Key Points
SUMOylatable-WASp trans-activates and non-SUMOylatable-WASp trans-represses NF-κB response genes mediating adaptive immunity.
HDAC inhibitors reverse the immunodeficient and proinflammatory phenotype caused by SUMOylation-deficient WASp in TH1 cells.
Abstract
In Wiskott-Aldrich syndrome (WAS), immunodeficiency and autoimmunity often comanifest, yet how WAS mutations misregulate chromatin-signaling in Thelper (TH) cells favoring development of auto-inflammation over protective immunity is unclear. Previously, we identified an essential promoter-specific, coactivator role of nuclear-WASp in TH1 gene transcription. Here we identify small ubiquitin-related modifier (SUMO)ylation as a novel posttranslational modification of WASp, impairment of which converts nuclear-WASp from a transcriptional coactivator to a corepressor of nuclear factor (NF)-κB response genes in human (TH)1-differentiating cells. V75M, one of many disease-causing mutations occurring in SUMO*motif (72-ψψψψKDxxxxSY-83) of WASp, compromises WASp-SUMOylation, associates with COMMD1 to attenuate NF-κB signaling, and recruits histone deacetylases-6 (HDAC6) to p300-marked promoters of NF-κB response genes that pattern immunity but not inflammation. Consequently, proteins mediating adaptive immunity (IFNG, STAT1, TLR1) are deficient, whereas those mediating auto-inflammation (GM-CSF, TNFAIP2, IL-1β) are paradoxically increased in TH1 cells expressing SUMOylation-deficient WASp. Moreover, SUMOylation-deficient WASp favors ectopic development of the TH17-like phenotype (↑IL17A, IL21, IL22, IL23R, RORC, and CSF2) under TH1-skewing conditions, suggesting a role for WASp in modulating TH1/TH17 plasticity. Notably, pan-histone deacetylase inhibitors lift promoter-specific repression imposed by SUMOylation-deficient WASp and restore misregulated gene expression. Our findings uncovering a SUMOylation-based mechanism controlling WASp’s dichotomous roles in transcription may have implications for personalized therapy for patients carrying mutations that perturb WASp-SUMOylation.
Introduction
Wiskott-Aldrich syndrome (WAS) is an X-linked immunodeficiency-cum-autoimmunity disorder arising from mutations in WAS that encode WASp, whose deficiency in hematopoietic cells results in the human disease.1 WASp polymerizes actin in the cytoplasm via the actin-related protein (ARP)2/3-complex and reprograms RNA polymerase II-dependent transcription in the nucleus via its effect on mixed lineage leukemia (MLL)- and switch/sucrose non-fermentable (SWI/SNF)-dependent chromatin remodeling of gene promoters.2-4 Like human WASp, other nucleation-promoting factors (NPFs) of the WASp family, WAVE1, WASH, JMY, and N-WASp, also support transcription or other nuclear functions in different organisms.5-11 Accordingly, the classical cytoplasmic NPFs are emerging as key players in the nucleus in roles that are mechanistically distinct from that in the cytosol. How then is the compartment-delimited role enabled for these dual-compartment, dual-function NPFs? For WASp, its cytosolic effect on ARP2/3-mediated actin polymerization is enabled by allosteric activation, wherein association with CDC42-GTP or Fyn-mediated tyrosine phosphorylation switches WASp conformation from auto-inhibitory to active.12,13 This structural change facilitates dimerization/oligomerization of WASp, which optimizes ARP2/3 activation and cytosolic F-actin nucleation.14 In contrast, the cotranscriptional activity of human-WASp or Xenopus-WAVE1 does not seem to critically depend on ARP2/3 or the verprolin, cofilin, acidic (VCA) domain.5,15 Accordingly, the regulatory inputs, including unique posttranslational modifications (PTMs) involved in instructing the transcriptional output of WASp family proteins, remain elusive, clarification of which we propose is central to understanding the differential effect of WASp on immunity vs autoimmunity/autoinflammation.
Given the substantial evidence for the involvement of small ubiquitin-related modifier (SUMO)ylation in transcriptional regulation,16-19 we asked whether this PTM, not heretofore reported for any WASp family members, enables the transcriptional role of WASp. Testing a SUMOylation-linked hypothesis is tempting because (1) WASp associates with RanBP2,15 an E3-SUMO-ligase,20 raising the possibility that RanBP2 might also SUMO modify WASp; (2) SUMOylation impacts subcellular localization and protein-protein interactions on chromatin,19,21 events that could modulate WASp’s role as a transcriptional cofactor; and (3) SUMOylation targets entire groups of physically interacting proteins,19 this raises the possibility that nuclear WASp could be SUMOylated as part of collective SUMOylation of trans-regulatory factors (RBBP5, signal transducer and activator of transcription [STAT]1, nuclear factor [NF]-κB), nucleoporin (NUP358/RanBP2), or actin-myosin complex (actin, MYL12A, MYL6, ARP2/3), all proteins known to associate with WASp3,4,15 and be SUMO modified.21-26
In SUMOylation, a SUMO1, 2, or 3 covalently attaches to the lysine(K) residue contained within a SUMO consensus, ψKxE/D, or phospho-SUMOyl consensus, ψKxExxS/T (ψ, hydrophobic-residue; x, any residue; D, aspartic acid; E, glutamic acid) in a multistep reaction involving E1-activating (SAE1/2), E2-conjugating (UBC9), and E3-ligating (RanBP2; PIAS) enzymes.17,18,26 SUMOylation typically modifies only a small fraction of the substrate (∼10%), and is reversible through the effects of multiple SUMO-specific proteases. Substrates can be modified by 1 SUMO at 1 K-residue (monoSUMOylation) or 1 SUMO at multiple K-residues (multi-monoSUMOylation) or can form an oligomeric SUMO chain at 1 or multiple K-residues (polySUMOylation). Both SUMO1 and SUMO2/3 can catalyze oligomeric SUMO chains in vivo,20,26,27 which fulfill multiple signaling functions including transcriptional regulation and genome organization.28 Accordingly, SUMOylation of transcription factors has been linked to either gene activation or gene repression in both yeast and mammalian cells29,30 via mechanisms that include altering chromatin dynamics.16,31,32 For peroxisome proliferator-activated receptor (PPAR)-γ and Ikaros that function either as a transcriptional coactivator or corepressor, this switch in transcriptional output is effected by SUMOylation.33,34 Beyond serving as a simple on-off switch, SUMOylation can also fine-tune gene expression as demonstrated for the nuclear receptor SF-1,35 a paradigm that is relevant to testing whether WASp-SUMOylation differentially regulates genes that pattern immunity vs inflammation.
We previously demonstrated that nuclear WASp is required for transcriptional activation.3,4 Here, we tested the hypothesis that WASp is a bi-functional transcriptional activator and repressor and investigated the mechanism by which WASp functions in transcriptional repression. Because SUMO1 coenriches with H3K4me3 at gene promoters to activate transcription36 and because WASp is required to inscribe H3K4me3 at promoters,3 we explored the WASp:SUMO link as a chromatin-based mechanism for transcriptional reprogramming in Thelper (TH) cells. We provide evidence for SUMO modification serving as a regulatory switch that controls activating vs repressive bi-functionality of WASp in transcription. Our study uncovers a mechanism by which SUMOylation-deficient WASp perturbs the transcriptional output of NF-κB response genes important in immunity and inflammation, identifies pathogenic mutation that co-opts this deranged pathway, and proposes potential therapeutic use of histone deacetylase (HDAC) inhibitors to restore inappropriate gene activation in WAS.
Materials and methods
Cells, transfection, and RNAi-mediated knockdown
Primary TH, B, natural killer (NK), and monocytoid dendritic cells generated from donor peripheral blood mononuclear cells, Jurkat TH cells, HeLa cells, and WASnull TH cells from a WAS patient carrying a nonsense mutation were used. WASp-SUMO sites were predicted using the group-based prediction system (GPS)-SUMO algorithm37 and single or combinatorial K>R mutations and disease-causing mutations introduced by QuickChangeIISite-Directed Mutagenesis (Stratagene) using primers shown in supplemental Table 1 available on the Blood Web site, and the constructs were stably expressed in the TH cells by AmaxaNucleofector (Lonza) or Lipofectamine (Life Technologies) as previously described.4,15 Transfected cells were cultured under TH1-skewing (recombinant human interleukin [rhIL]-12, anti-IL-4 antibody, rhIL-2), TH17-skewing (rhIL-6, rhTGFβ1, rhIL-2), or nonskewing TH0 conditions for 7 days as previously described.3 Stable expression of mutants was verified by flow cytometry and western blotting at day 7 after transfection, which showed >90% T cells expressing the constructs. NK cells were propagated in rIL2 (100 IU/mL), dendritic cells were matured with lipopolysaccharide (LPS), and B cells were activated with immunoglobulin (Ig)M crosslinking. Expression of endogenous SUMO1 was suppressed in primary human CD4+ TH cells by transfecting SUMO1 short hairpin RNA (shRNA) or scrambled shRNA (SantaCruz) (0.8 mL shRNA Transfection Medium, 200 μL shRNA solution A + solution B, incubated at 37°C for different time points). For HDAC inhibition assays, CD4+ T cells were incubated in the presence of 100 ng/mL trichostatin-A and 2 mM sodium butyrate for 2 hours.
Mass spectrometry
Multiple mass spectrometry (MS) assays on immunoprecipitated WASp-enriched protein complexes were performed as previously described.4,15 Briefly, lysates from micrococcal nuclease (MNase)-treated nuclei of human primary or Jurkat TH1-skewed cells expressing WASp-Flag/Myc dual-tagged protein were incubated with anti-WASp, or anti-Flag and anti-Myc, or their isotype-Ig antibodies. The recovered polypeptides were analyzed by nano-liquid chromatography (LC)-MS on an LTQ-Orbitrap Velos mass spectrometer using the SEQUEST database.
Coimmunoprecipitation, immunoblotting, and electrophoretic mobility shift assay
Coimmunoprecipitations and immunoblottings were done as previously described3,4,15 using the reagents and antibodies shown in supplemental Table 1. Electrophoretic mobility shift assay (EMSA) was carried out in 5 μg nuclear extract coincubated with double-stranded oligonucleotide probes for NF-κB, Octamer, and CCCTC-binding factor (CTCF) that were 5′end-labeled with [γ-32P]ATP. EMSA assays were performed as previously described.4
Flow cytometry and deconvolution microscopy
Intracellular staining of cytokines/proteins was performed as previously described.4,15 Briefly, cells were treated with protein transport inhibitors (GolgiPlug/GolgiStop), fixed, permeabilized, labeled with fluorochrome-conjugated antibodies (or control immunoglobulin antibody), and analyzed on a Becton-Dickinson LSRII using FACSDiva, and geometric mean (GM) fluorescence intensity was calculated. Deconvolution imaging of paraformaldehyde-fixed single TH1 cells was performed with Zeiss digital microscopy integrated with SlideBook software, as previously described.38
In vitro and in vivo SUMOylation
For in vitro SUMOylation, transfected FLAG immunoprecipitate (500 µg cell extract) from HeLa or T cells was coincubated with recombinant SUMO1 (or mutant SUMO1), SUMO2, SUMO3, or SUMO1 + 2 + 3 (1 µL) plus SUMOylation buffer (2 µL), and Mg-ATP (1 µL) in 20 µL total volume and incubated for 60 minutes at 37°C. Each assay was quenched by adding 20 µL of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel-loading buffer followed by heating to 95°C for 5 minutes before subjecting to western blotting (Enzo). To capture in vivo SUMOylated WASp, TH1-skewed cells were treated with 1 mM of N-ethylmaleimide (NEM; Sigma Aldrich) for 4 hours.
Chromatin immunoprecipitation-quantitative polymerase chain reaction and reverse transcriptase-polymerase chain reaction
Chromatin immunoprecipitation (ChIP) assays were performed with micrococcal nuclease (MNase)-digested chromatin derived from NEM-treated and -nontreated TH cells as described3,4,15 using antibodies/reagents shown in supplemental Table 1. Nonspecific signals obtained with control IgG-ChIP were subtracted. For reverse transcriptase-polymerase chain reaction (RT-PCR) assays, cDNA was synthesized from total RNA prepared using Quick-RNA MiniPrep (Zymo), and NF-κB transcriptome analysis was performed with the RT2ProfilerPCR Array (Qiagen). The calculated fold-change [2^(−ΔΔCt)] is the normalized gene expression [2^(−ΔCt)] in the test sample (TH1 skewed/TH17 skewed) divided by the normalized gene expression [2^(−ΔCt)] in the control sample (TH0 nonskewed). Normalization was performed using Ct values derived for GAPDH, ACTB, or RPLP0.
Results
WASp associates with SUMO pathway enzymes
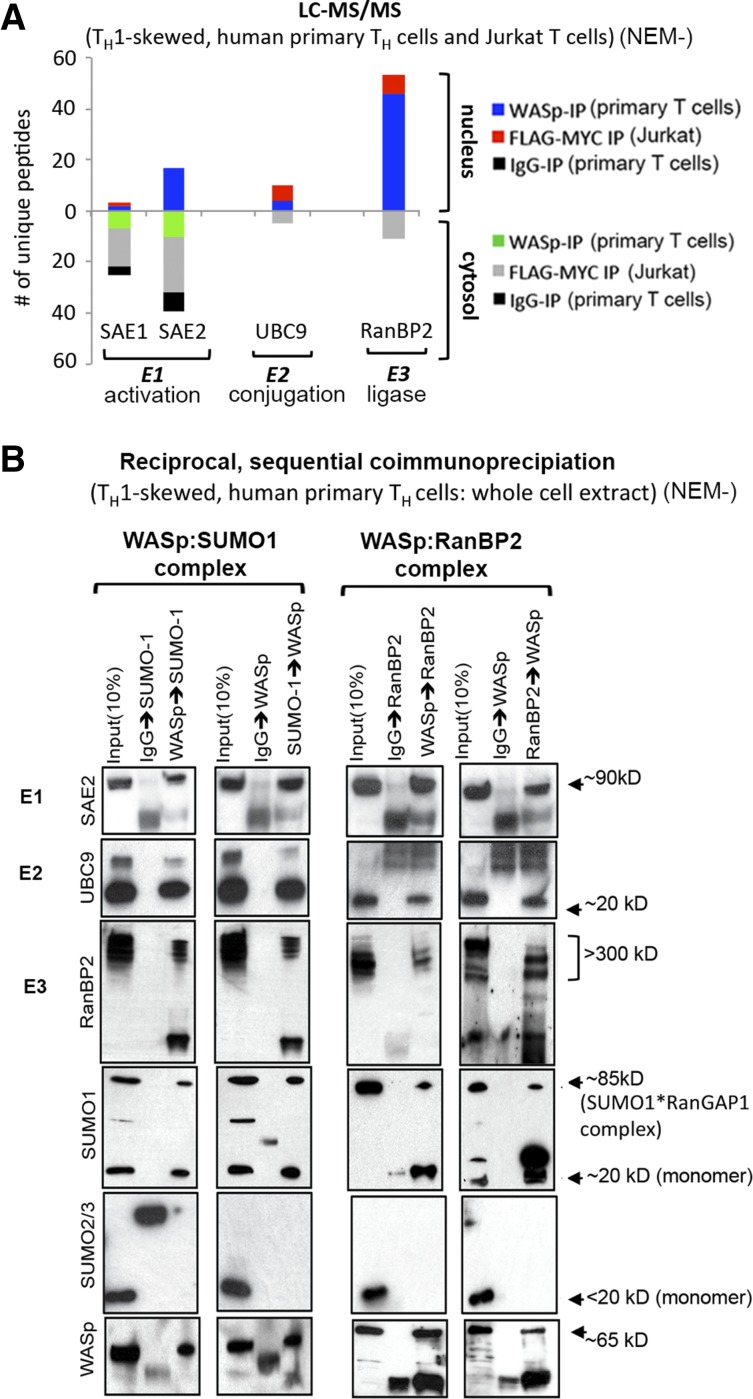
To investigate SUMO modification of WASp, we queried our LC-MS/MS dataset of WASp-associated proteins generated from human TH1-skewed cells, primary and Jurkat, with the latter transfected with Flag/Myc-tagged WASp.15 Polypeptides of SUMO enzymes involved in E1 activation (SAE1/2), E2 conjugation (UBC9), and E3 ligation (RanBP2) were recovered from cytosolic and nuclear WASp complexes, whereas those of other SUMO enzymes (PIAS1-4, NSE2, MAPL, PC2, HDAC4, TOPORs) were not (Figure 1A; supplemental Figure 1). The WASp:SUMO enzyme association yielded by MS was validated by reciprocal, sequential coimmunoprecipitation, which showed that the WASp:RanBP2 complex associates with SUMO1 but not SUMO2/3 in TH1 cells (Figure 1B). The collective presence of these SUMO enzymes raises the possibility that WASp is SUMO modified.
Figure 1.
WASp associates with SUMO-pathway proteins in vivo. (A) LC-MS/MS. Number of unique polypeptides of WASp-associated, SUMO-pathway proteins isolated from nuclear and cytosolic fractions of TH1-skewed, primary TH cells (reports on endogenous WASp-proteome) or Jurkat TH cells (reports on Flag/Myc-tagged transfected WASp-proteome) by immunoprecipitation (IP) with anti-WASp antibody (Ab) or anti-FLAG/Myc antibodies (2-step immunopurification, 1stIP: Flag Ab, 2ndIP: Myc Ab). MultiConsensus reports of peptides/proteins were generated from 3 to 4 biological replicates after applying the filtering criteria previously described15 (supplemental Figure 1). (B) Validation of MS-generated WASp-SUMO associations by coIP. Protein complexes isolated by reciprocal and sequential coIP (eg, WASp>RanBP2 sequence denotes 1stIP: WASp, 2ndIP: RanBP2) from whole cell extracts of TH1-skewed, human primary TH cells resolved by western blotting the same gel with indicated antibodies. NEM−, nontreated with NEM.
WASp is SUMOylated by SUMO1 in vivo and in vitro
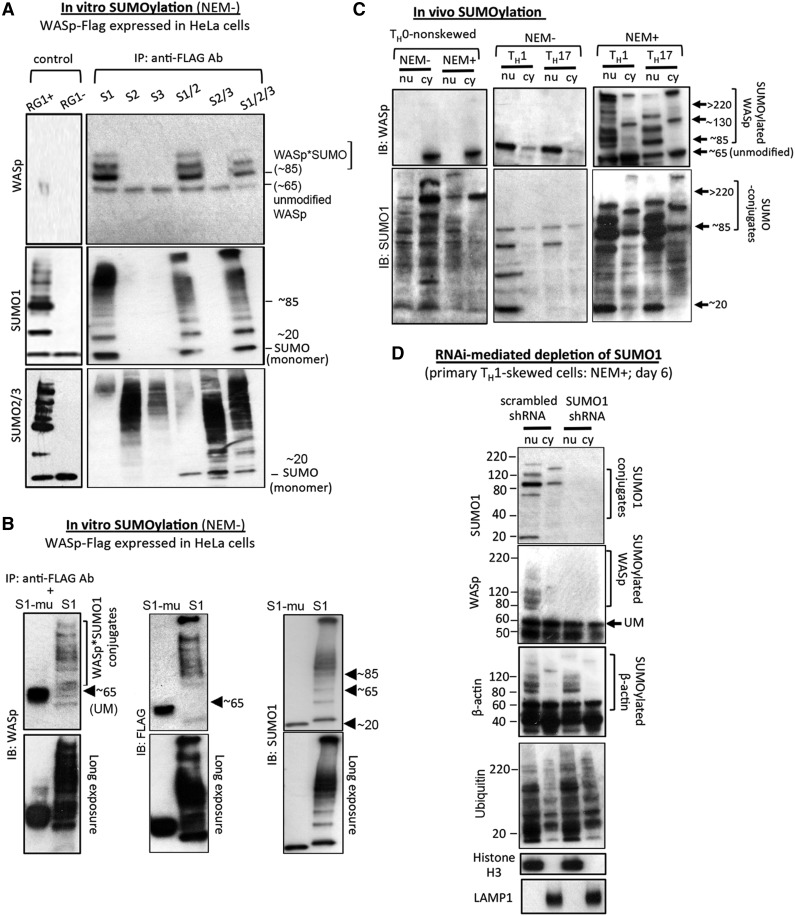
To investigate WASp-SUMOylation in vitro, FLAG/Myc-tagged WASp was expressed in HeLa cells, and the FLAG immunoprecipitate was coincubated with recombinant SUMO1 (or mutant SUMO1), SUMO2, SUMO3, or SUMO1 + 2 + 3. The presence of an ∼85 kDa band (∼20 kDa heavier than ∼65 kDa unmodified WASp) and multiple chain-like, slower-migrating bands (>85 kDa) suggests SUMOylated-WASp (multi-monoSUMOylated or polySUMOylated), observed with SUMO1 but not SUMO2, SUMO3, or mutant SUMO1 (Figures 2A-B).
Figure 2.
WASp is SUMO modified in vitro and in vivo. (A-B) In vitro SUMOylation using HeLa cells stably transfected with FLAG/Myc-tagged human full-length WASp construct. Immunoprecipitated WASp-FLAG incubated with SUMO-1, -2, and -3 (S1, S2, S3) individually or with the indicated combinations of SUMO-proteins sequentially probed with SUMO-1, -2/3, and WASp Abs in western blot assays (A). Purified RanGAP1 (RG1) protein, provided with the assay kit, served as a positive control and was coincubated with all 3 SUMO proteins (S1, S2, S3) together. RG1(+), RanGap1, E1, and E2, ATP included (+); RG(−), RanGap1, E1, E2 included but ATP excluded from the assay. WASp-FLAG samples incubated with SUMO1 (S1) or with the SUMO1 mutant lacking enzymatic activity (S1-mutated [mu]) (B). The data are representative of 3 biologic replicates for A and 2 biologic replicates for B. Numbers indicate the molecular masses (kDa) of key bands. SUMOylated-WASp is indicated as WASp*SUMO conjugates at ∼85 kDa and above. NEM−, assay performed in the absence of NEM, a chemical that inhibits de-SUMOylation. (C) In vivo SUMOylation. Nuclear (nu) and cytoplasmic (cy) extracts of NEM-treated, TH1- or TH17-skewed/TCR-activated or TH0-nonskewed/TCR-nonactivated primary TH-cells in the presence (+) or absence (−) of NEM sequentially reprobed with the indicated antibodies by western blot. (D) RNA interference-mediated depletion of endogenous SUMO1 was performed in primary TH1-skewed/TCR-activated cells using SUMO1-shRNA or scrambled-shRNA at day 6 after transfection and NEM treatment (supplemental Figure 2B for temporal progression of SUMO1 k/d) and western blots sequentially reprobed with the indicated Abs. (supplemental Table 1 for reagents). Data are representative of 2 independent experiments. NEM+/−, treated/nontreated with NEM.
Endogenous WASp is SUMOylated in vivo in the absence of exogenous SUMO enzymes in primary TH1-skewed and TH17-skewed but not in TH0-nonskewed cells treated with NEM, an inhibitor of SUMO isopeptidases (Figure 2C). SUMO*WASp conjugates are first captured at 4 hours in cytosol and 8 hours in the nucleus on TH1 skewing (supplemental Figure 2A). Besides unmodified WASp (∼65 kDa), multiple slower-migrating forms (>85 kDa) were captured in the nuclear fraction of TH1 and TH17 cells (Figure 2C). For reactions that did not include NEM, additional, slower-migrating WASp bands were undetected, upholding that these additional bands are SUMO*WASp conjugates. The cytosolic fraction, which lacked the chain-like patterning seen in the nucleus, displayed 2 prominent bands at ∼130 and >300 kDa in NEM-treated cells (Figure 2C), which are sizes that correspond to the WASp dimer/oligomer.14
Because NEM also inhibits deubiquitinases, it is possible that some of these slower-migrating bands could represent hybrid SUMO ubiquitin chains (∼30 kDa heavier; SUMO: ∼20 kDa, Ub: ∼10 kDa), especially given the cooperative roles of ubiquitin/SUMO dual modification in nuclear functions.39 Significantly, RNAi-mediated SUMO1 knockdown, which does not impair polyubiquitination, precludes WASp-SUMOylation without affecting β-actin SUMOylation (Figures 2D; supplemental Figure 2B); the latter is catalyzed by SUMO2/3.25 These results establish WASp as a physiologic SUMO1 substrate in TH1 and TH17 cells, including in activated B cells, NK cells, and mature dendritic cells (supplemental Figure 2C).
WASp contains the SUMO consensus and SUMO interaction motif
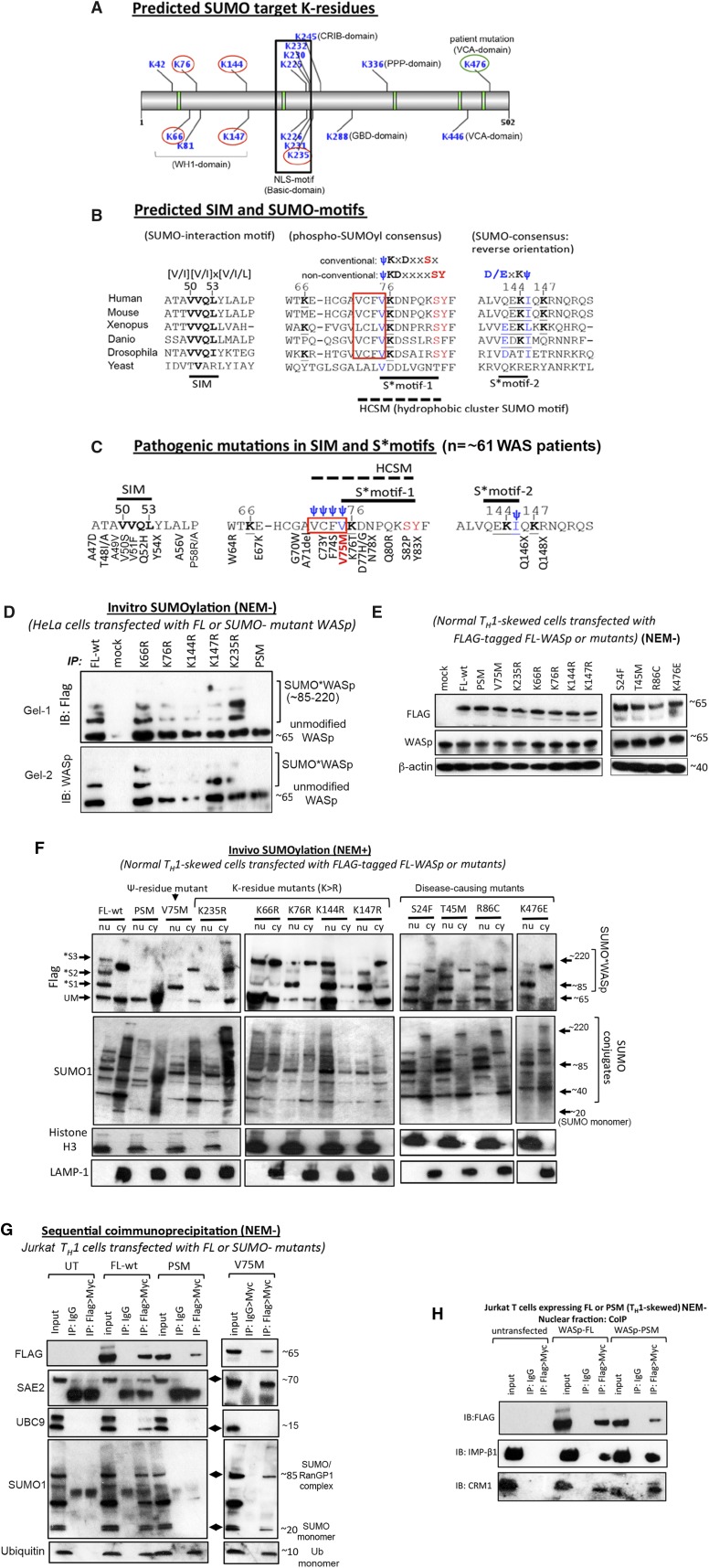
The GPS-SUMO algorithm37 predicted SUMO-acceptor K-residues in all WASp domains, although most occurred in WH1 and Basic domains (Figure 3A). Some of these K-residues occurred in the nonclassical SUMO consensus configurations previously identified in the global mapping of SUMOylation sites.40 These are S*motif-1, 75-ψKDxxxxSY-83, a variant of phospho:SUMOyl consensus (ψKxDxxSx) and S*motif-2, 142-D/ExKψ-145, an inverted SUMO consensus (Figure 3B). In both S*motifs, K-residues (K76, K144) and ψ-residues (V75, I145) are conserved through evolution and are in-frame with the SUMO consensus (ψKxD/E). In phospho:SUMOyl-consensus, the K76-residue is preceded by four evolutionarily conserved, hydrophobic (ψ) amino acids (75-VFCV-72), known as hydrophobic cluster SUMOylation motif (HCSM) and followed by Ser82 (+6 position) and Tyr83 (+7 position), phosphorylation of which functions to augment SUMOylation, as does the HCSM.41,42 Additionally, WASp contains the potential SUMO interaction motif (SIM) 50-VVQL-53, which follows the classical SIM consensus [V/I][V/I] × [V/I/L] that is also conserved through evolution (Figure 3B). Accordingly, WASp contains target sequences for both SUMOylation and SUMO interaction, the functional significance of which is implied in the clustering of disease-causing mutations (∼61 patients reported) within these sites (Figure 3C; supplemental Figure 3D).
Figure 3.
Identification and characterization of SUMO target lysine residues in WASp. (A) GPS-SUMO tool predicted SUMO-acceptor K-residues and SUMO-interaction motif (SIM) in different WASp domains (WH1, CRIB, Basic, GBD, PPP, VCA). K-residues circled in red were experimentally mutated (K>R), whereas K476 circled in green is a natural pathogenic mutation (K476E). These 6 K-residue mutants were included in SUMOylation assays. (B) Comparison of amino acid sequences of putative SUMO motifs (S*motif-1; S*motif-2) and SIM motif of WASp from yeast to human. Hydrophobic cluster SUMO motif (HCSM) preceding S*motif-1 is boxed in red. (C) Disease-causing mutations (n = ∼61 patients reported thus far) occurring in the SUMO, HCSM, and SIM motifs of WASp are shown (see supplemental Figure 3D for additional details). V75M (n = ∼22 patients) occurring in S*motif-1/HCSM that initially presents as XLT and in some patients progresses to serious disease (clinical severity grade 5 with autoimmunity and/or malignancy)72 is highlighted with red lettering. (D) In vitro SUMOylation assays were performed as described in legend to Figure 2A with full-length, wild-type WASp (FL-wt), K>R single mutants, penta-SUMO-site mutant (PSM), and mock (untransfected) as substrates immunoprecipitated with anti-FLAG Ab from HeLa cells transfected with the indicated constructs. Western blot with anti-FLAG or -WASp Ab is shown. The data are representative of 2 biologic replicates shown as Gel-1 and Gel-2 to highlight subtle interassay variations in the SUMOylation patterns (see supplemental Figure 3 for WASp*SUMO construct sequences and expression by flow cytometry). (E) The indicated FLAG-tagged mutants (PSM, K>R, and disease-causing) and FL-wt were stably transfected (or mock transfected) in the normal human TH-cell line, and the expression of the mutants on day 7 of TH1-skewing/TCR-activation was monitored by anti-WASp and -FLAG antibodies western blot. β-Actin served as loading control. (F) Transfected mutants immunoprecipitated with anti-FLAG antibody from nuclear (nu) and cytoplasmic (cy) fractions of NEM-treated, normal TH1-skewed/TCR-activated cells were used as substrate for the in vivo SUMOylation assays as described in the legend to Figure 2C. The same blot was sequentially reprobed with the indicated antibodies. Multiple SUMO*WASp conjugates are indicated as *S1/S2/S3 bands (molecular weight ≥ 85 kDa). The purity of nuclear and cytoplasmic fractions was monitored with Histone H3 and LAMP-1 immunoblotting, respectively. (G-H) Two-step sequential immunoprecipitation (1stIP: FLAG, 2nd IP: Myc) performed on the (G) whole cell extracts or (H) nuclear extract generated from TH1-skewed Jurkat cells stably transfected with the indicated mutants and immunoblotted with the indicated antibodies. Molecular mass of indicated bands is in kilodaltons. NEM+/−, treated/nontreated with NEM.
Lysine residues K66, K76, K144, K147, and K235 and hydrophobic residue V75 are important for WASp-SUMOylation in vivo
To test whether the predicted K-residues are SUMO targets, we generated K>R deletion/substitution mutants with FLAG/MYC dual-tag, stably expressed in HeLa, and immunoprecipitated WASp mutants with anti-FLAG antibody for in vitro SUMOylation (supplemental Figure 3). FL-WASp (wild-type) coincubated with recombinant SUMO1, E1/E2-enzymes, and ATP revealed multiple slower-migrating WASp forms (∼85 to ∼220 kDa), which is consistent with SUMO modification (multi-monoSUMOylation or polySUMOylation cannot be distinguished). In contrast, K>R single mutants demonstrated that K76, K144, and K147 residues are more critical than K66 or K235 for in vitro SUMOylation based on the intensity of SUMO-modified forms (Figure 3D). Notably, K235R results were mixed, displaying both preserved and partially compromised SUMO*WASp conjugates in separate assays (Figure 3D).
To verify the above findings in vivo, normal TH cells were stably transfected with FLAG-MYC-tagged FL-WASp or K>R single mutants with comparable expression (Figure 3E). After TH1 skewing/T-cell receptor (TCR) activation and NEM treatment, nuclear/cytosolic fractions were examined by western blot. Transfected FL-WASp (FL-wt) gave multiple slower-migrating bands in the nucleus that corresponded with those of SUMO1 (∼85-220 kDa) (Figure 3F). This implies that transfected FL-WASp is SUMOylated by endogenous SUMO enzymes, and the heavier bands are consequent to posttranslational modification rather than alternative splicing. Although neither K>R single mutant showed complete abrogation of SUMOylation, some gradation of SUMOylation defect was observed, wherein 1 or more slower-migrating form(s) was lost in nuclear and/or cytosolic fractions. This indicates alternative usage of various K-residues for SUMOylation. We therefore generated a penta-SUMO site mutant (PSM: lacking all 5 K-residues) (supplemental Figure 3C), which we show abrogates WASp-SUMOylation in vitro and in vivo (Figures 3D,F), implying that these lysines are required for WASp-SUMOylation.
In sequential coimmunoprecipitation assays, the PSM mutant does not bind SAE2 and UBC9, establishing that failed WASp-SUMOylation is consequent to impaired interaction of the PSM mutant with SUMO enzymes (Figure 3G). Although the PSM mutant does not associate with SUMO1, its association with ubiquitin is unaffected (Figure 3G), implying that the deleted K-residues are SUMO specific. This finding complies with the paradigm that only about one-quarter of SUMOylation sites are also ubiquitinated in the human proteome.41 It is possible, however, that some of these K-residues, upon SUMOylation, may become targets of ubiquitination, acetylation, or methylation. Notably, the PSM mutant binds importin-β1 and CRM1 and translocates to the TH1-cell nucleus (Figure 3H), assuming the same 4′,6-diamidino-2-phenylindole (DAPI)-dim distribution as normal-WASp (supplemental Figure 4),3 implying that SUMOylation and nuclear import are not linked events for WASp, as they are for actin.25
We next examined V75M, 1 of 3 disease-causing mutations occurring in HCSM (72-ψψψψKDxxxxSY-83) (Figure 3C) and found that, although the monoSUMOylated-WASp*S1 form (∼85 kDa) is present in the nucleus, multiple high-molecular-weight WASp*SUMO conjugates (>85 kDa) are deficient, suggesting a selective defect in SUMO chain (or SUMO:ubiquitin chain) formation, a finding also captured in K76R and K235R mutants (Figure 3F). Unexpectedly, the unmodified form (∼65 kDa) of V75M and K235R WASp mutants is also deficient in the nucleus. Why this might occur is unclear, although a similar finding was reported for another polySUMOylation-deficient mutant of yeast SMT3 (hSUMO1 homolog).28
Coimmunoprecipitation demonstrates that, although the binding of SAE2 to the V75M mutant is unaffected, that of UBC9 is lost, which is consistent with the paradigm that S*motifs are bound by UBC9.43 The finding that mutation of either lysine- or hydrophobic-residue perturbs WASp-SUMOylation lends further support to the functionality of S*motif-1. Significantly, in contrast to V75- and K76-residue mutations, other disease-causing mutations involving S24-, T45-, R86-, and K476-residues, although stably expressed (Figure 3E), do not impair WASp-SUMOylation (Figure 3F). This implies that not all pathogenic WAS mutations disrupt WASp-SUMOylation.
SUMOylation-deficient WASp favors COMMD1 recruitment to destabilize NF-κB at gene promoters in TH1 cells
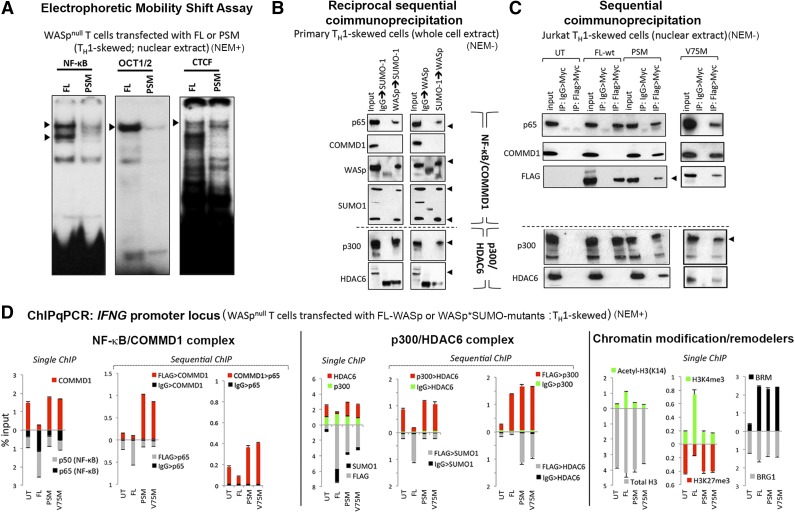
Because SUMO chain modification of both endogenous and transfected WASp is prominent in the nucleus and because SUMO chains are linked to chromatin signaling functions of its substrates,27,28 we sought to clarify its contribution to WASp’s role as a cotranscriptional factor. We focused on NF-κB signaling because of its relevance to TH1 differentiation44 and also because we previously showed that WASp deficiency impairs NF-κB activity in TH cells.4 EMSA assays demonstrate that, in PSM-expressing TH1 cells, DNA-binding activity of NF-κB and its downstream effector Octamer (OCT) proteins45 is diminished compared with that in TH cells expressing FL-WASp (Figure 4A). DNA-binding activity of CTCF is unaffected, which denotes the specificity of the WASp*SUMO effect on chromatin signaling.
Figure 4.
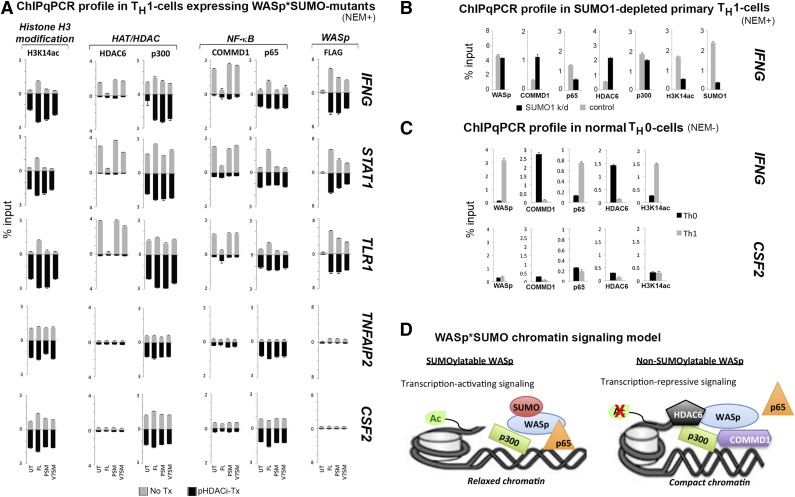
Molecular and functional characterization of SUMOylation-deficient WASp mutants. (A) EMSA with NF-κB, OCT1/2, and CTCF oligos performed on the nuclei isolated from TH1-skewed/TCR-activated WASp-deficient TH cells stably transfected with FL-WASp or PSM-WASp. Arrowheads indicate the location of the shifted bands. (B) Reciprocal, sequential immunoprecipitation (eg, WASp>SUMO1 sequence denotes 1stIP: WASp, 2ndIP: SUMO1) performed on the whole cell extract derived from primary TH1-skewed cells was resolved by western blotting the same gel with indicated antibodies. (C) The same gel shown in Figure 3G (Jurkat TH1-skewed cells, nuclear extract) was sequentially reprobed with the indicated antibodies. (D) MNase-ChIP. Chromatin fragmentation pattern and efficiency achieved with MNase treatment is shown in supplemental Figure 7. Chromatin enrichment profiles of the indicated proteins, at the 5′ untranslated region (−200 to −250 bp from first coding ATG, a region known to contain functional promoter elements) of the IFNG gene in TH1-skewed WASnull TH cells untransfected (UT) or stably transfected with Flag/Myc-tagged, full-length WASp (FL) or SUMOylation-deficient WASp mutants (PSM and V75M). For sequential ChIP, 2 rounds of conventional ChIPs were performed in the indicated sequence (FLAG>COMMD1, 1stChIP:FLAG, 2ndChIP:COMMD1; IgG>COMMD1, 1stChIP:IgG, 2ndChIP:COMMD1). The displayed ChIP values (mean ± standard error of the mean [SEM]) are percentage of total nuclear input chromatin and were derived after subtracting the background values obtained with isotype IgG antibody, the latter not shown for single ChIPs. Data were generated from 3 biologic replicates. NEM+/−, treated/nontreated with NEM.
To explicate the mechanism underlying impaired NF-κB:DNA-binding activity, we asked whether COMMD1, an inhibitor of NF-κB signaling at the DNA level,46,47 contributes to this defect. In sequential coimmunoprecipitation assays, we show that the WASp:SUMO1:NF-κB(p65) complex formed with endogenous WASp or transfected FL-WASp in TH1 cells does not contain COMMD1 (Figures 4B-C). Similarly, LC-MS/MS failed to recover COMMD1 polypeptides associated with normal WASp in primary TH1 cells.15 In contrast, SUMOylation-deficient WASp mutants (PSM, V75M) associate with COMMD1 and NF-κB(p65) (Figure 4C). In vivo, sequential ChIP assays showed increased coenrichment of COMMD1 with SUMOylation-deficient mutants compared with FL-WASp at the IFNG promoter in TH1 cells (Figure 4D). This finding was also captured at promoters of other WASp target genes, STAT1 and TLR1, but not TNFAIP2 or CSF2, which are both WASp nontarget genes (Figure 5A, no-Tx panel). Accordingly, single ChIP showed decreased promoter enrichment of NF-κB(p65) in TH1 cells expressing SUMOylation-deficient WASp, which is consistent with COMMD1’s function of removing NF-κB from DNA.46,47 Increased COMMD1 and decreased p65 enrichment at the IFNG promoter was also captured in normal TH1 cells depleted of endogenous SUMO1 by RNA interference (Figure 5B), as well as in normal TH0 cells (Figure 5C), where endogenous-WASp is non-SUMOylated and nonnuclear (Figure 2C). These findings propose a chromatin signaling function for WASp*SUMO1 in p65-dependent IFNG activation during TH0 to TH1 differentiation (Figure 5D).
Figure 5.
Characterizing the effect of SUMOylation-deficient WASp on the promoters of NF-κB response genes in the presence or absence of panHDAC inhibitors. (A) MNase-ChIP assays performed as described in the legend to Figure 4D in the absence (no Tx) or presence of panHDACi (pHDACi-Tx). The displayed ChIP values (mean ± SEM) are percentage of total nuclear input chromatin and were derived after subtracting the background values obtained with isotype IgG antibody; the latter is not shown. IFNG (TH1-signature cytokine), STAT1 (TH1-transcription factor), and TLR1 were selected as representative genes downregulated by SUMOylation-deficient WASp, whereas CSF2 (TH17-proinflammatory cytokine) and TNFAIP2 (proinflammatory cytokine) were representative genes ectopically upregulated by SUMOylation-deficient WASp. UT, untransfected WASp-null TH-cells; FL, same TH cells transfected with full-length WASp; PSM, transfected with penta-SUMO mutant; V75M, transfected with V75M disease-causing mutant. (B) MNase-ChIP assay performed on primary TH1-skewed cells from normal donor depleted of endogenous SUMO1 (by SUMO1-shRNA) or its control (scrambled shRNA) (see supplemental Figure 7 for chromatin-shearing efficiency and supplemental Figure 2B for temporal progression of SUMO1 depletion in these TH1 cells over 6 days following shRNA transfection). (C) MNase-ChIP assay performed on primary Th0 nonskewed and Th1-skewed cells with indicated antibodies at the indicated gene loci. (D) A working model of WASp*SUMO1 chromatin signaling at WASp target gene promoters proposing differential, activating vs repressive outputs of chromatin-located WASp that is informed by its SUMOylaton state. NEM+/−, treated/nontreated with NEM.
SUMOylation-deficient WASp favors HDAC6 recruitment and hypoacetylation of histone H3-lysine14 at p300-marked promoters of NF-κB response genes
Because histone acetyltransferase p300 regulates NF-κB-dependent gene activation48,49 including via impacting COMMD147 and HDAC6,50 we wondered whether a concomitant defect of p300 contributes to the NF-κB defect linked to non-SUMOylatable-WASp. Using sequential coimmunoprecipitation, we show that the endogenous WASp:SUMO1:p300 complex nucleated in primary TH1 cells does not contain HDAC6 (Figure 4B), suggesting that SUMOylatable-WASp associates with coactivator histone acetyltransferase activity. In contrast, SUMOylation-deficient WASp binds HDAC6 (Figure 4C), suggesting its ectopic association with corepressor HDAC activity.50 Furthermore, sequential ChIP assays showed HDAC6 coenriched with p300 at the IFNG promoter in PSM or V75M but not in FL-WASp-expressing TH1 cells, where this coassociation was nominal (Figure 4D). Similar findings were also captured at STAT1 and TLR1 promoters (Figure 5A, nontreated HAT/HDAC panel). Notably, promoter enrichment of p300 is unaffected (Figure 4D), suggesting that WASp-SUMOylation status does not impact p300 recruitment/retention but rather its functional output: activation vs repression.
We next asked whether the aberrant chromatin enrichment of HDAC6 impairs p300 HAT function of acetylating H3-lysine14 required for transcriptional activation.51 Sequential ChIP shows that increased HDAC6 enrichment occurs contemporaneously with decreased enrichment of acetylated H3K14 at IFNG, STAT1, and TLR1 promoters in PSM- and V75M-expressing TH1 cells (Figures 4D and 5A). Increased HDAC6 and decreased H3K14ac enrichment at the IFNG promoter was also captured in primary TH1 cells depleted of SUMO1 by RNA interference (Figure 5B) and in normal TH0 cells (Figure 5C). Because promoter occupancy/activity of NF-κB is attenuated by histone-H3 hypo-/deacetylation,52 together our data suggest that, for certain NF-κB response genes, HDAC6-mediated attenuation of the p300 HAT activity50 may contribute to NF-κB destabilization at chromatin in TH1 cells expressing SUMOylation-deficient WASp (Figure 5D).
SUMOylation-deficient WASp favors ectopic activation of proinflammatory genes while impeding activation of genes patterning TH1 immunity
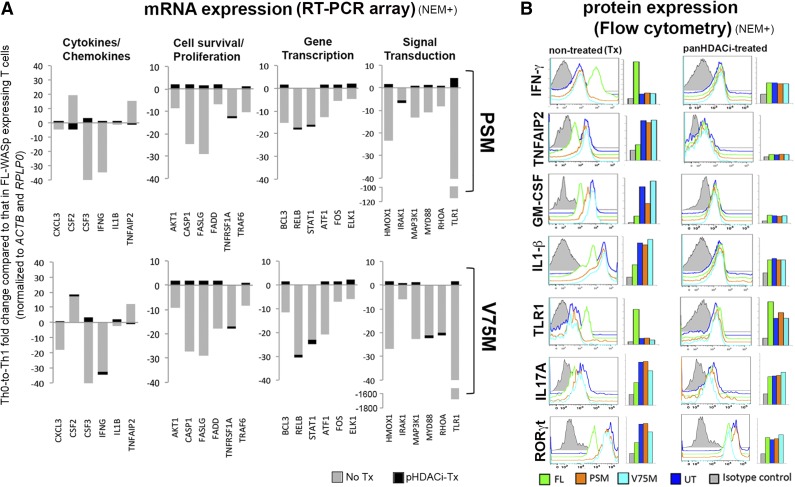
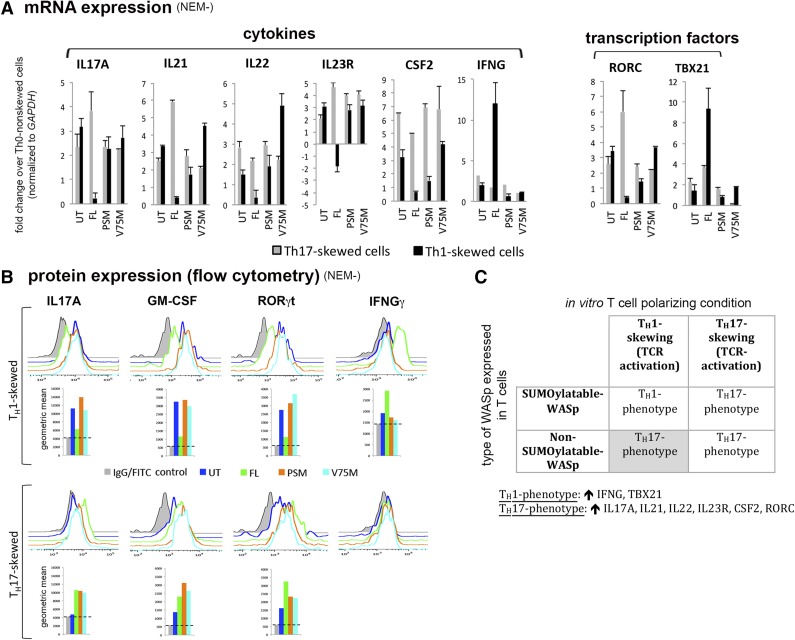
RT-PCR-based transcriptome profiling was used to broadly assess the effects of WASp-SUMOylation on the functional modulation of NF-κB response genes (n = 84; Figure 6A; supplemental Figure 5). Compared with FL-WASp, SUMOylation-deficient mutants showed decreased expression of most NF-κB response genes (n = 80) in TH1 cells. These genes fall into 4 gene ontology (GO) categories: (1) cytokines/chemokines, (2) cell survival/proliferation, (3) transcription, and (4) signal transduction. Whereas mRNA and protein expression of IFNG, a TH1 cytokine, is decreased, that of the proinflammatory cytokines granulocyte macrophage–colony-stimulating factor53 and tumor necrosis factor, alpha-induced protein 2 (TNFAIP2)54 is paradoxically increased (Figure 6, non-Tx panel). Similarly, expression of proinflammatory cytokine IL-1β (proIL-1β or mature IL-1β cannot be distinguished) is increased (Figure 6B, non-Tx panel) but without a concomitant mRNA increase (Figure 6A), a finding that could be linked to impaired NF-κB activity of preventing maturation of IL-1β.55 Furthermore, mRNA expression of BCL3 that imparts stability to the TH1 phenotype by preventing spontaneous conversion to the TH17 phenotype56 is low in TH1-skewed cells expressing SUMOylation-deficient WASp (Figure 6A). Accordingly, expression (mRNA and/or protein) of TH1 genes (IFNG, TBX21 STAT1, RELB57) is deficient, whereas that of TH17 genes (IL17A, IL21, IL22, IL23R, RORC, CSF2) is paradoxically increased in TH1-skewed cells expressing SUMOylation-deficient WASp (Figures 6B and 7). In contrast, upregulation of TH17 genes is unaffected in TH17-skewed cells expressing SUMOylation-deficient WASp (Figure 7). These finding imply that WASp-SUMOylation is necessary not only for activation of TH1 immunity but also for checking ectopic activation of proinflammatory pathways that destabilizes TH1 specification.
Figure 6.
Modulation of NF-κB response genes by SUMOylation-deficient WASp mutant. (A) The effect of SUMOylation-deficient WASp mutants (PSM and V75M) on mRNA expression levels of multiple NF-κB response genes in human TH1-skewed cells, in the presence (+) or absence (−) of panHDAC inhibitors (HDACi). Shown are some representative genes from the different gene ontology (GO) categories selected from the complete dataset including their heatmaps displayed in supplemental Figure 5. The mRNA expression, quantified by RT-PCR array, in SUMOylation-deficient WASp-expressing TH1 cells was compared with that in FL-WASp-expressing TH1 cells after normalizing to their respective control genes (ACTB and RPLP0) and then represented as fold upregulation (activated) or downregulation (repressed). (B) Flow cytometry showing histogram profiles for intracellular expression of indicated cytokines and transcription factors (after permeabilization) and cell surface expression of TLR1 (without permeabilization) along with their corresponding geometric means (GMs). Staining with isotype IgAb (gray histogram) is shown as a control. NEM+, treated with NEM.
Figure 7.
SUMOylation-deficient WASp favors paradoxical development of TH17-like phenotype in TH1-skewed cells. (A) mRNA expression profiles of TH1 and TH17 genes in WASnull TH cells reconstituted with SUMOylation-deficient mutants, normal wt-WASp (FL), or untransfected (UT) control quantified by RT-PCR. The displayed values (mean ± SEM) are fold change (up or down) in TH1 or TH17 cells compared with values obtained in TH0 cells after normalizing to GAPDH. The data are representative of 2 biologic replicates each with 3 samples assayed. (B) Flow cytometry displaying protein expression of some representative TH1/TH17 genes in the same cells and mutants described in A. Geometric mean (GM) is displayed underneath individual histograms. Horizontal dotted lines demarcate background fluorescence obtained with control immunoglobulin-fluorescein isothiocyanate/phycoerythrin staining. (C) A 2 × 2 table summarizing the key findings of this figure. ↑, increased expression compared to Th0 non-skewed cells; NEM−, NEM nontreated.
Pan-histone deacetylase inhibitors restore misregulated gene expression imposed by SUMOylation-deficient WASp
Because NF-κB activity is downmodulated by HDAC1, 2, 3, and 658-60 and because the pan-histone deacetylase inhibitor (pHDACi) was shown to displace HDAC661,62 and relieve p300 transcriptional repression,50 we tested the hypothesis that pHDACi may be sufficient to neutralize COMMD1- and HDAC6-mediated transcriptional repression imposed by SUMOylation-deficient WASp.
TH1-differentiating, WASpnull patient TH cells untransfected (UT) or transfected with FL-WASp, PSM-WASp, or V75M-WASp were cocultured with or without pHDACi (trichostatin A and Na butyrate), and their mRNA levels were quantified in the RT-PCR-based NF-κB array. TH1 cells expressing normal WASp showed that about two-thirds of the NF-κB response genes were neither up- nor downregulated (>2.0 fold) by pHDACi (supplemental Figure 5C, blue-shaded column), which agrees with prior reports demonstrating that <20% of genes become misregulated by pHDACi treatment.63,64 Of the genes downregulated by pHDACi in normal TH1 cells, most belonged to cell survival/proliferation and chemokines/cytokines GO categories, with CSF3 and CCL2 demonstrating marked suppression by pHDACi. In TH1 cells expressing SUMOylation-deficient WASp, however, pHDACi treatment displayed a more dramatic switch in mRNA expression patterns compared with their nontreated controls. Specifically, pHDACi restored to normal (or increased) mRNA expression of most NF-κB response genes, and correspondingly also some of the tested proteins, which were deficient in nontreated controls (Figure 6; supplemental Figure 5). Interestingly, pHDACi treatment normalized (or decreased) the expression of multiple proinflammatory factors (granulocyte macrophage–colony-stimulating factor, TNFAIP2, IL-1β, IL17A) that were aberrantly increased in nontreated controls (Figure 6; supplemental Figure 5C), a finding that aligns with the known inhibitory effect of pHDACi on proinflammatory cytokine signaling.63
pHDACi reverses promoter trans-repression imposed by SUMOylation-deficient WASp
Because pHDACi functions in the nucleus to either deacetylase histone tails or evict HDAC-containing corepressors from gene promoters,62,65,66 we asked whether the salutary effect of pHDACi on misregulated transcription is consequent to these epigenetic modes of actions. First, we show that the dose and duration of pHDACi treatment is sufficient to augment promoter enrichment of acetylated H3K14 in FL-WASp-expressing TH1 cells by ChIP (Figure 5A, histoneH3 modification column). Significantly, in TH1 cells expressing SUMOylation-deficient WASp, low H3K14ac and high HDAC6 enrichment observed in nontreated cells is restored to near-normal levels on pHDACi treatment at the IFNG, STAT1, and TLR1 promoters (Figure 5A, HAT/HDAC column). Contemporaneously, promoter occupancy of NF-κB is increased but that of its inhibitor COMMD1 is decreased by pHDACi (Figure 5A, NF-κB column), implying that acetylated H3K14 stabilizes NF-κB on gene promoters in TH1 cells, a paradigm that was previously established in the HeLa and SIRT6 murine model.52 Conversely, neither HDAC6 nor COMMD1 is enriched at the CSF2 and TNFAIP2 promoters, which could explain the chromatin “licensing” for its ectopic activation in TH1 cells expressing SUMOylation-deficient WASp. We conclude that HDAC-associated activity imposed by SUMOylation-defective WASp is required for trans-repression of a subset of NF-κB response genes in TH1 cells, the perturbation of which may underlie imbalanced activation of pathways supporting inflammation over immunity in WAS TH cells.
Discussion
A coactivator role for WAS family proteins in transcription is established for nucleus-located WASp, Wave1, N-WASp, WASH, and JMY in different organisms.3,5,8,11,67,68 The present study identifies a corepressive role for WASp and provides the initial insights into the molecular pathway that converts nuclear WASp from a transcriptional coactivator to a promoter-specific corepressor of NF-κB response genes in TH1 cells.
WASp bound to promoters instructs a dichotomous transcriptional outcome depending on whether it is SUMO modified or not. A model emerges wherein SUMOylatable-WASp transactivates and non-SUMOylatable-WASp transrepresses gene activation during TH1 differentiation (Figure 5D). Implicit in this model is the idea that WASp participates in the process that controls the exchange between coactivator and corepressor occupancy at target loci and that WASp-SUMOylation informs this outcome. Because coactivators and corepressors are continuously exchanged at transcriptionally active genes in TH cells,69 whether loss of WASp-SUMOylation actively recruits corepressors (repression model) or it functions to evict promoter-bound corepressors (derepression model)70 cannot be distinguished. The constitutive presence of corepressive complexes on the IFNG locus in normal TH0 cells, where endogenous WASp is physiologically non-SUMOylated, favors the derepression model. Furthermore, because the corepressive activity of WASp is manifest in a disease context where WASp is mutated, this begs the question of whether wild-type WASp is intrinsically a coactivator or corepressor. Our identification of SUMOylation as a physiologic PTM of WASp in multiple immune cells favors WASp to be inherently a coactivator. This idea, however, could be challenged if physiologic processes that disrupt WASp-SUMOylation converting normal (nonmutated) WASp from a coactivator to corepressor are uncovered.
Pending clarification of these working models, accumulating evidence proposes WASp family proteins (WASp, WASH/FAM21) as nuclear factors that modulate transcriptional output of NF-κB response genes,4,67 with the present study elucidating the molecular underpinnings of the WASp effect on NF-κB chromatin signaling. Although a detailed molecular analysis of all NF-κB response genes affected by disrupted WASp-SUMOylation is beyond the scope of this study, a picture emerges wherein TH1-skewed cells expressing SUMOylation-deficient WASp exhibit a proinflammatory signature in the face of deficient TH1 function. First, multiple TH17 genes are ectopically upregulated under TH1 conditions. Second, the expression of two proinflammatory genes CSF2 and TNFAIP2 is inappropriately increased. Third, the expression of the IL-1β protein is increased but without a concomitant increase in mRNA expression, a finding that is consistent with the enhanced processing of mature IL-1β from proIL-1β, a caspase-1-dependent process that is negatively regulated by NF-κB.55 Because these proinflammatory cytokines have been linked to autoimmunity/autoinflammation,53,54,71 our data propose that part of the cellular defect underling development of autoimmunity/autoinflammation in a subset of WAS patients expressing non-SUMOylatable-WASp is contributed by the ectopic activation of inappropriate immune function genes.
Regardless of how many pathway genes become misregulated by non-SUMOylatable-WASp, the linking of the WASp-SUMOylation defect to the imbalance between TH1 immunity and inflammation has immediate clinical relevance for WAS, because disease-causing mutations, many resulting in XLT-to-WAS clinical progression including V75M, reside in SUMO and SIM motifs of WASp (Figure 3C).72 Whether these mutations similarly disrupt WASp-SUMOylation to imbue proinflammatory qualities to TH1-deficient cells remains to be determined. We do, however, show that not all disease-causing mutations that result in XLT-to-WAS progression (eg, missense mutations of S24, T45, and R86 residues that do not reside in SUMO or SIM motifs) impair WASp-SUMOylation and yet impede gene activation, the latter via a SWI/SNF-linked mechanism.4 In contrast, V75M impairs WASp-SUMOylation but does not disrupt SWI/SNF promoter activity (Figure 4D). The SWI/SNF-disrupting T45M mutation that phenocopies V75M clinically (XLT-to-WAS progression) does not display ectopic enrichment of HDAC6 or COMMD1 at the IFNG promoter observed with V75M but yet impairs NF-κB enrichment (supplemental Figure 6). These observations imply that pathogenic mutations have differing proclivity to disrupt different steps of transcription along the DNA translational axis. Because we show that loss of WASp phenocopies non-SUMOylatable-WASp in causing TH1 immunity/inflammation imbalance, our findings are also relevant for some patient T cells lacking WASp expression.
Interestingly, of the disease-causing mutations that do impair WASp-SUMOylation (missense mutations of V75-, K76-, and K476-residues), not all display the identical SUMOylation defect. Mutations of V75- and K76-residues both impair polySUMO chains without affecting monoSUMOylation, a defect sufficient to recruit corepressors (COMMD1, HDAC6) at promoters of NF-κB response genes that are also H3K14 hypoacetylated. Certainly, besides HDAC6 and COMMD1, other HDAC-containing corepressors70 may also contribute to the repressive chromatin signaling module associated with deficient polySUMO chain modification of WASp. We also cannot exclude the possibility that V75M, besides disrupting SUMOylation, may introduce conformational defects, which may contribute to altered function. At the minimum, our findings linking a selective SUMO chain defect to aberrant gene repression suggest that polySUMO modification support a specific chromatin signaling function distinct from that of the monoSUMO modification of WASp. Although the causality of this association remains speculative, emerging evidence linking polySUMO chain defects to impaired chromatin-to-nucleolar targeting and DNA instability,73 altered chromatin organization and transcriptional repression,28 and Alzheimer’s disease74 together propose a physiologically relevant role for WASp polySUMO chains in immunity.
Future challenges in the WASp-SUMO field will be to determine (1) what fraction of these polymeric chains comprises a SUMO ubiquitin hybrid vs SUMO or ubiquitin alone and how their individual loss impairs function; (2) the effect of WASp-SUMOylation on immunological synapse formation; and (3) how WASp-SUMOylation influences regulatory networks controlling immunity/inflammation balance in health and disease. The latter is topical because many causal variants in autoimmune diseases map to NF-κB consensus motifs in the TH-cell genome,75 and SUMOylation-deficient WASp, we show, disturbs this NF-κB:DNA interaction. Notwithstanding, our identification of HDACi-mediated correction of gene misregulation in SUMOylation-impaired WASp TH cells offers chromatin alteration using panHDACis (or the selective HDAC6 inhibitor) as a new target for individualized therapy to restore immunodeficiency and attenuate autoimmunity in WAS patients, as previously entertained.76
Acknowledgments
The authors thank Jessica Hook of the University of Iowa for assistance with microscopy and imaging, M. Balasubramani of the University of Pittsburgh Proteomic Core Facility for analyzing the MS data, and the University of Iowa Dance Marathon for providing laboratory space.
This research was supported by National Institutes of Health, National Institute of Allergy and Infectious Diseases grants R01 AI073561 and R01 AI084957.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: K.S. performed the majority of the experiments; S.S. generated all SUMO mutants; S.-S.H. performed the EMSA assays; and Y.M.V. conceived the study, designed the experiments, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Current affiliation for S.S. is Department of Botany, Raiganj University, Raiganj 733134, West Bengal, India.
Correspondence: Yatin M. Vyas, Division of Pediatric Hematology-Oncology, Stead Family Department of Pediatrics, University of Iowa Children’s Hospital, Iowa City, IA 52242; e-mail: yatin-vyas@uiowa.edu.
References
- 1.Villa A, Notarangelo L, Macchi P, et al. X-linked thrombocytopenia and Wiskott-Aldrich syndrome are allelic diseases with mutations in the WASP gene. Nat Genet. 1995;9(4):414–417. doi: 10.1038/ng0495-414. [DOI] [PubMed] [Google Scholar]
- 2.Symons M, Derry JM, Karlak B, et al. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84(5):723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 3.Taylor MD, Sadhukhan S, Kottangada P, et al. Nuclear role of WASp in the pathogenesis of dysregulated TH1 immunity in human Wiskott-Aldrich syndrome. Sci Transl Med. 2010;2(37):37ra44. doi: 10.1126/scitranslmed.3000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarkar K, Sadhukhan S, Han SS, Vyas YM. Disruption of hSWI/SNF complexes in T cells by WAS mutations distinguishes X-linked thrombocytopenia from Wiskott-Aldrich syndrome. Blood. 2014;124(23):3409–3419. doi: 10.1182/blood-2014-07-587642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyamoto K, Teperek M, Yusa K, Allen GE, Bradshaw CR, Gurdon JB. Nuclear Wave1 is required for reprogramming transcription in oocytes and for normal development. Science. 2013;341(6149):1002–1005. doi: 10.1126/science.1240376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verboon JM, Rincon-Arano H, Werwie TR, et al. Wash interacts with lamin and affects global nuclear organization. Curr Biol. 2015;25(6):804–810. doi: 10.1016/j.cub.2015.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11(4):451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X, Yoo Y, Okuhama NN, Tucker PW, Liu G, Guan JL. Regulation of RNA-polymerase-II-dependent transcription by N-WASP and its nuclear-binding partners. Nat Cell Biol. 2006;8(7):756–763. doi: 10.1038/ncb1433. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez-Mesa E, Abreu-Blanco MT, Rosales-Nieves AE, Parkhurst SM. Developmental expression of Drosophila Wiskott-Aldrich Syndrome family proteins. Dev Dyn. 2012;241(3):608–626. doi: 10.1002/dvdy.23742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goley ED, Ohkawa T, Mancuso J, et al. Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science. 2006;314(5798):464–467. doi: 10.1126/science.1133348. [DOI] [PubMed] [Google Scholar]
- 11.Xia P, Wang S, Huang G, et al. WASH is required for the differentiation commitment of hematopoietic stem cells in a c-Myc-dependent manner. J Exp Med. 2014;211(10):2119–2134. doi: 10.1084/jem.20140169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404(6774):151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- 13.Badour K, Zhang J, Shi F, Leng Y, Collins M, Siminovitch KA. Fyn and PTP-PEST-mediated regulation of Wiskott-Aldrich syndrome protein (WASp) tyrosine phosphorylation is required for coupling T cell antigen receptor engagement to WASp effector function and T cell activation. J Exp Med. 2004;199(1):99–112. doi: 10.1084/jem.20030976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA. 2011;108(33):E472–E479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadhukhan S, Sarkar K, Taylor M, Candotti F, Vyas YM. Nuclear role of WASp in gene transcription is uncoupled from its ARP2/3-dependent cytoplasmic role in actin polymerization. J Immunol. 2014;193(1):150–160. doi: 10.4049/jimmunol.1302923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cubeñas-Potts C, Matunis MJ. SUMO: a multifaceted modifier of chromatin structure and function. Dev Cell. 2013;24(1):1–12. doi: 10.1016/j.devcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 18.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev. Mol Cell Biol. 2007;8(12):947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 19.Jentsch S, Psakhye I. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu Rev Genet. 2013;47:167–186. doi: 10.1146/annurev-genet-111212-133453. [DOI] [PubMed] [Google Scholar]
- 20.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108(1):109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 21.Nayak A, Viale-Bouroncle S, Morsczeck C, Muller S. The SUMO-specific isopeptidase SENP3 regulates MLL1/MLL2 methyltransferase complexes and controls osteogenic differentiation. Mol Cell. 2014;55(1):47–58. doi: 10.1016/j.molcel.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Ungureanu D, Vanhatupa S, Grönholm J, Palvimo JJ, Silvennoinen O. SUMO-1 conjugation selectively modulates STAT1-mediated gene responses. Blood. 2005;106(1):224–226. doi: 10.1182/blood-2004-11-4514. [DOI] [PubMed] [Google Scholar]
- 23.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2(2):233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 24.Hendriks IA, D’Souza RC, Yang B, Verlaan-de Vries M, Mann M, Vertegaal AC. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol. 2014;21(10):927–936. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofmann WA, Arduini A, Nicol SM, et al. SUMOylation of nuclear actin. J Cell Biol. 2009;186(2):193–200. doi: 10.1083/jcb.200905016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Werner A, Flotho A, Melchior F. The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol Cell. 2012;46(3):287–298. doi: 10.1016/j.molcel.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 27.Ulrich HD. The fast-growing business of SUMO chains. Mol Cell. 2008;32(3):301–305. doi: 10.1016/j.molcel.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Srikumar T, Lewicki MC, Costanzo M, et al. Global analysis of SUMO chain function reveals multiple roles in chromatin regulation. J Cell Biol. 2013;201(1):145–163. doi: 10.1083/jcb.201210019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosonina E, Duncan SM, Manley JL. SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev. 2010;24(12):1242–1252. doi: 10.1101/gad.1917910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terui Y, Saad N, Jia S, McKeon F, Yuan J. Dual role of sumoylation in the nuclear localization and transcriptional activation of NFAT1. J Biol Chem. 2004;279(27):28257–28265. doi: 10.1074/jbc.M403153200. [DOI] [PubMed] [Google Scholar]
- 31.Ouyang J, Gill G. SUMO engages multiple corepressors to regulate chromatin structure and transcription. Epigenetics. 2009;4(7):440–444. doi: 10.4161/epi.4.7.9807. [DOI] [PubMed] [Google Scholar]
- 32.Makhnevych T, Sydorskyy Y, Xin X, et al. Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol Cell. 2009;33(1):124–135. doi: 10.1016/j.molcel.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 33.Pascual G, Fong AL, Ogawa S, et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437(7059):759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gómez-del Arco P, Koipally J, Georgopoulos K. Ikaros SUMOylation: switching out of repression. Mol Cell Biol. 2005;25(7):2688–2697. doi: 10.1128/MCB.25.7.2688-2697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee FY, Faivre EJ, Suzawa M, et al. Eliminating SF-1 (NR5A1) sumoylation in vivo results in ectopic hedgehog signaling and disruption of endocrine development. Dev Cell. 2011;21(2):315–327. doi: 10.1016/j.devcel.2011.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu HW, Zhang J, Heine GF, et al. Chromatin modification by SUMO-1 stimulates the promoters of translation machinery genes. Nucleic Acids Res. 2012;40(20):10172–10186. doi: 10.1093/nar/gks819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Q, Xie Y, Zheng Y, et al. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014;42(Web Server issue):W325-30. doi: 10.1093/nar/gku383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang W, Ochs HD, Dupont B, Vyas YM. The Wiskott-Aldrich syndrome protein regulates nuclear translocation of NFAT2 and NF-kappa B (RelA) independently of its role in filamentous actin polymerization and actin cytoskeletal rearrangement. J Immunol. 2005;174(5):2602–2611. doi: 10.4049/jimmunol.174.5.2602. [DOI] [PubMed] [Google Scholar]
- 39.Praefcke GJ, Hofmann K, Dohmen RJ. SUMO playing tag with ubiquitin. Trends Biochem Sci. 2012;37(1):23–31. doi: 10.1016/j.tibs.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 40.Impens F, Radoshevich L, Cossart P, Ribet D. Mapping of SUMO sites and analysis of SUMOylation changes induced by external stimuli. Proc Natl Acad Sci USA. 2014;111(34):12432–12437. doi: 10.1073/pnas.1413825111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hendriks IA, D’Souza RC, Yang B, Verlaan-de Vries M, Mann M, Vertegaal AC. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol. 2014;21(10):927–936. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matic I, Schimmel J, Hendriks IA, et al. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell. 2010;39(4):641–652. doi: 10.1016/j.molcel.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 43.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276(24):21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 44.Aronica MA, Mora AL, Mitchell DB, et al. Preferential role for NF-kappa B/Rel signaling in the type 1 but not type 2 T cell-dependent immune response in vivo. J Immunol. 1999;163(9):5116–5124. [PubMed] [Google Scholar]
- 45.Mueller K, Quandt J, Marienfeld RB, et al. Octamer-dependent transcription in T cells is mediated by NFAT and NF-κB. Nucleic Acids Res. 2013;41(4):2138–2154. doi: 10.1093/nar/gks1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burstein E, Hoberg JE, Wilkinson AS, et al. COMMD proteins, a novel family of structural and functional homologs of MURR1. J Biol Chem. 2005;280(23):22222–22232. doi: 10.1074/jbc.M501928200. [DOI] [PubMed] [Google Scholar]
- 47.O’Hara A, Simpson J, Morin P, et al. p300-mediated acetylation of COMMD1 regulates its stability, and the ubiquitylation and nucleolar translocation of the RelA NF-κB subunit. J Cell Sci. 2014;127(Pt 17):3659–3665. doi: 10.1242/jcs.149328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perkins ND, Felzien LK, Betts JC, Leung K, Beach DH, Nabel GJ. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275(5299):523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 49.Gerritsen ME, Williams AJ, Neish AS, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA. 1997;94(7):2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Girdwood D, Bumpass D, Vaughan OA, et al. P300 transcriptional repression is mediated by SUMO modification. Mol Cell. 2003;11(4):1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 51.Luebben WR, Sharma N, Nyborg JK. Nucleosome eviction and activated transcription require p300 acetylation of histone H3 lysine 14. Proc Natl Acad Sci USA. 2010;107(45):19254–19259. doi: 10.1073/pnas.1009650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawahara TL, Michishita E, Adler AS, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136(1):62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El-Behi M, Ciric B, Dai H, et al. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol. 2011;12(6):568–575. doi: 10.1038/ni.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greten FR, Arkan MC, Bollrath J, et al. NF-kappaB is a negative regulator of IL-1beta secretion as revealed by genetic and pharmacological inhibition of IKKbeta. Cell. 2007;130(5):918–931. doi: 10.1016/j.cell.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang W, Wang H, Claudio E, et al. The oncoprotein and transcriptional regulator Bcl-3 governs plasticity and pathogenicity of autoimmune T cells. Immunity. 2014;41(4):555–566. doi: 10.1016/j.immuni.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corn RA, Hunter C, Liou HC, Siebenlist U, Boothby MR. Opposing roles for RelB and Bcl-3 in regulation of T-box expressed in T cells, GATA-3, and Th effector differentiation. J Immunol. 2005;175(4):2102–2110. doi: 10.4049/jimmunol.175.4.2102. [DOI] [PubMed] [Google Scholar]
- 58.Ashburner BP, Westerheide SD, Baldwin AS., Jr The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol. 2001;21(20):7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21(23):6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang W, Kone BC. NF-kappaB inhibits transcription of the H(+)-K(+)-ATPase alpha(2)-subunit gene: role of histone deacetylases. Am J Physiol Renal Physiol. 2002;283(5):F904–F911. doi: 10.1152/ajprenal.00156.2002. [DOI] [PubMed] [Google Scholar]
- 61.Fiskus W, Rao R, Fernandez P, et al. Molecular and biologic characterization and drug sensitivity of pan-histone deacetylase inhibitor-resistant acute myeloid leukemia cells. Blood. 2008;112(7):2896–2905. doi: 10.1182/blood-2007-10-116319. [DOI] [PubMed] [Google Scholar]
- 62.Akimova T, Beier UH, Liu Y, Wang L, Hancock WW. Histone/protein deacetylases and T-cell immune responses. Blood. 2012;119(11):2443–2451. doi: 10.1182/blood-2011-10-292003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joseph J, Mudduluru G, Antony S, Vashistha S, Ajitkumar P, Somasundaram K. Expression profiling of sodium butyrate (NaB)-treated cells: identification of regulation of genes related to cytokine signaling and cancer metastasis by NaB. Oncogene. 2004;23(37):6304–6315. doi: 10.1038/sj.onc.1207852. [DOI] [PubMed] [Google Scholar]
- 64.Peart MJ, Smyth GK, van Laar RK, et al. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2005;102(10):3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenetics. 2012;4(1):5. doi: 10.1186/1868-7083-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith KT, Martin-Brown SA, Florens L, Washburn MP, Workman JL. Deacetylase inhibitors dissociate the histone-targeting ING2 subunit from the Sin3 complex. Chem Biol. 2010;17(1):65–74. doi: 10.1016/j.chembiol.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deng ZH, Gomez TS, Osborne DG, Phillips-Krawczak CA, Zhang JS, Billadeau DD. Nuclear FAM21 participates in NF-κB-dependent gene regulation in pancreatic cancer cells. J Cell Sci. 2015;128(2):373–384. doi: 10.1242/jcs.161513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shikama N, Lee CW, France S, et al. A novel cofactor for p300 that regulates the p53 response. Mol Cell. 1999;4(3):365–376. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- 69.Wang Z, Zang C, Cui K, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138(50):1019-1031. [DOI] [PMC free article] [PubMed]
- 70.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11(2):109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 71.Noster R, Riedel R, Mashreghi MF, et al. IL-17 and GM-CSF expression are antagonistically regulated by human T helper cells. Sci Transl Med. 2014;6(241):241ra80. doi: 10.1126/scitranslmed.3008706. [DOI] [PubMed] [Google Scholar]
- 72.Albert MH, Bittner TC, Nonoyama S, et al. X-linked thrombocytopenia (XLT) due to WAS mutations: clinical characteristics, long-term outcome, and treatment options. Blood. 2010;115(16):3231–3238. doi: 10.1182/blood-2009-09-239087. [DOI] [PubMed] [Google Scholar]
- 73.Takahashi Y, Strunnikov A. In vivo modeling of polysumoylation uncovers targeting of Topoisomerase II to the nucleolus via optimal level of SUMO modification. Chromosoma. 2008;117(2):189–198. doi: 10.1007/s00412-007-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y, Wang H, Wang S, Quon D, Liu YW, Cordell B Li Y1. Positive and negative regulation of APP amyloidogenesis by sumoylation. Proc Natl Acad Sci USA. 2003;100(1):259–264. doi: 10.1073/pnas.0235361100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Farh KK, Marson A, Zhu J, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518(7539):337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Teitell MA. Alternative control: what’s WASp doing in the nucleus? Sci Transl Med. 2010;2(37):37ps31. doi: 10.1126/scitranslmed.3001336. [DOI] [PubMed] [Google Scholar]