In HLA-matched allogeneic hematopoietic stem cell transplantation (SCT), donor T cells can mediate graft-versus-leukemia/lymphoma (GvL) reactivity and graft-versus-host disease (GvHD) by recognition of minor histocompatibility antigens (MiHA).1–4 Only a minority of MiHA shows hematopoiesis-restricted expression, and donor T cells for these MiHA may induce beneficial GvL reactivity without GvHD. The number of well-characterized MiHA with therapeutic relevance based on hematopoiesis-restricted expression remains limited and only 25% and 40% of recipients transplanted with grafts from sibling and unrelated donors, respectively, are eligible for therapies targeting known hematopoietic MiHA.3,4 Thus, in order to increase the efficacy and applicability of cellular therapy for selective GvL induction, more hematopoiesis-restricted MiHA with balanced population frequencies in common HLA molecules must be identified. Here, we investigated the therapeutic significance of a MiHA encoded by ARHGDIB.5 We demonstrated hematopoiesis-restricted gene expression with the exception of intermediate mRNA expression in endothelial cells and showed that T cells recognized LB-ARHGDIB-1R presented by HLA-B*07:02 on primary leukemic cells, but not on [interferon-gamma (IFN-γ)]-treated fibroblasts and keratinocytes. To evaluate potential toxicity against endothelial cells, we tested T cell recognition of LB-ARHGDIB-1R on human umbilical vein endothelial cells (HUVEC) and found only limited reactivity under inflammatory conditions. Furthermore, we demonstrated in vivo targeting of LB-ARHGDIB-1R in eight out of ten patients who were screened for post-transplant specific T-cell responses. In one patient with relapsed lymphoma, high T-cell frequencies were induced after donor lymphocyte infusion (DLI), coinciding with long-lasting anti-lymphoma immunity without GvHD. Our data thus support the relevance of LB-ARHGDIB-1R as a therapeutic target with the potential to induce selective GvL reactivity.
We previously demonstrated that CD8 T cells specific for a MiHA (LB-ARHGDIB-1R) encoded by the ARHGDIB gene were induced in a patient with myelodysplastic syndrome who responded to DLI after HLA-matched allogeneic SCT.5 LB-ARHGDIB-1R is translated from the normal ARHGDIB transcript (NM_001175) in an alternative reading frame. Since ARHGDIB has been described to be expressed in hematopoietic cells,6,7 we investigated the therapeutic value of LB-ARHGDIB-1R to stimulate GvL reactivity after allogeneic SCT without GvHD. We first examined ARHGDIB expression by microarray gene expression analysis using Illumina HT-12 v3/4 BeadChips8 and compared gene expression between (malignant) hematopoietic and non-hematopoietic cells, which were cultured in the absence or presence of IFN-γ to mimic a state of inflammation. ARHGDIB showed strong overexpression in the majority of (malignant) hematopoietic versus (IFN-γ pre-treated) nonhematopoietic cells. The ARHGDIB expression profile was comparable to the strictly hematopoietic HMHA1 and MYO1G (Figure 1A and data not shown).
Figure 1.
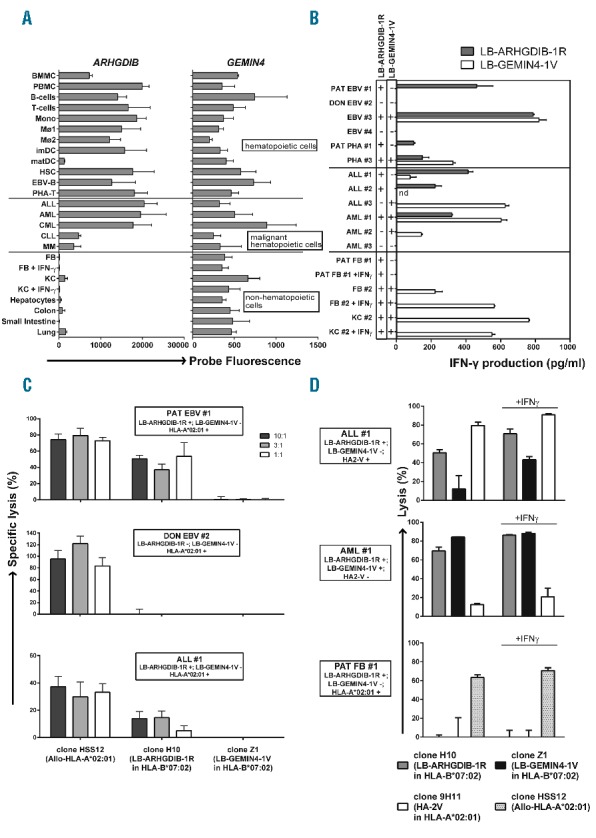
LB-ARHGDIB-1R as a target with therapeutic relevance for leukemia. (A) Expression profiles for ARHGDIB and GEMIN4 as determined by microarray gene expression analysis. GEMIN4 was selected as a representative gene for MiHA with ubiquitous expression on (non)-La by (non-)hematopoietic cells.5 The probe fluorescence, as measured on Illumina Human HT-12 v3/4 BeadChips, is indicated.8 Hematopoietic cells included bone marrow and peripheral blood mononuclear cells (BMMC and PBMC), B cells, T cells, monocytes (Mono), macrophages type I and II (MØ1 and MØ2), (im)mature DC (imDC and matDC), hematopoietic stem cells (HSC), Epstein-Barr virus-infected B (EBV-B) cells and phytohemagglutinin-stimulated T (PHA-T) cells. Malignant hematopoietic cells included acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic myeloid leukemia (CML), chronic lymphocytic leukemia (CLL) and multiple myeloma (MM). Non-hematopoietic cells included fibroblasts (FB) and keratinocytes (KC) cultured from skin biopsies in the absence or presence of IFN-γ and hepatocytes, colon and small intestine epithelial cells and lung epithelial cells. (B–D) Recognition and lysis of (non-)hematopoietic cells as mediated by T cells for LB-ARHGDIB-1R (clone H10).5 All samples were positive for HLA-B*07:02. Cell types included EBV-B and PHA-T cells from patient (PAT EBV #1 and PHA #1) or donor (DON EBV #2) origin or from third party individuals (EBV #3, EBV #4 and PHA #3) as well as primary ALL (ALL #1–3) and AML (AML #1–3) samples. One representative example of two independent experiments is shown. (B) Recognition of LB-ARHGDIB-1R in HLA-B*07:02 as expressed on (malignant) hematopoietic cells, but not on (IFN-γ pre-treated) FB and KC. Malignant cells were isolated by flow cytometry cell sorting based on CD19 (ALL) or CD33 (AML) expression. T cells for LB-GEMIN4-1V (clone Z1) were included as a positive control. Genotyping results (+ or −) for the single nucleotide polymorphisms encoding LB-ARHGDIB-1R (filled bars) and LB-GEMIN4-1V (open bars) are shown. Mean release of IFN-γ of duplicate wells as measured by ELISA after overnight co-incubation of T cells and stimulator cells at a ratio of 1:6 is depicted. (C) Cytolysis of primary leukemic blasts as mediated by T cells for LB-ARHGDIB-1R in a 51Cr-release assay. Target cells (1×103) labeled with Na251CrO4 (Perkin Elmer, Waltham, MA, USA) were co-incubated with T cells for 10 hours (h) at effector:target ratios of 10:1, 3:1 and 1:1. Mean specific lysis of triplicate wells is shown for patient and donor EBV-B (upper and middle graphs, respectively) and ALL #1 expressing LB-ARHGDIB-1R and HLA-B*07:02 (lower graph). Patient and donor EBV-B and ALL #1 were positive for HLA-A*02:01 and negative for LB-GEMIN4-1V. Allo-HLA-A*02:01 reactive T cells (clone HSS12) and T cells for LB-GEMIN4-1V were included as controls. (D) Cytolysis of primary leukemic blasts and patient’s FB as mediated by T cells for LB-ARHGDIB-1R in a 48 h flow cytometry-based cytotoxicity assay. T cells (2.5×104) were labeled with PKH67 (Sigma-Aldrich, St. Louis, MO, USA) and co-incubated for 48 h with AML or ALL samples or FB cultured from a skin biopsy of the patient in the absence or presence of IFN-γ (1×104). After co-incubation, cultures were stained with CD33-APC (AML), CD19-PE (ALL) or CD90-PE (FB). Sytox blue was added to gate on viable cells and flow count fluorospheres (Beckman Coulter, Brea, CA, USA) were added to calculate specific lysis. T cells recognizing HA-2 in HLA-A*02:01 (clone 9H11), T cells for LB-GEMIN4-1V and Allo-HLA-A*02:01 reactive T cells were included as controls. Mean lysis of triplicate wells is depicted.
Next, we investigated T-cell recognition of different leukemic samples and both primary fibroblasts and keratinocytes cultured from skin biopsies in the absence or presence of IFN-γ. Samples were collected from patients and healthy individuals after approval from the Leiden or Radboud UMC Institutional Review Board and informed consent according to the Declaration of Helsinki. Recognition of (non-)hematopoietic cell types by LB-ARHGDIB-1R-specific T cells was measured by IFN-γ enzyme-linked immunosorbent assay (ELISA) after overnight co-incubation. The data confirmed hematopoiesis-restricted T-cell recognition of LB-ARHGDIB-1R in HLA-B*07:02 on all MiHA-positive leukemic cells, but not on (IFN-γ pre-treated) fibroblasts and keratinocytes (Figure 1B). T cells for LB-ARHGDIB-1R were also shown to recognize healthy hematopoietic cell types, including Epstein-Barr virus-infected (EBV) B cells, phytohemagglutinin-stimulated T cells and dendritic cells (Figure 1B and data not shown), indicating that alternative translation of LB-ARHGDIB-1R is not restricted to malignant cells. T cells for LB-ARHGDIB-1R also showed specific lysis of patient’s, but not donor’s, EBV-B cells and specific lysis of an acute lymphoblastic leukemia sample (ALL #1) in a 10 h 51Cr-release assay (Figure 1C). Lysis of other acute lymphoblastic and myeloid leukemia samples was not detected by 51Cr-release, but could be measured after 48 h co-incubation in a flow cytometry-based cytotoxicity assay, while (IFN-γ pre-treated) patient’s fibroblasts were not lysed (Figure 1D and data not shown). The data showed that T cells for LB-ARHGDIB-1R can specifically lyse hematological malignancies of different origins.
In addition to hematopoietic cells, ARHGDIB can also be expressed in endothelial cells.9 Therefore, we addressed potential toxicity and measured T-cell reactivity against endothelial cells. We confirmed intermediate ARHGDIB gene expression in HUVEC under both steady-state and inflammatory conditions by microarray gene expression analysis (Online Supplementary Figure S1A, upper panel). Increased ARHGDIB mRNA expression in HUVEC as compared to fibroblasts was also detectable by quantitative polymerase chain reaction analysis (see Online Supplementary Methods) (Online Supplementary Figure S1A, lower panel). To investigate whether gene expression in HUVEC can lead to T-cell recognition, we measured reactivity against two LB-ARHGDIB-1R-positive HUVEC samples by IFN-γ ELISA. T cells for LB-ARHGDIB-1R were only capable of recognizing one HUVEC sample (#2) after IFN-γ pre-treatment, which is known to up-regulate HLA, co-stimulatory and adhesion molecules and to increase the antigen processing and presentation capacity (Online Supplementary Figure S1B).10,11 However, recognition of HUVEC #2 was low compared to EBV-B cells and (IFN-γ pre-treated) HUVEC #1 was not or hardly recognized by specific T cells. Altogether, we demonstrate that ARGHDIB gene expression in HUVEC leads to low surface presentation of LB-ARHGDIB-1R that triggers only minimal T-cell reactivity under inflammatory conditions. Our results thus support the value of LB-ARHGDIB-1R as a target for T-cell therapy to selectively augment GvL reactivity after allogeneic SCT with a limited risk of GvHD.
Finally, we determined the in vivo immunogenicity of LB-ARHGDIB-1R and investigated its relevance as a T-cell target in clinical responses after allogeneic SCT. The population frequency of LB-ARHGDIB-1R in Caucasians is 77% (www.hapmap.org), resulting in a disparity rate, in which a LB-ARHGDIB-1R-positive patient is transplanted with a negative donor, of 18%. HLA-B*07:02 is expressed in approximately 20% of Caucasians.12 In our cohort of 93 HLA-B*07:02 patient-donor pairs, 14 LB-ARHGDIB-1R-positive patients were transplanted with a MiHA-negative donor and samples at relevant time points were available for ten patients. Peripheral blood mononuclear cells were stained with APC- and PE-conjugated HLA-B*07:02 tetramers containing LB-ARHGDIB-1R directly ex vivo as well as after 7 days of in vitro peptide stimulation as previously described.13 Peripheral blood mononuclear cells obtained after allogeneic SCT (and DLI) were analyzed for tetramer-positive T cells, and LB-ARHGDIB-1R-specific T cells were detected in four patients ex vivo (Figure 2A) and in four additional patients after in vitro peptide stimulation (Figure 2B), resulting in eight positive patients (80%) out of a total of ten.
Figure 2.
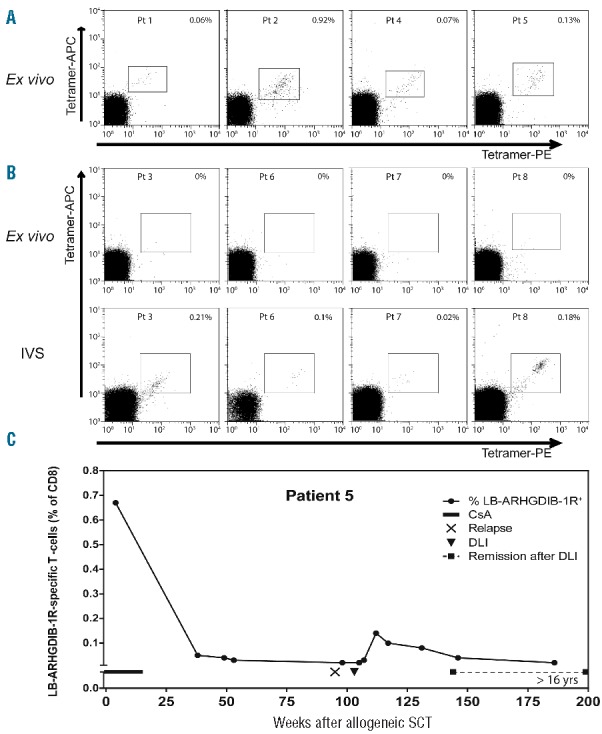
LB-ARHGDIB-1R as an immunogenic target with relevance for GvL reactivity. Peripheral blood mononuclear cells from ten LB-ARHGDIB-1R and HLA-B*07:02 positive patients transplanted with HLA-matched negative donors were screened for LB-ARHGDIB-1R-specific CD8 T cells directly ex vivo as well as after 1 week of in vitro peptide stimulation (IVS).13 (A) Patients with detectable LB-ARHGDIB-1R tetramer-positive T-cells ex vivo. Numbers indicate the percentage of CD8 T cells that are positive for both LB-ARHGDIB-1R tetramers (PE and APC) in the Sytox blueneg, CD8pos, CD4neg, CD14neg, CD16neg, and CD19neg T-cell population. (B) Dot plots of patients with detectable LB-ARHGDIB-1R tetramer-positive T cells after IVS: day 0 (upper panels) and day 7 (lower panels). Numbers indicate the percentage of CD8 T cells that are positive for both LB-ARHGDIB-1R tetramers (PE and APC). (C) Ex vivo frequencies of LB-ARHGDIB-1R specific T cells and clinical response in patient #5 who was treated with DLI in the absence of additional (chemo) therapy for relapsed lymphoma after partial T-cell-depleted allogeneic SCT.
All patients were treated with partial T-cell-depleted allogeneic SCT followed by at least 4 months of immunosuppression with cyclosporine A as GvHD prophylaxis. Six of ten patients received DLI after allogeneic SCT (Online Supplementary Table S1). In three of six patients treated with DLI, tetramer-positive T cells were detected ex vivo at frequencies between 0.06–0.92%. Five patients received prophylactic DLI, and a clinical response against (malignant) hematopoietic cells of patient origin could not therefore be monitored. Patient #5 with relapsed follicular lymphoma, which was confirmed by lymph node biopsy, received therapeutic DLI that in the absence of additional (chemo)therapy induced a long-lasting complete remission (>16 years) without any signs of GvHD. T cells for LB-ARHGDIB-1R were measured in three patients with GvHD after DLI (patients #2, 6 and 7). These T cells were detectable ex vivo (0.92%) in patient #2 and after in vitro stimulation in the other two patients. Although it cannot be excluded that T cells for LB-ARHGDIB-1R may have contributed to GvHD in these patients, we consider it more likely that T cells with other specificities mediated GvHD, since we previously demonstrated that a variety of MiHA are often targeted in patients with GvHD5,14 and that the majority of these MiHA are ubiquitously expressed on (non-)hematopoietic tissues. This is further supported by the observation that T cells for LB-ARHGDIB-1R were also measured ex vivo (0.13%) in patient #5. Induction of tetramer-positive T cells in this patient 2 months after DLI coincided with long-lasting GvL reactivity without GvHD. Dynamic analysis of LB-ARHGDIB-1R tetramer-positive T cells in this patient demonstrated high frequencies not only after DLI, but also within the first weeks after allogeneic SCT during immunosuppression with cyclosporine A (Figure 2C). Although the long-lasting GvL response in patient #5 suggests that LB-ARHGDIB-1R-specific T cells are capable of mediating strong anti-tumor immunity, T cells with specificities other than LB-ARHGDIB-1R may also be involved in the therapeutic effect of DLI. Systemic toxicity as a result of vascular damage has not been observed in any of the patients with circulating LB-ARHGDIB-1R-specific T cells. Thus, clinical observations support the therapeutic value of LB-ARHGDIB-1R and do not show evidence for specific attack of endothelial cells as might be suggested based on detectable ARHGDIB gene expression and low T-cell recognition of endothelial cells in vitro.
In conclusion, our data support the relevance of LB-ARHGDIB-1R as highly immunogenic and hematopoiesis-restricted MiHA with the potential to shift the delicate balance between GvL reactivity and GvHD in favor of desired anti-tumor reactivity. At the Radboud UMC, we have started a clinical trial in which transplanted patients are vaccinated with donor dendritic cells loaded with mRNA and included ARHGDIB as one of the transcripts for hematopoiesis-restricted MiHA (Dutch Trial Registry #NTR4128). Future clinical data will, therefore, provide definite evidence of whether T cells for LB-ARHGDIB-1R are capable of inducing selective GvL responses. In addition to hematopoietic cells, ARHGDIB has been reported to be expressed in several solid tumors correlating with advanced tumor stage and metastatic potential.15 As such, LB-ARHGDIB-1R may have broad value as a target for T-cell therapy to treat hematologic malignancies and solid tumors after allogeneic SCT.
Acknowledgments
The authors would like to thank Cynthia Kramer (Radboud UMC) for tetramer analyses and cell culture, and Martijn Dane (Department of Nephrology and the Einthoven Laboratory of Experimental Vascular Medicine, Leiden University Medical Center, Leiden, The Netherlands) for isolation of HUVEC
Footnotes
Funding: this work was supported by the Dutch Cancer Society (UL 2010-4748) and ZonMW (grant 95103004).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Kolb HJ. Graft-versus-leukemia effects of transplantation and donor lymphocytes. Blood. 2008;112(12):4371–4383. [DOI] [PubMed] [Google Scholar]
- 2.Bleakley M, Riddell SR. Exploiting T cells specific for human minor histocompatibility antigens for therapy of leukemia. Immunol Cell Biol. 2011;89(3):396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warren EH, Zhang XC, Li S, et al. Effect of MHC and non-MHC donor/recipient genetic disparity on the outcome of allogeneic HCT. Blood. 2012;120(14):2796–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spierings E, Hendriks M, Absi L, et al. Phenotype frequencies of autosomal minor histocompatibility antigens display significant differences among populations. PLoS Genet. 2007;3(6):e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Bergen CAM, Rutten CE, Van Der Meijden ED, et al. High-throughput characterization of 10 new minor histocompatibility antigens by whole genome association scanning. Cancer Res. 2010; 70(22):9073–9083. [DOI] [PubMed] [Google Scholar]
- 6.Lelias JM, Adra CN, Wulf GM, et al. cDNA cloning of a human mRNA preferentially expressed in hematopoietic cells and with homology to a GDP-dissociation inhibitor for the rho GTP-binding proteins. Proc Natl Acad Sci USA. 1993;90(4):1479–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scherle P, Behrens T, Staudt LM. Ly-GDI, a GDP-dissociation inhibitor of the RhoA GTP-binding protein, is expressed preferentially in lymphocytes. Proc Natl Acad Sci USA. 1993;90(16):7568–7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kremer AN, van der Meijden ED, Honders MW, et al. Endogenous HLA class II epitopes that are immunogenic in vivo show distinct behavior toward HLA-DM and its natural inhibitor HLA-DO. Blood. 2012;120(16):3246–3255. [DOI] [PubMed] [Google Scholar]
- 9.Theodorescu D, Sapinoso LM, Conaway MR, Oxford G, Hampton GM, Frierson HF., Jr Reduced expression of metastasis suppressor RhoGDI2 is associated with decreased survival for patients with bladder cancer. Clin Cancer Res. 2004;10(11):3800–3806. [DOI] [PubMed] [Google Scholar]
- 10.Fu H, Kishore M, Gittens B, et al. Self-recognition of the endothelium enables regulatory T-cell trafficking and defines the kinetics of immune regulation. Nat Commun. 2014;(3)5:3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma W, Lehner PJ, Cresswell P, Pober JS, Johnson DR. Interferon-gamma rapidly increases peptide transporter (TAP) subunit expression and peptide transport capacity in endothelial cells. J Biol Chem. 1997;272(26):16585–16590. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011; 39:D913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobo W, Broen K, van der Velden WJ, et al. Association of disparities in known minor histocompatibility antigens with relapse-free survival and graft-versus-host disease after allogeneic stem cell transplantation. Biol Blood Marrow Transplant. 2013;19(2):274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffioen M, Honders MW, van der Meijden ED, et al. Identification of 4 novel HLA-B*40:01 restricted minor histocompatibility antigens and their potential as targets for graft-versus-leukemia reactivity. Haematologica. 2012;97(8):1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho HJ, Baek KE, Yoo J. RhoGDI2 as a therapeutic target in cancer. Expert Opin Ther Targets. 2010;14(1):67–75. [DOI] [PubMed] [Google Scholar]


