Background: Artemis is a nuclease that is necessary for hairpin opening in V(D)J recombination.
Results: Artemis action on blunt DNA ends is dependent on DNA sequence (breathing) and Ku.
Conclusion: Breathing of blunt DNA ends into a transient ss/dsDNA boundary is needed for Artemis action.
Significance: Unification of Artemis nuclease action explains the features of NHEJ and V(D)J recombination.
Keywords: DNA, DNA binding protein, DNA endonuclease, DNA enzyme, DNA repair, endonuclease
Abstract
Artemis is a member of the metallo-β-lactamase protein family of nucleases. It is essential in vertebrates because, during V(D)J recombination, the RAG complex generates hairpins when it creates the double strand breaks at V, D, and J segments, and Artemis is required to open the hairpins so that they can be joined. Artemis is a diverse endo- and exonuclease, and creating a unified model for its wide range of nuclease properties has been challenging. Here we show that Artemis resects iteratively into blunt DNA ends with an efficiency that reflects the AT-richness of the DNA end. GC-rich ends are not cut by Artemis alone because of a requirement for DNA end breathing (and confirmed using fixed pseudo-Y structures). All DNA ends are cut when both the DNA-dependent protein kinase catalytic subunit and Ku accompany Artemis but not when Ku is omitted. These are the first biochemical data demonstrating a Ku dependence of Artemis action on DNA ends of any configuration. The action of Artemis at blunt DNA ends is slower than at overhangs, consistent with a requirement for a slow DNA end breathing step preceding the cut. The AT sequence dependence, the order of strand cutting, the length of the cuts, and the Ku-dependence of Artemis action at blunt ends can be reconciled with the other nucleolytic properties of both Artemis and Artemis·DNA-PKcs in a model incorporating DNA end breathing of blunt ends to form transient single to double strand boundaries that have structural similarities to hairpins and fixed 5′ and 3′ overhangs.
Introduction
In dividing mammalian cells, there are an estimated 10 DNA double-stranded breaks (DSBs)2 per day per cell (1–3). These pathological DSBs arise from ionizing radiation, oxygen free radicals, replication errors, and inadvertent cleavage by nuclear enzymes (4). Many of these pathological breaks and the physiological breaks in V(D)J recombination require the nonhomologous DNA end-joining (NHEJ) nuclease Artemis to repair the DNA. In NHEJ, the DSB is first recognized by the Ku heterodimer (Ku70/80), which acts as a “tool belt” to which other NHEJ proteins are recruited as needed. When DNA resection is required, a DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is recruited in complex with Artemis. DNA-PKcs undergoes autophosphorylation and activates Artemis endonuclease activity (5, 6). Activated Artemis then gains the ability to cut many DNA substrates at the single to double strand DNA boundaries (7) (Fig. 1).
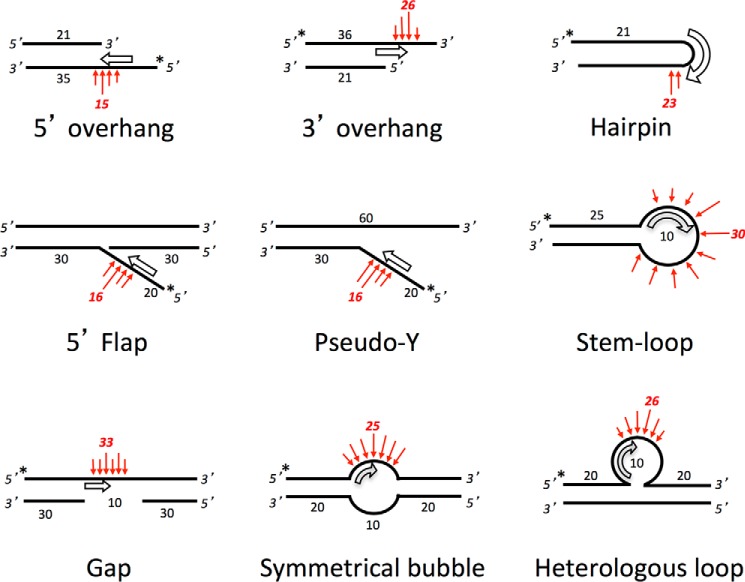
FIGURE 1.
Optimal substrates for Artemis have ss/dsDNA boundaries that direct Artemis to the point of preferred nuclease action. Shown are the known major substrates for Artemis (7, 11). The putative orientation and position of the Artemis enzyme are indicated by the open thick arrows, consistent with the preference of Artemis for ss/dsDNA boundaries and its propensity to cut on the 3′ side of an ∼4-nt ssDNA region at this boundary. The preferred cutting sites of Artemis are noted with red arrows. We propose that weaker activity may be seen on variants of these structures when conditions favor DNA breathing into one of the forms shown here. In addition to the substrates shown, Artemis has 5′ exonuclease activity at 5′ protruding DNA ends (9), which is the only activity that does not rely on DNA-PKcs (under Mg2+ conditions) (see text). Artemis also has very weak endonuclease activity on ssDNA, which is stimulated by DNA-PKcs and can be slightly stronger for ssDNA that can self-anneal or fold back at internal positions because of self-complementarity (6).
Artemis is in the metallo-β-lactamase family of nucleases, characterized by conserved metallo-β-lactamase and β-CASP domains. This family of nucleases is able to hydrolyze DNA or RNA in various configurations (8). Artemis alone has intrinsic 5′ exonuclease activity on single-stranded DNA (ssDNA) (9). On duplex DNA, Artemis, in complex with DNA-PKcs, has endonuclease activity on the 5′ and 3′ DNA overhangs that are often created by pathological DNA breaks and on DNA hairpins that are formed during V(D)J recombination. Therefore, patients lacking Artemis suffer from severe combined immunodeficiency because of a defect in antibody formation (10).
We have determined biochemically that Artemis, in complex with DNA-PKcs, resects 5′ and 3′ DNA overhangs to create DNA end structures that are able to be ligated by the DNA ligase IV-XRCC4 complex (11, 12). At 5′ overhangs, Artemis cuts directly at the ss/dsDNA boundary. However, when processing 3′ overhangs and DNA hairpins, it preferentially leaves a 4-nt overhang (7). Perfect DNA hairpins have sterically constrained tight turns and, therefore, have ss/ds boundaries at the last two base pairs (4 nt) (13). This may make them structurally similar to DNA overhangs. From these observations, we proposed that Artemis activity on duplex DNA can be unified under a model in which Artemis·DNA-PKcs binds to the ss/dsDNA boundary to occupy 4 nt along the single-stranded segment. This binding is followed by preferentially nicking the 3′ side of those 4 nt. Although the current model is extensive, it lacks information on whether Artemis can cut blunt-ended DNA that can be generated by chemotherapeutic agents, free radicals, or ionizing radiation (14).
Here we explore the action of Artemis at blunt DNA ends, which appears substantially weaker than its action at ss/dsDNA boundaries. We found that resection at blunt end depends on the DNA sequence of the duplex end, with AT-rich DNA ends being resected by Artemis alone but GC ends hardly at all. When Artemis is accompanied by DNA-PKcs, it can act at all blunt ends, but in a Ku-dependent manner. This is the first documentation showing that Ku can modulate Artemis activity. The sequence dependence and other features of Artemis action permit the unification of all nucleolytic properties of Artemis into a single model for its action at ss/dsDNA boundaries.
Experimental Procedures
Oligonucleotides and DNA Substrates
The Oligonucleotides used in this study were synthesized by Integrated DNA Technologies, Inc. (San Diego, CA). The oligonucleotides were purified using denaturing PAGE, and the concentrations were determined spectrophotometrically. 5′ end labeling of DNA substrates was done with [γ-32P]ATP (3000 Ci/mmol) (PerkinElmer Life Sciences) and T4 polynucleotide kinase (PNK) (New England Biolabs) according to the instructions of the manufacturer. Unincorporated radioisotope was removed by using Sephadex G-25 spin columns (Enzymax). To create dsDNA, the unlabeled complementary strand was added, and the sample was boiled for 5 min to inactivate T4 PNK. The DNA was allowed to slowly cool to room temperature and then incubated overnight at 4 °C to promote proper base pairing of the complementary strand.
3′ end labeling was performed by fill-in synthesis of [α-32P]dNTP (3000 Ci/mmol) (PerkinElmer Life Sciences) onto preannealed dsDNA with a 1-nt 5′ overhang with the Klenow fragment of DNA polymerase I (3′ → 5′ exo−) (New England Biolabs) to create a blunt-ended substrate. Unincorporated radioisotope was removed as stated above.
The sequences of the oligonucleotides used in this study were as follows: HC33, 5′-GCG GAG TGT CTG CAT CTT ACT TGA CGG ATG CAA TCG TCA CGT GCT AGA CTA CTG GTC AAG CGG ATC TTA GGG G-3′; HC34, 5′-CCC CTA AGA TCC GCT TGA CCA GTA GTC TAG CAC GTG ACG ATT GCA TCC GTC AAG TAA GAT GCA GAC ACT CCG C-3′; HC57, 5′-TTT TAG TGT CTG CAT CTT ACT TGA CGG ATG TTT T-3′; HC58, 5′-AAA ACA TCC GTC AAG TAA GAT GCA GAC ACT AAA A-3′; HC73, 5′-phosphate-AAA ACA TCC GTC AAG TAA GAT GCA GAC ACT AAA A-biotin-3′; HC74, 5′-TTT TAG TGT CTG CAT CTT ACT TGA CGG ATG TTT-3′; HC76, 5′-biotin-AAA ACA TCC GTC AAG TAA GAT GCA GAC ACT AAA A-3′; HC77, 5′-phosphate-A*A*A* A*C*A* T*C*C* G*TC AAG TAA GAT GCA GAC ACT AAA A-biotin-3′ (the asterisks represent phosphorothioate bonds); HC79, 5′-AAA AAG TGT CTG CAT CTT ACT TGA CGG ATG TTT T-3′; HC80, 5′-GGG GAG TGT CTG CAT CTT ACT TGA CGG ATG TTT T-3′; HC81, 5′-CCC CAG TGT CTG CAT CTT ACT TGA CGG ATG TTT T-3′; HC82, 5′-biotin-AAA ACA TCC GTC AAG TAA GAT GCA GAC ACT TTT T-3′; HC83, 5′-biotin-AAA ACA TCC GTC AAG TAA GAT GCA GAC ACT CCC C-3′; HC84, 5′-biotin-AAA ACA TCC GTC AAG TAA GAT GCA GAC ACT GGG G-3′; HC85, 5′-TTT TTT TTT TTT TAG TGT CTG CAT CTT ACT TGA CGG ATG TTT T-3′; HC86, 5′-TTT TAG TGT CTG CAT CTT ACT TGA CGG ATG AAA-3′; HC87, 5′-TTT TAG TGT CTG CAT CTT ACT TGA CGG ATG GGG-3′; HC88, 5′-TTT TAG TGT CTG CAT CTT ACT TGA CGG ATG CCC-3′; HC89, 5′-phosphate-TTT TCA TCC GTC AAG TAA GAT GCA GAC ACT AAA A-biotin-3′; HC90, 5′-phosphate-CCC CCA TCC GTC AAG TAA GAT GCA GAC ACT AAA A-biotin-3′; HC91, 5′-phosphate-GGG GCA TCC GTC AAG TAA GAT GCA GAC ACT AAA A-biotin-3′; HC98, 5′-CCC CCA TCC GTC AAG TAA GAT GCA GAC ACT AAA A-biotin-3′; and HC99, 5′-GGC CAG TGT CTG CAT CTT ACT TGA CGG ATG TTT T-3′.
Protein Expression and Purification
Wild-type Artemis and catalytically inactive ArtemisH115A (ARM14) were purified as described previously (9, 15). Briefly, Sf9 insect cells (Life Technologies, catalog no. 11496-015) were infected with baculovirus containing C-terminal His-tagged Artemis DNA. Cells were lysed and purified by nickel-nitrilotriacetic acid affinity chromatography, anion exchange chromatography, and size exclusion chromatography. The Ku70-Ku80 complex was purified as described previously (11). Briefly, Hi-5 insect cells (Invitrogen, catalog no. B855-02) were coinfected with baculoviruses containing the DNA for C-terminal His-tagged Ku70 and untagged Ku80. Cells were lysed and purified by nickel-nitrilotriacetic acid affinity chromatography, dsDNA (oligo) affinity chromatography, and anion exchange chromatography. The purification of endogenous DNA-PKcs from HeLa cells has been described previously (16). Ku and DNA-PKcs were confirmed to have no detectable nuclease activities. At least three entirely independent fresh preparations of Artemis and DNA-PKcs were used in the subsequent assays, and these preparations generated indistinguishable results from one another.
Biochemical Nuclease Assay
The in vitro DNA nuclease assays were performed in a volume of 10 μl with a buffer composition of 25 mm Tris-HCl (pH 8.0), 75 mm KCl, 10 mm MgCl2, and 1 mm DTT. The reactions consisted of 20 nm of 32P-labeled DNA incubated with 0.5 mm ATP (or 0.5 mm AMP-PNP), 50 nm Ku, 50 nm DNA-PKcs, and 50 nm Artemis (or 50 nm ARM14) at 37 °C for 30 min unless specified otherwise. The time course assays were performed in a volume of 12 or 15 μl. The ladders were created by incubating 40 nm 32P-labeled ssDNA with 0.5 milliunits/ml snake venom phosphodiesterase I (Sigma-Aldrich, catalog no. P3243) for 15 min at 37 °C. All reactions were stopped by the addition of an equal volume of 98% formamide, heated to 95 °C for 5 min, placed on ice for 5 min, and analyzed on a 12 or 14% denaturing PAGE. The gels were then dried and exposed to a phosphorimaging screen overnight. The screen was scanned, and quantification was performed in Quantity One® (Bio-Rad).
Results
Artemis Can Resect Blunt DNA Ends But at a Much Slower Rate Than at DNA Overhangs
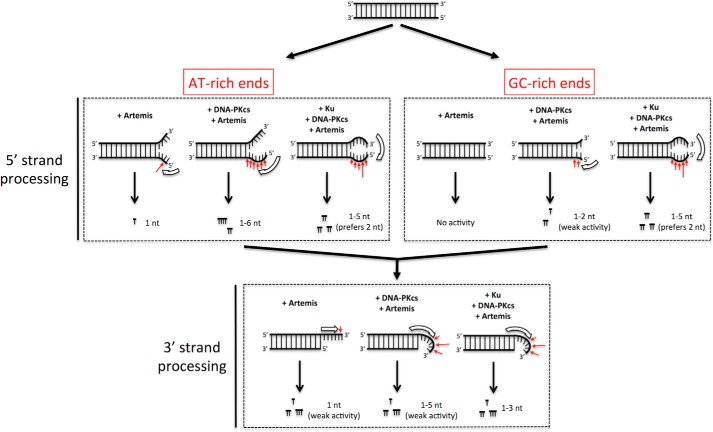
In Fig. 1, we show a summary of major Artemis substrates and preferred positions of Artemis cutting on those substrates. The open thick arrows indicate the putative orientation and position of the Artemis enzyme, and the red arrows indicate the preferred cutting sites. Artemis is known to have endonuclease activity on DNA hairpins and on 5′ and 3′ overhangs of duplex DNA, which is dependent on DNA-PKcs activity (Fig. 1, top row). Previous biochemical data have indicated that Artemis preferentially cuts on the 3′ side of a 4-nt ssDNA segment directly adjacent to a ss/dsDNA boundary (7, 11). The other known Artemis substrates listed in Fig. 1 (center and bottom rows) are physiologically relevant because they may arise from replication errors or by inadvertent action by nuclear enzymes. Nevertheless, we found that Artemis is able to cut these substrates on the ssDNA portion nearest the ss/dsDNA boundaries. We then wondered what would occur at blunt DNA ends when such ends may be able breathe open to form a transient pseudo-Y structure (Fig. 1, center row and center column).
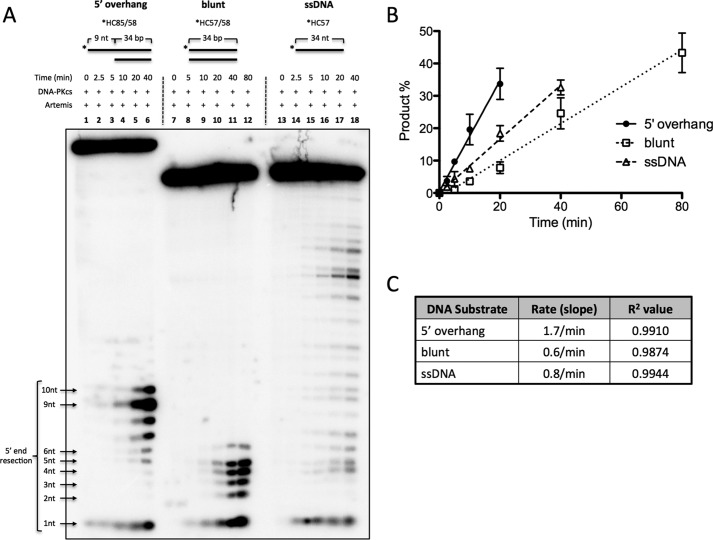
We initially performed a time course experiment with Artemis and DNA-PKcs on 9-nt 5′ overhang, blunt-ended, and ssDNA substrates (Fig. 2). Predictably, the predominant cut on the 5′ overhang substrate is at 9 nt, producing a blunt-ended product (Fig. 2A, lanes 1–6). The 34-nt ssDNA substrate (which matched the sequence of the top strand of the duplex portion of the other substrates) produced random endonucleolytic cuts (Fig. 2A, lanes 13–18). In addition, the apparent intense band at ∼18 nt may be explained by the palindromic 5′-TGCA-3′ on the 11th-14th nt, allowing the substrate to self-anneal. The 18th nt is then 4 nt on the 3′ side of the substrate, resulting in a more intense band. In contrast, the 34-bp blunt-ended substrate produced a distribution of cuts 1–6 nt into the duplex DNA end (Fig. 2A, lanes 7–12).
FIGURE 2.
Artemis activity comparison on blunt dsDNA and 5′ DNA overhangs. 100 nm 5′ 32P-radiolabeled substrate was incubated with 25 nm DNA-PKcs and 25 nm Artemis for the indicated times at 37 °C. The substrates were a 9-nt poly(dT) 5′ overhang substrate (*HC85/58); a 34-bp, blunt-ended substrate flanked by four dTs (*HC57/58); and a 34-nt ssDNA substrate (*HC57). The asterisks indicate [32P] phosphates. A, samples from time points were run on a 14% denaturing PAGE. B, cut products were quantified, and the average of two independent experiments was plotted as a function of time. Error bars indicate mean ± S.D. (Several additional similar time courses gave results indistinguishable from those shown here.). C, linear regression was used to calculate the slope and R2 values.
The cut products were then quantified as a percentage of the total substrate and plotted as a function of time (Fig. 2B). The rate of activity was determined by calculating the slope from the average of two entirely independent experiments (product percent per minute of reaction). (Several additional very similar experiments further supported these observations.) The 5′ overhang substrate was processed ∼3-fold faster than blunt-ended DNA and 2-fold faster than ssDNA (Fig. 2C). Although it is known that Artemis has endonuclease activity on ssDNA (6), it was surprising to observe that the activity was only 2-fold slower than at overhangs. Unlike homopolymeric ssDNA substrates, we suspect that the ssDNA substrate used here may have self-annealed to produce partial duplex DNA structures.
Artemis Endonuclease Activity on Blunt DNA Ends Is Strictly Dependent on DNA-PKcs and ATP and Is Modulated by Ku
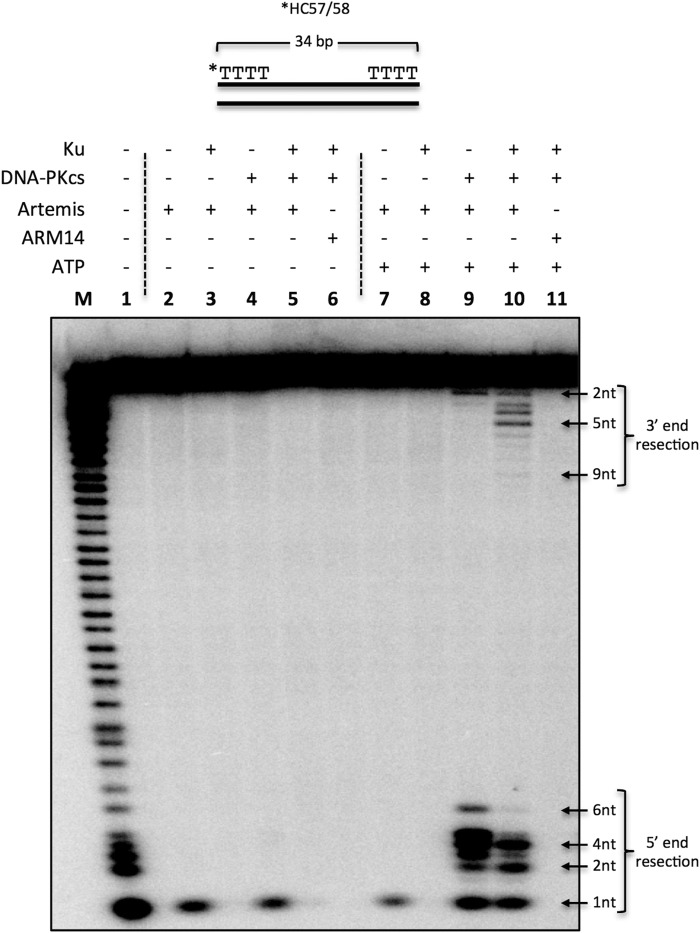
As mentioned, Artemis has intrinsic 5′ exonuclease activity and DNA-PKcs-dependent endonuclease activity. Therefore, we wondered whether this activity also applied to blunt-ended DNA. We incubated a 5′-radiolabeled 34-bp dsDNA substrate flanked with dTs in a reaction with Ku, DNA-PKcs, Artemis, and ARM14 (catalytically inactive ArtemisH115A) with or without ATP for 30 min at 37° (Fig. 3). Intrinsic Artemis 5′ exonuclease activity was able to resect the first 5′ nucleotide in the absence of DNA-PKcs and ATP (Fig. 3, lanes 2, 4, and 7). Furthermore, Artemis 5′ endonuclease activity predominately cut 1–6 nt from the 5′ end in the presence of DNA-PKcs (Fig. 3, lane 7 versus lane 9), and this activity was dependent on ATP (Fig. 3, lane 4 versus lane 9).
FIGURE 3.
Artemis activity on blunt dsDNA with dT ends. 20 nm 5′ 32P-radiolabeled, 34-bp dsDNA (*HC57/58) was incubated with 50 nm Ku, 50 nm DNA-PKcs, 50 nm Artemis, 50 nm ARM14, and 0.5 mm ATP in the respective reactions for 30 min at 37 °C, followed by resolving the sample on a 12% denaturing PAGE. Reactions included Artemis (lane 2); Ku and Artemis (lane 3); DNA-PKcs and Artemis (lane 4); Ku, DNA-PKcs, and Artemis (lane 5); and Ku, DNA-PKcs, and ARM14 (lane 6). ATP was included in lanes 7–11. M, marker lane; asterisk, [32P]phosphate. This is a representative gel of at least three identical experiments and at least two preparations of Artemis and DNA-PKcs. (Several additional similar experiments gave results indistinguishable from those shown here.)
Interestingly, Ku modulated Artemis endonuclease activity at blunt ends by restricting the cuts to 2 and 4 nt instead of the 1- to 6-nt distribution observed in Ku-independent reactions (Fig. 3, lane 9 versus lane 10, 5′ end resection). The addition of Ku also stimulated 3′ endonuclease activity by enabling Artemis to cut up to 9 nt into the 3′ end (Fig. 3, lane 9 versus lane 10, 3′ end resection). This 3′ activity remained DNA-PKcs-dependent (Fig. 3, lane 8 versus lane 10, 3′ end resection). Furthermore, the observed activities are not due to contaminating nucleases because the catalytically inactive Artemis point mutant, ARM14, which was purified in the same manner, did not show any 5′ or 3′ activity (Fig. 3, lanes 10 and 11). We also confirmed that, similar to Artemis endonuclease activity on overhangs and ssDNA (5, 6), Artemis phosphorylation is not required for its endonuclease activity on blunt-ended DNA (data not shown), although autophosphorylated DNA-PKcs is required for all ends.
Titration Reveals That DNA-PKcs Can Be Inhibitory at High Concentrations
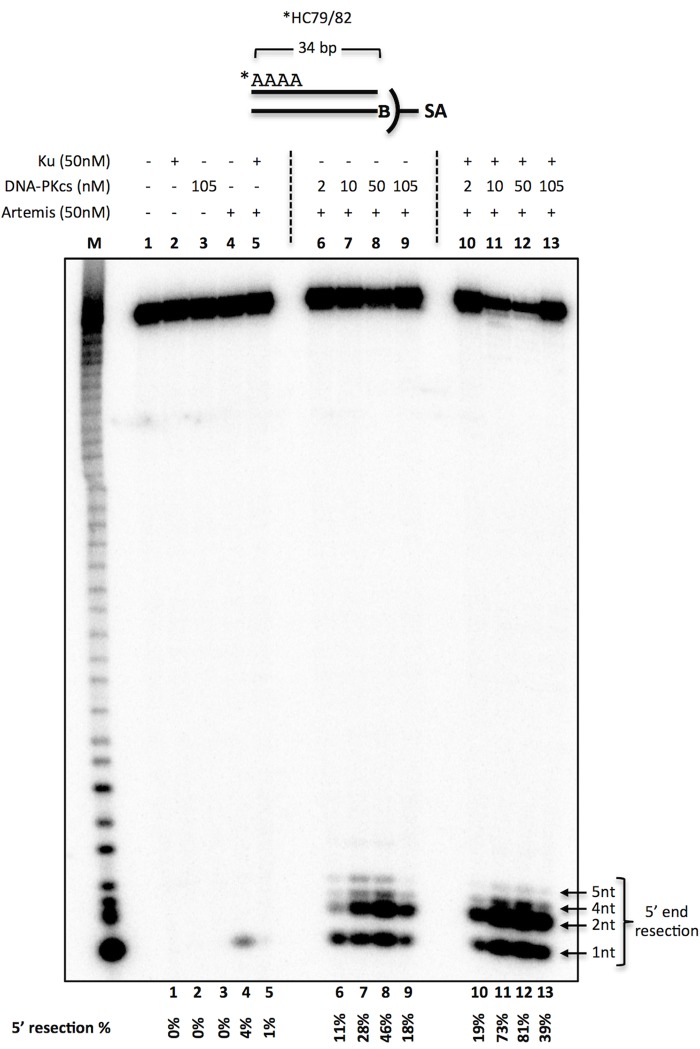
Next we examined the effect of the amount of DNA-PKcs on the observed activity of Artemis. We generated a 5′-labeled, 34-bp, blunt-ended DNA by annealing 5′-labeled ssDNA to its unlabeled complementary strand containing a 5′ biotin (Fig. 4). This substrate was then incubated along with streptavidin, which binds biotin tightly, to prevent/inhibit Artemis action on one end of the dsDNA to examine the activity on the labeled end only. Using a blunt-ended substrate flanked with four dAs, we titrated the amount of DNA-PKcs in the reaction from 2 to 105 nm (Fig. 4). Ku and DNA-PKcs alone did not have any detectable levels of nuclease activity (Fig. 4, lanes 2 and 3). Artemis activity was maximal at a 1:1 molar ratio with DNA-PKcs (Fig. 4, lanes 6–9).
FIGURE 4.
DNA-PKcs titration for Artemis activity on blunt ends. 20 nm 5′ 32P-radiolabeled, 34-bp dsDNA (*HC79/82) with four dAs was created by annealing the radiolabeled (asterisk) top strand with its complementary strand with a 5′ biotin (B). The substrate was incubated with 50 nm Ku, 50 nm Artemis, and various amounts of DNA-PKcs in the respective reactions for 30 min at 37 °C, followed by resolving the sample on a 14% denaturing PAGE. In addition, 0.5 mm ATP and 0.1 μm streptavidin (SA) were included in all reactions to suppress Artemis activity on the 3′ end. Reactions consisted of Ku (lane 2), DNA-PKcs (lane 3), Artemis (lane 4), Ku and Artemis (lane 5), Artemis with increasing amounts of DNA-PKcs (lanes 6–9), and Ku and Artemis with increasing amounts of DNA-PKcs (lanes 10–13). The quantified 5′ resection percentage is noted at the bottom of the gel. This is a representative gel of at least two identical experiments. (Several additional similar experiments gave results indistinguishable from those shown here.)
Furthermore, Artemis activity was stimulated ∼2- to 3-fold in the presence of Ku (Fig. 4, lanes 6–9 versus lanes 10–13). Interestingly, increasing the molar ratio of DNA-PKcs to Artemis to 2:1 reduced Artemis activity by ∼50% (Fig. 4, lane 8 versus lane 9 and lane 12 versus lane 13). These data support the view that Artemis acts in a complex with DNA-PKcs at a 1:1 molar ratio and that increasing the concentration of DNA-PKcs may block the DNA ends from Artemis activity.
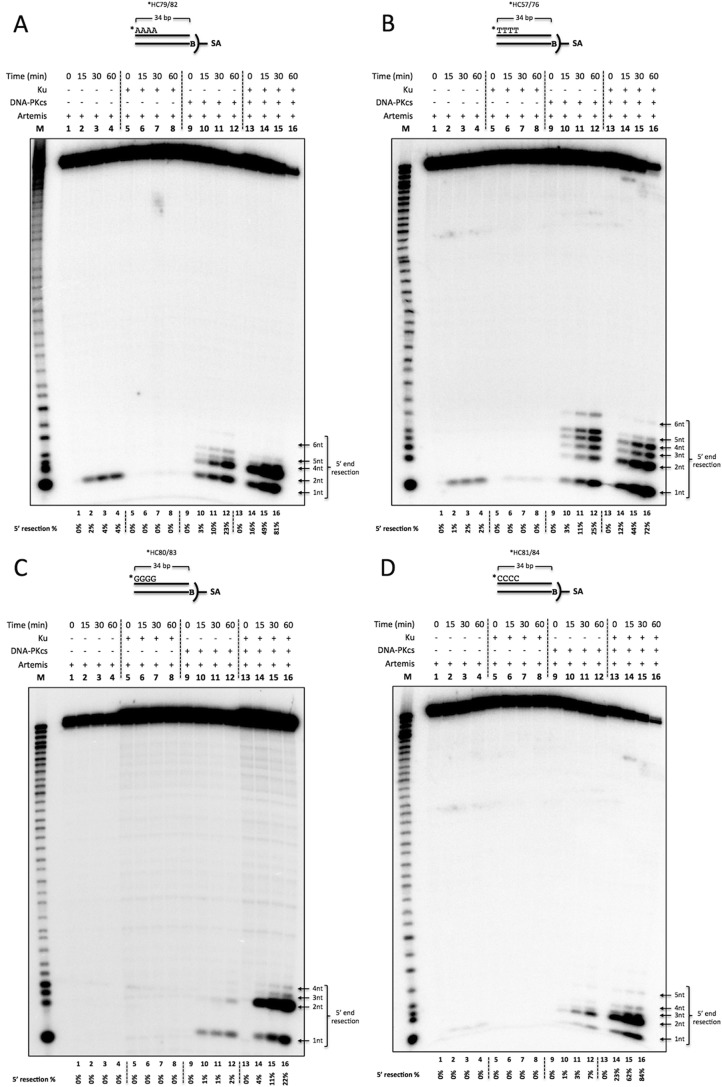
The Blunt End DNA Sequence Determines the Extent to Which Ku and DNA-PKcs Stimulate the 5′ Nuclease Action of Artemis
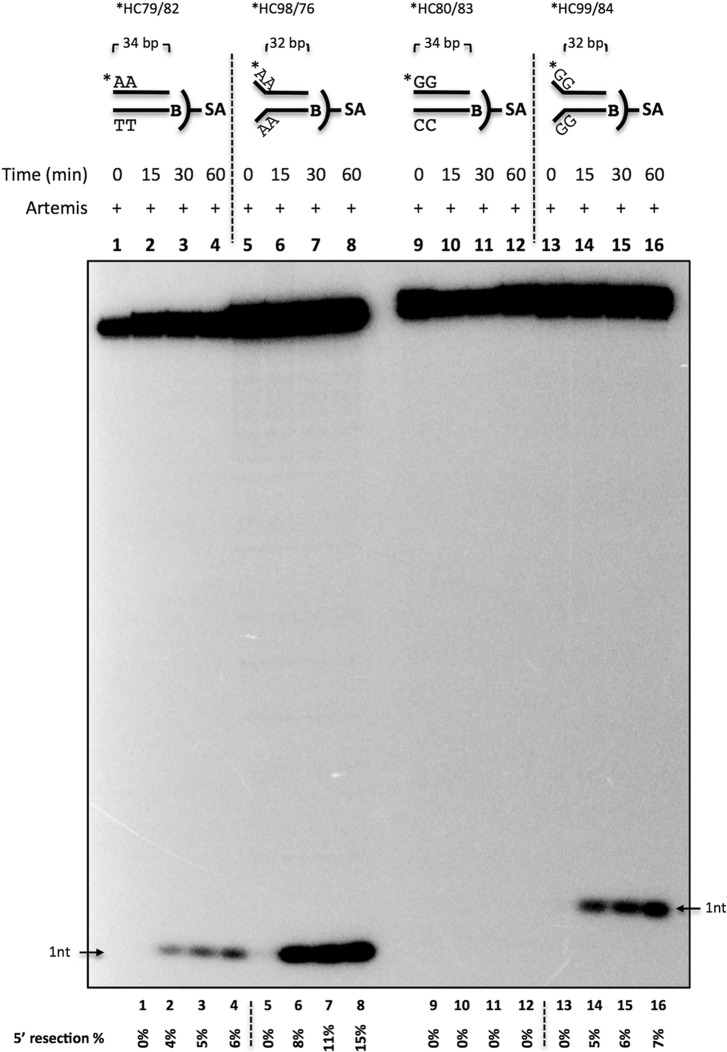
We then asked whether the observed Ku stimulation is sequence-dependent at 5′ blunt ends. We generated 5′-labeled, 34-bp, blunt-ended DNA with four dAs (Fig. 5A), four dTs (Fig. 5B), four dGs (Fig. 5C), or four dCs (Fig. 5D) by annealing 5′-labeled ssDNA with unlabeled 5′ biotinylated complementary strands. The substrate was then incubated with proteins as indicated, along with ATP and streptavidin. Artemis 5′ exonuclease activity was observed with substrates that are flanked by AT but not GC base pairs at the blunt DNA ends (Figs. 3, lanes 2, 4, and 7; 4, lane 4; and 5, A–D, lanes 1–4). The observed exonuclease activity was reduced when Ku was added (Figs. 3, lane 2 versus lane 3, lane 4 versus lane 5, and lane 7 versus lane 8; 4, lane 4 versus lane 5; and 5, A and B, lanes 1–4 versus lanes 5–8). We hypothesized that the weak base pairing in AT-rich ends compared with GC-rich ends allowed the DNA end to breathe and form transient ss/dsDNA boundaries to provide a substrate for Artemis. To examine the sequence-dependent variation further, we simulated breathing by creating a fixed 2-nt pseudo-Y structure with dA or dG unpaired ssDNA overhangs (Fig. 6). DNA with 5′ dGs was processed by Artemis when forced into a pseudo-Y structure (Fig. 6, lanes 9–12 versus lanes 13–16). Interestingly, Y-structure DNA with 5′ dAs was also stimulated 2- to 3-fold (Fig. 6, lanes 1–4 versus lanes 5–8). These data indicate that DNA breathing may allow AT-rich blunt DNA to be cleaved by the Artemis 5′ exonuclease activity in the absence of other proteins (DNA-PKcs and Ku).
FIGURE 5.
The Time course of 5′ endonuclease activity on blunt-ended DNA reveals that Artemis resects up to 6 nt, requires DNA-PKcs, and is modulated by Ku to preferentially cut 2 nt into the duplex. 20 nm 5′ 32P-radiolabeled, 34-bp biotinylated dsDNA (B) was incubated with 50 nm Ku, 50 nm DNA-PKcs, and 50 nm Artemis for 0, 15, 30, and 60 min at 37 °C in the lanes as indicated. In addition, 0.5 mm ATP and 0.1 μm streptavidin (SA) were included in all reactions to suppress Artemis activity on the 3′ end. The samples were resolved on a 14% denaturing PAGE. M, marker lane; asterisk, [32P]phosphate. The quantified 5′ resection percentage is noted at the bottom of the gel. This is a representative gel of at least three identical experiments and at least two preparations of Artemis and DNA-PKcs. (Several additional similar time courses gave results indistinguishable from those shown here.). A, 5′ dA substrate (*HC79/82). B, 5′ dT substrate (*HC57/76). C, 5′ dG substrate (*HC80/83). D, 5′ dC substrate (*HC81/84).
FIGURE 6.
DNA end breathing provides a substrate for Artemis 5′ exonuclease activity. Shown is the time course of Artemis activity on blunt-ended and fixed 2-nt Y-structure DNA. 20 nm 5′ 32P-radiolabeled, 34-bp biotinylated-dsDNA (B) was incubated with 50 nm Artemis and 0.1 μm streptavidin (SA) for 0, 15, 30, and 60 min at 37 °C. The samples were resolved on a 12% denaturing PAGE. Substrates were blunt-ended dA (*HC79/82, lanes 1–4), dA Y-structure (*HC98/76, lanes 5–8), blunt-ended dG (*HC80/83, lanes 9–12), and dG Y-structure (*HC99/84, lanes 13–16). Asterisk, [32P]-phosphate. The quantified 5′ resection percentage is noted at the bottom of the gel. This is a representative gel of at least two identical experiments.
In contrast, Artemis 5′ endonuclease activity is more complex. Artemis 5′ endonuclease activity was stimulated by DNA-PKcs with AT-rich blunt DNA substrates but only mildly stimulated with GC-rich blunt DNA substrates (Fig. 5, A–D, lanes 9–12). With blunt AT ends, up to 6 nt were resected in the presence of DNA-PKcs (Fig. 5, A and B, lanes 9–12). In addition, Ku stimulated the endonuclease activity ∼3- to 4-fold (Fig. 5, A and B, lanes 9–12 versus lanes 13–16). Conversely, for GC ends, DNA-PKcs only slightly activated 5′ exo- and endonuclease activities (Fig. 5, C and D, lanes 9–12). However, the addition of Ku stimulated Artemis activity more than 10-fold (Fig. 5, C and D, lanes 9–12 versus lanes 13–16). In summary, Ku could modulate the activity of Artemis·DNA-PKcs depending on the AT- or GC-richness of blunt ends. For AT-rich ends, Ku stimulated Artemis activity only 3- to 4-fold and diminished the diversity of resection. Specifically, 1- to 6-nt resection without Ku and 1 to 2 nt with Ku (Fig. 5, A and B, lanes 9–12 versus lanes 13–16). In contrast, Ku stimulated Artemis activity more than 10-fold in GC-rich ends overall, with the most frequent resection events at 1–2 nt (Fig. 5, C and D).
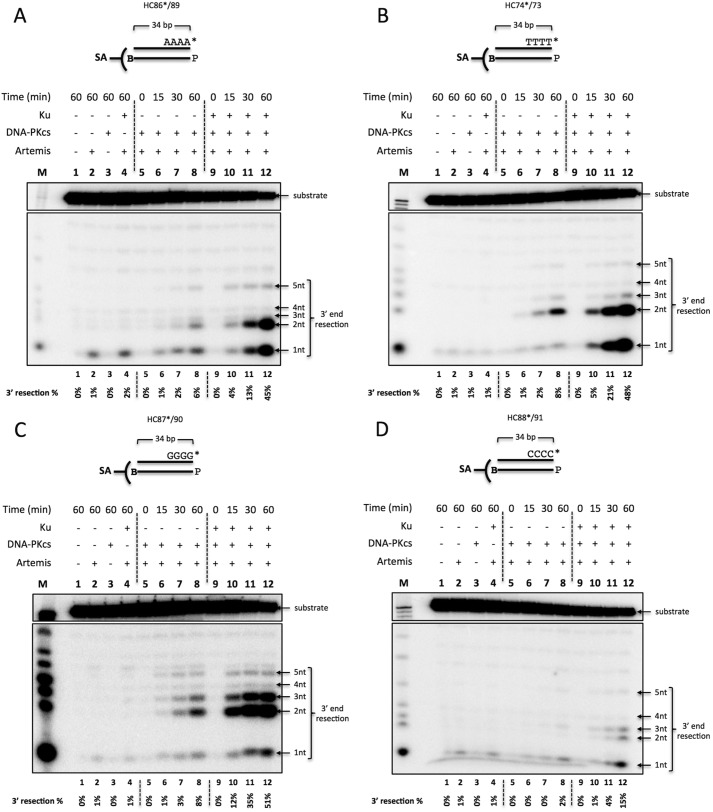
Artemis 3′ Endonuclease Resection Does Not Show Substantial Sequence Dependence but Is Stimulated by DNA-PKcs and Ku
We then asked whether the sequence-dependent activity is also observed on the 3′ end of blunt-ended DNA. 3′-labeled, 34-bp, blunt-ended DNA flanked by four dAs (Fig. 7A), four dTs (Fig. 7B), four dGs (Fig. 7C), or four dCs (Fig. 7D) was generated by fill-in synthesis with the Klenow fragment of DNA polymerase I (3′ → 5′ exo−) and [α-32P]dNTP. The substrate was then incubated with Ku, DNA-PKcs, and Artemis, as indicated, in the presence of ATP and streptavidin. The unlabeled complementary strand contained an unlabeled 5′ phosphate and a 3′ biotin to block Artemis action on the 3′ end. 3′ exonuclease activity was observed only on the 3′ dA DNA and slightly on 3′ dT, dG, and dC DNA (Fig. 7, A–D, lane 2). Interestingly, Ku did not block this 3′ exonuclease activity (Fig. 7, A–D, lane 2 versus lane 4). Artemis activity on the 3′ end was only mildly stimulated by DNA-PKcs and produced a distribution of cuts up to 5 nt into the duplex portion of the substrate (Fig. 7, A–D, lane 2 versus lane 8). Ku further stimulated Artemis activity 6- to 8-fold independent of sequence (Fig. 7, A–D, lane 8 versus lane 12).
FIGURE 7.
The Time course of 3′ endonuclease activity on blunt-ended DNA reveals that Artemis resects predominantly 1–3 nt, requires DNA-PKcs, and is stimulated by Ku. 20 nm 3′ 32P-radiolabeled, 34-bp biotinylated dsDNA (B) was incubated with 50 nm Ku, 50 nm DNA-PKcs, and 50 nm Artemis for 0, 15, 30, and 60 min at 37 °C in the lanes as indicated. In addition, 0.5 mm ATP and 0.1 μm streptavidin (SA) were included in all reactions to suppress Artemis activity on the 5′ end. The P on the bottom strand denotes a cold phosphate group on the 5′ end. The samples were resolved on a 14% denaturing PAGE. M, marker lane; asterisk, [32P]phosphate. The quantified 3′ resection percentage is noted at the bottom of the gel. This is a representative gel of at least three identical experiments and at least two preparations of Artemis and DNA-PKcs. (Several additional similar time courses gave results indistinguishable from those shown here.). A, 3′ dA substrate (HC86*/89). B, 3′ dT substrate (HC74*/73). C, 3′ dG substrate (HC87*/90). D, 3′ dC substrate (HC88*/91).
Interestingly, these results are in contrast to Artemis activity observed on the 5′ end. For one, the breathing requirement was not sequence-dependent on the 3′ end. The mild activity stimulation by DNA-PKcs on the 3′ end, in which only up to 8% of the substrate was cut after 60 min (Fig. 7, A–D, lane 8), was similar to the stimulation observed on GC-rich 5′ ends (Figs. 5, C and D, lane 12, and 7, A–D, lane 12). Secondly, the addition of Ku stimulated Artemis activity 6- to 8-fold on the 3′ end independent of sequence (Fig. 7, A–D, lane 8 versus lane 12), whereas Artemis activity on 5′ ends was sequence-dependent (a 3- to 4-fold increase for AT-rich and a more than 10-fold increase for GC-rich).
5′ Resection Occurs Prior to 3′ Resection on Blunt DNA Ends
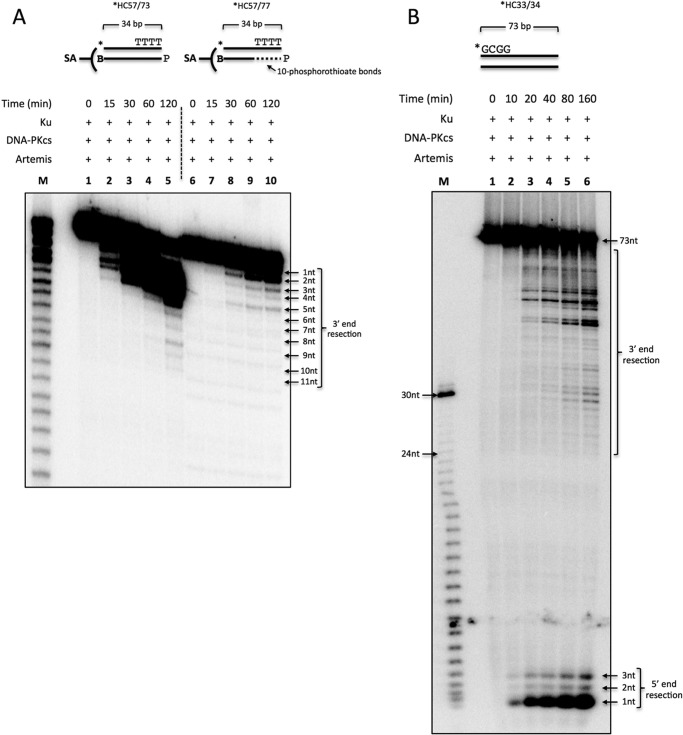
We determined that Artemis performs asymmetric resection for the 5′ versus 3′ strands. Resection on the 5′ strand at AT-rich blunt ends occurred in a largely Ku-independent manner. GC-rich 5′ ends were mildly stimulated by DNA-PKcs and stimulated strongly by Ku. Conversely, Artemis exhibited sequence-independent activity on 3′ ends that required Ku for robust activity. Because it appeared that 5′ end resection was more efficient, we asked whether 5′ end processing was required prior to 3′ end processing. We annealed a 5′-labeled, 34-nt oligo to its 3′ biotinylated complementary oligo with or without 10 phosphorothioate bonds on the 5′ end. A phosphorothioate bond, formed by the sulfurization of a nonbridging oxygen in the phosphodiester bond, suppresses the action of nucleases. We performed a time course of Artemis activity in the presence of DNA-PKcs and Ku. We observed that Artemis activity on the 3′ end of dsDNA substrate was reduced significantly when the complementary strand was unable to be cut first (Fig. 8A, lanes 1–5 versus lanes 6–10). Therefore, it appears that the 5′ strand resection occurs first, always leaving a 3′ ssDNA overhang that, by its ssDNA nature, does not have any breathing requirement. This allows the 3′ overhang resection to be sequence-independent.
FIGURE 8.
Artemis 5′ end resection occurs before 3′ end resection and progresses internally along the DNA with time. Shown is the time course of Artemis action. A, 20 nm 5′ 32P-radiolabeled, 34-bp dsDNA with four dTs on the 3′ end was created by annealing the radiolabeled (asterisk) top strand with its complementary strand with a cold 5′ phosphate (P) and 3′ biotin (B) (*HC57/73, lanes 1–5) or its complementary strand with 10 phosphorothioate bonds on the 5′ end (*HC57/77, lanes 6–10). The substrates were incubated with 50 nm Ku, 50 nm DNA-PKcs, and 50 nm Artemis for 0, 15, 30, 60, and 120 min at 37 °C and analyzed on a 12% denaturing PAGE. This is a representative gel of at least three identical experiments. (Several additional similar time courses gave results indistinguishable from those shown here.). B, 20 nm 5′ 32P-radiolabeled, 73-bp dsDNA (*HC33/34) was incubated with 50 nm Ku, 50 nm DNA-PKcs, 50 nm Artemis, and 0.5 mm ATP for 0, 10, 20, 40, 80, and 160 min at 37 °C and analyzed on a 12% denaturing PAGE. M, marker lane. This is a representative gel of at least 10 experiments using blunt-ended DNAs of various lengths.
Artemis Resection Progresses Internally along the DNA with Time
Because we established that Artemis first acts on the 5′ end prior to acting on the resulting 3′ overhang, we wondered whether this process would repeat until the DNA was processed completely. We initially showed that a 34-bp, blunt-ended DNA could be processed up to 11 nt after 2 h (Fig. 8A, lane 5), which would leave ∼23 bp remaining. We then incubated a 73-bp DNA with Ku, DNA-PKcs, and Artemis for up to 160 min (Fig. 8B, lanes 1–6). The resulting gel showed that Artemis endonucleolytically resected up to 4 nt on the 5′end (Fig. 8B, lane 6, 5′ end resection). Processing on the 3′ end proceeded until ∼25–30 nt remained (Fig. 8B, lane 6, 3′ end resection). Because we know that ∼26 bp are required for maximal Ku-dependent DNA-PKcs activity (17), it is likely that Artemis resection proceeds until the DNA is long enough for one Ku·DNA-PKcs complex to remain bound to the DNA.
Discussion
Under physiological conditions (Artemis plus DNA-PKcs in Mg2+ solutions), we found that resection at blunt DNA ends depends on the DNA sequence. However, when Ku is also present, the disparity is eliminated, and blunt ends of all sequences are resected efficiently. This is the first biochemical documentation showing that Ku can affect the nucleolytic behavior of Artemis. Further study of it has revealed major unifying features of the Artemis nuclease.
Unifying Kinetic Features for Artemis Action at DNA Ends
Previous work on Artemis has documented its action at ss/dsDNA boundaries, specifically at hairpins and overhangs and gaps and flaps (7, 11). One study also described endonuclease action by Artemis alone in Mg2+ solutions at blunt DNA ends (18). But this latter finding was curious because no other laboratory has found endonuclease activity of Artemis alone under Mg2+ conditions. We noted that numerous freeze and thaw cycles of stored Artemis enzyme preparations eventually cause such behavior, suggesting a denaturation-induced change in the enzyme that partially eliminates the requirement for DNA-PKcs. In addition, Artemis alone can cut overhangs in Mn2+ solutions, suggesting a permanent conformational change of the enzyme in the presence of this divalent cation (6, 9, 19). For this reason, evaluation of Artemis under conditions where it is dependent on DNA-PKcs was necessary to consider its function under physiologic conditions at blunt DNA ends and to integrate this into the spectrum of Artemis nuclease activities.
Our finding of resection by Artemis alone in Mg2+ solutions at AT-rich DNA ends but not at GC-rich DNA ends suggested that DNA end breathing is required for Artemis action. In other studies, we have documented the increased breathing of AT-rich DNA ends compared with GC-rich ends (20). When we “forced” breathing here by creating a fixed, 2-nt Y-structure in which the 2 nt arms of the Y cannot anneal, we found that, indeed, the DNA sequence of the arms no longer matters because the ends are in a permanent ssDNA configuration.
A requirement for end breathing for Artemis to act on a blunt end explains why such end resection by Artemis is slower than overhang resection. The breathing step is required, and the end is only in the available configuration (ss/dsDNA) for cutting for a fraction of the time.
A Unifying Physical Model for Artemis Nuclease Activity at DNA Ends
On the basis of the findings here and shown previously, it is possible to formulate a model that encompasses all of the nuclease activities of Artemis. These include hairpin opening (2 nt 3′ of the tip), 5′ overhang cutting (primarily at the ss/dsDNA boundary), 3′ overhang cutting (primarily 4 nt 3′ of the ss/dsDNA boundary), blunt end resection (slower than ss/dsDNA boundary substrates), and cutting at all DNA structures containing ss/dsDNA boundaries (gaps, flaps, heterologous loops, and Y-structures). We note that poly(dT) ssDNA can be cut by Artemis·DNA-PKcs but that this cutting is much slower than for DNA containing a ss/dsDNA boundary (6). The apparently efficient cutting of ssDNA shown in Fig. 2B is due to this ssDNA being a mixed sequence (not poly(dT)) and being capable of forming secondary structures.
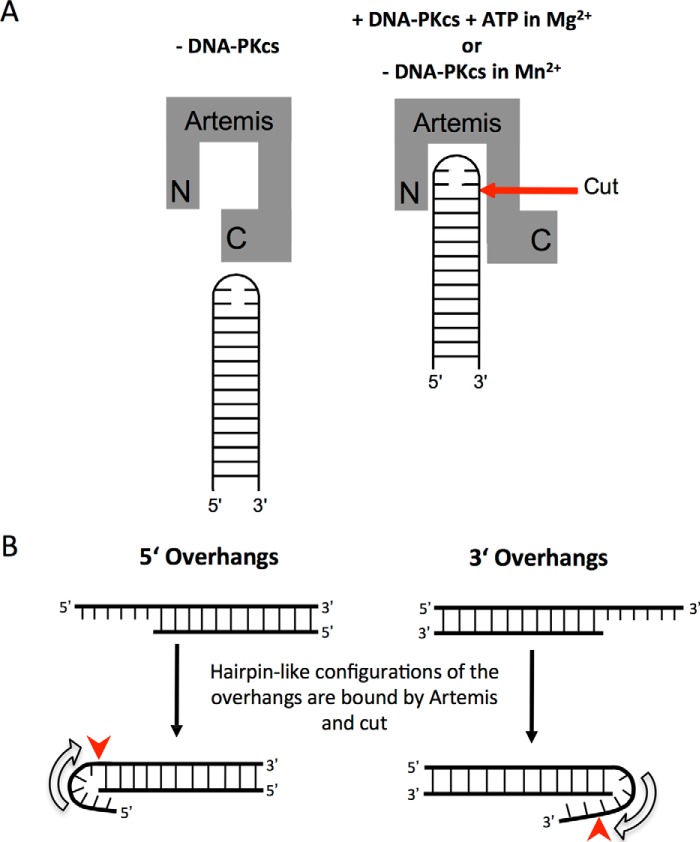
For optimal activity, we have proposed previously that the Artemis enzyme recognizes ss/dsDNA boundaries and cuts on the 3′ side of an ∼4-nt ssDNA region at this boundary. This explains the hairpin opening position, which is not at the tip but is located 2 nt 3′ of the tip (the turn in the perfect hairpin provides the 4 nt of ssDNA because it is largely unpaired) (11). This also explains why 5′ and 3′ overhangs yield different overhang products (blunt for 5′ overhangs but 4 nt 3′ overhangs for long 3′ overhangs) (7, 11). DNA bending by Artemis, and, more efficiently, by Artemis·DNA-PKcs, at a 5′ or 3′ overhang may generate a structure similar to a hairpin (21).
Fig. 9 shows how these same principles apply to Artemis action at blunt DNA ends. A blunt end can breathe into a Y-structure with short ssDNA arms. Artemis then resects the 5′ end first. The resulting 3′ overhang is then bent by Artemis or diffuses independently into a transient hairpin-like bend that is cut by Artemis. Because the resulting 3′ overhang may not be sufficiently long for efficient processing by Artemis, Ku is likely required to provide a scaffold for Artemis·DNA-PKcs to bind near the overhang. All of the structures in Fig. 1 are likely to require a similar bending of the ss/dsDNA boundary into a hairpin-like state (Fig. 10).
FIGURE 9.
DNA end breathing model for Artemis action at blunt DNA ends. The diagrams summarize the action of Artemis at blunt DNA ends (in 10 mm MgCl2 and 75 mm KCl). Consistent with the optimal substrate requirements of Artemis (Fig. 1), blunt DNA ends must first breathe to pseudo-Y structures before efficient nuclease action can begin. Artemis first acts on the 5′ strand of the DNA end and then on the 3′ strand (which, at that point, is a 3′ overhang). The result of 5′ and 3′ cuts is a blunt or near-blunt end that is then subject to iterative rounds of the same process, consistent with our results. The red arrows indicate the preferred Artemis cut sites. The open thick arrows represent the proposed Artemis binding site and orientation on the DNA substrates.
FIGURE 10.
Functional relationship between Artemis and ss/dsDNA ends. A, Artemis can only cut at DNA hairpins in Mn2+ solutions or in Mg2+ solutions that also contain DNA-PKcs and ATP. We propose that this allows the C-terminal portion (C) of Artemis to no longer suppress the endonuclease action by the active site of Artemis (N). The red arrow indicates the preferred Artemis cut site. B, we propose that 5′ and 3′ overhangs must adopt a folded structure similar to a hairpin to be cut by Artemis. Consistent with our previous proposals that Artemis recognizes ss/dsDNA boundaries and cuts on the 3′ side of a 4-nt length within the ssDNA portion, the 5′ overhangs are predominantly cut to initially generate a blunt DNA end, whereas the 3′ overhangs are initially cut predominantly to generate a 4-nt 3′ overhang. Of course, subsequent cuts into the DNA ends can occur in the manner shown in Figs. 1 and 9. The red arrows indicate the preferred Artemis cut sites.
The 5′ Exonuclease of Artemis
Even the 5′ exonuclease of Artemis can be incorporated into this model. We have noted previously that the 5′ exonuclease of Artemis is independent of DNA-PKcs at 5′ overhangs (7). However, for recessed 5′ overhangs or blunt ends, we find that AT-richness around such a non-protruding 5′ end is necessary for it to be cut by Artemis exonuclease activity (data not shown), implying that the 5′ exonuclease activity of Artemis also relies on ss/dsDNA transition structures in the substrate. Only the protruding 5′ DNA ends are targets of the 5′ exonuclease activity of Artemis without any effect on DNA sequence. This may arise because the protruding 5′ end can insert directly into the active site of Artemis without any more substantial substrate positioning by Artemis that is required for it to process all other DNA ends.
The Role of DNA-PKcs and Ku in Artemis Interaction with Its Substrate
We believe that the role of DNA-PKcs is to change the conformation of Artemis to a form that can more easily configure DNA ends into an optimal hairpin-like substrate (Fig. 10). As pointed out previously, Artemis under Mn2+ conditions does not require DNA-PKcs for any of the reactions shown in Fig. 1 (6, 9, 19).
DNA-PKcs and Ku might simply increase the length of time that Artemis remains bound so that, at the time of DNA breathing into a transient ss/dsDNA boundary structure, Artemis can hydrolyze the phosphodiester bond. Artemis in vivo exists predominantly in a very salt-stable complex with DNA-PKcs, whereas Ku only forms a complex with Artemis·DNA-PKcs when a DNA end is present (1, 11). DNA-PKcs autophosphorylation is required for DNA-PKcs to activate Artemis endonuclease activity (1). Ku improves the affinity of DNA-PKcs alone 100-fold, from 3 × 10−9 to 3 × 10−11 m (17), and we assume that this applies to the Artemis·DNA-PKcs complex as well. The Artemis·DNA-PKcs·Ku complex would be able to remain at a DNA end longer, giving Artemis more opportunity to cut.
Biological Implications of the Unified Model of Artemis Nuclease Action for NHEJ
The key in vivo roles of Artemis are diverse and have not been easy to reconcile with a single definition of the substrate configuration that is optimal for Artemis nuclease action. Among the key DNA end configurations at which Artemis is known to act are perfect DNA hairpins during the coding joint formation step of V(D)J recombination (11, 23, 24), repair of 10–40% of ionizing radiation-induced DSBs (25), and repair of DSBs produced by interruption of topoisomerase II action (26). All of the structures for in vivo and in vitro action of Artemis have ss/ds DNA boundaries. Importantly, the boundary for hairpins is due to the imperfect base pairing at the hairpin tip (11).
However, the action of Artemis at blunt DNA ends (and in a DNA-PKcs-independent manner) complicated this simple model (18). Here we confirmed the action at blunt ends, but we show that, like the other endonucleolytic actions of Artemis, this too is dependent on DNA-PKcs. Moreover, the DNA end-breathing requirement unifies this substrate configuration with the other known in vivo and in vitro substrate configurations.
Biological Implications of the Unified Model of Artemis Nuclease Action for V(D)J Recombination
This study is likely to explain why GC-rich coding ends suffer less end resection than AT-rich coding ends in human pre-B cell lines (27). GC-rich coding ends will breathe less than AT-rich coding ends. This curious but well documented finding on the sequence dependence of coding end resection has gone without explanation for nearly two decades, but the findings in this study provide the first explanation for how DNA end resection at coding ends in V(D)J recombination is affected by DNA end sequence.
A second point of in vivo relevance concerns the difference in resection of coding ends versus signal ends in V(D)J recombination. It has been unclear why signal ends suffer end resection much less often than coding ends. Signal ends are blunt, and they suffer end resection in wild-type human lymphoid cells but only at 5% of all signal joints (28). In contrast, coding ends are opened (by Artemis·DNA-PKcs) to give 3′ overhangs (usually 4 nt in length), and these coding ends suffer end resection in over 90% of coding joints in human pre-B cells (27). This disparity has never been explained, but one possible explanation is raised by our finding of a slower and weaker resection of Artemis at blunt DNA ends because of the requirement for DNA end breathing described here. Such an explanation would be consistent with the fact that end resection at signal ends decreases when Artemis is absent (29) and increases when the partner of Artemis, namely DNA-PKcs, is mutated—a point that has remained without any reasonable explanation until now (30).
A third point of possible in vivo relevance concerns our observation in human cells of inverted repeats at partially resected coding ends in the final coding joints formed in human pre-B cells (27). The Artemis resection in 2- to 6-nt increments at blunt ends, as in Fig. 2, may liberate these short oligonucleotides, allowing them to be religated via the single-strand ligation activity of the ligase IV complex (22). This would generate the short inverted repeats observed at partially resected coding ends (27), which have previously been difficult to explain mechanistically until now. Therefore, the findings here not only provide a unification of the enzymatic activities of an important structure-specific nuclease but also integrate several in vivo observations with this unified model.
Author Contributions
H. H. Y. C. and M. R. L. designed the experiments. H. H. Y. C. performed the experiments. G. W. purified and characterized DNA-PKcs. All authors wrote the paper.
Acknowledgments
We thank Dr. Ray Mosteller (USC) and members of the Lieber laboratory for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health (to M. R. L.). The authors declare that they have no conflicts of interest with the contents of this article.
- DSB
- double strand break
- NHEJ
- nonhomologous DNA end-joining
- β-CASP
- named after four representative metallo-β-lactamases: CPSF, Artemis, SNM1, PSO2
- CPSF
- cleavage and polyadenylation specificity factor
- SNM
- senstive to nitrogen mustard
- PSO
- psoralen
- RAG
- recombination activating gene
- DNA-PKcs
- DNA-dependent protein kinase catalytic subunit
- ss
- single-stranded
- ds
- double stranded.
References
- 1. Lieber M. R. (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lieber M. R., Karanjawala Z. E. (2004) Ageing, repetitive genomes and DNA damage. Nat. Rev. Mol. Cell Biol. 5, 69–75 [DOI] [PubMed] [Google Scholar]
- 3. Martin G. M., Smith A. C., Ketterer D. J., Ogburn C. E., Disteche C. M. (1985) Increased chromosomal aberrations in first metaphases of cells isolated from the kidneys of aged mice. Isr. J. Med. Sci. 21, 296–301 [PubMed] [Google Scholar]
- 4. Lieber M. R., Ma Y., Pannicke U., Schwarz K. (2003) Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 4, 712–720 [DOI] [PubMed] [Google Scholar]
- 5. Goodarzi A. A., Yu Y., Riballo E., Douglas P., Walker S. A., Ye R., Härer C., Marchetti C., Morrice N., Jeggo P. A., Lees-Miller S. P. (2006) DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 25, 3880–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gu J., Li S., Zhang X., Wang L. C., Niewolik D., Schwarz K., Legerski R. J., Zandi E., Lieber M. R. (2010) DNA-PKcs regulates a single-stranded DNA endonuclease activity of Artemis. DNA Repair 9, 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ma Y., Schwarz K., Lieber M. R. (2005) The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair 4, 845–851 [DOI] [PubMed] [Google Scholar]
- 8. Dominski Z. (2007) Nucleases of the metallo-β-lactamase family and their role in DNA and RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 42, 67–93 [DOI] [PubMed] [Google Scholar]
- 9. Li S., Chang H. H., Niewolik D., Hedrick M. P., Pinkerton A. B., Hassig C. A., Schwarz K., Lieber M. R. (2014) Evidence that the DNA endonuclease ARTEMIS also has intrinsic 5′-exonuclease activity. J. Biol. Chem. 289, 7825–7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moshous D., Callebaut I., de Chasseval R., Corneo B., Cavazzana-Calvo M., Le Deist F., Tezcan I., Sanal O., Bertrand Y., Philippe N., Fischer A., de Villartay J. P. (2001) Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105, 177–186 [DOI] [PubMed] [Google Scholar]
- 11. Ma Y., Pannicke U., Schwarz K., Lieber M. R. (2002) Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108, 781–794 [DOI] [PubMed] [Google Scholar]
- 12. Lu H., Shimazaki N., Raval P., Gu J., Watanabe G., Schwarz K., Swanson P. C., Lieber M. R. (2008) A biochemically defined system for coding joint formation in V(D)J recombination. Mol. Cell 31, 485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blommers M. J., Walters J. A., Haasnoot C. A., Aelen J. M., van der Marel G. A., van Boom J. H., Hilbers C. W. (1989) Effects of base sequence on the loop folding in DNA hairpins. Biochemistry 28, 7491–7498 [DOI] [PubMed] [Google Scholar]
- 14. Povirk L. F. (2012) Processing of damaged DNA ends for double-strand break repair in mammalian cells. ISRN Mol. Biol. 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pannicke U., Ma Y., Hopfner K. P., Niewolik D., Lieber M. R., Schwarz K. (2004) Functional and biochemical dissection of the structure-specific nuclease ARTEMIS. EMBO J. 23, 1987–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goodarzi A. A., Lees-Miller S. P. (2004) Biochemical characterization of the ataxia-telangiectasia mutated (ATM) protein from human cells. DNA Repair 3, 753–767 [DOI] [PubMed] [Google Scholar]
- 17. West R. B., Yaneva M., Lieber M. R. (1998) Productive and nonproductive complexes of Ku and DNA-dependent protein kinase at DNA termini. Mol. Cell. Biol. 18, 5908–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yannone S. M., Khan I. S., Zhou R. Z., Zhou T., Valerie K., Povirk L. F. (2008) Coordinate 5′ and 3′ endonucleolytic trimming of terminally blocked blunt DNA double-strand break ends by Artemis nuclease and DNA-dependent protein kinase. Nucleic Acids Res. 36, 3354–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang Y., Giblin W., Kubec M., Westfield G., St Charles J., Chadde L., Kraftson S., Sekiguchi J. (2009) Impact of a hypomorphic Artemis disease allele on lymphocyte development, DNA end processing, and genome stability. J. Exp. Med. 206, 893–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsai A. G., Engelhart A. E., Hatmal M. M., Houston S. I., Hud N. V., Haworth I. S., Lieber M. R. (2009) Conformational variants of duplex DNA correlated with cytosine-rich chromosomal fragile sites. J. Biol. Chem. 284, 7157–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Niewolik D., Pannicke U., Lu H., Ma Y., Wang L. C., Kulesza P., Zandi E., Lieber M. R., Schwarz K. (2006) DNA-PKcs dependence of Artemis endonucleolytic activity, differences between hairpins and 5′ or 3′ overhangs. J. Biol. Chem. 281, 33900–33909 [DOI] [PubMed] [Google Scholar]
- 22. Gu J., Lu H., Tsai A. G., Schwarz K., Lieber M. R. (2007) Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence. Nucleic Acids Res. 35, 5755–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rooney S., Alt F. W., Lombard D., Whitlow S., Eckersdorff M., Fleming J., Fugmann S., Ferguson D. O., Schatz D. G., Sekiguchi J. (2003) Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med. 197, 553–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rooney S., Sekiguchi J., Zhu C., Cheng H.-L., Manis J., Whitlow S., DeVido J., Foy D., Chaudhuri J., Lombard D., Alt F. W. (2002) Leaky SCID phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol. Cell 10, 1379–1390 [DOI] [PubMed] [Google Scholar]
- 25. Riballo E., Kühne M., Rief N., Doherty A., Smith G. C., Recio M.-J., Reis C., Dahm K., Fricke A., Krempler A., Parker A. R., Jackson S. P., Gennery A., Jeggo P. A., Löbrich M. (2004) A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell 16, 715–724 [DOI] [PubMed] [Google Scholar]
- 26. Kurosawa A., Koyama H., Takayama S., Miki K., Ayusawa D., Fujii M., Iiizumi S., Adachi N. (2008) The requirement of Artemis in double-strand break repair depends on the type of DNA damage. DNA Cell Biol. 27, 55–61 [DOI] [PubMed] [Google Scholar]
- 27. Gauss G. H., Lieber M. R. (1996) Mechanistic constraints on diversity in human V(D)J recombination. Mol. Cell. Biol. 16, 258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kulesza P., Lieber M. R. (1998) DNA-PK is essential only for coding joint formation in V(D)J recombination. Nucleic Acids Res. 26, 3944–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Touvrey C., Couedel C., Soulas P., Couderc R., Jasin M., de Villartay J. P., Marche P. N., Jouvin-Marche E., Candéias S. M. (2008) Distinct effects of DNA-PKcs and Artemis inactivation on signal joint formation in vivo. Mol. Immunol. 45, 3383–3391 [DOI] [PubMed] [Google Scholar]
- 30. Lieber M. R., Hesse J. E., Lewis S., Bosma G. C., Rosenberg N., Mizuuchi K., Bosma M. J., Gellert M. (1988) The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell 55, 7–16 [DOI] [PubMed] [Google Scholar]