Abstract
The original purification of the heterotrimeric eIF4F was published over 30 years ago (Grifo, J. A., Tahara, S. M., Morgan, M. A., Shatkin, A. J., and Merrick, W. C. (1983) J. Biol. Chem. 258, 5804–5810). Since that time, numerous studies have been performed with the three proteins specifically required for the translation initiation of natural mRNAs, eIF4A, eIF4B, and eIF4F. These have involved enzymatic and structural studies of the proteins and a number of site-directed mutagenesis studies. The regulation of translation exhibited through the mammalian target of rapamycin (mTOR) pathway is predominately seen as the phosphorylation of 4E-BP, an inhibitor of protein synthesis that functions by binding to the cap binding subunit of eIF4F (eIF4E). A hypothesis that requires the disassembly of eIF4F during translation initiation to yield free subunits (eIF4A, eIF4E, and eIF4G) is presented.
Keywords: eukaryotic initiation factor 4A (eIF4A), eukaryotic initiation factor 4B (eIF4B), eukaryotic translation initiation, mRNA, protein assembly, RNA helicase, translation initiation, 4E-BP, eIF4F, initiation factor recycling
The Biology of eIF4F
The initial findings in the study of natural mRNA translation reflected the newly discovered m7G cap at the 5′ end of eukaryotic mRNAs (2). mRNAs lacking this structure were translated with less efficiency than mRNAs that contained this structure (3). This unique structure allowed for specific assays or purifications, many established in the laboratory of Dr. Aaron Shatkin with assists from his colleagues. Two of note were the use of m7G-Sepharose for affinity purification (4, 5) and the crosslinking of periodate-oxidized mRNAs to proteins (6).
The initial application of these methodologies identified two different molecular weight species (about 25,000 and at least 200,000), although the high molecular weight protein contained the small molecular weight component, now known as eIF4E (7). Given the size of several other known translation factors, the question was whether these contained the small molecular weight subunit (notably eIF3 and eIF4B) (8–11). Ultimate purification of eIF4F indicated that neither of these were correct but that eIF4F would form stable complexes with either, thus being consistent with the eIF4E component tracking with them. At the same time, it was recognized that the 46,000 molecular weight subunit of eIF4F was eIF4A. By physical analysis, eIF4F was a heterotrimeric complex of eIF4A, eIF4E, and eIF4G (1).
The next studies were to attempt to identify the functions of the various proteins required specifically for natural mRNA translation (eIF4A, eIF4B, and eIF4F). The characteristics of these three proteins were very different. In the absence of ATP, binding to RNA could only be well demonstrated with eIF4F (Table 1). eIF4A and eIF4F could hydrolyze ATP in the presence of single-stranded RNA, and eIF4B would enhance both of these activities (12). In terms of mechanism, the coupling of the binding of ATP and RNA was realized in recognizing that eIF4A or eIF4F had the ability to unwind duplex RNA. As noted in Table 1, the “strength” of the helicase activity was greater with eIF4F (14).
TABLE 1.
Relative activities of translation initiation factors
| Initiation factor | Globin mRNA bindinga |
RNA-dependent ATPaseb |
Helicase activityc |
|||
|---|---|---|---|---|---|---|
| −ATP | +ATP | Kact | Vmax | ΔG = −17.9 | ΔG = −24.7 | |
| eIF4A | 130 | 1800 | 15,000 | 110 | 3.0 | 0.6 |
| eIF4B | 230 | 350 | –d | –d | –d | –d |
| eIF4F | 3610 | 3990 | 30 | 20 | 7.3 | 1.3 |
| eIF4A, eIF4B | 150 | 5170 | 60 | 135 | 10.3 | 3.6 |
| eIF4F, eIF4B | 3820 | 4260 | 30 | 30 | 9.6 | 3.6 |
a Radioactively labeled globin mRNA retention on nitrocellulose filters (cpm) (12).
b Hydrolysis of [γ-32P]ATP in response to added RNA (Kact in μm, Vmax in fmol of PO4 released per s per μg of either eIF4A or eIF4F) (13).
c Unwinding of duplex RNA with a 33-nucleotide single-stranded region or 29-nucleotide single-stranded region; duplexes were 11 and 15 bp, respectively (initial rate of unwinding) (14).
d — = not determined.
As the ability to determine amino acid sequence from RNA sequence advanced, it was found that there were numerous conserved amino acid motifs in eIF4A that could be found in other proteins, and this led to the establishment of the DEAD-box proteins (15). The DEAD-box proteins became the first group of well characterized RNA helicases.
This information was soon put into a variety of model initiation pathways in which the primary feature of the eIF4 proteins was their utilization for the unwinding of possible secondary structure to form a single-stranded RNA that could be bound to the 40S subunit (as the 43S preinitiation complex containing eIF1, eIF1A, eIF3, and the ternary complex eIF2·GTP·Met-tRNAi). A later finding that RNA helicases can displace protein from an RNA·protein complex adds a second element to the activation process as mRNAs exit the nucleus as mRNPs2 (16).
In the mid-90s, as efforts were continuing to define the biochemistry of the eIF4 family proteins, a very unique discovery was made. One of the major proteins to be phosphorylated in cells in response to insulin treatment was called PHAS-I (phosphorylated heat- and acid-stable protein regulated by insulin) (17). By a separate analytic procedure, this protein was also identified as a protein that bound to eIF4E and would later be discovered to be a major target of the mTOR pathway (mTORC1) (18). The phosphorylation of this protein (4E-BP) led to its inactivation (inability to bind to eIF4E). Because the binding of eIF4E by either eIF4G or 4E-BP is to overlapping sites on eIF4E, only one of the two can be bound to eIF4E at any point in time (19, 20).
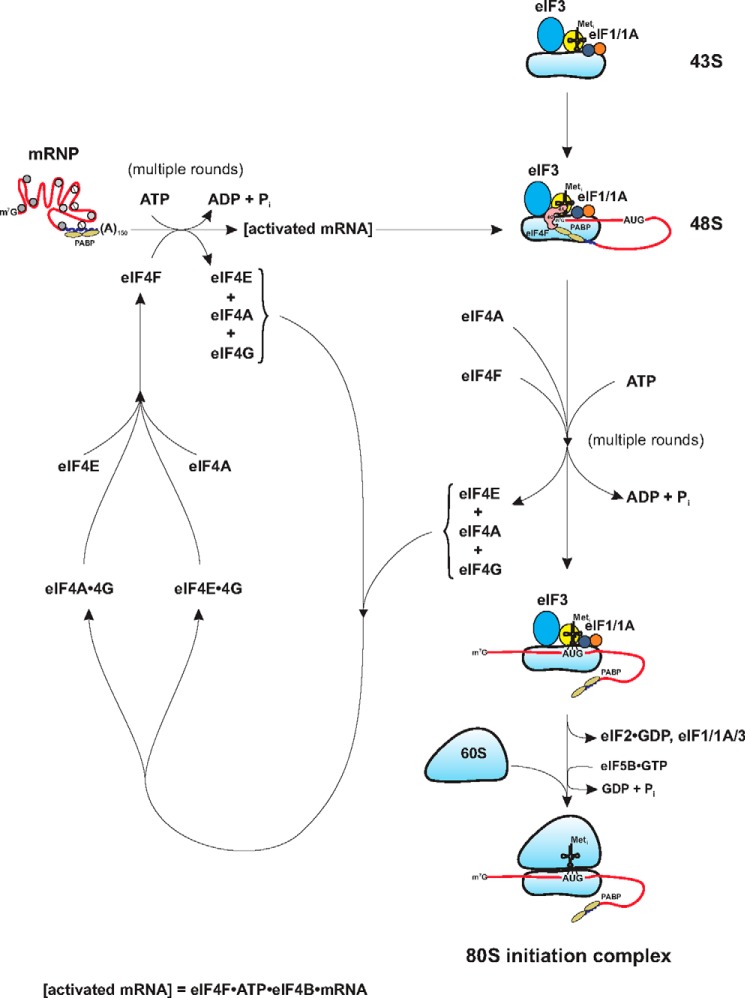
This finding added a second arm to the global regulation of eukaryotic protein synthesis. The first was the regulation of available initiator tRNA as the ternary complex eIF2·GTP·Met-tRNAi. The second was the restriction in the level of eIF4F activity due to the lowered availability of eIF4E. Consistent with the general pathway of 80S complex formation (Fig. 1) was that the binding of the ternary complex preceded the binding of mRNA. As a consequence, translation of most mRNAs was reduced equally when ternary complexes became limiting. In contrast, reduction of eIF4F activity forced mRNAs to compete for limiting eIF4F, and this resulted in “good” mRNAs being preferentially translated, whereas “poor” mRNAs were not. This was consistent with the mathematical modeling, initially by Lodish and colleagues (21, 22) and then refined by Godefroy-Colburn and Thach (23). Although a generalization, it would appear that the ability of an mRNA to compete for the translational apparatus was mostly determined by the availability of its m7G cap (24).
FIGURE 1.
Actions of eIF4F in translation initiation. Shown in this figure are the eIF4F steps in translation, the activation of mRNA, which converts an mRNP to an activated mRNA with bound eIF4F and scanning, and the movement in a 5′ to 3′ direction of the mRNA to the initiation codon AUG. Both of the steps are ATP-dependent. The degree wo which free eIF4A, eIF4B, or other RNA helicases may play a role in this process is not pictured. This figure is adapted from Merrick, W. C. (2010) J. Biol. Chem. 285, 21197–21201 (90).
A second protein that can influence the formation of eIF4F is the protein PDCD4, a tumor suppressor. This protein binds to eIF4A and can thereby limit the amount of eIF4A available to form eIF4F (25). PDCD4 is regulated through phosphorylation by the mTOR pathway (which inhibits its binding to eIF4A) (26–28). Based on the observation that mRNAs with structured 5′ ends require additional eIF4A for translation, the lack of PDCD4 phosphorylation could lead to reduced translation of these mRNAs, either as free eIF4A or as eIF4A to reconstitute eIF4F. In sum, mTOR coordinately regulates eIF4F formation by controlling the level of phosphorylation of both 4E-BP and PDCD4.
The Biochemistry of eIF4F
With the recognition that eIF4A was a subunit of eIF4F, much of the subsequent biochemistry focused on the difference between these two proteins and the differential affect of eIF4B on these proteins (it is noted that the relatively late discovery of eIF4H has resulted in more minimal studies in this comparison) (29). The standard assays initially used were: 1) RNA binding, an assay that monitors the retention of an RNA on nitrocellulose filters indicative of a protein·nucleic acid complex; 2) RNA-dependent ATPase, an assay that measures the hydrolysis of ATP in a reaction requiring the presence of RNA; and 3) duplex RNA unwinding, an assay that measures the ATP-dependent strand separation of an RNA duplex to yield two single strands. For a number of RNA helicases, there is a dependence on a single-stranded region being part of the duplex.
Data from these assays are shown in Table 1. Consistent with their behavior on phosphocellulose, in the absence of ATP, eIF4A failed to bind mRNA, whereas eIF4F bound significant levels of mRNA (30). However, the presence of ATP greatly enhanced the binding of mRNA by eIF4A (with or without eIF4B), whereas it only offered a slight stimulation with eIF4F. The kinetic data for the RNA-dependent ATPase assay indicate that the primary difference between eIF4A and eIF4F is the apparent affinity of the proteins for RNA, although the presence of eIF4B renders the ability of eIF4A to catalyze hydrolysis nearly equivalent to that seen with eIF4F (13). Similar trends are evident when the melting of RNA duplexes is considered; however, the -fold difference in the initial rates of unwinding is less dramatic with a 2–3-fold difference between eIF4A and eIF4F (± eIF4B) with a shorter duplex, but a greater difference with a more stable duplex (up to 6-fold) (14).
Yeast Just Aren't Human
A relatively early observation was that Saccharomyces cerevisiae eIF4F is different from the human protein and that some of these differences play out with eIF4A as well. Perhaps the most challenging difference is that a three-subunit eIF4F has not been purified from yeast, but rather only the two-subunit eIF4E·eIF4G complex has been isolated (31). Secondly, eIF4B is a monomer in yeast but a dimer in the mammalian system, which may have profound influences on the biochemical behavior of either eIF4A or eIF4F (32–34). In this light, eIF4B enhances the RNA-dependent ATPase activity of eIF4A in the mammalian system by reducing the Kact 1000-fold (13). In contrast, there is no stimulation observed in the yeast system. Thus, given these differences, the remaining discussion will focus on the mammalian eIF4F (and eIF4A and eIF4B), although it is anticipated that similar activities will be visualized for the yeast system as well.
Subunit Interactions in eIF4F
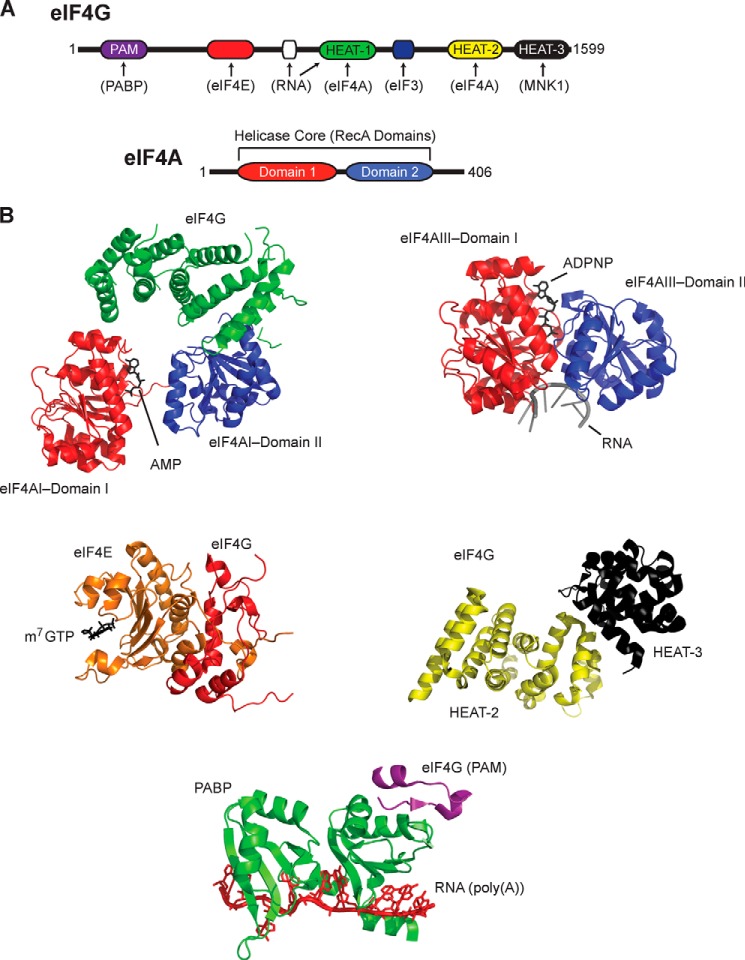
From numerous studies, interactive domains of eIF4G have been determined, and a graphic representation of these domains is seen in Fig. 2A. Because of the many interactions, eIF4G is able to coordinate functions related to m7G cap binding (eIF4E), RNA helicase unwinding (eIF4A), binding to the 40S subunit (eIF3), and coordinating initiation using freshly terminated ribosomes via the interaction with the poly(A)-binding protein (PABP) as the “circular” mRNA.
FIGURE 2.
High resolutions structures of eIF4F components. A, a graphic representation of the interactive domains of eIF4F and the two-domain structure of eIF4A. PAM, PABP-interacting motif. B, clockwise, a selection of a few high resolution structures that represent eIF4A·AMP·eIF4FG HEAT domain in the open confirmation (Protein Data Bank (PDB): 2VSO (40)), the closed confirmation of eIF4AIII as it exists in the exon junction complex with ADPNP (PDB: 2HYI (86)), the structure of the C-terminal HEAT-2 and HEAT-3 domains in eIF4G (PDB: 1UG3 39)), the N-terminal PABP binding domain of eIF4G with PABP·poly(A) (PDB 4F02 (87)), and a solution structure of m7GTP·eIF4E bound to eIF4G (PDB: 1RF8 (88)). Note that the colors of the domains in the structures are coded to those as seen in the graphic in panel A. This figure is used with the generous permission of Dr. Christopher Fraser, University of California at Davis, and with permission from Elsevier Publishing. This figure can be found in Ref. 89.
At the level of crystallographic studies, individual structures for eIF4A, eIF4E, and the HEAT domains of eIF4G have been determined (35–45). The only catalytically active co-crystal structure is for eIF4A complexed with the middle domain of eIF4G (amino acids 572–853), and in this structure, both the N-terminal and the C-terminal domains make contact with the eIF4G HEAT1 domain (Fig. 2B) (37–40). This interaction would appear to restrict the flexibility of the eIF4A molecule around the 11-amino acid flexible linker that joins the two domains. Of particular interest is the effect that eIF4G has on orienting the subdomains of eIF4A into a more active (open) confirmation in contrast to the variety of confirmations theoretically possible with free eIF4A (38, 40). At present, the mechanism for the observed activation of eIF4A activity by eIF4B is unknown, but it would not be surprising if it were similar. This activation appears to reflect primarily differences in binding nucleic acid as the apparent Km for ATP is relatively unaffected (13).
An interesting side note comes from the structural analysis of eIF4E complexed with the inhibitor peptide, 4EGI-1 (42, 43). In contrast to the 4E-BPs, the inhibitor peptide binds to an allosteric site on eIF4E, leading to the displacement of eIF4G from eIF4E (43).
Unfortunately, further crystal structure analysis has been limited by either the low level of protein in normal cells or the inability to readily express human full-length eIF4G (although see Feoktistova et al. (45)). This is accompanied by the concern that the post-translational modifications known to occur on both eIF4E and eIF4G may be important for function, and the degree of modification obtained with expression in either Escherichia coli or insect cells may be limiting. Also, as noted for eIF2, it is possible that a cellular protein might be required for the correct assembly of the complete complex (46). Thus, it will be important to compare both biochemically and physically native protein purified from actively translating systems (i.e. HeLa or reticulocytes) with recombinant proteins.
“Modern” Biochemistry with eIF4F Subunits/Reconstitution
The availability of molecular biological techniques and protein expression has allowed for the preparation of numerous translation initiation factors, either as subunits or as individual proteins. For the most part, these expressed proteins have demonstrated activity either as individual components or as added to a reconstitution assay. The most useful of these are when the expressed protein can be independently assessed for activity, as is the case for eIF4A. With respect to the “mRNA-specific initiation factors,” whereas eIF4E can be assessed for binding to m7GTP-Sepharose and eIF4B and eIF4G can be assessed for binding to nucleic acid, for most of the “functional assays” (RNA-dependent ATPase, RNA duplex unwinding, toe printing, protein synthesis), it is the effect that the added protein (or protein subunits) has on the activity of eIF4A that is most often measured.
Using expressed eIF4A, eIF4B, and either fragments or full-length eIF4G, Ozes et al. (47) found that eIF4B stimulated the unwinding activity of eIF4A about 10-fold, whereas eIF4G stimulated the unwinding less than 2-fold. However, the combination of eIF4A, eIF4B, and eIF4G provided for a 100-fold stimulation over free eIF4A (Table 1 of Ref. 47). Additional studies have examined the helicase activity of eIF4A in the presence of various eIF4G fragments with and without eIF4B and/or eIF4E. In part, these studies utilized a series of truncated (or full-length) forms of eIF4G (amino acids 682–1166, 557–1137, 557–1600, and 1–1600) (45). When a larger eIF4G fragment was used (or the complete protein) that contained the eIF4E binding site (amino acids 557–681), a surprising result was found. The 682–1105 fragment provided more stimulation of activity than either the 557–1137 or 557–1600 fragments or the full-length protein. However, the lost activity of the larger fragments containing the eIF4E binding site was recovered upon the addition of eIF4E. This led to an autoinhibitory model (Fig. 5 of Ref. 45) where the presence of eIF4E directly influences eIF4F activity by removing this inhibition. This observation may provide a partial answer as to how the eIF4F/mRNA interaction might be stabilized to allow kinetically for the interaction with eIF3 of the 43S preinitiation complex, perhaps something similar to the clamping activity of eIF4AIII when it is part of the exon junction complex (48).
From the above (and other) results, it is clear that both eIF4B and eIF4G stimulate the helicase activity of eIF4A, results consist with earlier observations where eIF4F was found to be more active than eIF4A and the eIF4A unwinding activity was stimulated by eIF4B (see Table 1). However, none of these studies addressed the root cause for these observations. Did the additional proteins enhance the catalytic rate, or did they enhance the affinity of eIF4A for substrate? Based upon the results from the RNA-dependent ATPase assay, the primary effect would appear to be an improvement in substrate affinity (13). However, it is clear that a secondary effect appears to be related to the stability of the RNA duplex and the helicase activity of eIF4A. Using a duplex with a ΔG of −24.7 kcal/mol, the stimulation of eIF4A activity by eIF4B in unwinding is 7-fold, whereas for a less stable duplex (ΔG = −17.9 kcal/mol), the stimulation was 3-fold (14). The suggestion here is that beyond the presumed hydrolysis of ATP and the partial strand separation of the RNA duplex, the other proteins are likely stabilizing the partially melted intermediate, allowing additional time for the duplex to unwind (and more complicated mechanisms could easily be envisaged). Thus, the roles played by both eIF4B and eIF4G may also relate to activities not directly related to eIF4A activity, but dependent upon it.
Very recently, using single molecule FRET, Garcia-Garcia et al. (49) have shown that the combination of eIF4A, eIF4B, and eIF4G can lead to the processive unwinding of a 70-bp hairpin. The processive character was dependent on eIF4B, and “hints” of this processivity were also seen previously with native eIF4F (14). Currently, it is not clear what the contribution of eIF4E might be in this processive unwinding. The unexpected finding was that a 70-bp hairpin (ΔG > 100 kcal/mol) could be completely unwound despite the observation that a hairpin this stable will usually block translation completely.
Hints at the Instability of eIF4F
The original purification of eIF4F took 5–8 days, suggesting that the complex of eIF4E, eIF4A, and eIF4G was quite stable (1). However, in the process of numerous purifications, my colleagues and I noted that the level of purity varied from one preparation to another. As a result, different alterations to the purification scheme were tried. The first effort was the use of m7GTP-Sepharose a second time, but with more extensive washing to remove contaminating proteins. Much to our dismay, the use of extensive washing of the bound eIF4F led to the purification of eIF4E only.3 It appears that the additional time taken to wash the column destabilized the interaction between eIF4E and eIF4G. In retrospect, this is consistent with the early purification efforts using m7GTP-Sepharose that resulted in the isolation of eIF4E (50, 51).
As an alternate, gradient elution from phosphocellulose was attempted as the early step in the purification scheme used batch elution from phosphocellulose. When this effort was made, again my colleagues and I were surprised. We found that we had separated eIF4A from eIF4F and that the remaining eIF4E-eIF4G complex was also purified (52). The eIF4F activity could be restored by adding back purified eIF4A to the eIF4E·eIF4G complex. The inference here is that the binding of eIF4F to phosphocellulose was a “poor man's RNA affinity column” and that the interaction of eIF4F with RNA was leading to the separation of eIF4A from the eIF4G·eIF4E complex.
Further studies with purified eIF4A and eIF4F indicated that free eIF4A could exchange with bound eIF4A in the eIF4F complex (53). In part, this observation provided an explanation for the dominant negative affects of a mutant eIF4A (R362Q) in translation (54). Although the exchange of the mutant eIF4A for the wild type eIF4A in eIF4F accounted for much of the inhibition observed, it did not account for the dominant negative aspect. At the time, this was taken as evidence that multiple rounds of eIF4F action were necessary for the utilization of a single mRNA via the cap-dependent pathway, likely to be 4–5 rounds of eIF4F utilization (54). However, it was not clear whether these multiple rounds of eIF4F utilization reflected the initial mRNA activation step or scanning or both.
The eIF4F Disassembly Hypothesis
The one biologic necessity of eIF4F would appear to be the requirement for the release of eIF4E. Otherwise, what would 4E-BP ever bind to? Although many of the above data/observations describe the differential activities depending on whether different combinations of subunits were added to the test tube, none looked at the actual formation or possible dissolution of the eIF4F complex. However, it is possible to piece together a hypothesis that builds upon the observations related to the instability elements of eIF4F noted above. 1) Based upon the observations above, it appears that the eIF4E/eIF4G interaction is destabilized upon binding the m7GTP cap. 2) Release of eIF4E might then stabilize the eIF4G·eIF4A·eIF4B complex on the mRNA with the postulated autoinhibitory activity seen with the loss of eIF4E (45). This might provide for an RNA clamping activity. 3) In what is likely an ATP-dependent event, eIF4A is released from eIF4G in a manner reflected in the phosphocellulose-induced release of eIF4A from eIF4F (52). This step could also be the source for loading of eIF4A onto the 43S preinitiation complex as there is generally 5–10 times more eIF4A on 40S subunits than eIF2, eIF3, or eIF4F (55, 56). 4) Additional eIF4F and/or eIF4A molecules are required for the ATP-dependent scanning of the mRNA. This step could also lead to the additional release of subunits from eIF4F. 5) As a result of mRNA activation and scanning, the products of the initiation pathway are the individual subunits of eIF4F (eIF4A, eIF4E, and eIF4G) as shown in Fig. 1.
Our initial observation that eIF4F purified as a complex with eIF4B does raise the question as to whether its association would enhance, hinder, or have no effect on the process of disassembly. As in general, eIF4B stimulates the activity of both eIF4A and eIF4F, it is anticipated that it might enhance the disassembly process as well in the context of the m7GTP cap and the RNA body of the mRNA or the RNA of the 40S subunit. Given the observations with 4EGI-1, it is also possible that a yet unidentified compound or factor may have an allosteric affect in triggering the release of subunits (44).
With the release of subunits, to perform the next round of initiation, eIF4F needs to reassemble its three subunits. The pathway below is suggested, but in the absence of real data. 1) eIF4G binds to eIF4A to form the eIF4G·4A complex. This is seen as the preferred first step as the concentration of eIF4A is roughly 10-fold greater than the concentration of eIF4E. Additionally, a functional complex of eIF4G·4A has been seen in the experiments of Fraser and colleagues (45, 47), and evidence from the yeast system also suggests that this association might be facilitated by eIF4B (47, 57). 2) The eIF4G·4A complex binds eIF4E to form eIF4F. Besides forming eIF4F, the binding of eIF4E also relieves the postulated “autoinhibition” from the N-terminal region of eIF4G (45). 3) eIF4F binds eIF4B to form the eIF4F·4B complex, which is seen as the “real” initiation factor that begins the mRNA activation process.
The combination of the disassembly process and the reformation of eIF4F are depicted in Fig. 1. The reformation process could also begin with the binding of eIF4G to eIF4E followed by the binding of eIF4A. Also, by that same token, it is possible that in the disassembly process, eIF4A might be released prior to the release of eIF4G. It should be noted that an equivalent thermodynamic pathway has been traced for the yeast proteins (see Fig. 1 in Ref. 58). Although the Kd values for the various associations would appear to favor eIF4E binding to eIF4G as the first step, the observation that in yeast eIF4A exceeds eIF4E concentration by about a factor of 3 would make either reassembly pathway possible (56). One of the supporting pieces of evidence for the postulated existence of the eIF4G·4A complex is that this is the experimentally functional component required for internal ribosomal entry site-mediated translation, and thus, there is a biological assay for (and use of) the two different forms of eIF4G complexes, eIF4F and eIF4G·4A (58, 59).
Other Complications
This minireview has focused on eIF4F as the primary driving force in the activation and utilization of mRNAs in cap-dependent protein synthesis. However, numerous articles have indicated that for mRNAs with highly structured 5′ UTRs, additional proteins may be required. One of these is additional eIF4A for which one might imagine a mechanism (60). The others represent different RNA helicases including DDX3, DDX41, DHX9, and DHX29 (61–64). These alternate possibilities are extensively reviewed in Parsyan et al. (65). Using either in vitro or in vivo conditions, researchers were able to show through the addition of more or less (depletion via siRNA) of these proteins that expression from a reporter mRNA was influenced. What is not clear is whether these RNA helicases are truly part of the normal initiation process, although not required for efficiently translated mRNAs, or whether they are specific for mRNAs with highly structured 5′-UTRs. One suggestion is that these “extra” helicases might be the first binders to the mRNP and that their subsequent action is influenced by the downstream actions of eIF4A, eIF4B, and eIF4F (52). These observations are relatively recent, and clearly more experimentation will be required to determine the exact mechanism for the utilization of these additional RNA helicases.
For the mammalian factors, an added complication is the number of isoforms for each of the eIF4F subunits (well reviewed in Hernández and Vazquez-Pianzola (66)) with three isoforms for eIF4A, three isoforms for eIF4E, and two isoforms for eIF4G. With no additional information (concentration, tissue-specific expression, relative affinities for other subunits), the number of combinations is 18. To date, most of the combined biochemical, biological, and structural data have focused on one of the possible eIF4F forms (eIF4E1, eIF4A1, eIF4G1). It remains to be seen whether dramatic differences in biologic function in the other possible combinations will emerge (i.e. such as the difference between eIF4A1 and eIF4A3, the former an active RNA helicase, the latter an RNA clamp in the exon junction complex) (48).
Two of the major proteins implicated in eIF4F function are eIF3 and PABP, both of which form complexes with eIF4F (67–69). Those formed with eIF3 are sufficiently stable to be isolated by either gel filtration or sucrose density gradients, whereas the PABP interaction has been seen by cryoelectron microscopy and pulldown experiments. It is anticipated that the eIF3 interaction is required for each initiation event, whereas the interaction of PABP with eIF4F may be most important in polysomes in assisting in a three-dimensional, intramolecular reaction that facilitates initiation events from a freshly terminated ribosome.
Although much of this review has focused on mammalian eIF4F, it does need to be recognized how different the yeast and mammalian proteins are. A small sampling is provided in Table 2 for yeast and mammalian eIF4A and eIF4F. From this sampling, the yeast and mammalian proteins are very different. The yeast eIF4A shows greater stimulation of its ATPase activity with double-stranded RNA than with single-stranded RNA by more than a factor of 10, whereas the reverse is true for the mammalian protein. At the same time, the yeast eIF4A is much more dependent on a single-stranded overhang for duplex unwinding, whereas the mammalian protein is only slightly stimulated by a single-stranded overhang.
TABLE 2.
Comparison of yeast and mammalian initiation factor activities
| Assaya | y4A | m4A | y4F | m4F |
|---|---|---|---|---|
| RNA-dependent ATPase | ||||
| Single-stranded RNA/poly(U) | 0.003 | 35 | 6 | 5 |
| Double-stranded RNA | 0.12 | 3 | 0.08 | NDb |
| RNA duplex unwinding | ||||
| +5′-Single-stranded region | 9.5 | 4.5 | 113 | 60 |
| +3′-Single-stranded region | 6.5 | 4.5 | 4.9 | 60 |
| Blunt end duplex | 1.2 | 3.3 | 1.2 | 5 |
a The indicated values are to be compared for relative differences and not for absolute level of activity and are taken from tables where direct comparisons between substrates were made; the yeast and mammalian values were reported in different publications. Direct comparisons should only be made within columns (i.e. all the y4A values), but relative comparisons can be made between columns. The values used in this table are from Refs. 12, 14, 70, and 71. Some values represent rates (yeast eIF4A and eIF4F, mammalian eIF4A), whereas others represent extent of reaction (mammalian eIF4A- and eIF4F-directed duplex unwinding).
b ND, not determined.
In contrast, the ATPase activity of yeast eIF4F is much better stimulated by single-stranded RNA than dsRNA as was seen with mammalian eIF4A and eIF4F. Both the yeast and the mammalian eIF4F show a marked dependence for a single-stranded overhang for duplex unwinding, with a requirement of at least 20+ nucleotides. However, there is a very dramatic difference in the yeast and mammalian proteins in that the yeast eIF4F shows an extreme preference for a 5′ single-stranded region over a 3′ single-stranded region, whereas the stimulation for mammalian eIF4F is equivalent with either 5′ or 3′ single-stranded regions.
The caution in relating the mechanism of action of the yeast and mammalian initiation factors reflects not just differences in methodologies used to assess function, but also in the physical and biochemical differences that have been noted. That said, there does tend to be general agreement on the pathway for forming an initiation complex. However, the most poorly described steps in translation initiation are in fact mRNA activation and scanning, two steps that are highly dependent on eIF4F and for which much better information is required to more accurately assess the individual steps catalyzed by this protein, either in solution (mRNA activation) or on the surface of the 40S subunit (scanning).
It should be recognized that this retrospective focused on eIF4F, and as such, is not a review of translation initiation in general, nor does it probe the various aspects that might call for other proteins, either as RNA helicases or as proteins that might allow for non-standard initiation events. To obtain a fuller appreciation of where eIF4F fits into the grand scheme of things, the reader may find one or more of the closing references to reviews useful (72–85).
Acknowledgments
I thank Dr. Anton Komar for assisting with the development and review of this article and generating Fig. 1 and Dr. Christopher Fraser for the contribution of Fig. 2 to this article.
This work was supported in part by the School of Medicine, Case Western Reserve University. The author declares that he has no conflicts of interest with the contents of this article.
W. Merrick, unpublished results.
- mRNP
- messenger ribonucleoprotein
- mTOR
- mammalian target of rapamycin
- PABP
- poly(A)-binding protein
- ADPNP
- 5′-adenylyl-β,γ-imidodiphosphate
- 4E-BP
- eIF4E-binding protein.
References
- 1. Grifo J. A., Tahara S. M., Morgan M. A., Shatkin A. J., Merrick W. C. (1983) New initiation factor activity required for globin mRNA translation. J. Biol. Chem. 258, 5804–5810 [PubMed] [Google Scholar]
- 2. Both G. W., Furuichi Y., Muthukrishnan S., Shatkin A. J. (1975) Ribosome binding to reovirus mRNA in protein synthesis requires 5′ terminal 7-methylguanosine. Cell 6, 185–195 [DOI] [PubMed] [Google Scholar]
- 3. Zan-Kowalczewska M., Bretner M., Sierakowska H., Szczesna E., Filipowicz W., Shatkin A. J. (1977) Removal of the 5′-terminal m7G from eukaryotic mRNAs by potato nucleotide pyrophosphatase and its effect on translation. Nucleic Acids Res. 4, 3065–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sonenberg N., Rupprecht K. M., Hecht S. M., Shatkin A. J. (1979) Eukaryotic mRNA cap binding protein: purification by affinity chromatography on Sepharose-coupled m7GDP. Proc. Natl. Acad. Sci. U.S.A. 76, 4345–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Webb N. R., Chari R. V., DePillis G., Kozarich J. W., Rhoads R. E. (1984) Purification of the messenger RNA cap-binding protein using a new affinity medium. Biochemistry 23, 177–181 [DOI] [PubMed] [Google Scholar]
- 6. Sonenberg N., Shatkin A. J. (1977) Reovirus mRNA can be covalently crosslinked via the 5′ cap to proteins in initiation complexes. Proc. Natl. Acad. Sci. U.S.A. 74, 4288–4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tahara S. M., Morgan M. A., Shatkin A. J. (1981) Two forms of purified m7G-cap binding protein with different effects on capped mRNA translation in extracts of uninfected and poliovirus-infected HeLa cells. J. Biol. Chem. 256, 7691–7694 [PubMed] [Google Scholar]
- 8. Padilla M., Canaani D., Groner Y., Weinstein J. A., Bar-Joseph M., Merrick W., Shafritz D. A. (1978) Initiation factor eIF-4B (IF-M3)-dependent recognition and translation of capped versus uncapped eukaryotic mRNAs. J. Biol. Chem. 253, 5939–5945 [PubMed] [Google Scholar]
- 9. Hansen J., Etchison D., Hershey J. W., Ehrenfeld E. (1982) Association of cap-binding protein with eukaryotic initiation factor 3 in initiation factor preparations from uninfected and poliovirus-infected HeLa cells. J. Virol. 42, 200–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Etchison D., Milburn S. C., Edery I., Sonenberg N., Hershey J. W. (1982) Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eukaryotic initiation factor 3 and a cap-binding protein complex. J. Biol. Chem. 257, 14806–14810 [PubMed] [Google Scholar]
- 11. Trachsel H., Sonenberg N., Shatkin A. J., Rose J. K., Leong K., Bergmann J. E., Gordon J., Baltimore D. (1980) Purification of a factor that restores translation of vesicular stomatitis virus mRNA in extracts from poliovirus-infected HeLa cells. Proc. Natl. Acad. Sci. U.S.A. 77, 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abramson R. D., Dever T. E., Lawson T. G., Ray B. K., Thach R. E., Merrick W. C. (1987) The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J. Biol. Chem. 262, 3826–3832 [PubMed] [Google Scholar]
- 13. Abramson R. D., Dever T. E., Merrick W. C. (1988) Biochemical evidence supporting a mechanism for cap-independent and internal initiation of eukaryotic mRNA. J. Biol. Chem. 263, 6016–6019 [PubMed] [Google Scholar]
- 14. Rogers G. W. Jr., Richter N. J., Lima W. F., Merrick W. C. (2001) Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276, 30914–30922 [DOI] [PubMed] [Google Scholar]
- 15. Linder P., Lasko P. F., Ashburner M., Leroy P., Nielsen P. J., Nishi K., Schnier J., Slonimski P. P. (1989) Birth of the D-E-A-D box. Nature 337, 121–122 [DOI] [PubMed] [Google Scholar]
- 16. Jankowsky E., Gross C. H., Shuman S., Pyle A. M. (2001) Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 291, 121–125 [DOI] [PubMed] [Google Scholar]
- 17. Hu C., Pang S., Kong X., Velleca M., Lawrence J. C. Jr (1994) Molecular cloning and tissue distribution of PHAS-I, an intracellular target for insulin and growth factors. Proc. Natl. Acad. Sci. U.S.A. 91, 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pause A., Belsham G. J., Gingras A. C., Donzé O., Lin T. A., Lawrence J. C. Jr., Sonenberg N. (1994) Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371, 762–767 [DOI] [PubMed] [Google Scholar]
- 19. Lukhele S., Bah A., Lin H., Sonenberg N., Forman-Kay J. D. (2013) Interaction of the eukaryotic initiation factor 4E with 4E-BP2 at a dynamic bipartite interface. Structure 21, 2186–2196 [DOI] [PubMed] [Google Scholar]
- 20. Peter D., Igreja C., Weber R., Wohlbold L., Weiler C., Ebertsch L., Weichenrieder O., Izaurralde E. (2015) Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol. Cell 57, 1074–1087 [DOI] [PubMed] [Google Scholar]
- 21. Temple G., Lodish H. F. (1975) Competition between α and β globin messenger RNA. Biochem. Biophys. Res. Comm. 63, 971–979 [DOI] [PubMed] [Google Scholar]
- 22. Bergmann J. E., Lodish H. F. (1979) A kinetic model of protein synthesis: application to hemoglobin synthesis and translational control. J. Biol. Chem. 254, 11927–11937 [PubMed] [Google Scholar]
- 23. Godefroy-Colburn T., Thach R. E. (1981) The role of mRNA competition in regulating translation. IV Kinetic model. J. Biol. Chem. 256, 11762–11773 [PubMed] [Google Scholar]
- 24. Godefroy-Colburn T., Ravelonandro M., Pinck L. (1985) Cap accessibility correlates with the initiation efficiency of alfalfa mosaic virus RNAs. Eur. J. Biochem. 147, 549–552 [DOI] [PubMed] [Google Scholar]
- 25. Yang H. S., Jansen A. P., Komar A. A., Zheng X., Merrick W. C., Costes S., Lockett S. J., Sonenberg N., Colburn N. H. (2003) The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell. Biol. 23, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Suzuki C., Garces R. G., Edmonds K. A., Hiller S., Hyberts S. G., Marintchev A., Wagner G. (2008) PDCD4 inhibits translation initiation by binding to eIF4A using both its MA3 domains. Proc. Natl. Acad. Sci. U.S.A. 105, 3274–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang J. H., Cho Y. H., Sohn S. Y., Choi J. M., Kim A., Kim Y. C., Jang S. K., Cho Y. (2009) Crystal structure of the eIF4A-PDCD4 complex. Proc. Natl. Acad. Sci. U.S.A. 106, 3148–3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dennis M. D., Jefferson L. S., Kimball S. R. (2012) Role of p70S6K1-mediated phosphorylation of eIF4B and PDCD4 proteins in the regulation of protein synthesis. J. Biol. Chem. 287, 42890–42899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richter N. J., Rogers G. W. Jr., Hensold J. O., Merrick W. C. (1999) Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H. J. Biol. Chem. 274, 35415–35424 [DOI] [PubMed] [Google Scholar]
- 30. Merrick W. C. (1979) Purification of protein synthesis initiation factors from rabbit reticulocytes. Methods Enzymol. 60, 101–108 [DOI] [PubMed] [Google Scholar]
- 31. Lanker S., Müller P. P., Altmann M., Goyer C., Sonenberg N., Trachsel H. (1992) Interactions of the eIF4F subunits in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 267, 21167–21171 [PubMed] [Google Scholar]
- 32. Altmann M., Wittmer B., Méthot N., Sonenberg N., Trachsel H. (1995) The Saccharomyces cerevisiae translation initiation factor Tif3 and its mammalian homologue, eIF4B, have RNA annealing activity. EMBO J. 14, 3820–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grifo J. A. (1982) Eukaryotic initiation factors which recognize and bind mRNA. Ph. D. thesis, Case Western Reserve University, Cleveland, OH [Google Scholar]
- 34. Méthot N., Song M. S., Sonenberg N. (1996) A region rich in aspartic acid, arginine, tyrosine and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol. Cell. Biol. 16, 5328–5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Caruthers J. M., Johnson E. R., McKay D. B. (2000) Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. U.S.A. 97, 13080–13085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meng H., Li C., Wang Y., Chen G. (2014) Molecular dynamics simulation of the allosteric regulation of eIF4A protein from the open to closed state, induced by ATP and RNA substrates. PLoS One 9, e86104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marcotrigiano J., Lomakin I. B., Sonenberg N., Pestova T. V., Hellen C. U. T., Burley S. K. (2001) A conserved HEAT domain with eIF4G directs assembly of the translation initiation machinery. Mol. Cell 7, 193–203 [DOI] [PubMed] [Google Scholar]
- 38. Oberer M., Marintchev A., Wagner G. (2005) Structural basis for the enhancement of the eIF4A helicase activity by eIF4G. Genes Dev. 19, 2212–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bellsolell L., Cho-Park P. F., Poulin F., Sonenberg N., Burley S. K. (2006) Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1. Structure 14, 913–923 [DOI] [PubMed] [Google Scholar]
- 40. Schütz P., Bumann M., Oberholzer A. E., Bieniossek C., Trachsel H., Altmann M., Baumann U. (2008) Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 105, 9564–9569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marcotrigiano J., Gingras A. C., Sonenberg N., Burley S. K. (1997) Co-crystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89, 951–961 [DOI] [PubMed] [Google Scholar]
- 42. Matsuo H., Li H., McGuire A. M., Fletcher C. M., Gingras A. C., Sonenberg N., Wagner G. (1997) Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 4, 717–724 [DOI] [PubMed] [Google Scholar]
- 43. Brown C. J., Verma C. S., Walkinshaw M. D., Lane D. P. (2009) Crystallization of eIF4E complexed with eIF4GI peptide and glycerol reveals distinct structural differences around the cap-binding site. Cell Cycle 8, 1905–1911 [DOI] [PubMed] [Google Scholar]
- 44. Papadopoulos E., Jenni S., Kabha E., Takrouri K. J., Yi T., Salvi N., Luna R. E., Gavathiotis E., Mahalingam P., Arthanari H., Rodriguez-Mias R., Yefidoff-Freedman R., Aktas B. H., Chorev M., Halperin J. A., Wagner G. (2014) Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G. Proc. Natl. Acad. Sci. U.S.A. 111, E3187–E3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Feoktistova K., Tuvshintogs E., Do A., Fraser C. S. (2013) Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc. Natl. Acad. Sci. U.S.A. 110, 13339–13344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Perzlmaier A. F., Richter F., Seufert W. (2013) Translation initiation requires cell division cycle 123 (Cdc123) to facilitate biogenesis of the eukaryotic initiation factor 2 (eIF2). J. Biol. Chem. 288, 21537–21546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Özeş A. R., Feoktistova K., Avanzino B. C., Fraser C. S. (2011) Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B. J. Mol. Biol. 412, 674–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nielsen K. H., Chamieh H., Andersen C. B., Fredslund F., Hamborg K., Le Hir H., Andersen G. R. (2009) Mechanism of ATP turnover inhibition in the EJC. RNA 15, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. García-García C., Frieda K. L., Feoktistova K., Fraser C. S., Block S. M. (2015) Factor-dependent processivity in human eIF4A DEAD-box helicase. Science 348, 1486–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sonenberg N., Morgan M. A., Merrick W. C., Shatkin A. J. (1978) A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc. Natl. Acad. Sci. U.S.A. 75, 4843–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hellmann G. M., Chu L.-Y., Rhoads R. E. (1982) A polypeptide which reverses cap analog inhibition of cell-free protein synthesis: Purification and binding to capped oligonucleotides. J. Biol. Chem. 257, 4056–4062 [PubMed] [Google Scholar]
- 52. Ray B. K., Lawson T. G., Kramer J. C., Cladaras M. H., Grifo J. A., Abramson R. D., Merrick W. C., Thach R. E. (1985) ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J. Biol. Chem. 260, 7651–7658 [PubMed] [Google Scholar]
- 53. Yoder-Hill J., Pause A., Sonenberg N., Merrick W. C. (1993) The p46 subunit of eukaryotic initiation factor (eIF)-4F exchanges with eIF4A. J. Biol. Chem. 268, 5566–5573 [PubMed] [Google Scholar]
- 54. Pause A., Méthot N., Svitkin Y., Merrick W. C., Sonenberg N. (1994) Dominant negative mutants of mammalian translation initiation factor eIF4A define a critical role for eIF4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13, 1205–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Duncan R., Hershey J. W. (1983) Identification and quantitation of levels of protein synthesis initiation factors in crude HeLa cell lysates by two-dimensional polyacrylamide gel electrophoresis. J. Biol. Chem. 258, 7228–7235 [PubMed] [Google Scholar]
- 56. von der Haar T., McCarthy J. E. G. (2002) Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol. Microbiol. 46, 531–544 [DOI] [PubMed] [Google Scholar]
- 57. Park E. H., Walker S. E., Zhou F., Lee J. M., Rajagopal V., Lorsch J. R., Hinnebusch A. G. (2013) Yeast eukaryotic initiation factor 4B (eIF4B) enhances complex assembly between eIF4A and eIF4G. J. Biol. Chem. 288, 2340–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mitchell S. F., Walker S. E., Algire M. A., Park E.-H., Hinnebusch A. G., Lorsch J. R. (2010) The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol. Cell 39, 950–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Svitkin Y. V., Herdy B., Costa-Mattioli M., Gingras A. C., Raught B., Sonenberg N. (2005) Eukaryotic initiation factor 4E availability controls the switch between cap-dependent and internal ribosome entry site-mediated translation. Mol. Cell. Biol. 25, 10556–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Svitkin Y. V., Pause A., Haghighat A., Pyronnet S., Witherell G., Belsham G. J., Sonenberg N. (2001) The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7, 382–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chuang R. Y., Weaver P. L., Liu Z., Chang T. H. (1997) Requirement of the DEAD-box protein Ded1p for messenger RNA translation. Science 275, 1468–1471 [DOI] [PubMed] [Google Scholar]
- 62. Hilliker A., Gao Z., Jankowsky E., Parker R. (2011) The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol. Cell 43, 962–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Soto-Rifo R., Rubilar P. S., Limousin T., de Breyne S., Décimo D., Ohlmann T. (2012) DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 31, 3745–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pisareva V. P., Pisarev A. V., Komar A. A., Hellen C. U., Pestova T. V. (2008) Translation initiation on mammalian mRNAs with structured 5′ UTRs requires DExH-box protein DHX29. Cell 135, 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Parsyan A., Svitkin Y., Shahbazian D., Gkogkas C., Lasko P., Merrick W. C., Sonenberg N. (2011) mRNA helicases: the tacticians of translational control. Nat. Rev. Mol. Cell Biol. 12, 235–245 [DOI] [PubMed] [Google Scholar]
- 66. Hernández G., Vazquez-Pianzola P. (2005) Functional diversity of the eukaryotic translation factors belong to eIF4 families. Mech. Dev. 122, 865–876 [DOI] [PubMed] [Google Scholar]
- 67. Hui D. J., Terenzi F., Merrick W. C., Sen G. C. (2005) Mouse p56 blocks a distinct function of eukaryotic initiation factor 3 in translation initiation. J. Biol. Chem. 280, 3433–3440 [DOI] [PubMed] [Google Scholar]
- 68. Otero L. J., Ashe M. P., Sachs A. B. (1999) The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 18, 3153–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wells S. E., Hillner P. E., Vale R. D., Sachs A. B. (1998) Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2, 135–140 [DOI] [PubMed] [Google Scholar]
- 70. Rajagopal V., Park E.-H., Hinnebusch A. G., Lorsch J. R. (2012) Specific domains in yeast translation initiation factor eIF4G strongly bias RNA unwinding activity of the eIF4F complex toward duplexes with 5′-overhangs. J. Biol. Chem. 287, 20301–20312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Grifo J. A., Abramson R. D., Satler C. A., Merrick W. C. (1984) RNA-stimulated ATPase activity of eukaryotic initiation factors. J. Biol. Chem. 259, 8648–8654 [PubMed] [Google Scholar]
- 72. Lorsch J. R., Dever T. E. (2010) Molecular view of 43S complex formation and start site selection in eukaryotic translation initiation. J. Biol. Chem. 285, 21203–21207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jackson R. J., Hellen C. U., Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ranji A., Boris-Lawrie K. (2010) RNA helicases: Emerging roles in viral replication and the host innate response. RNA Biol. 7, 775–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jankowsky E. (2010) RNA Helicases, The Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 76. Komar A. A., Hatzoglou M. (2011) Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 10, 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Malys N., McCarthy J. E. (2011) Translation initiation: variations in the mechanism can be anticipated. Cell. Mol. Life Sci. 68, 991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hershey J. W., Sonenberg N., Mathews M. B. (2012) Principles of translational control: an overview. Cold Spring Harb. Perspect. Biol. 4, a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Komar A. A., Mazumder B., Merrick W. C. (2012) A new framework for understanding IRES-mediated translation. Gene 502, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hinnebusch A. G., Lorsch J. R. (2012) The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 4, a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fonseca B. D., Smith E. M., Yelle N., Alain T., Bushell M., Pause A. (2014) The ever-evolving role of mTOR in translation. Semin. Cell Dev. Biol. 36, 102–112 [DOI] [PubMed] [Google Scholar]
- 82. Hinnebusch A. G. (2014) The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 83, 779–812 [DOI] [PubMed] [Google Scholar]
- 83. Browning K. S., Bailey-Serres J. (2015) Mechanism of cytoplasmic mRNA translation. Arabidopsis Book 13, e0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pelletier J., Graff J., Ruggero D., Sonenberg N. (2015) Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 75, 250–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bhat M., Robichaud N., Hulea L., Sonenberg N., Pelletier J., Topisirovic I. (2015) Targeting the translational machinery in cancer. Nat. Rev. Drug Discov. 14, 261–278 [DOI] [PubMed] [Google Scholar]
- 86. Andersen C. B., Ballut L., Johansen J. S., Chamieh H., Nielsen K. H., Oliveira C. L., Pedersen J. S., Séraphin B., Le Hir H., Andersen G. R. (2006) Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313, 1968–1972 [DOI] [PubMed] [Google Scholar]
- 87. Safaee N., Kozlov G., Noronha A. M., Xie J., Wilds C. J., Gehring K. (2012) Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol. Cell 48, 375–386 [DOI] [PubMed] [Google Scholar]
- 88. Gross J. D., Moerke N. J., von der Haar T., Lugovskoy A. A., Sachs A. B., McCarthy J. E., Wagner G. (2003) Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115, 739–750 [DOI] [PubMed] [Google Scholar]
- 89. Fraser C. S. (2015) Quantitative studies of mRNA recruitment to the eukaryotic ribosome. Biochimie 114, 58–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Merrick W. C. (2010) Eukaryotic protein synthesis: still a mystery. J. Biol. Chem. 285, 21197–21201 [DOI] [PMC free article] [PubMed] [Google Scholar]