Background: Heterotrimeric type IV collagen has breaks in the triple-helix repeating Gly-X-Y sequence.
Results: NMR studies on a type IV peptide model show heterotrimeric chain selection and register, with new hydrogen bonds formed at an interruption site.
Conclusion: Interruption sites may provide a driving force for self-assembly and chain register.
Significance: Interruptions can have a positive effect on triple-helix continuity in non-fibrillar heterotrimer collagens.
Keywords: circular dichroism (CD), collagen, extracellular matrix, nuclear magnetic resonance (NMR), structure-function, interruption, heterotrimer
Abstract
All non-fibrillar collagens contain interruptions in the (Gly-X-Y)n repeating sequence, such as the more than 20 interruptions found in chains of basement membrane type IV collagen. Two selectively doubly labeled peptides are designed to model a site in type IV collagen with a GVG interruption in the α1(IV) and a corresponding GISLK sequence within the α2(IV) chain. CD and NMR studies on a 2:1 mixture of these two peptides support the formation of a single-component heterotrimer that maintains the one-residue staggering in the triple-helix, has a unique chain register, and contains hydrogen bonds at the interruption site. Formation of hydrogen bonds at interruption sites may provide a driving force for self-assembly and chain register in type IV and other non-fibrillar collagens. This study illustrates the potential role of interruptions in the structure, dynamics, and folding of natural collagen heterotrimers and forms a basis for understanding their biological role.
Introduction
Collagens present a large family of triple-helical, extracellular matrix molecules that provide structural integrity for the human body (1–3). Twenty-eight distinct types of collagen have been identified, including those that form fibrils with a 67-nm axial period and others classified as non-fibrillar collagens. Some of the collagens are homotrimers, such as fibrillar type II and type III collagens, whereas others are heterotrimers, such as the fibrillar type I and non-fibrillar basement membrane type IV collagens (4, 5). Collagen displays a unique (Gly-X-Y)n repetitive sequence pattern, which leads to the formation of the characteristic triple-helix structure (6). The strict requirement of Gly as every third residue is perfectly maintained throughout the triple helical domain of fibrillar collagens, such as type I, II, and III collagens; even the replacement of one Gly by a larger residue leads to disease (7). In contrast, interruptions in the repetitive (Gly-X-Y)n sequences have been observed within the triple helical domain of all non-fibrillar collagens (8). There are more than 350 interruptions found in non-fibrillar collagen chains, such as the ∼20 breaks in the Gly-X-Y repeating pattern seen in the triple-helix domain of type IV collagen chains in basement membranes.
These natural interruptions in the collagen triple-helix are suggested to play important roles in molecular structure, self-association, binding, and degradation (9–14). A number of flexible regions seen in type IV collagen by rotary shadowing electron microscopy correspond to interruption sites in the sequence and are considered to be important for the network-like structure seen in basement membrane collagen (12). One interruption in type IV collagen appears to play a role in binding to an integrin cell receptor in tumor cell adhesion, whereas two interruptions within type X collagen were shown to serve as specific cleavage sites of matrix metalloproteinases (10, 13, 14).
Basement membranes contain three different forms of type IV collagen, which are all heterotrimers: [α1(IV)]2α2(IV), α3(IV)α4(IV)α5(IV), and [α5(IV)]2α6(IV). Interruptions are often found at corresponding sites in the different chains within one heterotrimeric triple-helix, but the lengths of the interruptions usually differ within the two or three different chains at a given site. It is poorly understood how interruptions in three different chains are accommodated collectively in the self-assembly of heterotrimer type IV collagen.
Triple-helical peptides have been extensively utilized as valuable models to investigate the stability, folding, dynamics, and molecular structure of collagen (15–19). Although the large majority of studies have been done on homotrimer collagen model peptides, a heterotrimer platform is needed to model collagens with more than one type of chain, such as type IV collagen. In some studies, covalent linkages have been employed to force the selection of three different chains and their alignment for heterotrimer peptide models of collagen, although challenges in purity are significant (20–22). More recently, important progress has been achieved in the design of heterotrimer peptide models using electrostatic interaction strategies (23–27) to create defined compositions of heterotrimeric triple-helices, introduce mutations in one, two, or three chains, and create sticky ends to promote association. In addition, a computational approach has been used to optimize interactions to produce heterotrimers that are more stable than homotrimeric peptides (28, 29).
We have previously reported a study on a heterotrimer composed of two peptides designed to model a natural interruption site in the triple helical domain of the most abundant isoform, [α1(IV)]2α2(IV) of type IV collagen (30). One peptide models the GVG interruption at residue 292 of the α1(IV) chain, whereas the other peptide contains the corresponding GISLKG interruption at residue 288 of the α2(IV) chain at the same site (30). This earlier study indicated the formation of a heterotrimeric triple-helix, but structural details of the interruption and surrounding triple-helix could not be defined because only a single chain was labeled. Here, two 15N/13C selectively labeled peptides were synthesized with sequences corresponding to the human α1(IV) and α2(IV) chains at this GVG/GISLKG interruption site. Application of a number of NMR techniques demonstrated that each peptide forms a homotrimer, with an absence of hydrogen bonding throughout the interruption. When the two peptides were mixed in a 2:1 ratio, a single-component heterotrimer was observed with a defined register α1α1α2(IV) and with interchain hydrogen bonds at the interruption site.
Experimental Procedures
Notation
The number of residues between the repeating Gly-X-Y tripeptides can categorize the interruptions. Interruptions in the form of Gly-X-Y-Gly-(AA)n-Gly-X-Y are denoted as GnG type interruptions (8).
Sample Preparation
Peptides A and B were synthesized by Tufts University Core Facility (Boston, MA) (see Table 1). Peptide B was identical to that previously reported in Ref. 30, synthesized with 15N/13C-labeled amino acids at positions Ile13, Ser14, Leu15, and Ala25. The sequence of peptide A studied here had four minor changes outside of the interruption site when compared with that studied in Ref. 30. These minor changes in flanking residues were designed to try to improve overall triple-helix stability, but as reported below, the A homotrimer was very similar to that found previously. Importantly, the A peptide now contains selectively 15N/13C doubly labeled residues at four positions, Gly14, Val15, Gly16, and Gly25. Samples of homotrimer peptides A and B were prepared in 10% D2O, 90% H2O with a concentration of 6 mm. Peptides A and B were mixed in solution at a molar ratio of 2A:1B (4 mm:2 mm) to look for formation of heterotrimeric molecules. The mixtures were heated to 50 °C, cooled slowly to 0 °C, and incubated for 30 h to prepare heterotrimer samples for measurements.
TABLE 1.
Triple-helix content (MRE225) and thermal stability for peptides A, B, and a mixture with a molar ratio of 2A:1B
Underlined residues are sequences from the human α1(IV) and α2(IV) chains. 15N/13C doubly labeled residues in A and B are colored in red. Note that an attempt was made to create more blunt ends by having N-terminal OG for peptide A and N-terminal POG for peptide B.

Circular Dichroism Spectroscopy
CD spectra were recorded on an Aviv model 62DS spectrophotometer with a Peltier thermoelectric temperature controller (Aviv Biomedical Inc., Lakewood, NJ). Samples were prepared at a concentration of 0.28 mm in 20 mm PBS (10 mm NaH2PO4, 10 mm Na2HPO4, 150 mm NaCl) at pH 7. Peptide concentrations were determined by tyrosine absorbance at 275 nm using ϵ275 = 1400 m−1 cm−1. Wavelength scans of the peptides were carried out by collecting the signal from 260 to 215 nm at 0.5-nm intervals at 0 °C.
CD was applied to determine the thermal stability by monitoring the amplitude of the peak at 225 nm as a function of increasing temperature with an average heating rate of 0.1 °C/min (31). The peptides or mixtures were equilibrated for at least 40 h at 0 °C prior to the melting experiments. The melting temperature (Tm) is defined as the temperature at which the fraction folded is equal to 0.5 in the curve fitted to the trimer to monomer transition.
NMR Spectroscopy
NMR experiments were performed on a Varian INOVA 600-MHz spectrometer with a cold probe or a Varian VNMRS 800-MHz spectrometer. To accomplish sequential assignments, 1H-15N heteronuclear single quantum coherence (HSQC)3 (32) and three-dimensional HNCA experiments (33) were carried out at 5 °C. Three-dimensional HNCA experiments consisted of 100(t1) × 30(t2) × 1024(t3) complex points and were recorded with spectral widths of 8000(F1), 2800(F2), and 8000(F3) Hz. Three-dimensional 15N-edited NOESY-HSQC experiments (34–36) were performed with a mixing time of 50 or 60 ms at 5 °C and 10 °C. Relaxation R1, R2, and heteronuclear NOE measurements (37–39) were done at 10 °C as described previously. To obtain residue-specific melting curves and calculate amide proton temperature gradients, 1H-15N HSQC experiments were performed at 0–29 °C for peptides A and AAB, and at 3–19 °C for peptide B. All data were processed using the FELIX 2004 software package (MSI, San Diego, CA) and analyzed with FELIX 2004 or NMRView.
Results
Peptide Design
Peptides were designed to model one natural interruption in the [α1(IV)]2α2(IV) isoform of type IV collagen (Fig. 1). Peptide A includes residues 289–299 from the sequence of the human α1(IV) chain with a G1G interruption (underlined) (Table 1). Residues GVG at the interruption site were selectively 15N/13C-labeled together with a labeled Gly25 within a Gly-Pro-Hyp triplet at the C terminus. Peptide B includes residues 288–295 from the human α2(IV) chain at the corresponding GISLKG interruption (underlined) with selectively 15N/13C-labeled residues ISL at the interruption site and a labeled Ala25 near the C terminus (Table 1). All collagen sequences are flanked by stabilizing triplets and a terminal Tyr for concentration determination.
FIGURE 1.
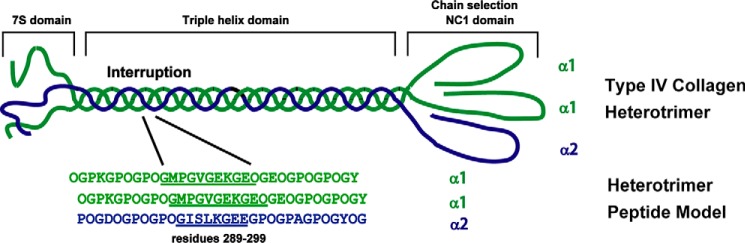
Heterotrimer peptide models of interruptions in type IV collagen. The major isoform of type IV collagen [α1(IV)]2α2(IV) consists of two chains with identical amino acid sequences (green) and one chain with a different sequence (blue). All type IV collagens contain three domains: an N-terminal 7S domain, a central triple helical domain, and a C-terminal globular non-collagenous (NC1) domain. Heterotrimer peptides are designed to model a GVG interruption in the α1(IV) chains corresponding with a GISLKG interruption in the α2(IV) chain at sites 289–302 of type IV collagen, showing sequences from the human α1(IV) and α2(IV) chains (underlined).
CD Characterization of Peptides A and B and the Mixture of 2A:1B
CD studies were performed on peptides A and B separately, as homotrimers. Dissolving peptide A in solution leads to formation of a stable triple-helix with a characteristic positive CD peak at 225 nm (MRE225 = 2611 deg cm2/dmol−1) and a melting temperature of 14.9 °C (Table 1, Fig. 2). Dissolving peptide B in solution only leads to partial formation of homotrimers, with a lower MRE225 value (1150 deg cm2/dmol−1) and a partial thermal transition (Fig. 2). Mixing peptides A and B at a molar ratio of 2A:1B to model the type IV [α1(IV)]2α2(IV) composition leads to the formation of triple-helix structure with a characteristic CD peak at 225 nm (MRE225 = 2218 deg cm2/dmol−1) and a single-mode transition with a melting temperature of 14.4 °C (Fig. 2). The mixture shows a stability comparable with homotrimer A, but is much more stable than homotrimer B (Table 1).
FIGURE 2.
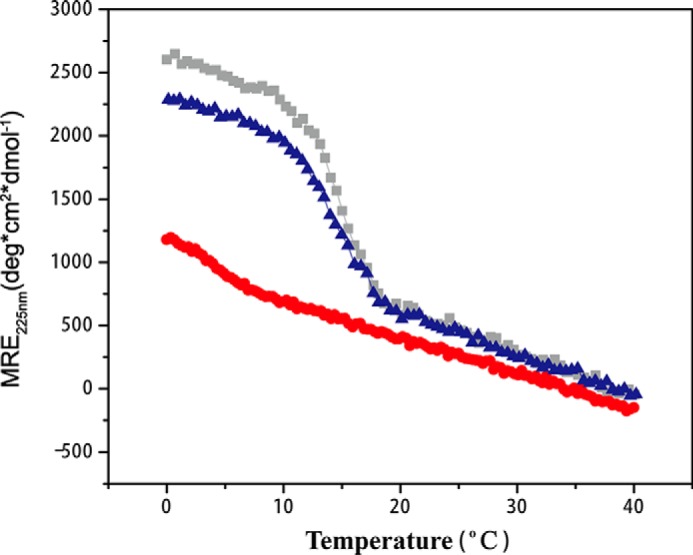
CD thermal transitions of peptide A (gray) and B (red) and a mixture with molar ratio of 2A:1B (blue). CD measurements were recorded at a concentration of 0.28 mm, whereas the NMR experiments were carried out at a higher concentration of 6 mm.
NMR Shows That the 2A:1B Peptide Mixture Forms a Single-component Heterotrimer
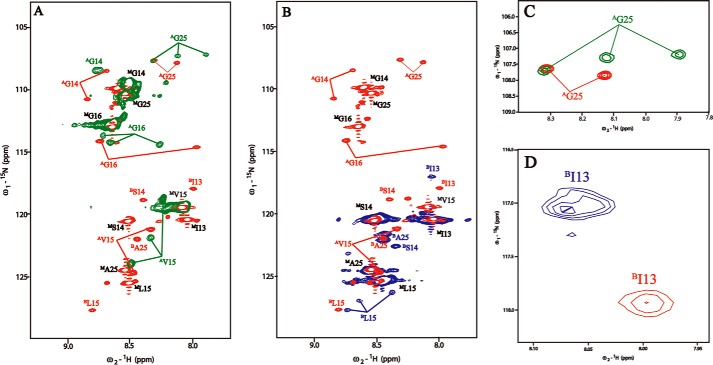
NMR studies were performed on the 2A:1B peptide mixture, as well as A homotrimers and B homotrimers, to evaluate whether heterotrimers had formed (Fig. 3). The 1H-15N HSQC spectra of peptides A and B separately and a 2A:1B peptide mixture were obtained, and the chain-specific sequential assignments were based on three-dimensional HNCA experiments. The peaks corresponding to monomer states are denoted with superscript M, whereas the trimer resonances are denoted with superscript A and B, indicating the chain composition of the trimer species (Fig. 3). Minor resonances in the HSQC spectrum arise due to cis-trans isomerization of Gly-Pro and Pro-Hyp bonds in the peptide in the unfolded states (40). The HSQC spectrum of homotrimer peptide A (green) shows three trimer peaks for labeled residues Val15, Gly16, and Gly25, indicating distinct chemical environments in three different chains (Fig. 3A). The one-residue stagger of the three chains within the triple-helix leads to a different environment for residues in each chain. Only two trimer peaks are seen for Gly14; the expected third peak may be overlapped with other resonances (Fig. 3A). The HSQC spectrum of the homotrimer peptide B (blue) shows three trimer peaks for labeled residue Leu15 (BLeu15, superscript B for trimer resonances), indicating different environments for the Leu residues located in the interruption region in each of the three chains of the homotrimer (Fig. 3B). The presence of only one or two trimer resonances for Ile (BIle13), Ser (BSer14), and Ala (BAla25) is likely due to overlapping trimer resonances (Fig. 3B).
FIGURE 3.
A, overlapped 1H-15N HSQC spectra of AAB (red) and A (green). B, overlapped 1H-15N HSQC spectra of AAB (red) and B (blue). C, zoomed-in spectra of AGly25 (colored red in AAB and green in A). D, zoomed-in spectra of BIle13 (colored red in AAB and blue in B). Superscript A and B indicate the trimer resonances for peptide chains A and B, respectively, whereas superscript M represents the monomer resonances.
The HSQC spectrum of the 2A:1B peptide mixture is well resolved (Fig. 4a). In the spectrum for the mixture, all labeled residues in peptide A have two trimer resonances (superscript A), whereas the labeled residues in peptide B all show a single trimer resonance (superscript B), which is consistent with the composition of the heterotrimer as two A chains and one B chain (Fig. 4a). The difference in the chemical shifts of trimer peaks in the homotrimers when compared with the 2A:1B mixture indicates that the residues in polypeptide chains A and B have different chemical environments in homotrimers versus heterotrimers (Fig. 3, A and B). These differences are clearly illustrated by the trimer resonances of Gly25 (AGly25) and Ile13 (BIle13) in Fig. 3, C and D. Three trimer resonances of Gly25 (AGly-25) in homotrimer peptide A become two peaks in the 2A:1B mixture (Fig. 3C), whereas the trimer resonance of Ile13 (BIle13) in homotrimer peptide B is significantly shifted in the 2A:1B mixture (Fig. 3D). One monomer resonance is also observed for each residue (indicated by superscript M), supporting an equilibrium of trimers and monomers in the 2A:1B peptide mixture, which is also seen for homotrimers (Fig. 3, A and B). No trimer resonance corresponding to the chemical shifts seen for homotrimers is found for the labeled residues in the peptide mixture, indicating that no homotrimer is formed. These results indicate that the 2A:1B peptide mixture forms a single-component heterotrimer without any observable homotrimers.
FIGURE 4.
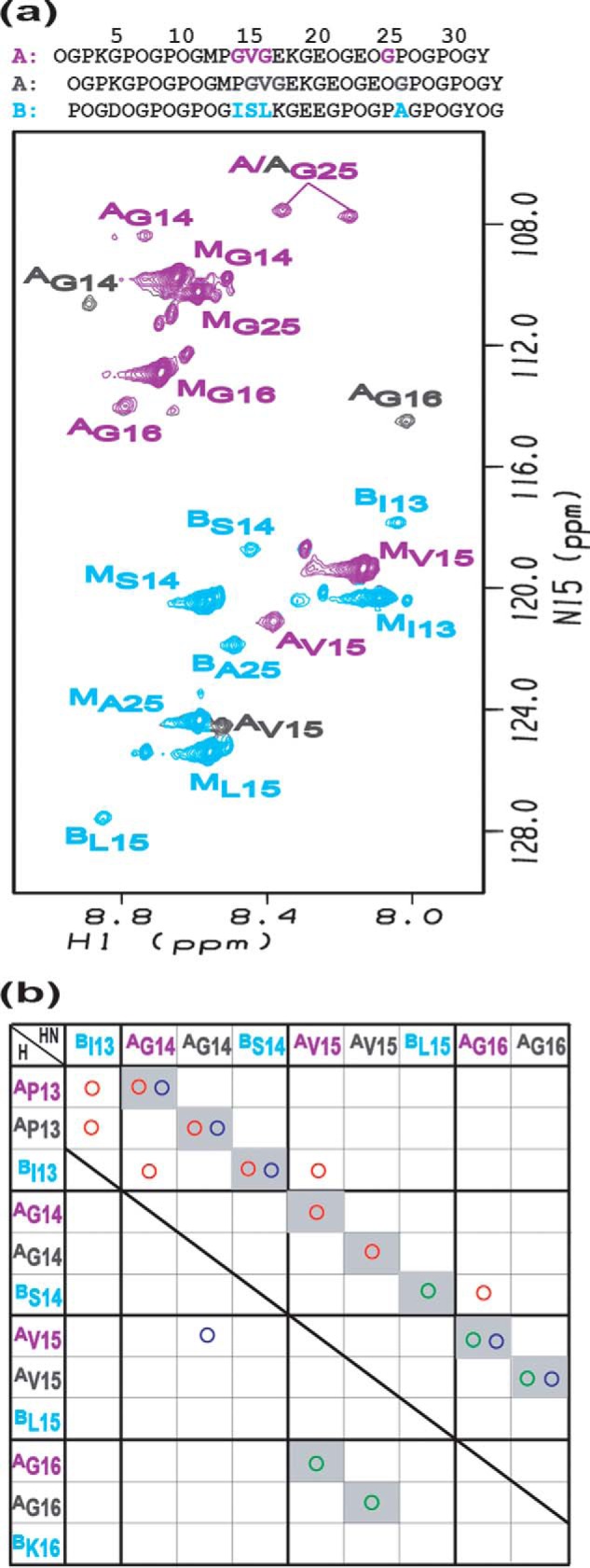
a, 1H-15N HSQC spectrum of peptide mixture 2A:1B. Violet and gray colors are used to distinguish two different A chains in the heterotrimer, and chain B is colored in azure. Labeled residues are colored violet, gray, and azure in the first A chain, the second A chain, and chain B, respectively. The peaks corresponding to the trimer states are denoted with a superscript A or B, respectively, whereas those for the monomer state are denoted with a superscript M. b, contact map generated from NH-H experimental NOEs of peptide mixture 2A:1B. Experimental NOEs are represented by circles: HN-HN (green circle); HN-HA (red circle); and HN-side chain protons (blue circle). The shaded squares in the background denote intrachain NOEs.
Chain Register of Heterotrimer 2A:1B
The heterotrimer formed by the peptide mixture 2A:1B has three possible registers, AAB, ABA, and BAA, with B as the lagging, middle, or leading chain, respectively. Chain assignments for 2A:1B were based on interchain NOEs, which are summarized in the experimental contact map (Fig. 4b) (41). Violet and gray colors are used to distinguish two different A chains in the heterotrimer, and chain B is colored in azure. The colored circles were utilized to represent experimentally observed NOEs (HN-HN (green circles); HN-HA (red circles); and HN-side chain protons (blue circles)). The shaded squares in the background denote intrachain NOEs. Only a few interchain NOEs could be observed including AGly14 NH-BIle13 αH, AVal15 NH-BIle13 αH, AGly16 NH-BSer14 αH, AGly14 NH-AVal15 γH, and BIle13 NH-A/APro13 αH. The interchain NOEs between AGly14 NH-BIle13 αH and AGly14 NH-AVal15 γH are only consistent with having the A chain (violet) as the leading chain, whereas the B chain is the lagging chain (Fig. 4a). All other interchain NOEs supported these chain assignments. Thus, the peptide 2A:1B mixture forms a single-component heterotrimer with chain register AAB.
Hydrogen Bonding of Heterotrimer Versus Homotrimer
Amide proton temperature gradients were measured for homotrimers of A3 and B3 and the heterotrimer AAB (Fig. 5). Amide protons showing the temperature gradients with a more positive value than a −4.6 threshold are considered to be involved in hydrogen bonding (42, 43), and this value is used as the cut-off in our analysis. For homotrimer A, Gly25 in all three chains of triple-helix has amide proton temperature gradients ∼−2, indicating the formation of hydrogen bonds in this stable C-terminal Gly-Pro-Hyp region. Residues Gly14-Val15-Gly16 at the interruption site all show temperature gradients more negative than −6, indicating that the hydrogen bonds of Gly, which are preserved for continuous Gly-X-Y region, have been broken (Fig. 5a). For homotrimer B, all labeled residues including Ile13, Ser14, and Leu15 at the interruption sites and Ala25 at the Tyr position in the Gly-X-Y repeating triplets show temperature gradients more negative than −6, indicating the absence of hydrogen bonds (Fig. 5b).
FIGURE 5.
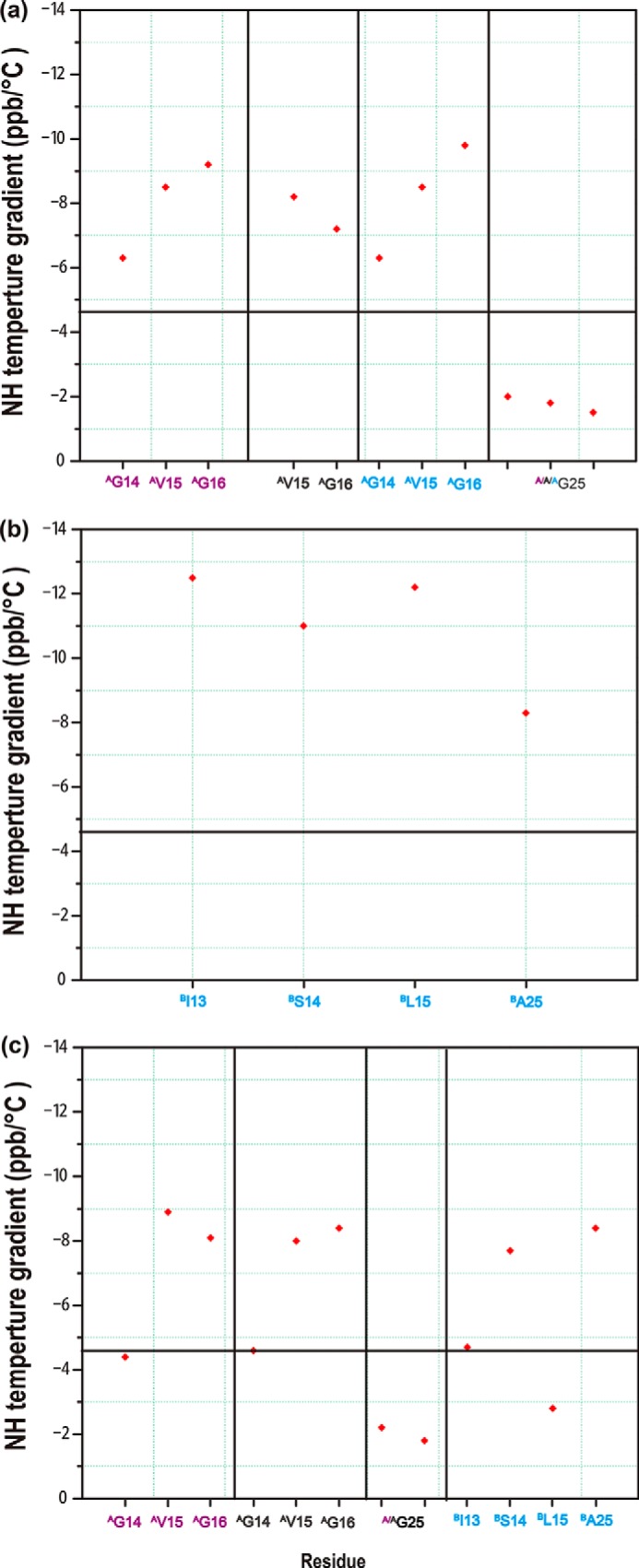
Amide proton temperature gradients of homotrimer A (a), homotrimer B (b), and heterotrimer AAB (c). The black horizontal line corresponds to a value of −4.6, the cut-off for hydrogen bonding, with less negative values indicative of hydrogen bonding.
In the heterotrimer AAB, the amide proton temperature gradient values indicate the formation of new hydrogen bonds at the interruption site (Fig. 5c). Leu15 in the B chain now has a temperature gradient with a value of −2.8, indicating involvement of its NH in hydrogen bonding. In addition, Gly14 residues in the two A chains have temperature gradients with values of −4.4 and −4.6, near the cut-off value, indicating the potential hydrogen bonds. When compared with the A chain Gly14 residues in heterotrimer AAB, the A chain Gly16 residues have temperature gradients more negative than −6, indicating the absence of hydrogen bonding. Gly25 in the Gly-X-Y C terminus of the heterotrimer AAB forms hydrogen bonds, as indicated by its temperature gradients with values more positive than −3.
Dynamics of Heterotrimer AAB on the Fast, Picosecond Timescale
The flexibility of residues within the heterotrimer AAB was estimated by measuring the 1H-15N NOE values (Fig. 6). Heteronuclear 1H-15N NOE measurements provide useful information on the fast, picosecond timescale motion, such as bond vibration and side-chain rotation. Smaller NOE values indicate higher flexibility on this time scale (44). Among the labeled residues of the A chains, Gly14-Val15-Gly16, Gly14 is the most rigid, whereas Gly16 is the most flexible, in both chains. Gly25 residues in the C-terminal region of both chains are very rigid. For the B chain, the labeled residues Ile13-Ser14-Leu15 at the interruptions sites as well as Ala25 in the non-disrupted C-terminal region all have NOE values larger than 0.6, indicating that all of the labeled residues in the B chain are rigid. Overall, most residues at the interruption sites are rigid except for the two Gly16 at the C terminus of the GVG interruption sites in the A chains.
FIGURE 6.
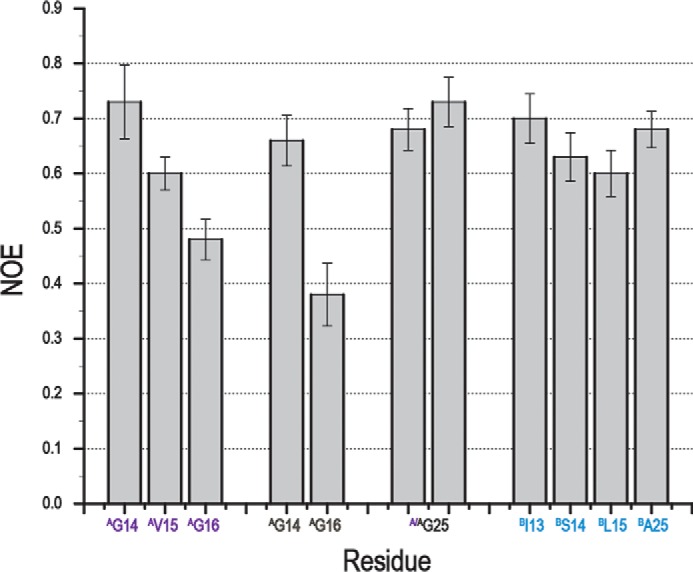
Heteronuclear 1H-15N NOE values of heterotrimer peptide AAB. Superscript A and B indicate the trimer resonances for peptide chains A and B, respectively. Violet and gray colors represent the leading and middle chain A, respectively, whereas the azure color represents the lagging chain B. Gly25 residues in two A chains cannot be distinguished from each other. Decreasing NOE values indicate increasing mobility on the fast, picosecond time scale. Gly16 residues in two A chains have the smallest NOE values, suggesting that they are the most flexible among all labeled residues. Error bars indicate means ± S.E.
Discussion
Although collagen is distinguished by its repeating (Gly-X-Y)n amino acid sequence, the triple-helix domains of non-fibrillar collagens all include a number of sites where this repeating tripeptide sequence is interrupted. For the heterotrimeric type IV collagen, ∼20 of these sequence discontinuities are distributed throughout the triple-helix, and they may play significant roles in the organization and mechanical integrity of basement membranes. Knowledge about the effect of heterotrimeric interruptions on the triple-helix is important for understanding their biological role. NMR studies are reported here on a heterotrimeric peptide that models an interruption in both chains at residues ∼289–299 of the most widespread type IV collagen isoform [α1(IV)]2α2(IV), and the results provide direct experimental evidence on chain composition, hydrogen bonding, register, and interactions between disparate length interruptions.
The 1450-residue-long triple-helix of type IV collagen has a C-terminal globular domain (NC1) responsible for chain selection, which likely also promotes triple-helix nucleation and the register in which the three chains are staggered by one residue (e.g. AAB, ABA, or BAA). In type IX collagen, the non-collagenous NC2 domain has been shown to be crucial to control of the chain register (45). Some non-fibrillar collagens, such as type X collagen, are homotrimers, with the same interruptions in all three chains, so that chain register can be preserved on both sides. However, in heterotrimeric type IV collagens, there are almost always disparate lengths of interruptions in different chains at a given site (Fig. 7a). These disparate lengths can be either commensurate, maintaining the three chains in the same register at either side of the interruption, or non-commensurate, where the chain register is not maintained (46). The interruption in our peptide model, with a G1G interruption (GVG) in two chains and a G4G interruption (GISLKG) in the third chain, would be categorized as commensurate, because Gly residues could be maintained in the same register on both sides (46).
FIGURE 7.
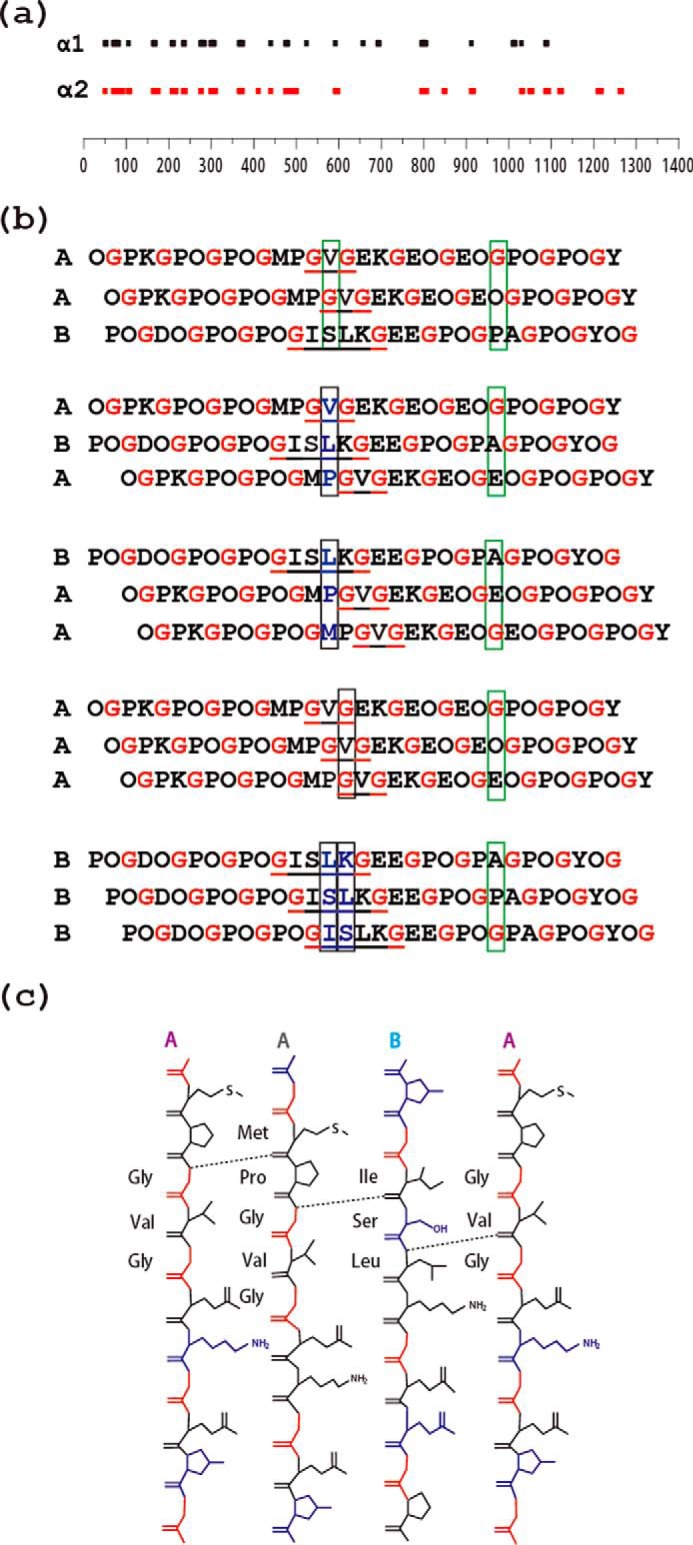
a, schematic diagram of the location of interruptions along the α1(IV) (top) and α2(IV) (bottom) chains of human type IV collagen. The lengths of the bars are proportional to the number of amino acids in the interruption sequence. The alignment illustrates that many, but not all, interruptions are at the same sites in the two chains within a [α1(IV)]2α2 triple-helix molecule, whereas the lengths of the interruptions often differ between the two chains at a given site. b, schematic illustration of the effect of the staggering between the three chains within a triple-helix on interchain hydrogen bonding at the interruption site. The three possible registers for the A and B peptides are shown on the top (AAB, ABA, BAA), whereas homotrimers AAA and BBB are shown at the bottom. The presence of a one-residue GVG interruption in two chains and a four-residue GISLKG interruption in the other two chains allows maintenance of the stagger on both sides of the interruption (commensurate). Gly residues are shown in red to visualize the stagger, and interruptions are underlined. Black boxes are used to designate an axial level where there is no Gly and the typical stabilizing interchain Gly NH … CO bonds cannot be formed. Green boxes are used to exemplify a regular axial level that contains one and only one Gly with the expected formation of a hydrogen bond. Only AAB allows hydrogen bonding at all axial positions, as proposed by Bella (46). c, hydrogen bonding at the interruption site GVG/GVG/ISLK in heterotrimer AAB deduced from amide temperature-dependent NMR studies, with hydrogen bonds formed between the amide group of Gly14/Leu16 and the carbonyl group of nearby amino acids indicated as dashed lines.
The CD spectrum and thermal transition indicate that the 2A:1B peptide mixture is forming a largely triple-helical structure, consistent with triple-helix forming on both sides of the interruption. NMR studies indicated, surprisingly, that the 2A:1B peptide mixture formed only one type of triple-helical molecule, with composition A2B. No A3 homotrimers were observed, although they have a slightly higher stability than the 2A:1B mixture, nor were B3 homotrimers, a much less stable species, or AB2 molecules observed. Within the triple-helix structure, the three chains are staggered by one residue with respect to each other, and the NOEs showed the register for this heterotrimeric peptide to be AAB, or α1α1α2(IV) (leading, middle, trailing). Interestingly, the register for the type IV [α1(IV)]2α2(IV) molecule was experimentally determined for one other site in type IV collagen (47). Golbik et al. (47) used FRET on a tryptic fragment to define the stagger of type IV (Gly-X-Y)n residues 460–480 (which includes the α1β1 integrin binding site) and found it to be α1α1α2(IV), the same observed in our heterotrimeric peptide. It was unexpected to find the same register for these two different sites separated by ∼200 residues and by four additional interruptions, including three that are non-commensurate. The observation of the same register at two distinct and separate locations raises the possibility that the α1α1α2(IV) register is maintained throughout the entire type IV triple-helix, despite the many non-commensurate interruption sites. The ability of the small A and B peptides studied here to self-assemble into a heterotrimer triple-helix with a unique chain composition and register suggests that this particular G1G/G1G/G4G (α1/α1/α2) interruption plays a role in preserving the type IV collagen register and may facilitate triple-helix propagation.
The current study suggests that hydrogen bonding may play a key role in defining chain composition and the precise chain register within this heterotrimer peptide. Interchain hydrogen bonding involving Gly NH of one chain and O=C in the X residue on another chain is known to be a main driving force for triple-helix stabilization (48). The homotrimer A3, which forms a stable triple-helix structure (Tm ∼15 °C), has amide proton gradient values reflecting hydrogen bonding for the Gly NH in the C-terminal Gly-Pro-Hyp region, but no hydrogen bonding for any residues in the Gly-Val-Gly interruption. This is surprising because there is at least one Gly residue at every axial level for the A3 homotrimer, and direct H-bonds might be expected (Fig. 7b) (46). For the weakly stable homotrimer B3 peptide, no residues in the ISLK interruption site form hydrogen bonds. This may be due to the lack of Gly residues at two axial levels for the B3 homotrimer (see boxed axial triplets for B3 in Fig. 7b). In contrast, within the heterotrimer context of AAB, Gly14(A) in the leading and middle chains form hydrogen bonds, and Leu15(B) in the lagging chain does as well. A recent analysis by Bella (46) proposed that the most favorable register for a heterotrimeric interruption consisting of two G1G and one G4G would be AAB (α1α1α2) because the presence of a Gly at every axial level would allow maximum hydrogen bonding. This predicted favorable register is observed for the AAB peptide (Fig. 7b), but the expected H-bonds are not observed for Gly16.
An unexpected hydrogen bond was observed between the amide group of Leu15 in chain B and the carbonyl in an adjacent chain A (Fig. 7c). The Leu within the α2(IV) interruption sequence GISLKG occupies the position normally occupied by a Gly residue, and this residue is predominantly hydrophobic within all such G4G interruptions (11, 41). Previous NMR studies showed that a typical G4G interruption GAAVMG within a homotrimer context allowed the preservation of the one-residue stagger and close packing of the three chains, with a very local alteration of typical triple-helix dihedral angles leading to a small hydrophobic cluster in the center of the triple-helix (41). In the heterotrimer, the Leu from the α2(IV) chain may have such hydrophobic interactions with the Val chains from α1(IV) chains, with the buried Leu forming a backbone hydrogen bond to another chain as well.
Although the appearance of hydrogen bonds in the heterotrimer that are absent in homotrimers is striking, there may well be contributions from Gly-X-Y sequences surrounding the interruption determining heterotrimer chain composition and stagger. It has been reported that atypical sequences are found adjacent to interruptions in type IV collagen (47), which could play a role. In addition, electrostatic interactions have the capacity to control chain selection (48), and the presence of oppositely charged residues at two sites in A versus B chains may make some contribution to stability.
The dynamics at interruption sites is of interest because interruptions are thought to be associated with looseness or flexibility based in part on rotary shadowing electron microscopy and on the known cleavage of two G4G sites in type X collagen by MMP-9 (12, 14). All labeled residues within the G1G/G1G/G4G interruption studied here appear to be rigid on the fast timescale, reflecting bond vibration and side-chain rotation, except for Gly16 in the two A chains (Fig. 6). The Gly16 residues do not show hydrogen bonding and are the most flexible residues when compared with the more rigid Gly14 on the N-terminal side of the Gly14-Val15-Gly16 sequence, which does form hydrogen bonds. This is consistent with the high resolution x-ray structure of a homotrimer peptide with a GPG interruption, where the Gly N-terminal to the Pro shows standard hydrogen bonding, whereas the Gly C-terminal to the Pro does not (49). It is possible that the mobility seen for Gly16 could have some functional significance in binding or cleavage.
Although type IV collagen contains more than 20 interruptions in each chain, it has a similar melting temperature as type I collagen without any breaks. Peptides, such as those studied in this report, magnify the destabilizing effect of such interruptions and allow biophysical studies to characterize distinctive molecular features. Our studies suggest that complementary interruptions in different chains may provide specificity and selectivity within the triple-helix as well as serving as external recognition sites for MMPs or receptors. MMP-9 cleaves a Gly-Leu or Gly-Ile in a G4G interruption in type X collagen (14). Type X is a homotrimer with G4G interruptions in all three chains, and it cannot have H-bonds at three axial levels. The NMR studies reported here suggest that the structure in the homotrimer is looser and therefore may be more exposed than the G1G/G4G case in heterotrimer.
Jalan and Hartgerink (1) raised the possibility that the triple-helix itself has the capacity for chain selection and chain registration, and have employed an electrostatic strategy to promote formation of heterotrimers in (Gly-X-Y)n peptides. Here, we have shown that heterotrimers can be produced by peptide models with natural interruptions in collagen and that hydrogen bonding, as well as electrostatic interactions, may provide a driving force for heterotrimer formation. We suggest that the data presented here reframe our understanding of at least some interruptions in non-fibrillar collagens. Previously, interruptions have been viewed as “obstacles” to folding, which may either passively allow the chain register to continue (commensurate) or alter the register (non-commensurate). We propose that the interactions between interruptions in different chains, together with atypical surrounding sequences, may play a role in facilitating folding around these interruptions. The heterotrimeric interruption studied here does not represent an “obstacle” to folding and a “disruption” of register, but rather a stable, relatively rigid site whose interactions favor this register and chain interaction. We hypothesize that heterotrimeric interruptions in type IV collagen contain important information that supports the establishment and maintenance of chain register. Definition of the basement membrane collagen structure is important for understanding the normal functioning of basement membranes and their role in kidney diseases and tumor metastases, as well as forming substrates for stem cell growth in tissue engineering.
Author Contributions
J. B. and B. B. conceived and coordinated the study and wrote the paper. J. X. designed, performed, and analyzed the experiments. X. S. contributed to the analysis of data and the preparation of the figures. M. B. performed and analyzed the experiments shown in Table 1. All authors reviewed the results and approved the final version of the manuscript.
This work was supported by National Institutes of Health Grants GM45302 (to J. B.) and GM60048 (to B. B.); National Science Foundation Grants DBI-0403062 and DBI-0320746 (to J. B.); and National Natural Science Foundation of China Grant 21305056 and Fundamental Research Funds for the Central Universities Grant lzujbky-2014-73) (to J. X.). The authors declare that they have no conflicts of interest with the contents of this article.
- HSQC
- heteronuclear single quantum coherence
- deg
- degrees
- MMP
- matrix metalloproteinase
- MRE
- mean residue ellipticity.
References
- 1. Jalan A. A., Hartgerink J. D. (2013) Pairwise interactions in collagen and the design of heterotrimeric helices. Curr. Opin. Chem. Biol. 17, 960–967 [DOI] [PubMed] [Google Scholar]
- 2. Shoulders M. D., Raines R. T. (2009) Collagen structure and stability. Annu. Rev. Biochem. 78, 929–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kielty C. M., Grant M. E. (2002) The collagen family: structure, assembly, and organization in the extracellular matrix. in Connective Tissue and Its Heritable Disorders, Molecular, Genetic and Medical Aspects (Royes P. M., Steinmann B. U., eds), pp. 159–222, Wiley Liss, New York [Google Scholar]
- 4. Fields G. B. (2013) Interstitial collagen catabolism. J. Biol. Chem. 288, 8785–8793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Myllyharju J., Kivirikko K. I. (2001) Collagens and collagen-related diseases. Ann. Med. 33, 7–21 [DOI] [PubMed] [Google Scholar]
- 6. Brodsky B., Persikov A. V. (2005) Molecular structure of the collagen triple helix. Adv. Protein Chem. 70, 301–339 [DOI] [PubMed] [Google Scholar]
- 7. Marini J. C., Forlino A., Cabral W. A., Barnes A. M., San Antonio J. D., Milgrom S., Hyland J. C., Körkkö J., Prockop D. J., De Paepe A., Coucke P., Symoens S., Glorieux F. H., Roughley P. J., Lund A. M., Kuurila-Svahn K., Hartikka H., Cohn D. H., Krakow D., Mottes M., Schwarze U., Chen D., Yang K., Kuslich C., Troendle J., Dalgleish R., Byers P. H. (2007) Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum. Mutat. 28, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thiagarajan G., Li Y., Mohs A., Strafaci C., Popiel M., Baum J., Brodsky B. (2008) Common interruptions in the repeating tripeptide sequence of non-fibrillar collagens: sequence analysis and structural studies on triple-helix peptide models. J. Mol. Biol. 376, 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brazel D., Oberbäumer I., Dieringer H., Babel W., Glanville R. W., Deutzmann R., Kühn K. (1987) Completion of the amino acid sequence of the α1 chain of human basement membrane collagen (type IV) reveals 21 non-triplet interruptions located within the collagenous domain. Eur. J. Biochem. 168, 529–536 [DOI] [PubMed] [Google Scholar]
- 10. Miles A. J., Knutson J. R., Skubitz A. P., Furcht L. T., McCarthy J. B., Fields G. B. (1995) A peptide model of basement membrane collagen α 1 (IV) 531–543 binds the α3β1 integrin. J. Biol. Chem. 270, 29047–29050 [DOI] [PubMed] [Google Scholar]
- 11. Mohs A., Popiel M., Li Y., Baum J., Brodsky B. (2006) Conformational features of a natural break in the type IV collagen Gly-X-Y repeat. J. Biol. Chem. 281, 17197–17202 [DOI] [PubMed] [Google Scholar]
- 12. Hofmann H., Voss T., Kühn K., Engel J. (1984) Localization of flexible sites in thread-like molecules from electron micrographs. Comparison of interstitial, basement membrane and intima collagens. J. Mol. Biol. 172, 325–343 [DOI] [PubMed] [Google Scholar]
- 13. Miles A. J., Skubitz A. P., Furcht L. T., Fields G. B. (1994) Promotion of cell adhesion by single-stranded and triple-helical peptide models of basement membrane collagen α 1(IV)531–543: evidence for conformationally dependent and conformationally independent type IV collagen cell adhesion sites. J. Biol. Chem. 269, 30939–30945 [PubMed] [Google Scholar]
- 14. Furcht L. T., Skubitz A. P., Fields G. B. (1994) Tumor cell invasion, matrix metalloproteinases, and the dogma. Lab. Invest. 70, 781–783 [PubMed] [Google Scholar]
- 15. Brodsky B., Thiagarajan G., Madhan B., Kar K. (2008) Triple-helical peptides: an approach to collagen conformation, stability, and self-association. Biopolymers 89, 345–353 [DOI] [PubMed] [Google Scholar]
- 16. Bella J., Eaton M., Brodsky B., Berman H. M. (1994) Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution. Science 266, 75–81 [DOI] [PubMed] [Google Scholar]
- 17. Baum J., Brodsky B. (1999) Folding of peptide models of collagen and misfolding in disease. Curr. Opin Struct. Biol. 9, 122–128 [DOI] [PubMed] [Google Scholar]
- 18. Xiao J., Addabbo R. M., Lauer J. L., Fields G. B., Baum J. (2010) Local conformation and dynamics of isoleucine in the collagenase cleavage site provide a recognition signal for matrix metalloproteinases. J. Biol. Chem. 285, 34181–34190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiao J., Cheng H., Silva T., Baum J., Brodsky B. (2011) Osteogenesis imperfecta missense mutations in collagen: structural consequences of a glycine to alanine replacement at a highly charged site. Biochemistry 50, 10771–10780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fields G. B., Prockop D. J. (1996) Perspectives on the synthesis and application of triple-helical, collagen-model peptides. Biopolymers 40, 345–357 [DOI] [PubMed] [Google Scholar]
- 21. Fiori S., Saccà B., Moroder L. (2002) Structural properties of a collagenous heterotrimer that mimics the collagenase cleavage site of collagen type I. J. Mol. Biol. 319, 1235–1242 [DOI] [PubMed] [Google Scholar]
- 22. Raynal N., Hamaia S. W., Siljander P. R., Maddox B., Peachey A. R., Fernandez R., Foley L. J., Slatter D. A., Jarvis G. E., Farndale R. W. (2006) Use of synthetic peptides to locate novel integrin α2β1-binding motifs in human collagen III. J. Biol. Chem. 281, 3821–3831 [DOI] [PubMed] [Google Scholar]
- 23. Jalan A. A., Jochim K. A., Hartgerink J. D. (2014) Rational design of a non-canonical “sticky-ended” collagen triple helix. J. Am. Chem. Soc. 136, 7535–7538 [DOI] [PubMed] [Google Scholar]
- 24. Fallas J. A., Lee M. A., Jalan A. A., Hartgerink J. D. (2012) Rational design of single-composition ABC collagen heterotrimers. J. Am. Chem. Soc. 134, 1430–1433 [DOI] [PubMed] [Google Scholar]
- 25. O'Leary L. E., Fallas J. A., Hartgerink J. D. (2011) Positive and negative design leads to compositional control in AAB collagen heterotrimers. J. Am. Chem. Soc. 133, 5432–5443 [DOI] [PubMed] [Google Scholar]
- 26. Gauba V., Hartgerink J. D. (2007) Self-assembled heterotrimeric collagen triple helices directed through electrostatic interactions. J. Am. Chem. Soc. 129, 2683–2690 [DOI] [PubMed] [Google Scholar]
- 27. Xu F., Li J., Jain V., Tu R. S., Huang Q., Nanda V. (2012) Compositional control of higher order assembly using synthetic collagen peptides. J. Am. Chem. Soc. 134, 47–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu F., Zahid S., Silva T., Nanda V. (2011) Computational design of a collagen A:B:C-type heterotrimer. J. Am. Chem. Soc. 133, 15260–15263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu F., Zhang L., Koder R. L., Nanda V. (2010) De novo self-assembling collagen heterotrimers using explicit positive and negative design. Biochemistry 49, 2307–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Madhan B., Xiao J., Thiagarajan G., Baum J., Brodsky B. (2008) NMR monitoring of chain-specific stability in heterotrimeric collagen peptides. J. Am. Chem. Soc. 130, 13520–13521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Persikov A. V., Pillitteri R. J., Amin P., Schwarze U., Byers P. H., Brodsky B. (2004) Stability related bias in residues replacing glycines within the collagen triple helix (Gly-Xaa-Yaa) in inherited connective tissue disorders. Hum. Mutat 24, 330–337 [DOI] [PubMed] [Google Scholar]
- 32. Kay L. E., Keifer P., Saarinen T. (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114, 10663–10665 [Google Scholar]
- 33. Ikura M., Kay L. E., Bax A. (1990) A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry 29, 4659–4667 [DOI] [PubMed] [Google Scholar]
- 34. Fesik S. W., Zuiderweg E. R. (1988) Heteronuclear three-dimensional NMR spectroscopy. A strategy for the simplification of homonuclear two-dimensional NMR spectra. J. Magn. Reson 78, 588–593 [Google Scholar]
- 35. Messerle B. A., Wider G., Otting G., Weber C., Wüthrich K. (1989) Solvent suppression using a spin-lock in 2D and 3D NMR spectroscopy with H2O solutions. J. Magn. Reson. 85, 608–613 [Google Scholar]
- 36. Marion D., Kay L. E., Sparks S. W., Torchia D. A., Bax A. (1989) Three-dimensional heteronuclear NMR of 15N labeled proteins. J. Am. Chem. Soc. 111, 1515–1517 [Google Scholar]
- 37. Fan P., Li M. H., Brodsky B., Baum J. (1993) Backbone dynamics of (Pro-Hyp-Gly)10 and a designed collagen-like triple-helical peptide by 15N NMR relaxation and hydrogen-exchange measurements. Biochemistry 32, 13299–13309 [DOI] [PubMed] [Google Scholar]
- 38. Palmer A. G., 3rd. (1993) Dynamic properties of proteins from NMR spectroscopy. Curr. Opin. Biotechnol. 4, 385–391 [DOI] [PubMed] [Google Scholar]
- 39. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 40. Buevich A. V., Dai Q. H., Liu X., Brodsky B., Baum J. (2000) Site-specific NMR monitoring of cis-trans isomerization in the folding of the proline-rich collagen triple helix. Biochemistry 39, 4299–4308 [DOI] [PubMed] [Google Scholar]
- 41. Li Y., Brodsky B., Baum J. (2007) NMR shows hydrophobic interactions replace glycine packing in the triple helix at a natural break in the (Gly-X-Y)n repeat. J. Biol. Chem. 282, 22699–22706 [DOI] [PubMed] [Google Scholar]
- 42. Baxter N. J., Williamson M. P. (1997) Temperature dependence of 1H chemical shifts in proteins. J. Biomol. NMR 9, 359–369 [DOI] [PubMed] [Google Scholar]
- 43. Cierpicki T., Otlewski J. (2001) Amide proton temperature coefficients as hydrogen bond indicators in proteins. J. Biomol. NMR 21, 249–261 [DOI] [PubMed] [Google Scholar]
- 44. Xu Y., Hyde T., Wang X., Bhate M., Brodsky B., Baum J. (2003) NMR and CD spectroscopy show that imino acid restriction of the unfolded state leads to efficient folding. Biochemistry 42, 8696–8703 [DOI] [PubMed] [Google Scholar]
- 45. Boudko S. P., Bächinger H. P. (2012) The NC2 domain of type IX collagen determines the chain register of the triple helix. J. Biol. Chem. 287, 44536–44545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bella J. (2014) A first census of collagen interruptions: collagen's own stutters and stammers. J. Struct. Biol. 186, 438–450 [DOI] [PubMed] [Google Scholar]
- 47. Golbik R., Eble J. A., Ries A., Kühn K. (2000) The spatial orientation of the essential amino acid residues arginine and aspartate within the α1β1 integrin recognition site of collagen IV has been resolved using fluorescence resonance energy transfer. J. Mol. Biol. 297, 501–509 [DOI] [PubMed] [Google Scholar]
- 48. Rich A., Crick F. H. (1955) The structure of collagen. Nature 176, 915–916 [DOI] [PubMed] [Google Scholar]
- 49. Bella J., Liu J., Kramer R., Brodsky B., Berman H. M. (2006) Conformational effects of Gly-X-Gly interruptions in the collagen triple helix. J. Mol. Biol. 362, 298–311 [DOI] [PubMed] [Google Scholar]