Sherman H, Frumin I, Gutman R, Chapnik N, et al. Long-term restricted feeding alters circadian expression and reduces the level of inflammatory and disease markers. Journal of cellular and molecular medicine. 2011;15:2745–2759. doi: 10.1111/j.1582-4934.2010.01160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 68.Sherman H, Genzer Y, Cohen R, Chapnik N, et al. Timed high-fat diet resets circadian metabolism and prevents obesity. Faseb J. 2012;26:3493–3502. doi: 10.1096/fj.12-208868. [DOI] [PubMed] [Google Scholar]
- 69.Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 2009;17:2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garaulet M, Gomez-Abellan P, Alburquerque-Bejar JJ, Lee YC, et al. Timing of food intake predicts weight loss effectiveness. Int J Obes (Lond) 2013;37:604–611. doi: 10.1038/ijo.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng HY, Papp JW, Varlamova O, Dziema H, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nagel R, Clijsters L, Agami R. The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. The FEBS journal. 2009;276:5447–5455. doi: 10.1111/j.1742-4658.2009.07229.x. [DOI] [PubMed] [Google Scholar]
- 73.Na YJ, Sung JH, Lee SC, Lee YJ, et al. Comprehensive analysis of microRNA-mRNA co-expression in circadian rhythm. Experimental & molecular medicine. 2009;41:638–647. doi: 10.3858/emm.2009.41.9.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.