Background: hmtLeuRS has a degenerate CP1 domain that lacks post-transfer editing.
Results: We identified the function of the hmtLeuRS's CP1 domain.
Conclusion: CP1 contributes to aminoacylation efficiency, and hmtLeuRS lacks discrimination toward Nva.
Significance: We elucidated the significance of a defunct CP1 domain.
Keywords: amino acid, aminoacyl tRNA synthetase, enzyme catalysis, transfer RNA (tRNA), translation, CP1 domain, aminoacylation, editing, human mitochondrial leucyl-tRNA synthetase
Abstract
The connective polypeptide 1 (CP1) editing domain of leucyl-tRNA synthetase (LeuRS) from various species either harbors a conserved active site to exclude tRNA mis-charging with noncognate amino acids or is evolutionarily truncated or lost because there is no requirement for high translational fidelity. However, human mitochondrial LeuRS (hmtLeuRS) contains a full-length but degenerate CP1 domain that has mutations in some residues important for post-transfer editing. The significance of such an inactive CP1 domain and a translational accuracy mechanism with different noncognate amino acids are not completely understood. Here, we identified the essential role of the evolutionarily divergent CP1 domain in facilitating hmtLeuRS's catalytic efficiency and endowing enzyme with resistance to AN2690, a broad-spectrum drug acting on LeuRSs. In addition, the canonical core of hmtLeuRS is not stringent for noncognate norvaline (Nva) and valine (Val). hmtLeuRS has a very weak tRNA-independent pre-transfer editing activity for Nva, which is insufficient to remove mis-activated Nva. Moreover, hmtLeuRS chimeras fused with a functional CP1 domain from LeuRSs of other species, regardless of origin, showed restored post-transfer editing activity and acquired fidelity during aminoacylation. This work offers a novel perspective on the role of the CP1 domain in optimizing aminoacylation efficiency.
Introduction
In the three domains of life, genetic information is delivered from DNA to mRNA and is subsequently translated into protein. The fidelity of the whole process is vital for maintaining cellular physiology. Aminoacyl-tRNA synthetases (aaRSs)3 are a family of canonical and diverse enzymes that catalyze the esterification of the amino acids to the cognate tRNAs in a two-step reaction, supplying the building blocks, aminoacyl-tRNAs (aa-tRNAs) for protein biosynthesis. First, the corresponding amino acid is activated with the hydrolysis of ATP, forming an enzyme-aminoacyl·adenylate complex, and subsequently, the activated aminoacyl moiety is transferred to the terminal adenine (A76) of the 3′ end of tRNA (1–3). Based on sequence alignments and structural and functional analyses, the 20 aaRSs are divided exclusively into class I and class II, each consisting of 10 enzymes (4–6). Class I aaRSs utilize a Rossmann dinucleotide-binding fold, characterized by two conserved motifs, HIGH and KMSKS, as the framework to perform their tRNA charging activities (4, 5). However, class II aaRSs use an anti-parallel β-sheet active-site architecture with signature motifs, motifs 1–3, to dimerize and to bind ATP and amino acids (4, 6). During evolution, half of the aaRSs have gradually incorporated an editing domain to remove the wrong amino acid charged on the cognate tRNA, possessing a “double-sieve” mechanism to correct errors in aminoacylation and guarantee the accuracy of protein synthesis. Amino acids larger than the cognate amino acid are excluded via the aminoacylation active site, i.e. “coarse sieve”, and the editing site performs as the “fine sieve” to hydrolyze the incorrect but isosteric amino acid, based on size and structure (3, 7).
Leucyl-tRNA synthetase (LeuRS), along with isoleucyl-tRNA synthetase (IleRS) and valyl-tRNA synthetase (ValRS), belongs to sub-group Ia, which shares a homologous connective polypeptide 1 (CP1) domain tethered to the halves of the Rossmann fold by two flexible β-strands. LeuRS could mis-activate a series of analogs, including valine (Val), isoleucine (Ile), methionine (Met), and intermediate metabolites, like norvaline (Nva) and l-α-aminobutyric acid (ABA) (8, 9). The high fidelity in discriminating the cognate substrate from the large pool of structurally similar noncognate amino acids is largely maintained by proofreading (editing). Based on rectifying errors in the aminoacyl-adenosine monophosphate (aa-AMP) level or the aa-tRNA level, total editing can be divided into pre-transfer editing and post-transfer editing. In addition, pre-transfer editing is further classified as tRNA-dependent and tRNA-independent pre-transfer editing, depending on whether the tRNA can trigger the hydrolytic reaction significantly (2, 3, 10). In class I aaRSs, the catalytic site for post-transfer editing is harbored in the CP1 domain, which has an important role in ensuring the fidelity of aminoacylation (9, 11). Moreover, the existence of a functionally active CP1 domain also increased the aminoacylation activity of Escherichia coli LeuRS (EcLeuRS) (12, 13). Residues that are essential for the hydrolytic role of the CP1 domain have been identified by structural and biochemical analyses, and these include a Thr-rich peptide and a GTG motif (constituting the editing catalytic pocket that can perform the hydrolytic role) and an absolutely conserved Asp residue (such as Asp345 in EcLeuRS), which forms a salt bridge with the α-amino group of the noncognate amino acid) (14–16).
LeuRSs from various species differ in the presence pattern and editing activity of the CP1 domain (Fig. 1). For most LeuRSs, such as LeuRSs from E. coli, Aquifex aeolicus (AaLeuRS) to Saccharomyces cerevisiae cytoplasm (ScLeuRS), and Homo sapiens cytoplasm (HsLeuRS), the active site residues are highly homologous and conserved, possessing the conserved Asp residue, the GTG and Thr-rich regions, and are functionally active in post-transfer editing. These active CP1 domains provide a direct safeguard to guarantee accuracy in tRNALeu aminoacylation. Nevertheless, the CP1 domains of some LeuRSs from Mycoplasma species have been severely truncated, and these LeuRSs are thought to possess no post-transfer editing activity. As a particular example, Mycoplasma mobile encodes a minimized LeuRS (MmLeuRS), which completely lacks a CP1 domain, having a nonapeptide linker (MmLinker, 227KEEIDGKIT235) instead. It is thought that LeuRSs in Mycoplasma parasites gradually lost the important residues in the editing domain to achieve translational inaccuracy and consequent phenotypic plasticity to accommodate host defense (13, 17). Thus, truncation or loss of the CP1 domain is evolutionarily beneficial for their growth or survival. However, LeuRSs from the mitochondria of some higher eukaryotes (such as Mus musculus and H. sapiens) are unique because they have inherent substitutions at key residues in the hydrolytic active sites (18).
FIGURE 1.
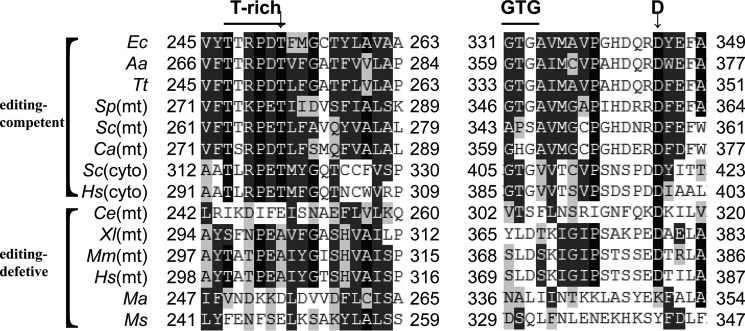
Alignment of active sites in CP1 domain from various LeuRSs. The alignment shows that LeuRSs from different origins have variations in the conserved regions of the CP1 domain. Residues that are conserved or similar are shaded in black or dark gray, respectively. Cyto, cytoplasmic LeuRSs; mt, mitochondrial LeuRSs. The abbreviations used for each species are as follows: Ec, E. coli; Aa, A. aeolicus; Tt, T. thermophiles; Sp, S. pombe; Sc, S. cerevisiae; Ca, C. albicans; Hs, H. sapiens; Ce, C. elegans; Xl, X. laevis; Mm, M. musculus; Ma, M. agalactiae; Ms, M. synoviae.
hmtLeuRS is encoded by the nuclear LARS2 gene and is then imported into the mitochondria. By contrast, human mitochondrial tRNALeus (including tRNALeu(CUN) and tRNALeu(UUR)) are encoded by the mitochondrial genome. The precursor of hmtLeuRS, with a full length of 903 residues, is transported with the guidance of an N-terminal signal peptide and cleaved between Ser39 and Ile40 (19). Although hmtLeuRS preserves the key conserved Asp residue in the CP1 domain, the GTG region and Thr-rich region have variations to different extents (Fig. 1), leading to a degenerated CP1 that has lost its post-transfer editing activity (18). Compared with the truncation or complete loss of the CP1 domain in Mycoplasma parasites, which favors their survival, the reason why hmtLeuRS retains this degraded domain is unclear. Furthermore, a previous study by Lue and Kelley (18) compared the initial velocity of ATP-PPi exchange among Ile, Val, Met, Thr, Ser, Ala, and Gly and found that among them Ile is the most efficiently activated by hmtLeuRS, albeit much more weakly compared with EcLeuRS. They indicated that hmtLeuRS could discriminate Ile efficiently via a precise synthetic core (18, 20). However, it was suggested recently that the main targets of LeuRS editing include Nva (21). Thus, whether hmtLeuRS is able to rigorously discriminate Nva and other Leu analogs (such as ABA) during aminoacylation needs to be further explored.
Inhibition of aaRSs would halt protein translation and stall the growth of organisms; thus, they have long been pursued as anti-parasitic drug targets. Benzoxaborole 5-fluoro-1,3-dihydro-1-hydroxy-2,1-benzoxaborole (AN2690) is in phase 3 clinical trials for the treatment of onychomycosis, an infection of nails caused by fungi (22). AN2690 inhibits LeuRSs from diverse species, including prokaryotic EcLeuRS, eukaryotic Candida albicans LeuRS (CaLeuRS), ScLeuRS, and HsLeuRS, by forming an adduct with the 2′- and 3′-OH group of the terminal adenosine (A76) of non-aminoacylated tRNALeu in the editing-active site (23). It blocks the translocation of tRNA and the turnover of the enzyme. AN2690 mostly interacts with the main chains and side chains of some residues in the CP1 domain, including the Thr-rich region, the GTG region, and the region around the conserved Asp residue (22, 23). To date, two LeuRSs have been proven to be insensitive to this compound, eukaryotic Giardia lamblia LeuRS (GlLeuRS) and prokaryotic AaLeuRS (22, 24). Based on the degeneration of the CP1 domain of hmtLeuRS, we wanted to clarify whether hmtLeuRS is a target of AN2690 or not.
In this study, we systematically studied the role of the editing-defunct CP1 domain in aminoacylation by comparing the activities of hmtLeuRS and CP1 domain-deleted/substituted chimeric variants. The CP1 domain in hmtLeuRS improved its catalytic rate and made it resistant to AN2690. As revealed by others, Ile could be effectively discriminated at the synthetic core of hmtLeuRS as is the case for EcLeuRS; however, our study showed that Nva and Val could be mis-activated by hmtLeuRS and may pose threats to the programmed insertion of Leu in newly synthesized peptides. Moreover, we found hmtLeuRS only retains tRNA-independent pre-transfer editing, which is insufficient to remove mis-activated Nva in vitro. The substitution of CP1 domains from other LeuRSs could assist chimeric enzymes to regain their post-transfer editing activities and correct the tendency to mis-incorporate noncognate Leu analogs. Taken together, these results emphasized the crucial role of the hmtLeuRS CP1 domain in enhancing the catalytic rate and revealed the mis-activation and editing features of hmtLeuRS for noncognate amino acids.
Experimental Procedures
Materials
l-Leu, l-Nva, l-Val, l-ABA, l-Met, Tris-HCl, MgCl2, NaCl, KCl, ATP, tetrasodium pyrophosphate (Na4PPi), inorganic pyrophosphate, dithiothreitol (DTT), sodium acetate (pH 5.2), and activated charcoal were purchased from Sigma. l-[14C]Leu, [32P]Na4PPi, and [α-32P]ATP were obtained from PerkinElmer Life Sciences. Isopropyl 1-thio-β-d-galactopyranoside and AN2690 were purchased from Amescro (Solon, OH) and Milestone Pharmatech (Hangzhou, China), respectively. Pyrophosphatase was purchased from Roche Diagnostics (Basel, Switzerland). Nitrocellulose membranes (0.22 μm), polyethyleneimine cellulose plates, and Amicon ultra-15 filters were obtained from Merck (Darmstadt, Germany). Nucleoside triphophate (NTP) and deoxynucleoside triphosphate (dNTP) mixtures were purchased from Sangon (Shanghai, China). The DNA fragment rapid purification kit and plasmid extraction kit were obtained from Tiangen (Beijing, China). Oligonucleotide primers were synthesized by Invitrogen. The KOD-plus mutagenesis kit and KOD-plus Neo enzyme were purchased from TOYOBO (Osaka, Japan); and variants were confirmed by DNA sequencing performed by Biosune (Shanghai, China). T4 ligase, inorganic pyrophosphatase, and restriction endonucleases were obtained from Thermo Scientific (Waltham, MA). The nickel-nitrilotriacetic acid Superflow was purchased from Qiagen (Hilden, Germany). SuperdexTM 75 and DEAE-Sepharose CL-6B were purchased at GE Healthcare. Competent E. coli Top10 and BL21 (DE3) cells were prepared in our laboratory. T7 RNA polymerase and E. coli CCA-adding enzyme were purified from an overproducing E. coli BL21 (DE3) strain.
Gene Cloning, Mutagenesis, Expression, and Protein Purification
The definition of the CP1 domain of hmtLeuRS and two prokaryotic LeuRSs (EcLeuRS and AaLeuRS) was based on the crystal structure of Thermus thermophilus LeuRS (TtLeuRS, Ser227–Arg418, PDB code 2BTE) and sequence alignment and that of two eukaryotic LeuRSs (HsLeuRS and ScLeuRS) was based on the crystal structure of Pyrococcus horikoshii LeuRS (PhLeuRS, Val204–Asn447, PDB code 1WKB) (Fig. 2A).
FIGURE 2.
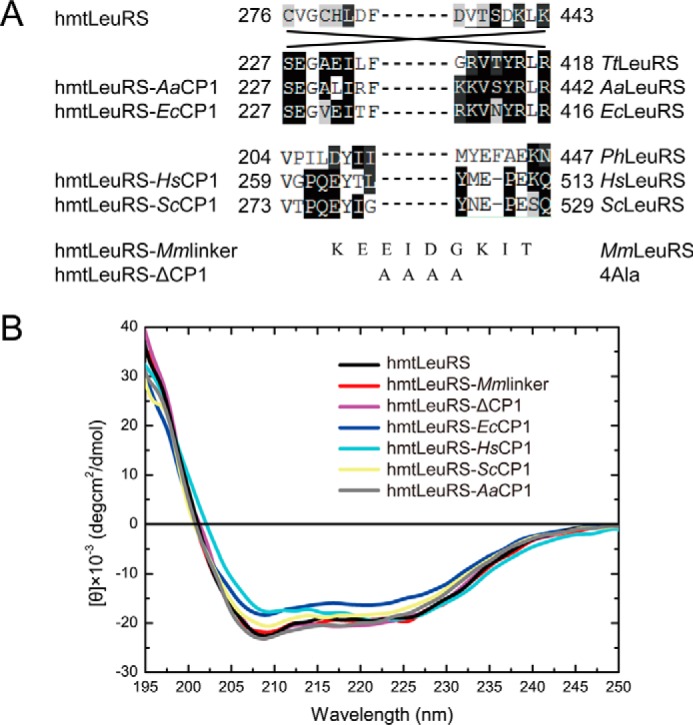
Construction of clones encoding mosaic enzymes and CD spectroscopic analysis of these proteins. A, schematic demonstration of detailed fusion sites of the chimeric proteins. The definition of CP1 domain was based on the crystal structure of TtLeuRS (PDB code 2BTE) and PhLeuRS (PDB code 1WKB) and sequence alignment. Numbers represent the beginning and end of each CP1 domain in the context of full-length enzyme. The abbreviations are the same as Fig. 1, except for two species (Ph, P. horikoshii; Mm, M. mobile). B, CD spectra suggesting the proper secondary structure of hmtLeuRS and its derived mosaic enzymes.
The plasmid pET22b(+)-hmleuS-40, encoding the mature form of hmtLeuRS with an N-terminal His6 tag, was constructed previously in our laboratory (25). The CP1 domain (Cys276–Lys443) of hmtLeuRS was substituted by a four-alanine peptide and by a nonapeptide MmLinker (227KEEIDGKIT235) (13) to obtain the variants hmtLeuRS-ΔCP1 and hmtLeuRS-MmLinker, respectively (Fig. 2A).
Other mosaic enzyme constructs were obtained in a stepwise process. First, we deleted the gene segment encoding four Ala residues and introduced SacII and BspTI sites into the hmtLeuRS-ΔCP1 construct in one-round mutagenesis, creating plasmid pHMBS. The CP1 domains of HsLeuRSs (Val259–Gln513), ScLeuRS (Val273–Gln529), EcLeuRS (Ser227–Arg416), and AaLeuRS (Ser227–Arg442) were amplified separately from plasmids encoding the corresponding LeuRSs constructed previously (11, 13, 26, 27). We digested the PCR products and pHMBS with SacII and BspTI and ligated them to form chimeric constructs of hmtLeuRS-HsCP1, hmtLeuRS-ScCP1, hmtLeuRS-EcCP1, and hmtLeuRS-AaCP1 (Fig. 2A). To inactivate the post-transfer editing activity of the CP1 domain of hmtLeuRS-EcCP1, the Asp residue essential to the activity (Asp345 in EcLeuRS) was mutated to Ala to mimic the corresponding EcLeuRS deficient in editing, generating hmtLeuRS-EcCP1-D345A (16, 28).
hmtLeuRS and its derived variants were overexpressed in E. coli BL21 (DE3) cells (25). Proteins were purified by nickel-nitrilotriacetic acid affinity and gel filtration chromatography with SuperdexTM 75. Purity was confirmed by SDS-PAGE, and their final concentrations were determined by active site titration, as described previously (29).
Circular Dichroism (CD) Spectroscopy
The secondary structure of the chimeric LeuRSs was determined by CD spectroscopy, as reported previously (9). Enzyme samples at 0.2 mg/ml were analyzed on Jasco J-715 spectropolarimeter with a nitrogen purge at room temperature. Spectra were accumulated over three scans, using a 1-mm path length cuvette.
Acquisition of tRNAs
The tRNA constructs of hmtRNALeu, which has a hammerhead ribozyme sequence between a T7 RNA polymerase promoter sequence and the tRNA sequence, were constructed previously (30, 31). In vitro transcriptions were performed as reported previously (32, 33). E. coli tRNALeu (CAG) (EctRNALeu) was isolated from an overexpression strain constructed in our laboratory (34). hmtLeuRS can leucylate EctRNALeu efficiently (19, 25); therefore, EctRNALeu was used to assay the aminoacylation activity of hmtLeuRS.
ATP-PPi Exchange Reaction
The ATP-PPi exchange reaction was performed at 37 °C in buffer containing 2 mm [32P]Na4PPi (22 cpm/pmol), 100 mm HEPES-KOH (pH 7.8), 10 mm MgCl2, 10 mm KF, 2 mm ATP and different amino acids. The reaction was initiated by the addition of 300 nm LeuRS. For Km determinations, Leu (0.02–0.5 mm), Nva (0.5–100 mm), ABA (5–160 mm), Val (5–100 mm), Ile (10–150 mm), and Met (5–150 mm) were used. At regular time intervals, samples were quenched by adding 200 μl of a solution containing 2% activated charcoal, 3.5% HClO4, and 50 mm Na4PPi. The concentration of Leu used to assay activation time curves of hmtLeuRS and chimeric enzymes was 5 mm.
[32P]Labeling at 3′ Terminus of tRNA
EctRNALeu was labeled with [α-32P]ATP, as described previously (10). Briefly, 1000 pmol of tRNA was added to a 50-μl reaction mixture containing 60 mm Tris-HCl (pH 8.0), 12 mm MgCl2, 50 μm Na4PPi, 15 μm ATP, 1 mm DTT, 0.2 μm [α-32P]ATP, and 10 μm E. coli CCA-adding enzyme at 37 °C for 5 min. Thereafter, 0.1 unit or pyrophosphatase was added to the mixture for a further 5 min. The solution was extracted with phenol/chloroform and then precipitated in 3 volumes of ethanol at −20 °C overnight. The product was dissolved in buffer containing 5 mm MgCl2. By liquid scintillation counting of samples that were washed with and without 5% trichloroacetic acid, the ratio of 32P-labeled tRNA was determined.
Aminoacylation, Mis-acylation, and De-acylation
Aminoacylation activities of hmtLeuRS and its various variants were measured in a reaction mixture containing 50 mm HEPES-KOH (pH 7.6), 25 mm KCl, 10 mm MgCl2, 2.5 mm ATP, 1 mm spermidine, 100 μg/ml BSA, 20 μm l-[14C]Leu, and 5 μm EctRNALeu with 300 nm enzymes at 37 °C. EctRNALeu from 0.5 to 30 μm was used for Km determination. AN2690 was added to the reaction buffer to reach a final concentration of 1 mm to determine its effect on hmtLeuRS and hmtLeuRS-EcCP1.
The mis-acylation activities of hmtLeuRS and its variants, hmtLeuRS-EcCP1 and hmtLeuRS-EcCP1-D345A in the presence of [32P]EctRNALeu with various amino acids (20 mm Leu, 20 mm Nva, and 100 mm Val) were assayed. The reaction was initiated by adding 300 nm hmtLeuRS or 600 nm hmtLeuRS-EcCP1 and hmtLeuRS-EcCP1-D345A, respectively. Aliquots were taken and quenched at various time intervals with NaAc and ethanol and precipitated at −20 °C overnight. Samples were centrifuged (12,000 × g) for 30 min at 4 °C, dried at room temperature for 1 h, and subsequently digested by nuclease S1 (25 units). 1.5 μl of the digestion mixture was loaded on polyethyleneimine cellulose plates. Thin layer chromatography (TLC) was then performed and developed with 0.1 m NH4Ac and 5% acetic acid to separate [32P]ATP, [32P]AMP, and aa-[32P]AMP. A series of known amounts of [α-32P]ATP was applied on the TLC plates for quantification of the produced aa-[32P]AMP (from Nva-[32P]EctRNALeu). Data were collected using phosphorimaging and analyzed using MultiGauge Version 3.0 software (Fujifilm).
In the de-acylation assay, mis-acylated Nva-[32P]EctRNALeu was obtained with 5 μm EctRNALeu containing 4.64 nm [32P]EctRNALeu, 50 mm Nva, and 3 μm hmtLeuRS. De-acylation of Nva-[32P]EctRNALeu was carried out with hmtLeuRS in a reaction mixture containing 100 mm Tris-HCl (pH 7.5), 30 mm KCl, 12 mm MgCl2, and 0.5 mm DTT. 500 nm enzymes were used to initiate the reaction, and samples were treated with the same procedure as mis-acylation of [32P]EctRNALeu. After nuclease S1 digestion, TLC was performed on de-acylated samples, similarly to the procedure described for mis-acylation.
AMP Formation
The net effect of the editing reaction is the consumption of ATP. Therefore, editing could be measured by monitoring the breakdown of ATP (to AMP and PPi) in the presence of noncognate amino acids. The AMP formation assay was performed in a reaction mixture containing 100 mm Tris-HCl (pH 7.5), 30 mm KCl, 12 mm MgCl2, 0.5 mm DTT, 3 mm ATP, 5 units/ml pyrophosphatase, 20 nm [32P]ATP (3000 Ci/mm), and 20 mm Nva (or 100 mm Val, 1 mm Leu), with or without 5 μm transcribed hmtRNALeus at 37 °C. Reactions were initiated by the addition of 3 μm hmtLeuRS. 1.5-μl aliquots at various time intervals were quenched in 6 μl of 200 mm NaAc, and then 1.5 μl of the quenched aliquots were spotted on pre-activated polyethyleneimine cellulose plates. A solution containing 0.1 m NH4Ac and 5% acetic acid was used as the flowing phase to separate aa-[32P]AMP, [32P]AMP, and [32P]ATP on the TLC plates.
Nonenzymatic Spontaneous Hydrolysis of Adenylates
The rate of nonenzymatic hydrolysis of adenylates was measured by a pulse-chase experiment in which a large excess of unlabeled ATP was added to the reaction buffer following the initiation of Nva-AMP and Val-AMP. The excess of unlabeled ATP induces the release of the noncognate aminoacyl-adenylate from the active site into solution, where its spontaneous hydrolysis is monitored (11, 35). A solution of 3 μm hmtLeuRS was first incubated with 20 mm Nva, 100 μm ATP, and 0.25 μm [α-32P]ATP for 15 min at 37 °C to prepare Nva-[32P]AMP, in a buffer containing 100 mm Tris-HCl (pH 7.5), 30 mm KCl, 12 mm MgCl2, 0.5 mm DTT, 5 units/ml pyrophosphatase. A relatively large amount of unlabeled ATP was then added such that the final concentration of unlabeled ATP was a 500-fold molar excess of the labeled ATP. The hydrolysis activity was quenched at various time points (2–10 min) by mixing 1.5 μl of the reaction mixture with 6 μl of 200 mm NaAc and 0.1% SDS. Separation of Nva-[32P]AMP and [32P]AMP by TLC was then performed, and the reactions were quantified as described above.
Results
Significance of the Degenerate CP1 Domain in Amino Acid Activation and Aminoacylation
To determine functional significance of the degenerate CP1 of hmtLeuRS, we obtained the two variants of hmtLeuRS, hmtLeuRS-ΔCP1, and hmtLeuRS-MmLinker by cloning and expressing their genes. The two hmtLeuRS variants had the same CD spectrogram as hmtLeuRS, indicating that they have a stable secondary structure (Fig. 2B).
We assayed and compared the kinetic constants of the two variants with those of hmtLeuRS in the amino acid activation reaction (Table 1). The Km values of both variants for Leu were decreased by less than 2-fold after CP1 truncation, showing the affinity between the variant and Leu is slightly increased; however, the kcat values of hmtLeuRS-ΔCP1 and hmtLeuRS-MmLinker sharply decreased to 3.3 × 10−2 and 12.1 × 10−2 s−1, respectively, when compared with that of hmtLeuRS (1.15 s−1). The catalytic efficiency of hmtLeuRS-ΔCP1 (1.7 s−1 mm−1) or hmtLeuRS-MmLinker (7.6 s−1 mm−1) only retained ∼4 or 20% compared with that of the wild-type enzyme (37.8 s−1 mm−1).
TABLE 1.
Kinetic parameters of hmtLeuRS, hmtLeuRS-ΔCP1, and hmtLeuRS-MmLinker toward Leu in amino acid activation
Data were obtained from three independent experiments. The catalytic efficiency of hmtLeuRS was designated as 100%.
| Km | kcat | kcat/Km | Relative | |
|---|---|---|---|---|
| μm | ×10−2 s−1 | s−1 mm−1 | % | |
| hmtLeuRS | 30.4 ± 4.2 | 115 ± 25 | 37.8 | 100 |
| hmtLeuRS-ΔCP1 | 19.3 ± 0.2 | 3.3 ± 0.4 | 1.7 | 4 |
| hmtLeuRS-MmLinker | 15.9 ± 0.2 | 12.1 ± 0.9 | 7.6 | 20 |
In subsequent aminoacylation assays, both variants had almost an unchanged Km for tRNA; however, hmtLeuRS-ΔCP1 and hmtLeuRS-MmLinker displayed obvious decreases in kcat values (0.40 × 10−2 and 1.46 × 10−2 s−1, respectively), accounting for only 9 and 33% that of the native enzyme (4.51 × 10−2 s−1). Although in aminoacylation the catalytic efficiency of hmtLeuRS-MmLinker (8.8 s−1 mm−1) was much higher than that of hmtLeuRS-ΔCP1 (1.9 s−1 mm−1), they only retained 9 and 42% of that of native enzyme (21.0 s−1 mm−1), respectively (Table 2). The data also showed that the nonapeptide MmLinker was a good substitute for the degenerate CP1 domain of hmtLeuRS in synthesis of aminoacyl-tRNA.
TABLE 2.
Kinetic parameters of hmtLeuRS, hmtLeuRS-ΔCP1, and hmtLeuRS-MmLinker toward tRNALeu in aminoacylation
Data were obtained from three trials. The catalytic efficiency of hmtLeuRS was designated as 100%.
| Km | kcat | kcat/Km | Relative | |
|---|---|---|---|---|
| μm | ×10−2 s−1 | s−1 mm−1 | % | |
| hmtLeuRS | 2.14 ± 0.32 | 4.51 ± 0.24 | 21.0 | 100 |
| hmtLeuRS-ΔCP1 | 2.06 ± 0.22 | 0.40 ± 0.02 | 1.9 | 9 |
| hmtLeuRS-MmLinker | 1.65 ± 0.19 | 1.46 ± 0.26 | 8.8 | 42 |
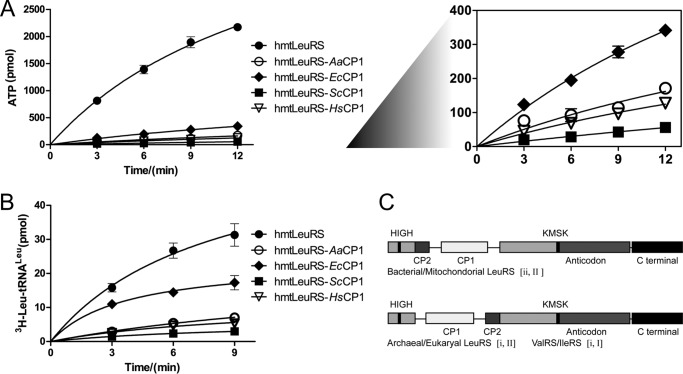
To further explore the importance of the degenerate CP1 domain, we replaced it with its counterparts from both bacterial and eukaryotic LeuRSs (LeuRSs of E. coli, A. aeolicus, and S. cerevisiae cytoplasmic form and H. sapiens cytoplasmic form), and we obtained four chimeric variants, including hmtLeuRS-EcCP1, hmtLeuRS-AaCP1, hmtLeuRS-ScCP1, and hmtLeuRS-HsCP1. Their proper secondary structures were confirmed by CD spectrograms (Fig. 2B). We compared Leu activation and tRNA charging activities of wild-type hmtLeuRS and these chimeras. The variants showed reduced catalytic activities to different extents (Fig. 3, A and B). Further analysis of the activities of these variants revealed that the activity was related to the origin of the CP1 domain. Chimeric variants with a CP1 domain from bacterial LeuRSs showed slightly more efficient catalysis than those with CP1 domains from eukaryotic LeuRSs (especially in Leu activation). Sequence, insertion point, and orientation variation between CP1 domains from prokaryotic and eukaryal/archaeal LeuRSs when substituting for hmtLeuRS's CP1 might affect the activity of the chimeras (Fig. 3C) (36).
FIGURE 3.
Leu activation and aminoacylation time curves of hmtLeuRS and corresponding CP1 chimeric enzymes. A, Leu activation with 5 mm Leu using 300 nm hmtLeuRS and its chimeras. The right panel compares the Leu activation activities of hmtLeuRS chimeras. B, aminoacylation of 5 μm EctRNALeu using 300 nm hmtLeuRS and chimeras. C, discrepancy of CP1 domain insertion points between bacterial/mitochondrial LeuRSs and archaeal/eukaryal LeuRSs. Insertion point type is distinguished by i and ii; rotational state type is distinguished by I and II. ValRS/IleRS (i, I) and archaeal/eukaryal LeuRS (i, II) have the same insertion point but differ in their editing domain rotational state because the insertion of the long variable loop in tRNALeu. Although bacterial/mitochondrial LeuRSs (ii, II) have a different insertion point; they also change the rotational state to accommodate tRNALeu (35).
The above data showed that the presence of the CP1 domain in hmtLeuRS, despite the degeneracy in sequence and deficiency in post-transfer editing (18), is necessary for the amino acid activation and aminoacylation activities of hmtLeuRS. Its deletion or replacement by other CP1 domains led to severe impairment of enzyme function. Therefore, hmtLeuRS must be under evolutionary pressure to keep its CP1 domain.
AN2690 Does Not Affect hmtLeuRS
AN2690 is a broad-spectrum LeuRS inhibitor, targeting the active site of the CP1 editing domain of various LeuRSs. Indeed, our previous analysis revealed that it inhibits the aminoacylation and post-transfer editing activity of HsLeuRS (26). However, does the degeneracy of hmtLeuRS's CP1 domain affect its response to AN2690? To address this question, we compared the aminoacylation activity of hmtLeuRS in the absence or presence of AN2690. The data showed that AN2690 did not affect the aminoacylation activity of hmtLeuRS (Fig. 4A). To further reveal the significance of degeneracy of the CP1 domain in the AN2690 resistance of hmtLeuRS, we assayed the aminoacylation activity of hmtLeuRS-EcCP1, which has a functional and AN2690-sensitive CP1, in the absence or presence of AN2690 (Fig. 4B). Indeed, addition of the EcCP1 domain restored sensitivity to AN2690. Furthermore, post-transfer editing of hmtLeuRS-EcCP1 to Nva-tRNALeu was obviously impaired by AN2690 (the restoration of post-transfer editing of hmtLeuRS-EcCP1 is described later in the text). However, the effect of AN2690 on the post-transfer editing of hmtLeuRS could not be determined because hmtLeuRS naturally lacks post-transfer editing of Nva-tRNALeu (Fig. 4C), which is consistent with the results of Lue and Kelley (18). Thus, the above data showed that AN2690 could not inhibit the activity of hmtLeuRS because of its degenerate CP1 domain. However, the presence of a functional CP1 domain restored sensitivity to AN2690.
FIGURE 4.
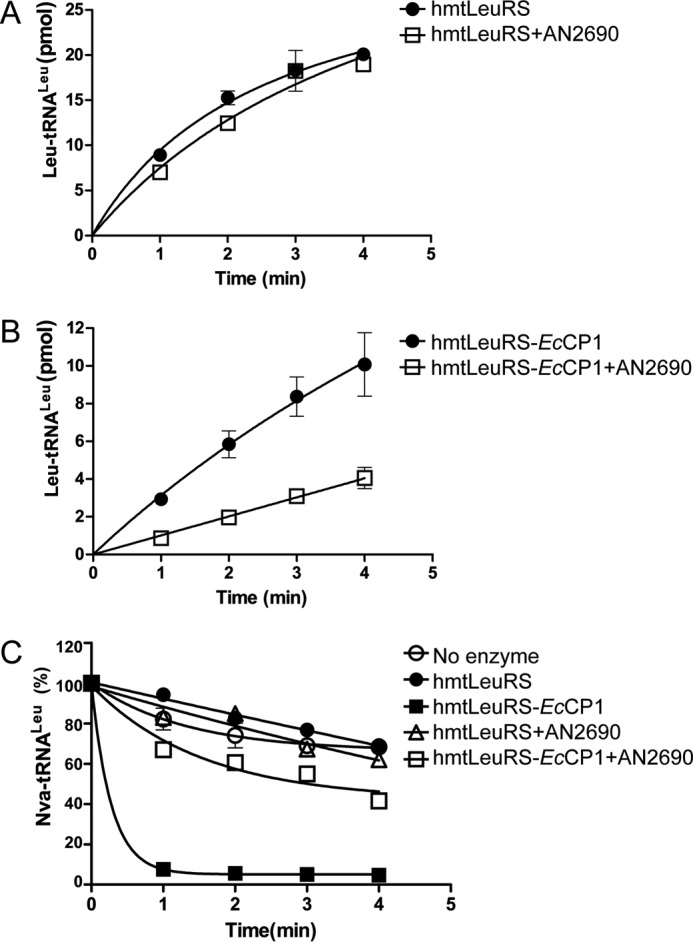
Degenerate CP1 domain endows hmtLeuRS insensitivity toward AN2690. A and B, aminoacylation by 300 nm hmtLeuRS and hmtLeuRS-EcCP1 in the presence and absence of 1 mm AN2690. C, effect of AN2690 on the post-transfer editing activities of hmtLeuRS and hmtLeuRS-EcCP1. 500 nm enzymes were used in the deacylation assay.
HmtLeuRS Could Obviously Mis-activate Nva and Val
Lue and Kelley (18, 20) assayed and compared the initial rates of activation of several proteinic amino acids by hmtLeuRS and found that, among them, Ile could be mis-activated at the highest rate and the canonical active core of hmtLeuRS could more efficiently discriminate Ile at the activation step compared with EcLeuRS. However, the kinetic parameters for other Leu analogs and nonprotein amino acids, such as Nva and ABA, have not been determined.
To further understand the mis-activation features of hmtLeuRS, we assayed its ATP-PPi exchange activity and measured the kinetic parameters for several analogs of Leu, including Nva and ABA (Table 3). The results showed that hmtLeuRS could effectively discriminate ABA and Met at the first step of aminoacylation. The catalytic rate constants (kcat) of hmtLeuRS for ABA and Met were about 11% (0.13 s−1) and 5% (0.061 s−1) that for Leu (1.15 s−1), respectively; however, the Km values for ABA and Met increased by about 530-fold (16.2 mm) and 670-fold (20.5 mm) that for Leu (30.4 μm), respectively (Table 3). The mis-activation discrimination factors were 4730 and 12,600 for ABA and Met, which are much higher than the threshold of the general error level in protein biosynthesis (3300) (37). These results indicated ABA and Met should not pose a threat to the catalytic fidelity of hmtLeuRS.
TABLE 3.
Amino acid activation kinetic parameters of hmtLeuRS for Leu and its analogs
Discrimination factor represents the loss of catalytic efficiency relative to Leu.
| Amino acids | Km | kcat | kcat/Km | Discrimination factor |
|---|---|---|---|---|
| μm | s−1 | s−1 m−1 | ||
| Leu | 30.4 ± 4.2 | 1.15 ± 0.25 | 37.8 | 1 |
| Nva | 6200 ± 130 | 1.31 ± 0.17 | 2.1 ×10−1 | 180 |
| Val | 19600 ± 900 | 0.62 ± 0.09 | 3.2 ×10−2 | 1180 |
| ABA | 16200 ± 1100 | 0.13 ± 0.02 | 8.0 ×10−3 | 4730 |
| Met | 20500 ± 1600 | 0.061 ± 0.008 | 3.0 × 10−3 | 12600 |
Meanwhile, although Nva and Val, the other two analogs of Leu, had weaker affinity with the enzyme (higher Km values, 6.2 mm for Nva and 19.6 mm for Val), they were activated with comparable catalytic rate constants (1.31 s−1 for Nva and 0.62 s−1 for Val), compared with that for Leu (1.15 s−1) (Table 3). They could be activated by hmtLeuRS efficiently. The discrimination factors for Nva and Val were 180 and 1180, respectively, which were much lower than the mistranslation frequency (3300); therefore, hmtLeuRS might have evolved a pre-transfer proofreading pathway to remove mis-activated amino acids and maintain the catalytic fidelity or allow the generation of mis-charged tRNA without proofreading.
hmtLeuRS Has No tRNA-dependent Pre-transfer Editing
Editing is used to ensure a precise recognition between aaRS and its cognate amino acid, and it minimizes errors in mis-incorporation of amino acids in peptides or proteins. Theoretically, the net effect of the editing reaction is the consumption of ATP. Both hydrolysis of mis-aminoacyl-adenylate in pre-transfer editing and removal of mis-charged tRNAs after aminoacylation in post-transfer editing contribute to the formation of AMP (24). By monitoring AMP formation by TLC in the presence of noncognate amino acids, we could characterize the editing activity of hmtLeuRS.
hmtLeuRS can mis-activate Nva and Val and has lost its post-transfer editing activity; therefore, we determined whether it possesses tRNA-dependent pre-transfer editing and the contributions of tRNA-independent and -dependent pre-transfer editing to total editing in the presence of these two Leu analogs.
First, we determined the AMP formation rate of hmtLeuRS in the presence of Leu. The observed AMP formation rate (kobs) was 1.41 × 10−3 s−1 without tRNAs and 1.88 × 10−3 s−1 or 1.79 × 10−3 s−1 in the presence of hmttRNALeu (CUN) or hmttRNALeu (UUR) transcripts (Table 4). Considering the cognate substrate cannot trigger hmtLeuRS's editing reaction, AMP should come from the hydrolysis of ATP in Leu activation. Then we monitored the editing of hmtLeuRS toward noncognate Nva and Val in the absence and presence of hmttRNALeu transcripts. For Nva, the kobs value without tRNA was 2.79 × 10−2 s−1, ∼20-fold greater than that for Leu (1.41 × 10−3 s−1); and in the presence of hmttRNALeu (CUN) or hmttRNALeu (UUR), it was 3.29 × 10−2 or 2.78 × 10−2 s−1, respectively (Table 4 and Fig. 5). tRNA does not affect the pre-transfer editing of hmtLeuRS; therefore, hmtLeuRS only has tRNA-independent pre-transfer editing and has no tRNA-dependent pre-transfer editing for Nva. Compared with EcLeuRS (the kobs of EcLeuRS for Nva was 3.3 × 10−1 and 3.42 s−1 in the absence and presence of 5 μm EctRNALeu, respectively) (24), hmtLeuRS possesses weak editing function.
TABLE 4.
Observed rate constants for AMP synthesis in the presence of Nva or Val by hmtLeuRS
Rates are the average of two independent repeats, with the standard deviations indicated.
| Amino acids | Rate of AMP formation kobs (s−1) |
||
|---|---|---|---|
| No tRNA | hmttRNALeu (CUN) | hmttRNALeu (UUR) | |
| Leu | (1.41 ± 0.08) ×10−3 | (1.88 ± 0.88) ×10−3 | (1.79 ± 0.22) ×10−3 |
| Nva | (2.79 ± 0.14) ×10−2 | (3.29 ± 0.19) ×10−2 | (2.78 ± 0.52) ×10−2 |
| Val | (6.34 ± 2.86) ×10−4 | (5.42 ± 1.68) ×10−4 | (5.21 ± 1.64) ×10−4 |
FIGURE 5.
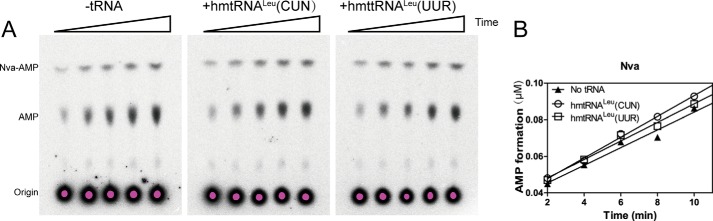
hmtLeuRS only possesses tRNA-independent pre-transfer editing activity. A, representative diagram of AMP formation assay conducted with 20 mm Nva by 3 μm hmtLeuRS in the absence and presence of 5 μm hmtRNALeu(CUN) and hmtRNALeu(UUR). B, corresponding graphical representations of the AMP formation rate using a TLC-based assay.
For Val, in the absence of tRNA, the kobs was 6.34 × 10−4 s−1, only 45% that for Leu (1.41 × 10−3 s−1) (Table 4), and we suggested the accumulated AMP might be derived from mis-activation rather than editing. hmttRNALeu (CUN) (5.42 × 10−4 s−1) or hmttRNALeu (UUR) (5.21 × 10−4 s−1) triggered no more AMP formation, and the kobs value was still lower than that for Leu in the absence of tRNA (1.41 × 10−3 s−1) (Table 4). Therefore, hmtLeuRS has no pre-transfer editing (including tRNA-independent pre-transfer editing and tRNA-dependent pre-transfer editing) for Val.
AMP could be produced by hmtLeuRS catalysis or by spontaneous hydrolysis of Nva-AMP. To measure the rate of possible spontaneous hydrolysis, we carried out a pulse-chase experiment (11, 35). The kobs value of the conversion of Nva-AMP to AMP was 1.18 × 10−4 s−1 (Fig. 6), which was 236-fold lower than the kobs determined for AMP formation (2.79 × 10−2 s−1 for no tRNA) by hmtLeuRS in the presence of Nva (Table 4), indicating that the spontaneous hydrolysis of Nva-AMP was negligible. The above data showed that hmtLeuRS is deficient in tRNA-dependent editing, including tRNA-dependent pre-transfer and post-transfer editing, and it only retains tRNA-independent pre-transfer editing for Nva.
FIGURE 6.
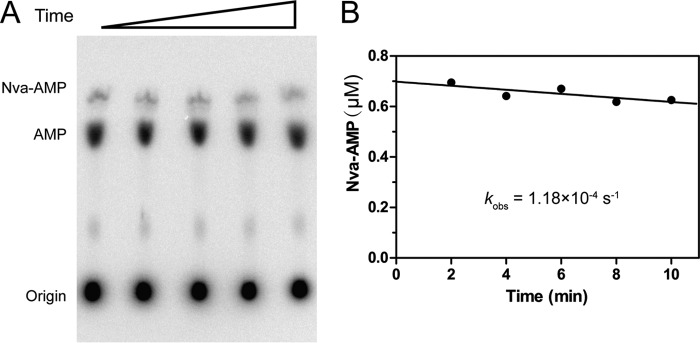
Nonenzymatic hydrolysis of Nva-AMP by hmtLeuRS. A, representative diagram of nonenzymatic hydrolysis of Nva-AMP. B, quantification of remaining Nva-AMP at different time points and the kobs of nonenzymatic hydrolysis of Nva-AMP calculated from three independent trials.
tRNA-independent Pre-transfer Editing Is Insufficient to Remove Mis-activated Nva
To understand whether hmtLeuRS could achieve fidelity toward Nva and Val, we examined the mis-acylation of hmtLeuRS using a TLC-based assay. By labeling the 3′ end of EctRNALeu with [α-32P]ATP, we could monitor the formation of Nva-[32P]AMP and Val-[32P]AMP from aminoacyl-tRNAs by nuclease S1 digestion. The data showed that hmtLeuRS catalyzes the formation of similar amounts of Nva-tRNALeu and Leu-tRNALeu, suggesting that tRNA-independent pre-transfer editing from the synthetic domain in hmtLeuRS could not remove Nva efficiently (Fig. 7). For Val, there was only a trace amount of Val mis-charged to tRNALeu by hmtLeuRS, although its concentration (100 mm) was 5-fold higher than those of Nva and Leu (20 mm) (Fig. 7). Considering Val was mis-activated by hmtLeuRS (Table 3) and hmtLeuRS possessed no tRNA-independent pre-transfer editing toward it (Table 4), it is suggested that the mis-activated Val was not efficiently transferred to tRNA. In summary, the tRNA-independent pre-transfer editing of hmtLeuRS was insufficient to ensure specificity in the aminoacylation reaction toward Nva.
FIGURE 7.
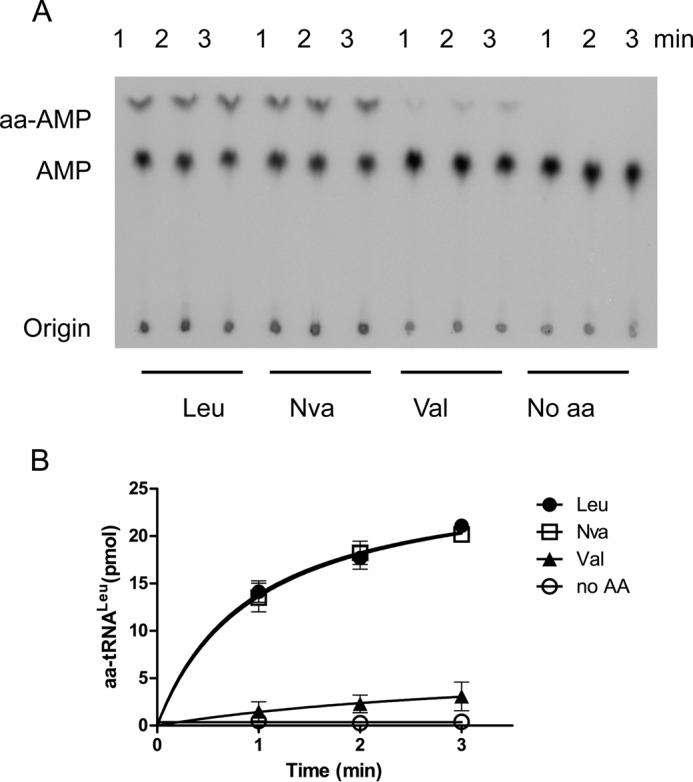
Mis-charging behavior of hmtLeuRS toward Val and Nva. A, representative diagram of aminoacylation assays using 300 nm hmtLeuRS in the presence of 5 μm mixture of 32P-labeled and -unlabeled EctRNALeu at 37 °C. The concentrations of Leu, Nva, and Val used here were 20, 20, and 100 mm, respectively. B, quantification of aminoacylated tRNA at various time points by hmtLeuRS for Leu, Nva, and Val.
Restoration of hmtLeuRS's Post-transfer Editing Activity
The homology in the CP1 domain of hmtLeuRS and many other LeuRSs showed that the deficient CP1 domain of hmtLeuRS might be an evolutionary remnant of an active editing domain. We performed reverse mutations of the degenerate Thr-rich region and GTG regions in hmtLeuRS to generate an editing-active hmtLeuRS. However, despite extensive efforts, we failed to obtain such a post-transfer editing-active enzyme. Subsequently, we utilized the above CP1 domain chimeras and determined their de-acylation activity, together with wild-type hmtLeuRS. The data showed that all of them could significantly restore the post-transfer editing activities toward Nva-tRNALeu (Fig. 8).
FIGURE 8.
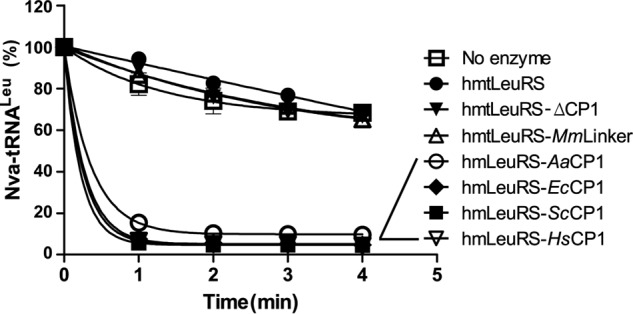
Restoration of post-transfer editing activity to Nva-tRNALeu by 500 nm hmtLeuRS and its chimeras.
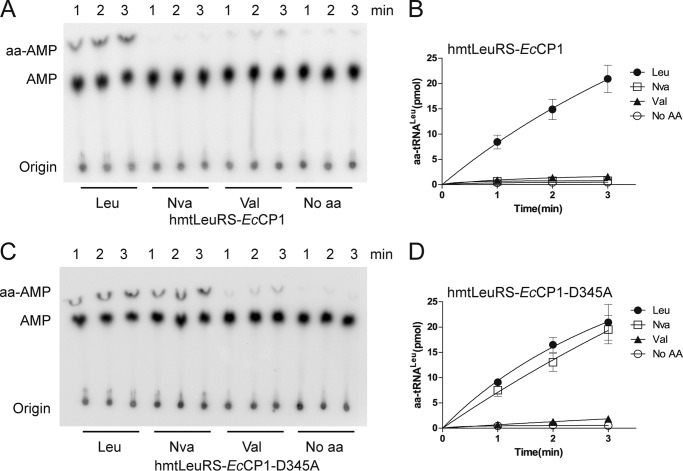
To determine whether an editing-functional CP1 domain could improve fidelity during aminoacylation, we examined the mis-acylation behavior of hmtLeuRS-EcCP1, because EcCP1 is evolutionarily close to CP1 of hmtLeuRS. In the presence of tRNALeu and Nva, the formation of Nva-tRNALeu of hmtLeuRS-EcCP1 was considerably weak compared with that of hmtLeuRS, because of the prevention of accumulation of Nva-AMP by post-transfer editing conferred by an active editing domain of EcLeuRS (Fig. 9, A and B). When the conserved Asp residue was mutated to Ala in hmtLeuRS-EcCP1 (hmtLeuRS-EcCP1-D345A), the production of Nva-tRNALeu dramatically increased (Fig. 9, C and D). hmtLeuRS-EcCP1 catalyzed the synthesis of Val-tRNALeu at a relatively lower rate compared with that of hmtLeuRS; hmtLeuRS-EcCP1-D345A produced Val-tRNALeu at a slightly higher rate than hmtLeuRS-EcCP1. In conclusion, the fusion of a functional CP1 domain endowed accuracy on an error-prone hmtLeuRS during aminoacylation.
FIGURE 9.
Restoration of translational fidelity of hmtLeuRS. A, graphical representations showing aminoacylation assays using 600 nm hmtLeuRS-EcCP1. The concentrations of Leu, Nva, and Val used here were 20, 20, and 100 mm, respectively. B, quantification of aminoacylated tRNA at various time points by hmtLeuRS-EcCP1 for Leu, Nva, and Val. C, graphical representations showing aminoacylation assays by 600 nm hmtLeuRS-EcCP1-D345A. The concentrations of Leu, Nva, and Val used here were 20, 20, and 100 mm, respectively. D, quantification of aminoacylated tRNA at various time points using hmtLeuRS-EcCP1-D345A for Leu, Nva, and Val.
Discussion
CP1 Domain Benefits hmtLeuRS in Catalytic Rate
The CP1 editing module found in class Ia aaRSs is present in nearly all species and is highly conserved throughout evolution. The editing domains of LeuRS and many other aaRSs in Mycoplasma parasites naturally have mutations or deletions (17). It causes mis-translation, phenotype diversity, and plasticity, facilitating the pathogens' escape from host resistance (13, 17). Compared with the retrogressive CP1 domain of LeuRSs in the mitochondria of several species, such as H. sapiens, Xenopus laevis, and Caenorhabditis elegans (18), some fungal mitochondrial LeuRSs (C. albicans and Schizosaccharomyces pombe) still remain intact in the essential residues of the CP1 domain. An exception is the LeuRS from the mitochondria of S. cerevisiae (ScmtLeuRS), which has variation in the GTG region; but it retains post-transfer editing activity (38).
Although defunct in editing, the reason why hmtLeuRS retains a CP1 domain is unknown. In this study, we confirmed the function of hmtLeuRS's CP1 domain in the aminoacylation reaction rather than editing. By comparison of hmtLeuRS and CP1 deletion/substitution enzymes, we found that the CP1 domain contributes significantly to the amino acid activation and tRNA charging activities. Complete removal of the CP1 domain (hmtLeuRS-ΔCP1) resulted in considerable loss of aminoacylation activity, whereas substitution of this domain with the Mmlinker (hmtLeuRS-MmLinker) could partly assist the enzyme in retaining the aminoacylation activity. Our results suggested that the existence of a CP1 domain is beneficial for hmtLeuRS to optimize activation and aminoacylation catalytic efficiency, as observed in EcLeuRS (12). We also validated that the MmLinker can functionally compensate for the CP1 domain of LeuRSs in aminoacylation activity, to some extent (13). It is possible that hmtLeuRS still retains this domain to benefit the catalytic function under environmental stress. Likewise, prolyl-tRNA synthetases (ProRSs) show divergence in the editing domain through evolution. For most bacterial ProRSs, a domain (INS) is inserted between the canonical core to perform cis-editing, and in lower eukaryotes, there is a homologous domain appended to its N terminus. Previous work showed that the N-terminal extension of S. cerevisiae ProRS (ScProRS) has low homology with the INS domain, and it is deprived of some key residues, which may be the relics of a previously functional domain. Although editing-deficient, it still contributes to the catalytic rate of the whole enzyme (39).
The endosymbiotic theory states that a mitochondrion is generated by the capture of an independent bacterium by a cell. Not only did early lives adapt themselves with increasing oxygen, but the host cells could also be provided with much energy via oxidative phosphorylation (40). Mammalian mitochondrial aaRSs, although they have various evolutionary origins, are mainly of bacterial type (41). From an evolutionary point of view, the CP1 domains of bacterial and mitochondrial LeuRSs have the same insertion point and orientation, which are different from archaeal/eukaryal LeuRSs, which may account for the discrepancy in the catalytic activities between mosaic enzymes, according to the origins of the CP1 domain.
AN2690 Resistance of LeuRSs
AN2690 is an antibiotic used to treat leukonychia. Rock et al. (23) isolated S. cerevisiae spontaneous and ethyl-methanesulfonate-induced AN2690-resistant variants and found all the AN2690-resistant mutations were in the CP1 domain or the editing-active site of ScLeuRS. They demonstrated that AN2690 inhibits LeuRS by forming a stable tRNALeu-AN2690 adduct in the editing site, mediated via tRNA's A76 and AN2690's boron atom (23). According to the crystal structure of TtLeuRS with tRNALeu and AN2690 (PDB number 2V0G), the configuration between AMP-AN2690 and the CP1 domain is facilitated by hydrogen bonds to the conserved Thr-rich peptide (Thr247 and Thr248 in TtLeuRS) and a water molecule (23).
Here, we found that the degenerate CP1 domain confers resistance to AN2690 on hmtLeuRS during aminoacylation. Sequence alignment showed that, in addition to alteration to Ala of the counterpart of TtLeuRS Thr248 in hmtLeuRS, hmtLeuRS also diverges in some residues whose mutations led to the insensitivity to AN2690 in ScLeuRS (Fig. 10). Therefore, we hypothesized that the degeneracy of the hmtLeuRS CP1 domain, especially in the binding site with AN2690, inhibits AN2690 from forming an appropriate adduct with A76 of the tRNA. We could not exclude the possibility that the 3′ end of the tRNA cannot shift between the synthetic active site and the defective CP1 domain of hmtLeuRS, as is found in EcLeuRS (where the 3′ end of tRNA translocates between the synthetic active site and the functional CP1 domain of EcLeuRS) (42).
FIGURE 10.
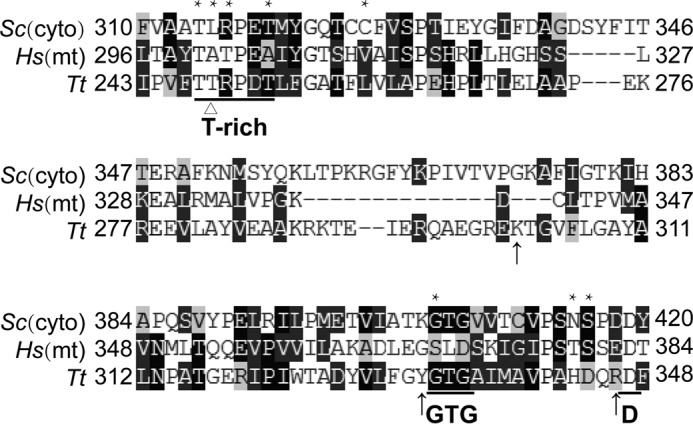
Sequence alignment showing the degeneracy of hmtLeuRS's CP1 domain in regions or sites that may be relevant to the resistance to AN2690 and the loss of tRNA-dependent pre-transfer editing in ScLeuRS and TtLeuRS. The abbreviations used are as follows: Sc(cyto), S. cerevisiae cytoplasmic LeuRS; Hs(mt), H. sapiens mitochondrial LeuRS; Tt, T. thermophilus LeuRS. Asterisks and triangles indicate residues whose mutation leads to resistance to AN2690 in ScLeuRS (T314M, L315V, R316I, T319I, C326R, C326F, G405V, N415D, and S416L) (23) and Thr248 in TtLeuRS, respectively. Arrows indicate certain essential residues (Lys302, Tyr332, and Arg346) in TtLeuRS that interact with tRNA in the post-transfer editing conformation.
It is notable that human cytosolic LeuRS is readily inhibited by AN2690, which affects the external use of AN2690 in disease treatment. Combined with the previously identified GlLeuRS and AaLeuRS, and hmtLeuRS here, three LeuRSs have been found to be insensitive to AN2690 despite bearing a CP1 domain (22). However, hmtLeuRS is the only enzyme located in an organelle. Indeed, the detailed resistance mechanism of hmtLeuRS to AN2690 remains to be determined.
hmtLeuRS Only Has tRNA-independent Pre-transfer Editing for Nva
Recent studies showed that Met40 in EcLeuRS plays a key part in discriminating Ile; prevention of incorrect Nva incorporation is the major biological editing activity of EcLeuRS (21). Our study determined systematically the catalytic efficiency of hmtLeuRS toward different analogs of Leu. We found that Nva and Val could be mis-activated by hmtLeuRS; however, the presence of tRNALeus does not affect its editing activity. The data showed that hmtLeuRS has neither tRNA-dependent pre-transfer editing nor post-transfer editing, only retaining tRNA-independent pre-transfer editing for Nva.
The tertiary structure of TtLeuRS-tRNALeu in the post-transfer editing conformation (PDB number 2BYT) showed that Lys302 and Arg346 of TtLeuRS interact with the C74 base and C75 phosphate of tRNA, whereas Thr332 (Thr in the GTG region) makes contact with A76 phosphate (43). Previous studies showed that a proper binding between the CCA end of a tRNA and the editing domain, rather than an active site of editing, is a prerequisite for LeuRS to hydrolyze noncognate adenylate intermediates and perform tRNA-dependent pre-transfer editing (24, 44), which is likely to occur in the aminoacylation active site. The communication between the aminoacylation and CP1 domain could modulate tRNA-dependent pre-transfer editing (45). Sequence alignments showed that hmtLeuRS has deletions in regions corresponding to the TtLeuRS's regions containing Lys302 and mutations in residues corresponding to Tyr332 and Arg346 (Fig. 10). Therefore, hmtLeuRS shows divergence in the conserved residues essential for binding tRNA, which might explain why hmtLeuRS possesses no tRNA-dependent pre-transfer editing.
As for hmtLeuRS, in vitro assays showed that it could produce considerable mis-charged product Nva-tRNALeu, indicating that tRNA-independent pre-transfer editing was not sufficient to clear mis-activated Nva. This is consistent with previous results (13, 24), which showed that pre-transfer editing alone is insufficient to prevent generation of mis-charged tRNALeu in vitro. For Val, it seems that mis-activated Val cannot be efficiently transferred to tRNALeu, and thus only a trace amount of Val-tRNALeu is synthesized.
Nva is a side product of the Leu biosynthetic pathway and differs from Leu only by the absence of a side chain methyl group (46). It may pose a significant threat to the fidelity of protein synthesis when accumulated to a concentration capable of jeopardizing the accuracy of Leu-tRNALeu synthesis. A recent study showed that intracellular accumulation of Nva could come from the downshift of free oxygen, although free Nva is not accumulated in E. coli W3110 in aerobic cultures (47). Ross-Inta et al. (48) analyzed the profiles of free amino acids and some specific derivatives in rat liver mitochondria and found that the levels of most of them (including Leu, 5.16 mm in mitochondria) were higher than in the cytosol or serum. Free amino acids in mitochondria come from the degradation of mtDNA-encoded proteins, rather than the fulfillment of an amino acid profile to suit mitochondrial protein synthesis. However, the amount of Nva in any mitochondrial system has not been reported. We hypothesized that normally the higher eukaryotic mitochondria contain Nva at a low concentration such that the mis-activated Nva could be removed completely by tRNA-independent pre-transfer editing, and tRNA-dependent editing might be redundant. Therefore, hmtLeuRS does not need to retain its tRNA-dependent editing function. However, it is possible that under specific circumstances, Nva would accumulate or be imported into mitochondria from the cytoplasm, where it would damage the quality control in mitochondrial protein synthesis. Otherwise, mitochondria may endure a considerable level of tRNA mis-charging, as revealed by the mitochondrial phenylalanyl-tRNA synthetase system (49). Quality control of protein synthesis in mitochondria might be focused in the co- or post-translational steps, and the incorporation of noncognate amino acids might lead to the production of misfolded proteins, which would be rapidly degraded by the mitochondrial proteolytic system (48).
hmtLeuRS Chimeras Gain Post-transfer Editing Function and Translational Accuracy
We found that hmtLeuRS chimeras with a fusion of the CP1 domain from different species gained post-transfer editing activities and fidelity during aminoacylation, just as ScProRS regained post-transfer editing function by replacement of N-terminal domain from the EcProRS INS domain (39). Additionally, the gain of post-editing activity was not dependent on the origin of the fused CP1 domain; both eukaryotic and prokaryotic LeuRSs were functional. EcLeuRS is close to hmtLeuRS in evolution; therefore, we specifically inspected the mis-acylation characteristics of hmtLeuRS-EcCP1 and hmtLeuRS-EcCP1-D345A. hmtLeuRS-EcCP1 prohibited the mis-charging of tRNA with Nva, although hmtLeuRS-EcCP1-D345A catalyzed the formation of a large amount of Nva-tRNALeu. The editing function of the hmtLeuRS-EcCP1 chimera to Nva also depended on the conserved D345A residue in the CP1 domain. Moreover, hmtLeuRS-EcCP1 and hmtLeuRS-EcCP1-D345A only produced a trace amount of Val-tRNALeu.
Concluding Remarks
Our study presents a new viewpoint on the effect of the editing-inactive CP1 domain on the aminoacylation function of hmtLeuRS. We showed that hmtLeuRS cannot function as a single sieve for Nva or Val. It exhibits only tRNA-independent pre-transfer editing for Nva, which is not sufficient to remove mis-activated Nva. In addition, the editing-inactive CP1 domain confers resistance to AN2690 on the enzymes. By fusing with the editing-active CP1 domain from other LeuRSs, hmtLeuRS chimeras regained post-transfer editing activities and displayed aminoacylation fidelity.
Author Contributions
Q. Y., X. L. Z., and E. D. W. designed the experiments, analyzed the data, and wrote the manuscript. M. W. assisted in the gene cloning and expression of mosaic enzymes. Z. P. F., Z. R. R., and Q. Q. J. assisted in obtaining tRNAs. Q. Y. performed all the other experiments. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We are grateful to Drs. Min Tan and Wei Yan for their valuable suggestions.
This work was supported by National Key Basic Research Foundation of China Grant 2012CB911000, Natural Science Foundation of China Grants 31130064 and 91440204, and Committee of Science and Technology in Shanghai Grants 12JC1409700 and 15ZR1446500. The authors declare that they have no conflicts of interest with the contents of this article.
- aaRS
- aminoacyl-tRNA synthetase
- CP1
- connective peptide 1
- LeuRS
- leucyl-tRNA synthetase
- hmtLeuRS
- human mitochondrial leucyl-tRNA synthetase
- Nva
- norvaline
- Val
- Valine
- PDB
- Protein Data Bank
- aa
- aminoacyl
- AN2690
- 5-fluoro-1,3-dihydro-1-hydroxy-2,1-benzoxaborole.
References
- 1. Ibba M., Söll D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 [DOI] [PubMed] [Google Scholar]
- 2. Ling J., Reynolds N., Ibba M. (2009) Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 63, 61–78 [DOI] [PubMed] [Google Scholar]
- 3. Zhou X., Wang E. (2013) Transfer RNA: a dancer between charging and mis-charging for protein biosynthesis. Sci. China Life Sci. 56, 921–932 [DOI] [PubMed] [Google Scholar]
- 4. Eriani G., Delarue M., Poch O., Gangloff J., Moras D. (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347, 203–206 [DOI] [PubMed] [Google Scholar]
- 5. Rould M. A., Perona J. J., Söll D., Steitz T. A. (1989) Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 Å resolution. Science 246, 1135–1142 [DOI] [PubMed] [Google Scholar]
- 6. Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. (1990) A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 Å. Nature 347, 249–255 [DOI] [PubMed] [Google Scholar]
- 7. Fersht A. R., Dingwall C. (1979) Evidence for the double-sieve editing mechanism in protein synthesis. Steric exclusion of isoleucine by valyl-tRNA synthetases. Biochemistry 18, 2627–2631 [DOI] [PubMed] [Google Scholar]
- 8. Zhou X. L., Fang Z. P., Ruan Z. R., Wang M., Liu R. J., Tan M., Anella F. M., Wang E. D. (2013) Aminoacylation and translational quality control strategy employed by leucyl-tRNA synthetase from a human pathogen with genetic code ambiguity. Nucleic Acids Res. 41, 9825–9838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen J. F., Guo N. N., Li T., Wang E. D., Wang Y. L. (2000) CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry 39, 6726–6731 [DOI] [PubMed] [Google Scholar]
- 10. Fang Z. P., Wang M., Ruan Z. R., Tan M., Liu R. J., Zhou M., Zhou X. L., Wang E. D. (2014) Coexistence of bacterial leucyl-tRNA synthetases with archaeal tRNA binding domains that distinguish tRNA(Leu) in the archaeal mode. Nucleic Acids Res. 42, 5109–5124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu B., Yao P., Tan M., Eriani G., Wang E. D. (2009) tRNA-independent pretransfer editing by class I leucyl-tRNA synthetase. J. Biol. Chem. 284, 3418–3424 [DOI] [PubMed] [Google Scholar]
- 12. Boniecki M. T., Vu M. T., Betha A. K., Martinis S. A. (2008) CP1-dependent partitioning of pretransfer and posttransfer editing in leucyl-tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 105, 19223–19228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tan M., Yan W., Liu R. J., Wang M., Chen X., Zhou X. L., Wang E. D. (2012) A naturally occurring nonapeptide functionally compensates for the CP1 domain of leucyl-tRNA synthetase to modulate aminoacylation activity. Biochem. J. 443, 477–484 [DOI] [PubMed] [Google Scholar]
- 14. Xu M. G., Li J., Du X., Wang E. D. (2004) Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination of amino acids and cell viability. Biochem. Biophys. Res. Commun. 318, 11–16 [DOI] [PubMed] [Google Scholar]
- 15. Lee K. W., Briggs J. M. (2004) Molecular modeling study of the editing active site of Escherichia coli leucyl-tRNA synthetase: two amino acid binding sites in the editing domain. Proteins 54, 693–704 [DOI] [PubMed] [Google Scholar]
- 16. Lincecum T. L. Jr., Tukalo M., Yaremchuk A., Mursinna R. S., Williams A. M., Sproat B. S., Van Den Eynde W., Link A., Van Calenbergh S., Grøtli M., Martinis S. A., Cusack S. (2003) Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol. Cell 11, 951–963 [DOI] [PubMed] [Google Scholar]
- 17. Li L., Boniecki M. T., Jaffe J. D., Imai B. S., Yau P. M., Luthey-Schulten Z. A., Martinis S. A. (2011) Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc. Natl. Acad. Sci. U.S.A. 108, 9378–9383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lue S. W., Kelley S. O. (2005) An aminoacyl-tRNA synthetase with a defunct editing site. Biochemistry 44, 3010–3016 [DOI] [PubMed] [Google Scholar]
- 19. Yao Y. N., Wang L., Wu X. F., Wang E. D. (2003) The processing of human mitochondrial leucyl-tRNA synthetase in the insect cells. FEBS Lett. 534, 139–142 [DOI] [PubMed] [Google Scholar]
- 20. Lue S. W., Kelley S. O. (2007) A single residue in leucyl-tRNA synthetase affecting amino acid specificity and tRNA aminoacylation. Biochemistry 46, 4466–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cvetesic N., Palencia A., Halasz I., Cusack S., Gruic-Sovulj I. (2014) The physiological target for LeuRS translational quality control is norvaline. EMBO J. 33, 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou X. L., Tan M., Wang M., Chen X., Wang E. D. (2010) Post-transfer editing by a eukaryotic leucyl-tRNA synthetase resistant to the broad-spectrum drug AN2690. Biochem. J. 430, 325–333 [DOI] [PubMed] [Google Scholar]
- 23. Rock F. L., Mao W., Yaremchuk A., Tukalo M., Crépin T., Zhou H., Zhang Y. K., Hernandez V., Akama T., Baker S. J., Plattner J. J., Shapiro L., Martinis S. A., Benkovic S. J., Cusack S., Alley M. R. (2007) An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 316, 1759–1761 [DOI] [PubMed] [Google Scholar]
- 24. Tan M., Zhu B., Zhou X. L., He R., Chen X., Eriani G., Wang E. D. (2010) tRNA-dependent pre-transfer editing by prokaryotic leucyl-tRNA synthetase. J. Biol. Chem. 285, 3235–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yao Y. N., Wang L., Wu X. F., Wang E. D. (2003) Human mitochondrial leucyl-tRNA synthetase with high activity produced from Escherichia coli. Protein Expr. Purif. 30, 112–116 [DOI] [PubMed] [Google Scholar]
- 26. Chen X., Ma J. J., Tan M., Yao P., Hu Q. H., Eriani G., Wang E. D. (2011) Modular pathways for editing non-cognate amino acids by human cytoplasmic leucyl-tRNA synthetase. Nucleic Acids Res. 39, 235–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yao P., Zhou X. L., He R., Xue M. Q., Zheng Y. G., Wang Y. F., Wang E. D. (2008) Unique residues crucial for optimal editing in yeast cytoplasmic leucyl-tRNA synthetase are revealed by using a novel knockout yeast strain. J. Biol. Chem. 283, 22591–22600 [DOI] [PubMed] [Google Scholar]
- 28. Englisch S., Englisch U., von der Haar F., Cramer F. (1986) The proofreading of hydroxy analogues of leucine and isoleucine by leucyl-tRNA synthetases from E. coli and yeast. Nucleic Acids Res. 14, 7529–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou X. L., Zhu B., Wang E. D. (2008) The CP2 domain of leucyl-tRNA synthetase is crucial for amino acid activation and post-transfer editing. J. Biol. Chem. 283, 36608–36616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hao R., Yao Y. N., Zheng Y. G., Xu M. G., Wang E. D. (2004) Reduction of mitochondrial tRNALeu(UUR) aminoacylation by some MELAS-associated mutations. FEBS Lett. 578, 135–139 [DOI] [PubMed] [Google Scholar]
- 31. Hao R., Zhao M. W., Hao Z. X., Yao Y. N., Wang E. D. (2005) A T-stem slip in human mitochondrial tRNALeu(CUN) governs its charging capacity. Nucleic Acids Res. 33, 3606–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fechter P., Rudinger J., Giegé R., Théobald-Dietrich A. (1998) Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: production and activity. FEBS Lett. 436, 99–103 [DOI] [PubMed] [Google Scholar]
- 33. Sohm B., Frugier M., Brulé H., Olszak K., Przykorska A., Florentz C. (2003) Towards understanding human mitochondrial leucine aminoacylation identity. J. Mol. Biol. 328, 995–1010 [DOI] [PubMed] [Google Scholar]
- 34. Yong L., Enduo W., Yinglai W. (1998) Overproduction and purification of Escherichia coli tRNALeu. Sci. China C Life Sci. 41, 225–231 [DOI] [PubMed] [Google Scholar]
- 35. Gruic-Sovulj I., Uter N., Bullock T., Perona J. J. (2005) tRNA-dependent aminoacyl-adenylate hydrolysis by a nonediting class I aminoacyl-tRNA synthetase. J. Biol. Chem. 280, 23978–23986 [DOI] [PubMed] [Google Scholar]
- 36. Fukunaga R., Yokoyama S. (2005) Crystal structure of leucyl-tRNA synthetase from the archaeon Pyrococcus horikoshii reveals a novel editing domain orientation. J. Mol. Biol. 346, 57–71 [DOI] [PubMed] [Google Scholar]
- 37. Loftfield R. B., Vanderjagt D. (1972) The frequency of errors in protein biosynthesis. Biochem. J. 128, 1353–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sarkar J., Poruri K., Boniecki M. T., McTavish K. K., Martinis S. A. (2012) Yeast mitochondrial leucyl-tRNA synthetase CP1 domain has functionally diverged to accommodate RNA splicing at expense of hydrolytic editing. J. Biol. Chem. 287, 14772–14781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. SternJohn J., Hati S., Siliciano P. G., Musier-Forsyth K. (2007) Restoring species-specific posttransfer editing activity to a synthetase with a defunct editing domain. Proc. Natl. Acad. Sci. U.S.A. 104, 2127–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zimorski V., Ku C., Martin W. F., Gould S. B. (2014) Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 22, 38–48 [DOI] [PubMed] [Google Scholar]
- 41. Fender A., Gaudry A., Jühling F., Sissler M., Florentz C. (2012) Adaptation of aminoacylation identity rules to mammalian mitochondria. Biochimie 94, 1090–1097 [DOI] [PubMed] [Google Scholar]
- 42. Palencia A., Crépin T., Vu M. T., Lincecum T. L. Jr., Martinis S. A., Cusack S. (2012) Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat. Struct. Mol. Biol. 19, 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tukalo M., Yaremchuk A., Fukunaga R., Yokoyama S., Cusack S. (2005) The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat. Struct. Mol. Biol. 12, 923–930 [DOI] [PubMed] [Google Scholar]
- 44. Zhou X. L., Du D. H., Tan M., Lei H. Y., Ruan L. L., Eriani G., Wang E. D. (2011) Role of tRNA amino acid-accepting end in aminoacylation and its quality control. Nucleic Acids Res. 39, 8857–8868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan M., Zhu B., Liu R. J., Chen X., Zhou X. L., Wang E. D. (2013) Interdomain communication modulates the tRNA-dependent pre-transfer editing of leucyl-tRNA synthetase. Biochem. J. 449, 123–131 [DOI] [PubMed] [Google Scholar]
- 46. Kisumi M., Sugiura M., Chibata I. (1976) Biosynthesis of norvaline, norleucine, and homoisoleucine in Serratia marcescens. J. Biochem. 80, 333–339 [DOI] [PubMed] [Google Scholar]
- 47. Soini J., Falschlehner C., Liedert C., Bernhardt J., Vuoristo J., Neubauer P. (2008) Norvaline is accumulated after a down-shift of oxygen in Escherichia coli W3110. Microb. Cell Fact. 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ross-Inta C., Tsai C. Y., Giulivi C. (2008) The mitochondrial pool of free amino acids reflects the composition of mitochondrial DNA-encoded proteins: indication of a post-translational quality control for protein synthesis. Biosci. Rep. 28, 239–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Roy H., Ling J., Alfonzo J., Ibba M. (2005) Loss of editing activity during the evolution of mitochondrial phenylalanyl-tRNA synthetase. J. Biol. Chem. 280, 38186–38192 [DOI] [PubMed] [Google Scholar]