Background: Diverse tristetraprolin family proteins exhibit sequence differences in their tandem zinc finger (TZF) RNA binding domains.
Results: Replacement of the S. pombe Zfs1 TZF domain with those from distant species fully complements the Zfs1 deletion phenotype.
Conclusion: Activities intrinsic to the TZF domain are interchangeable among distant eukaryotes.
Significance: Accumulated sequence changes in TZF domains during evolution still permit high-affinity RNA binding.
Keywords: mRNA decay, RNA binding protein, RNA turnover, yeast genetics, zinc finger
Abstract
Members of the tristetraprolin (TTP) family of proteins participate in the regulation of mRNA turnover after initially binding to AU-rich elements in target mRNAs. Related proteins from most groups of eukaryotes contain a conserved tandem zinc finger (TZF) domain consisting of two closely spaced, similar CCCH zinc fingers that form the primary RNA binding domain. There is considerable sequence variation within the TZF domains from different family members within a single organism and from different organisms, raising questions about sequence-specific effects on RNA binding and decay promotion. We hypothesized that TZF domains from evolutionarily distant species are functionally interchangeable. The single family member expressed in the fission yeast Schizosaccharomyces pombe, Zfs1, promotes the turnover of several dozen transcripts, some of which are involved in cell-cell interactions. Using knockin techniques, we replaced the TZF domain of S. pombe Zfs1 with the equivalent domains from human TTP and the single family member proteins expressed in the silkworm Bombyx mori, the pathogenic yeast Candida guilliermondii, and the plant Chromolaena odorata. We found that the TZF domains from these widely disparate species could completely substitute for the native S. pombe TZF domain, as determined by measurement of target transcript levels and the flocculation phenotype characteristic of Zfs1 deletion. Recombinant TZF domain peptides from several of these species bound to an AU-rich RNA oligonucleotide with comparably high affinity. We conclude that the TZF domains from TTP family members in these evolutionarily widely divergent species are functionally interchangeable in mRNA binding and decay.
Introduction
Tristetraprolin (TTP)2 is a well characterized CCCH tandem zinc finger (TZF) domain RNA binding protein that can promote the decay of target mRNAs after binding to AU-rich elements (AREs) found in their 3′UTRs (1–3). TTP family members contain two highly conserved CCCH zinc fingers of the CX8CX5CX3H type that are separated by an 18-amino acid linker. TTP family proteins from mammals, insects, and yeasts have been shown to bind to ARE-containing target mRNAs and promote their decay (1, 4–8). The preferred binding site for these proteins is thought to be UUAUUUAUU (9–11).
A structure of the TZF domain from the human TTP family member ZFP36L2 (TIS11d) bound to the 9-mer UUAUUUAUU revealed the nature of the interactions between the unstructured RNA binding site and key amino acid residues within the TZF domain (12). A synthetic peptide comprising the TZF domain of human TTP bound to this 9-mer sequence with a Kd of 3.2 nm at 24 °C, as shown by fluorescence anisotropy (11). Remarkably similar affinities have been demonstrated for the analogous TZF domains from TTP family members from a variety of other species, including the single family members expressed in Drosophila melanogaster (5), the fission yeast Schizosaccharomyces pombe (4, 7), and the human pathogenic yeast Candida albicans (6). In the experimental situations tested to date, mutation of any one of the cysteines or histidines that comprise the two zinc coordination centers of the TZF domain will abrogate RNA binding. However, the contributions of the other residues in the TZF domain are less well understood (2, 13–16). These could potentially alter the specificity and affinity of the RNA binding as well as the binding of potential protein cofactors, such as members of the AUF1 protein family, which can bind to TTP family members at their TZF domains and apparently increase their affinity for its RNA binding site (17).
TTP family members with characteristic TZF domains have been found in all major groups of eukaryotes, including plants and protists (2). This suggests that a common ancestor expressed one or more proteins containing this type of binding site more than 1.6 billion years ago, leaving considerable evolutionary time for sequence divergence to occur. Because of uncertainties regarding the roles of various residues within the TZF domains of different species, we addressed the hypothesis that TZF domains from evolutionarily distant species are functionally interchangeable. To do this, we analyzed RNA binding affinity using purified components and fluorescence anisotropy and performed in vivo functional analyses in S. pombe. In this species, loss of the single TTP family member Zfs1 results in the abnormal accumulation of several dozen transcripts, many of which have been shown to be bound directly to Zfs1 in coimmunoprecipitation analyses and which decayed more slowly in zfs1 deletion cells (4, 7). Many of these target transcripts encode proteins involved in cell-cell adhesion, and deletion of zfs1 caused a severe flocculation defect. The molecular phenotype characteristic of zfs1Δ mutants as well as the flocculation phenotype make this a convenient in vivo system for assaying the function of modified Zfs1 proteins.
We designed a method to replace the TZF domain of S. pombe Zfs1 at its normal chromosomal locus, allowing us to maintain endogenous levels of expression of proteins containing each domain replacement. We then assayed the mutant strains for expression of known Zfs1 target transcripts and measured the effects on calcium-induced flocculation. Finally, we attempted to replace endogenous S. pombe Zfs1 with the full-length proteins from a related member of the Schizosaccharomyces genus, Schizosaccharomyces octosporus, as well as the corresponding proteins from the more distantly related fungus Candida guilliermondii and full-length TTP from Mus musculus. We found that replacement of the S. pombe TZF domains by the analogous domains from a distantly related fungus, insect, mammal, or plant completely complemented the zfs1 null phenotype. However, with the exception of the protein from the closely related species S. octosporus, the full-length protein replacements did not fully complement the zfs1 deletion phenotype, suggesting that protein regions outside of the TZF domain are critical for full activity in this species.
Experimental Procedures
Yeast Strains and Media
All strains were made using standard yeast molecular genetics techniques. The deletion of zfs1 has been described previously (4). The wild-type control strain (referred to as Zfs1:FLAG, Sp TZF L1, and Sp Zfs1) was generated using the pFa6a-3×FLAG-6×Gly-KanMX vector (18). The open reading frame of zfs1 was cloned into the pFa6a-3×FLAG-6×Gly-KanMX vector using XmaI/PacI. The PacI site was removed using site-directed mutagenesis to allow the Zfs1:FLAG construct to be in-frame for protein expression. In addition, the alcohol dehydrogenase 1 (ADH) terminator in the pFa6a-Zfs1–3×FLAG-6×Gly-KanMX vector was replaced by subcloning 800 bases of the S. pombe genomic sequence (bases 3482848–3483648, GenBank accession no. NC_003423) into the ADH terminator site. TZF domain point mutants (C347R and C370R, using the residue numbering of GenBank accession no. NC_596453) were created using site-directed mutagenesis of the pFa6a-Zfs1–3×FLAG-6×Gly-KanMX vector. The TZF replacement strains were generated by cloning the TZF domain sequence from S. pombe Zfs1 (NP_596453), Homo sapiens TTP (NP_003398.2), Bombyx mori Tis11 (XP_00492873.1), the TTP family member protein from Chromolaena odorata (translation of Unigene mRNA sequence GACH01022939.1), and C. guilliermondii Zfs1 (XP_001482883), beginning at YKTE(L/P) to the final histidine in the second zinc finger, into the pFa6a-Zfs1–3×FLAG-6×Gly-KanMX vector. The mutation of Leu-341 (using the residue numbering in GenBank accession no. NP_596453) to Arg in each TZF replacement was created using site-directed mutagenesis of the pFa6a-Zfs1–3×FLAG-6×Gly-KanMX vector. Full-length S. octosporus Zfs1 (EPX71053), C. guilliermondii Zfs1 (XP_001482883), and M. musculus TTP (NP_035886.1) protein-coding regions were cloned into the pFa6a-Zfs1–3×FLAG-6×Gly-KanMX vector as described for S. pombe Zfs1. Primers were then used to amplify the cassette with sequences homologous to the endogenous zfs1 locus and transformed into strain PR110. Proper integration and expression were assessed by PCR, sequencing, and Western blot analysis. These genetic manipulations resulted in replacements of either the TZF domain or the full-length protein in the endogenous zfs1 locus in this haploid organism, leaving the promoter region, 3′UTR, and flanking regions intact.
NanoString Assays
NanoString assays for mRNA quantitation (19) were performed using 75 ng of total RNA isolated from four independent isolates each of the indicated strains using the RiboPure Yeast RNA Purification kit. Probe sets included 46 transcripts that were chosen on the basis of our previous mRNA sequencing analysis (4). The raw counts were adjusted for technical variability using positive and negative controls included in the code sets and then normalized to internal controls identified previously. The normalized RNA counts, standard deviation, and -fold change for each experiment are shown in supplemental Tables S1–S4.
Flocculation Assays
Flocculation in mid-log phase yeast was measured as described previously (4). Briefly, 5 ml of logarithmically growing cultures were centrifuged at 4500 × g for 5 min at 4 °C. Cells were then washed twice in 250 mm EDTA, once in 250 mm NaCl (pH 2), and, finally, in 250 mm NaCl (pH 4.5). Washed cells were resuspended to a final concentration of 1 × 108 cells/ml in 25 ml of 250 mm NaCl (pH 4.5) and placed into a 25-ml graduated cylinder. The cell suspension was adjusted to 4 mm CaCl2 with 100 mm CaCl2 (pH 4.5) and inverted 18 times. At defined intervals, 100 μl of cell suspension was removed from a fixed position in the graduated cylinder and diluted with 900 μl of water. Absorbance was then measured immediately at an optical density of 600 nm. All values were normalized to 100% cells in suspension at time 0.
Expression and Purification of Recombinant Zfs1 and TTP TZF Domains
The DNA encoding the TZF domain, beginning at LYKTEP, of Zfs1 from S. pombe was cloned using the SspI site of a modified pET28/30 vector (Novagen) that encodes six histidine residues preceding a maltose binding protein (MBP), followed immediately by a tobacco etch virus protease site at the N terminus (a gift from the laboratory of Dr. John Sondek, University of North Carolina at Chapel Hill). The TZF domain point mutant (C370R, using the residue numbering in GenBank accession no. NP_596453) was created using site-directed mutagenesis of the Zfs1-containing pET28/30 vector. The cDNAs encoding the TZF domain of TTP, beginning at RYKTEL, from H. sapiens and full-length Zfs1 from C. guilliermondii were cloned using the NotI/NheI sites of a modified pMAL-c5x vector (New England Biolabs) that encodes a maltose binding protein at the N terminus, as described previously (20). The recombinant fusion proteins were expressed in BL21 (DE3) cells after induction with 0.3 mm isopropyl 1-thio-β-d-galactopyranoside for 16 h at 20 °C. Cells were lysed by sonication and cleared by centrifugation for 35 min at 35,000 × g. The supernatants containing the TZF domain fusion peptides from S. pombe Zfs1 were incubated with nickel-nitrilotriacetic acid-agarose (Qiagen), and the supernatants containing the MBP fusions with the TZF domain of H. sapiens TTP and full-length Zfs1 from C. guilliermondii were incubated with amylose resin (New England Biolabs) for 1 h at 4 °C, washed 5 times, and then eluted with either 400 mm imidazole or 40 mm maltose, respectively. Fractions containing the S. pombe Zfs1 fusion peptides were incubated with tobacco etch virus protease overnight to cleave the affinity tag. The cleaved protein was subsequently applied to a Superdex 200 size exclusion column (GE Healthcare), followed by further purification on a HiTrap Q (GE Healthcare) anion exchange column. Fractions containing the purified S. pombe Zfs1 TZF L1 peptide, S. pombe Zfs1 TZF L1R peptide, S. pombe Zfs1 C370R peptide, C. guilliermondii Zfs1:MBP fusion protein, or H. sapiens TTP TZF:MBP fusion peptide were pooled and concentrated using a 30,000 Vivaspin 20 MWCO 30 000 centrifugal filtering device (GE Healthcare) and stored at −80 °C after flash-freezing in liquid nitrogen.
Determination of RNA Binding Affinity by Fluorescence Anisotropy
Fluorescence anisotropy experiments were carried out using 5′ fluorescein-labeled RNA probes as described previously (6). These probes consisted of a 13-base fluorescein-labeled RNA target substrate (5′-FL-UUUUAUUUAUUUU-3′, FL-ARE13) or a control polyU probe (5′-FL-UUUUUUUUUUUUU-3′) (11). Total measured anisotropy was measured over a range of purified S. pombe Zfs1 TZF L1 peptide, mutant S. pombe Zfs1 TZF L1R peptide, mutant S. pombe Zfs1 C370R peptide, C. guilliermondii Zfs1:MBP fusion protein, and H. sapiens TTP TZF:MBP fusion peptide concentrations. Binding constants were calculated by the nonlinear regression algorithm in PRISM, version 6.0 (GraphPad Software) using Equation 1 in Ref. 6.
Western Blot Analysis
Total protein lysates were prepared by tricholoracetic acid extraction. Briefly, cells were grown to A600 = 0.8, and 1.5 ml of culture was pelleted by centrifugation at 15,700 × g for 3 min at room temperature. Pellets were resuspended in 200 μl of 20% tricholoracetic acid. 100 μl of 0.7-mm, acid-washed glass beads was added, and cells were lysed by vortexing for 10 min. Proteins were precipitated on ice for 5 min and centrifuged at 15,700 × g for 10 min at 4 °C. Pellets were resuspended in 2× SDS-PAGE sample buffer (2.5% (w/v) SDS, 25% (v/v) glycerol, 125 mm Tris-Cl (pH 6.8), 0.01% (w/v) bromphenol blue, and 100 mm fresh 2-mercaptoethanol) and boiled for 5 min. Blots were probed with anti-FLAG (Sigma, catalog no. A8592) or anti-actin (Abcam, catalog no. 8224).
Solution Structure Modeling of the RNA-bound TZF Domains
The RNA-bound structure of the TZF domain of ZFP36L2 (TIS11d, PDB code 1RGO) (12) was used to construct the initial models of RNA-bound TZF domains from human TTP and from the Zfs1 proteins from S. pombe and C. guilliermondii. As a reference, the RNA-bound ZFP36L2 from PDB code 1RGO was also subjected to a similar molecular dynamics simulation as the other peptides of interest. Mutations were introduced into the sequence of ZFP36L2 using Coot (21) to generate starting models for each peptide in its RNA-bound form, with both CCCH coordinated Zn2+ ions present in their appropriate positions. Initial models were energy-minimized using the program, version 14 (22). Each of the RNA-peptide complexes was solvated in a box of water (H. sapiens, 9399 water molecules; ZFP36L2, 9764 water molecules; S. pombe, 10,666 water molecules; C. guilliermondii, 10,230 water molecules). Appropriate amounts of count ions were added to neutralize the central water box. Prior to equilibration, all systems were subjected to 100-ps belly dynamics runs with a fixed peptide; minimization; low-temperature, constant pressure dynamics at a fixed protein concentration to assure a reasonable starting density; minimization; stepwise heating molecular dynamics at a constant volume; and constant-volume molecular dynamics for 1 ns. All final unconstrained trajectories were calculated at 300 K under a constant volume (25 ns; time step, 1 fs) using the Particle Mesh Ewald Molecular Dynamics module of Amber.14 (22) to accommodate long-range interactions. The parameters were taken from the FF14SB force field for both the protein and the RNA. The charge on zinc ions was +2, whereas cysteine coordinated with zinc carried a −1 charge. Histidine coordinated to zinc remained neutral. No specific constraints were applied to maintain the zinc coordination. Images of molecular models were created using Chimera (23).
Results
Transcript Levels in zfs1Δ TZF Domain Mutant Strains
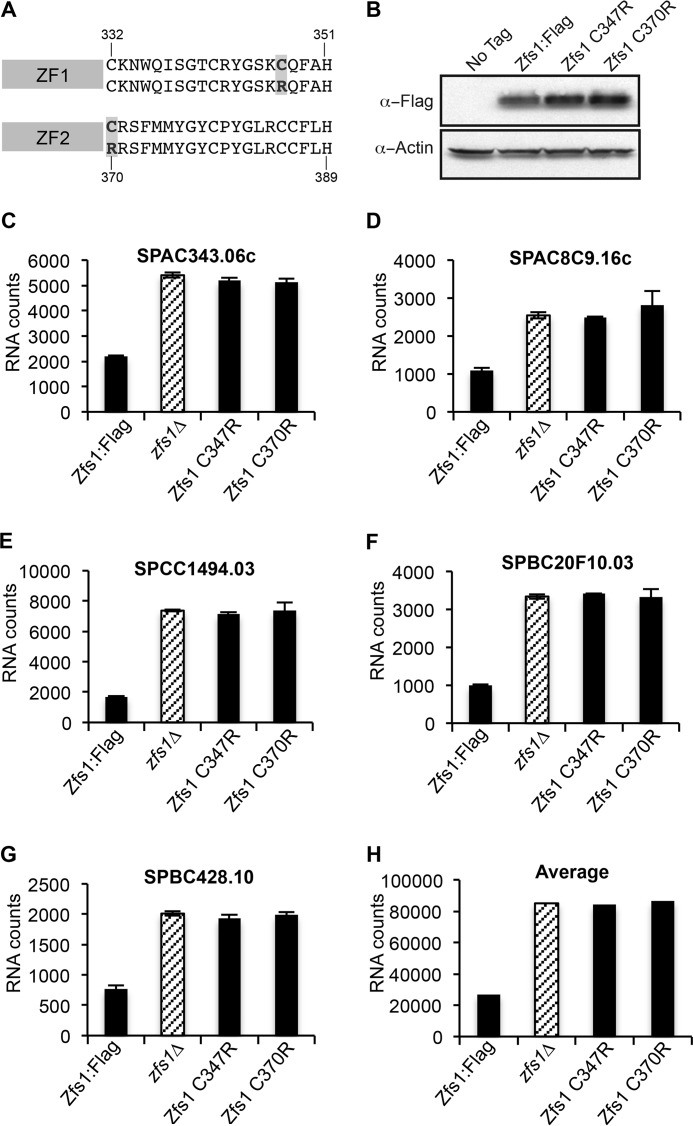
Our previous study used deep sequencing to identify 185 transcripts that were increased at least 1.5-fold in zfs1Δ mutants (4). Of those, 119 transcripts contained at least one potential TTP family member optimal binding site 7-mer (UAUUUAU) in their 3′UTR. Several of these transcripts were identified as direct Zfs1 targets by RNA coimmunoprecipitation studies and mRNA decay measurements. A subset of these transcripts encode proteins involved in cell-cell adhesion, including the transcription factor Cbf12. Our previous studies demonstrated that increases in several potential Zfs1 target transcripts were likely to be due to Cbf12 overexpression (4). To confirm that an intact TZF domain is required for the activity of Zfs1 in S. pombe, we developed a NanoString code set to analyze the top 28 transcripts identified by mRNA sequencing that were up-regulated in zfs1Δ mutants compared with the wild type. This code set included transcripts that were both dependent and independent of Cbf12 and the Cbf12 mRNA itself. Previous studies have shown that mutations of any of the CCCH residues within the TZF domain of human TTP result in loss of binding (13). Therefore, we mutated cysteine 347 in the first zinc finger (ZF1) and cysteine 370 in the second finger (ZF2) to arginine (Fig. 1A) and performed NanoString analyses. Western blot analysis, followed by densitometric quantitation, revealed a slight (∼1.6-fold) increase in Zfs1 expression in the Zfs1 C347R and Zfs1 C370R mutation strains compared with the WT (Fig. 1B). Focusing on the top five Cbf12-independent, ARE-containing, up-regulated target transcripts, we found that the C347R or C370R mutations had the same effect on target mRNA levels as the zfs1Δ mutation, indicating that these residues are required for the binding and eventual turnover of target transcripts (Fig. 1, C–G). The normalized RNA counts and -fold changes for all 28 transcripts are shown in supplemental Table S1). When we averaged the number of counts of transcripts that were increased at least 2-fold, which included 14 of the 28 transcripts used in the assay, in the zfs1Δ mutants, we found that the C347R and C370R mutant strains behaved almost identically to the zfs1Δ mutation (Fig. 1H).
FIGURE 1.
Analysis of target transcripts in strains expressing Zfs1 TZF domain point mutations. A, the sequence of the first (ZF1) and second (ZF2) zinc finger from S. pombe Zfs1. The residues highlighted in gray indicate the mutations within each zinc finger. B, Western blot analysis of whole-cell lysates isolated from the indicated strains and blotted using anti-FLAG or anti-actin antisera. C—G, data from five of the 28 transcripts that were analyzed by NanoString in the WT, zfs1Δ, Zfs1 C347R, and Zfs1 C370R strains. Diagonally striped columns represent the zfs1Δ deletion strain. Shown are the means ± S.D. from at least three independent isolates. The y axis represent normalized RNA counts for the indicated transcript. H, the averages for all transcripts increased 2-fold or more in the zfs1Δ strain for the indicated strains. These averages include 14 of the 28 transcripts from the NanoString analysis.
TZF Domain Complementation Strains
Certain amino acids within the RNA binding TZF domain are highly conserved in TTP family members from plants and yeasts to humans, but many are different (2, 13). For the this study, we compared the TZF domain sequences from S. pombe Zfs1 (Ref. 24 and NP_596453), human TTP (Ref. 25 and NP_003398.2), B. mori Tis11 (Ref. 5 and XP_00492873.1), the TTP family member protein from C. odorata (Ref. 2 and translation of the Unigene mRNA sequence GACH01022939.1), and the yeast CTG clade family member C. guilliermondii Zfs1 (Ref. 6 and XP_001482883). The B. mori and C. guilliermondii proteins were chosen for this analysis because they are the smallest TZF domain-containing proteins we found to date among insects and fungi, respectively. The C. odorata protein was chosen because it contains, in addition to the TZF domain, a C-terminal putative NOT1 binding domain characteristic of the mammalian and insect TTP family members (2). An alignment of the TZF domains from these species shows the perfect conservation of the cysteines and histidines in the two CCCH zinc fingers. These were separated by 18 amino acids in all cases, with the conserved intra-finger spacing of CX8CX5CX3H (Fig. 2A, yellow). In addition, the alignment shows the relative conservation of the lead-in sequence (L/R)YKTE(P/L) preceding the CCCH motifs (Fig. 2A). However, there are many differences among the proteins at other sites within the TZF domains, some of which might be expected to affect binding activity.
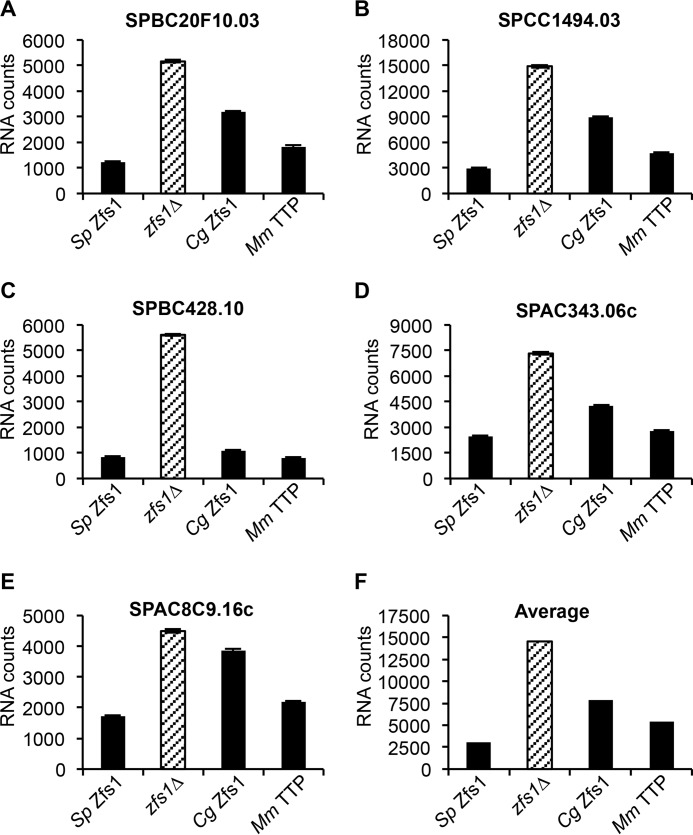
FIGURE 2.
Design and expression of Zfs1 TZF domain complementation constructs. A, alignment of TZF domains from the indicated species. The numbering corresponds to the position within the TZF domain. Sequences were aligned by ClustalW2, and colors were assigned by ClustalW2 on the basis of their physicochemical properties. The conserved CCCH is highlighted in yellow. Asterisks indicate the amino acid identity at that site, colons indicate a conserved substitution, and dots indicate a semiconserved substitution at that site. B, schematic of the construct used to substitute the TZF domains from various species into the endogenous S. pombe locus. In addition, a 3× FLAG tag was integrated into the endogenous locus of S. pombe Zfs1. Bm, B. mori; Cg, C. guilliermondii; Co, C. odorata; Hs, H. sapiens; Sp, S. pombe). C, Western blot analysis of whole-cell lysates isolated from the indicated strains and blotted using anti-FLAG or anti-actin antisera. TZF L1 and TZF L1R indicate the amino acid residue at the beginning of the highly conserved TZF domain lead in the sequence (L/R)YKTE(P/L), where position 1 is the first amino acid in the TZF domain. D, flocculation analysis of TZF domain replacement strains. Flocculation of the indicated strains was initiated by the addition of CaCl2 and determined using the Helm assay (13) as described previously (4). The percentages of cells in suspension were measured on the basis of optical density at 600 nm. Shown for each strain are the means of values from three independent experiments.
To address the hypothesis that TZF domains from these evolutionarily distant species are functionally interchangeable, we used knockin techniques to replace the TZF domain in S. pombe with the analogous domains from human TTP, silkworm Tis11, the TTP family member protein from C. odorata, and C. guilliermondii Zfs1 (Fig. 2B). The cognate domains, starting at the highly conserved (L/R)YKTE(P/L) sequence, were cloned into the endogenous locus of S. pombe zfs1 along with a C-terminal 3× FLAG epitope (Fig. 2, A and B). We also used similar techniques to knock in the same epitope tag into the S. pombe zfs1 locus. The endogenous zfs1 promoter and 3′UTR elements remained in their original genetic sites to ensure normal expression and regulation of this transcript. In addition, we mutated L1 from the S. pombe LYKTEP first zinc finger lead-in sequence to an arginine (Arg-1), which is the more common initial residue in proteins from higher eukaryotes (2). Strains expressing these mutant proteins were grown to mid-log phase, and protein extracts were prepared. Western blotting and densitometry measurements demonstrated essentially equal expression of the mutant proteins in all strains containing the TZF domain replacements (Fig. 2C). No apparent differences in growth rates were observed among the replacement strains.
Flocculation of Complementation Strains
Previous studies have shown that the loss of zfs1 in S. pombe causes an increase in transcripts and, presumably, proteins involved in cell-cell adhesion, resulting in increased flocculation in response to added calcium (4). To determine whether the TZF domain substitutions could complement the S. pombe TZF domain in Zfs1, we analyzed sedimentation rates in the substitution strains in response to calcium. We found that, upon the addition of calcium, the strains with the substituted TZF domains did not flocculate and remained in suspension, similar to a wild-type strain (Fig. 2D). We also found that the Leu-to-Arg mutations in all of the TZF domains did not affect the ability of these substituted proteins to prevent flocculation (Fig. 2D).
Target Transcript Levels in Complementation Strains
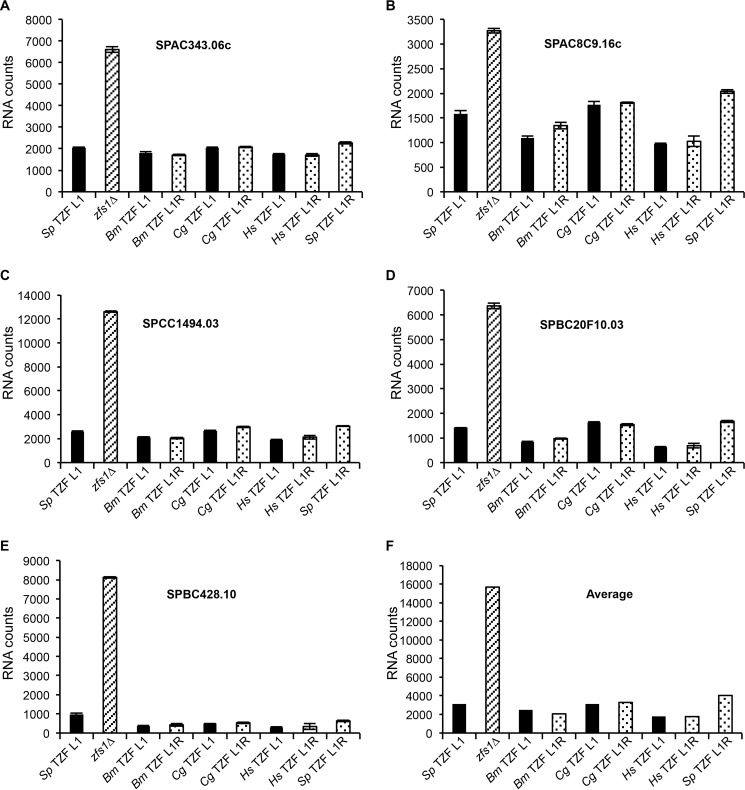
To determine whether the TZF domains from the human, insect, and other fungal proteins could complement the TZF domain of S. pombe Zfs1 in its function as an mRNA-destabilizing protein, we quantitated the levels of expression of the previously identified 28 Zfs1 target transcripts. When we focused on the five most up-regulated transcripts, we found that the TZF domain substitutions from the indicated species functioned almost exactly like the wild-type protein (Fig. 3, A–E, and supplemental Table S2). In addition, the Arg-1 mutation in the constructs had no effect on the levels of the target transcripts, compared with results with the Leu-1 constructs (Fig. 3, A–E, and supplemental Table S2). As an index of the overall effect of these complementations, we averaged the concentrations of all transcripts that were increased at least 2-fold, which included 17 of the 28 transcripts, in the zfs1Δ mutants, and found that all of the TZF domain-substituted proteins, including the Leu-to-Arg mutants, functioned almost identically to the wild-type S. pombe protein, indicating that the TZF domain is functionally interchangeable among these species (Fig. 3F), at least at the level of target transcript accumulation.
FIGURE 3.
Expression of Zfs1 target transcripts in the TZF domain complementation strains. Shown is a NanoString analysis of target transcripts in the S. pombe TZF domain Leu-1, zfs1Δ, and the complementation strains in which the S. pombe TZF domain was replaced with the indicated TZF domain. L1/L1R indicates the amino acid residue at the beginning of the TZF domain lead-in sequence (L/R)YKTE(P/L). Diagonally striped columns represent zfs1Δ, and dotted columns represent the L1R mutants. The y axes represent normalized RNA counts for the indicated transcript. Bm, B. mori; Cg, C. guilliermondii; Hs, H. sapiens; Sp, S. pombe. A—E, five of the 28 transcripts that were analyzed by NanoString. The y axes indicate normalized counts for the indicated transcripts, which are shown as the mean ± S.D. from at least four independent isolates. F, averages for all transcripts increased 2-fold or more in the zfs1Δ strain for the indicated substitution strains. These averages include 17 of the 28 transcripts from the NanoString analysis.
Replacement of the S. pombe TZF Domain with the TZF Domain from a Plant, C. odorata
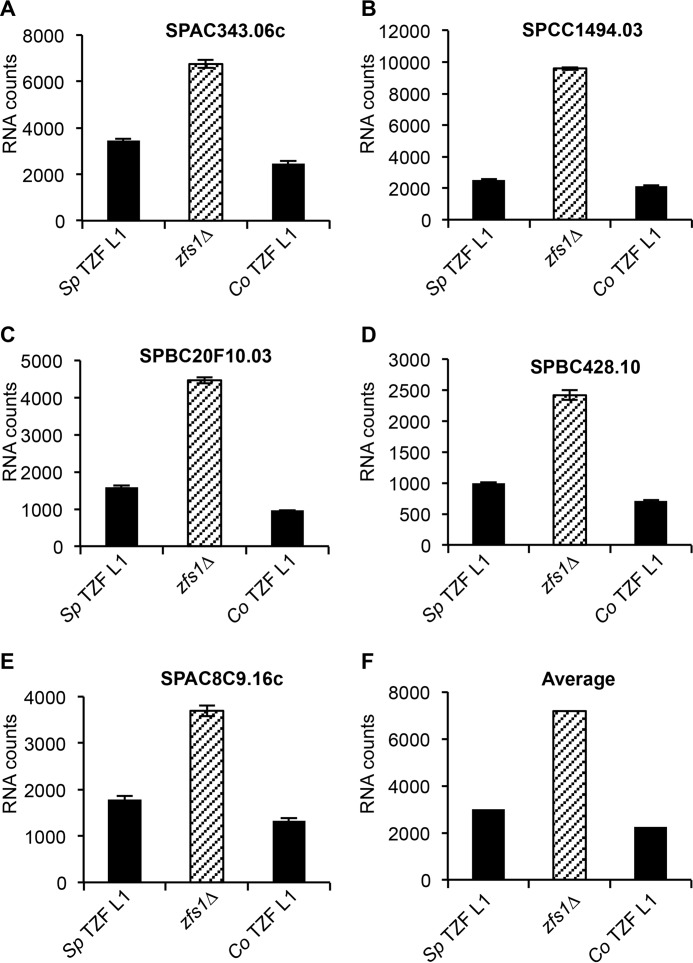
To examine a TTP family member protein more distantly related to S. pombe, we attempted similar complementation analyses with the TZF domain from a plant, C. odorata, which expresses a protein that contains both the TZF domain and a predicted C-terminal NOT1 binding domain (Fig. 2A) (2). As mentioned above, this complementation strain behaved like a wild-type strain in its resistance to calcium-induced flocculation (Fig. 2D). As with the other TZF domains, the substitution of the TZF domain from C. odorata into S. pombe Zfs1 completely normalized expression of the top five mRNA targets (Fig. 4, A–E, and supplemental Table S3). We also found that the TZF domain from C. odorata could functionally complement the TZF domain in S. pombe Zfs1, as indicated by the average of the transcripts that were increased 2-fold, which included 14 of the 28 transcripts, in the zfs1Δ mutants (Fig. 4F).
FIGURE 4.
Expression of Zfs1 target transcripts in the plant TZF domain substitution strain. Shown are the results of the NanoString analysis of target transcripts in the S. pombe TZF Leu-1, zfs1Δ, and C. odorata TZF L1 strains as described in the legend for Fig. 3. Sp TZF L1 indicates the wild-type S. pombe TZF domain sequence, and Co TZF L1 indicates the wild-type TZF domain sequence from C. odorata. Diagonally striped columns represent zfs1Δ. The y axes represent normalized RNA counts for the indicated transcript. A—E, shown are five of the 28 transcripts that were analyzed by NanoString. Shown are the means ± S.D. from at least four independent isolates. F, averages for all transcripts increased 2-fold or more in the zfs1Δ strain for the indicated strains. These averages include 14 of the 28 transcripts from the NanoString analysis.
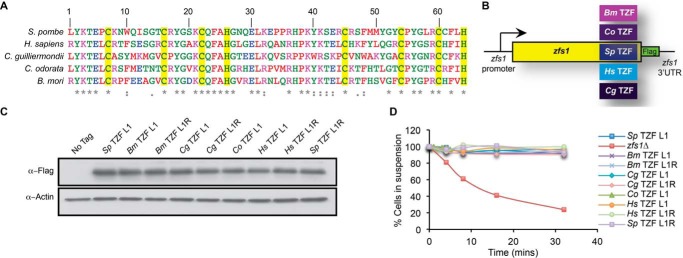
Binding of TZF Domains to an AU-rich Element RNA Oligonucleotide
Previous studies have shown high-affinity binding of a synthetic peptide containing the TZF domain from human TTP and full-length C. albicans Zfs1 to RNA targets with the optimal TTP binding site (UUAUUUAUU) (6, 11). To compare the RNA-binding characteristics of the TZF domain from the S. pombe protein Zfs1 with those of the human peptide, we expressed constructs encoding the TZF domain of Zfs1 from S. pombe fused to MBP:His in E. coli, purified the peptide, cleaved the MBP:His tag using tobacco etch virus protease, and performed binding measurements to a 13-base fluorescein-labeled RNA target (5′-FL-UUUUAUUUAUUUU-3′, FL-ARE13) using fluorescence anisotropy (11). We also tested a version of the S. pombe TZF domain peptide that contained the Leu-1 to Arg-1 mutation. Under these conditions, the wild-type Leu-1 S. pombe peptide exhibited a Kd of 1.2 + 0.1 nm at 24 °C (Fig. 5A), whereas there was no detectable binding of the S. pombe peptide to the labeled polyU oligonucleotide under these conditions (Fig. 5A). In addition, we determined whether the Sp Zfs1 C370R mutant peptide could bind to a probe containing the optimal TTP family member binding site. We found no detectable binding of the Sp Zfs1 C370R mutant to the probe (Fig. 5A). Using the same techniques, the Leu-1 to Arg-1 mutant S. pombe protein exhibited a Kd of 1.6 + 0.1 nm at 24 °C (Fig. 5B). We performed similar analyses with the TZF domain of human TTP fused to MBP and the MBP-fused, full-length Zfs1 protein from C. guilliermondii. The human fusion peptide bound to the same probe with a Kd of 0.7 + 0.07 nm under these conditions (Fig. 5B), whereas the C. guilliermondii fusion protein bound with a Kd of 0.9 + 0.1 nm (Fig. 5B). These values were very similar as those found with the TZF domain of C. albicans Zfs1 (Kd of 1.8 nm at 24 °C) (6).
FIGURE 5.
Measurement of Zfs1 TZF domain binding to RNA using fluorescence anisotropy. A, binding reactions contained the 13-base fluorescein-labeled RNA target (5′-FL-UUUUAUUUAUUUU-3′) or a polyU control (5′-FL-UUUUUUUUUUUUU-3′). Various concentrations of recombinantly expressed, purified peptide containing the TZF domain of Zfs1 from S. pombe (Sp Zfs1 TZF) or the purified peptide containing the Sp Zfs1 C370R mutant were added to the indicated probe, and fluorescence intensity was monitored. A nonlinear regression algorithm in PRISM was used to calculate TZF domain-dependent changes in anisotropy. B, binding reactions were performed as described in A using various concentrations of the analogous peptide containing the Sp Zfs1 TZF L1R mutant as well as fusion peptides containing the H. sapiens TTP TZF R1:MBP fusion peptide and the full-length C. guilliermondii Zfs1 L1:MBP fusion protein. A nonlinear regression algorithm in PRISM was used to calculate protein-dependent changes in anisotropy.
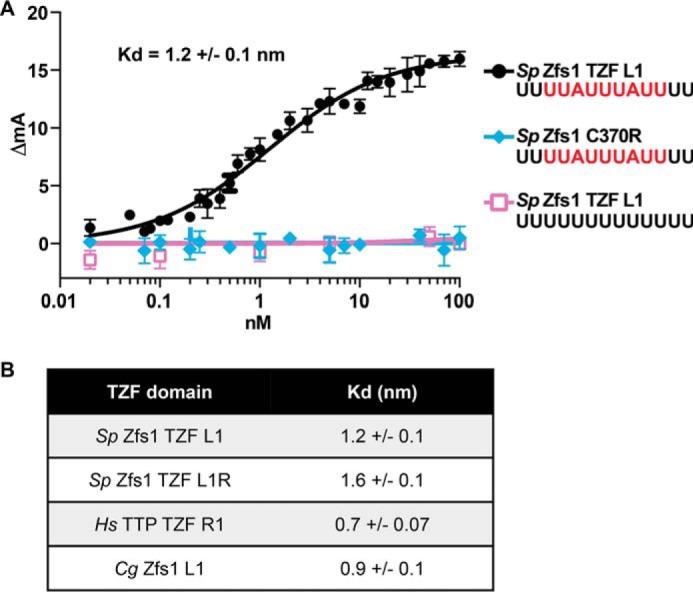
Replacement of S. pombe Zfs1 with Full-length TTP Family Member Proteins
We next asked whether similar functional complementation could be achieved with the full-length proteins. We cloned open reading frames for the complete proteins from a closely related member of the Schizosaccharomyces genus, S. octosporus, the CTG clade member C. guilliermondii, and TTP from M. musculus, into the endogenous zfs1 locus in S. pombe (Fig. 6A). Full-length S. octosporus Zfs1 is ∼51% identical to S. pombe Zfs1 at the level of amino acid sequence (4). Again, we preserved the endogenous S. pombe zfs1 promoter and 3′UTR in each construct, in an attempt to replicate endogenous levels of expression (Fig. 6A). As before, these intact protein replacements did not exhibit any obvious growth abnormalities. Western blotting showed that both C. guilliermondii and S. octosporus Zfs1 proteins were expressed at similar levels as S. pombe Zfs1 (Fig. 6B). However, the TTP protein from M. musculus was considerably more highly expressed compared with S. pombe Zfs1 (Fig. 6B). When we examined transcript levels for each construct, we found that C. guilliermondii zfs1 and S. octosporus zfs1 mRNAs were expressed at levels similar to the S. pombe zfs1 transcript, whereas the M. musculus TTP transcript increased ∼10-fold over S. pombe zfs1 mRNA levels (data not shown).
FIGURE 6.
Construction and expression of full-length Zfs1 replacement strains. A, schematic of the construct used to substitute the full-length ORFs from the TTP family member proteins from the indicated species into the endogenous S. pombe locus. In addition, a 3× FLAG tag was integrated into the endogenous locus of the S. pombe Zfs1 protein in-frame at with the C terminus. Cg, C. guilliermondii; Mm, M. musculus; So, S. octosporus; Sp, S. pombe. B, Western blot analyses of whole-cell lysates isolated from the indicated strains and blotted using anti-FLAG or anti-actin antibodies. The lysate from the M. musculus TTP strain was diluted 1:10 to obtain comparable expression levels to the proteins from the other species. C, flocculation analysis of full-length replacement strains. Flocculation of the indicated strains was performed as described in the legend for Fig. 2. Shown are the means of values from three independent experiments.
Flocculation of Full-length Complementation Strains
We next examined calcium-induced flocculation of the strains with the full-length Zfs1 replacements. The protein from the related species S. octosporus fully complemented S. pombe zfs1 in the flocculation analysis (Fig. 6C). However, the replacement of S. pombe Zfs1 with full-length M. musculus TTP or C. guilliermondii Zfs1 resulted in intermediate flocculation phenotypes compared with the wild-type strains (Fig. 6C).
Target Transcript Levels in the Full-length Complementation Strains
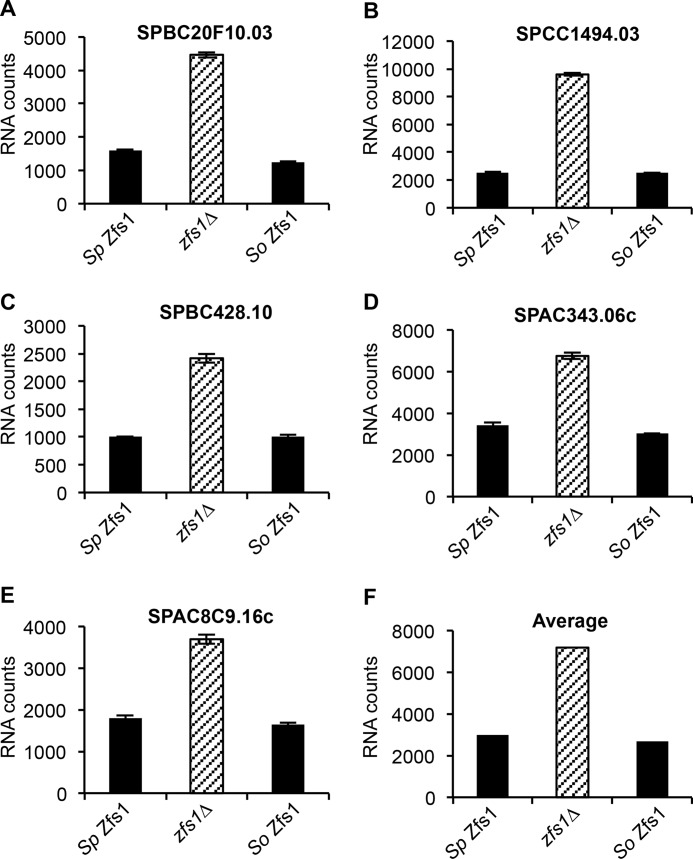
Finally, we examined expression of the 28 Zfs1 targets discussed above in the full-length complementation strains using NanoString analysis of total RNA. In the case of the S. octosporus Zfs1, we found that this protein affected target transcript levels almost identically to S. pombe Zfs1 when we focused on the top five up-regulated ARE-containing transcripts (Fig. 7, A–E, and supplemental Table S3). We also found that S. octosporus Zfs1 behaved like S. pombe Zfs1 when we averaged the levels of all transcripts that were increased at least 2-fold, which included 14 of the 28 transcripts, in the zfs1Δ mutants (Fig. 7F). However, replacement of S. pombe Zfs1 with the full-length TTP family member from another fungal organism, C. guilliermondii, or from mouse TTP did not fully complement S. pombe Zfs1 in regulating the top five targets (Fig. 8, A–E, and supplemental Table S4). The averages of all transcripts that were increased at least 2-fold, which included 18 of the 28 transcripts, in the zfs1Δ mutants showed incomplete complementation in both cases (Fig. 8F). However, it was noteworthy that neither full-length complementation strain exhibited levels of target transcripts similar to those seen in the zfs1Δ mutants, suggesting that at least some degree of functional complementation had taken place.
FIGURE 7.
Expression of Zfs1 target transcripts in the Sp Zfs1 replacement with So Zfs1. Shown are the NanoString analyses of target transcripts in strains containing S. pombe Zfs1, zfs1Δ, and S. octosporus Zfs1, in which the entire Zfs1 ORF from S. pombe was replaced with the corresponding ORF from S. octosporus (So Zfs1). Diagonally striped columns represent zfs1Δ. The y axes represent normalized RNA counts for the indicated transcript. A–E, five of the 28 transcripts that were analyzed by NanoString. Shown for the indicated transcripts are the means ± S.D. from at least four independent isolates. F, shown are the averages for all transcripts increased 2-fold or more in the zfs1Δ strain for the indicated strains. These averages include 14 of the 28 transcripts from the NanoString analysis.
FIGURE 8.
Expression of Zfs1 target transcripts in the Sp Zfs1 full-length replacement strains. Shown are the NanoString analyses of target transcripts in S. pombe Zfs1, zfs1Δ, and the full-length replacement strains, in which the ORF of S. pombe Zfs1 was replaced with the indicated ORFs. Diagonally striped columns represent zfs1Δ. The y axes represent normalized RNA counts for the indicated transcript. Cg, C. guilliermondii; Mm, M. musculus; Sp, S. pombe. A–E, five of the 28 transcripts that were analyzed by NanoString. Shown for the indicated transcripts are the means ± S.D. from at least four independent isolates. F, averages for all transcripts increased 2-fold or more in the zfs1Δ strain for the indicated strains. These averages include 18 of the 28 transcripts from the NanoString analysis.
Discussion
In this study, we tested the hypothesis that the putative RNA binding domains from TTP family members from evolutionarily distant species are functionally interchangeable. Using the Time-Tree calculator, the estimated date of a common ancestor of H. sapiens and the plant C. odorata is greater than 1.6 billion years, suggesting that the amino acid composition of this ancient protein domain has been subjected to a very long period of evolutionary pressure.
To test this hypothesis, we replaced the endogenous TZF domain of S. pombe Zfs1 with TZF domains from several evolutionarily distant species, including a mammal, a plant, an insect, and a distantly related fungus. This technique allowed the expression of the tagged chimeric proteins under the control of the endogenous zfs1 promoter, 3′UTR, and flanking region. We confirmed normal levels of expression of the chimeric Zfs1 proteins in these TZF domain complementation strains. By measuring the expression levels of known Zfs1 targets in this species and examining the calcium-induced flocculation phenotype characteristic of the zfs1Δ mutant, we found that the TZF domains from these evolutionarily very diverse species were functionally interchangeable in S. pombe. We also tested several of these peptides in quantitative RNA binding assays and found that they bound to a typical TTP family member binding site RNA oligonucleotide with similar low-nanomolar affinities. Therefore, despite the changes in amino acid sequence that have accumulated over millennia in these species, these TZF domains appear to behave essentially identically to the native S. pombe sequence in RNA binding and in promoting target mRNA decay in this species.
Some of the sequence changes observed in these comparisons are surprising. For example, in a mutational analysis of the human TTP TZF domain, the tyrosine residue at position 49 in the human TZF domain (Fig. 2A) has been shown to play an important role in the positioning of neighboring residues that interact with RNA (13). Mutation of this residue to an alanine resulted in the inability of the human TZF domain to bind RNA and promote its deadenylation. The previous model suggested that this mutation would create a slight overwinding of the helix, resulting in a shift of His-46 from its normal location and apparent interference with RNA binding.
However, as shown in Fig. 2A, there is a naturally occurring alanine at that site in the C. guilliermondii protein and a methionine at that site in the S. pombe protein. Nonetheless, the binding of the C. guilliermondii and S. pombe peptides to the AU-rich RNA oligonucleotide occurred with a similar high affinity to the other peptides, and their TZF domains were able to complement the zfs1 deficiency phenotype perfectly well. To investigate this paradox, we generated and compared simulation solution models of the various TZF domains on the basis of the original ZFP36L2 NMR structure (12) (Fig. 9, A–C). Analysis of this region of the human TTP TZF domain (Fig. 9A) suggests that the hydrophobic residues Leu-44, Phe-48, and Ile-63 (not shown) cluster to allow the zinc-coordinating His-64 to be in close proximity to the zinc ion. Tyr-49 interacts with Phe-48 to make a strong hydrogen bond with the backbone carbonyl oxygen of Ile-63 (Fig. 9A). In S. pombe Zfs1, the corresponding residue to human TTP Tyr-49 is Met-49 (Fig. 9B). Despite the difference in hydrophobicity between these two residues, it appears that several neighboring residues, including Arg-44 and Leu-63 (not shown), can compensate for the Tyr-to-Met change to facilitate zinc coordination and, ultimately, RNA binding (Fig. 9B). In this case, Met-49 is in contact with the hydrophobic part of the Arg-44 side chain. This interaction may contribute to the formation of a hydrophobic patch, which allows the correct folding for Zn2+ binding. A similar phenomenon was observed in the second zinc finger of C. guilliermondii Zfs1 (Fig. 9C). Trp-48, which corresponds to Phe-48 in human TTP, makes a strong hydrogen bond with His-64, allowing coordination of the Zn2+ ion. It is possible that this hydrogen binding compensates for the lack of the hydrophobic patch observed in C. guilliermondii.
FIGURE 9.
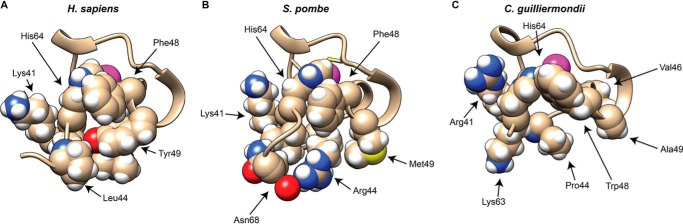
Comparison of portions of the second zinc fingers from H. sapiens TTP, S. pombe Zfs1, and C. guilliermondii Zfs1. Portions of the solution structure models are shown from H. sapiens TTP (A), S. pombe Zfs1 (B), and C. guilliermondii Zfs1 (C) TZF domains. Shown are the backbone structures of a portion of the second zinc finger from the TZF domains, including selected side chains. The indicated residues are thought to form hydrophobic patches that help fold the terminal histidine into position to facilitate both zinc and RNA binding. The magenta spheres represent the Zn2+ ions. The peptide backbone ribbon and side chain carbons are shown in a wheat color, and the atoms of the side chain residues are represented by colored spheres: white, hydrogen; red, oxygen; blue, nitrogen; yellow, sulfur. See the text for further details.
Our sample size of organisms tested is too small to conclude that putative TZF domains from other proteins and species that have identical spacing and certain invariant residues will work as well as the ones tested here. However, we suspect that will be the case. It might be interesting to use this experimental system to analyze “TZF domain-like” sequences that differ in key residues or in internal spacing. For example, in Caenorhabditis elegans, there is one typical TTP family member protein with a well conserved TZF domain and a C-terminal potential NOT1 binding site (CCCH-1, GenBank accession no. NP_505926). However, this species also expresses ∼15 other proteins with somewhat related tandem zinc finger domains, differing from CCCH-1 at many internal amino acid residues as well as internal spacing within the TZF domains. Similarly, species within the most recently evolved subdivision of fungi, the Pezizomycotina, have generally “lost” the typical TZF domains present in most other groups of fungi, apparently because of changes in internal spacing within the presumed TZF domains (2). In these and other cases, it may be useful to use the S. pombe complementation assay to determine the potential RNA binding specificities of these and other modified domains.
Somewhat less clear were the results of our attempts to complement the zfs1Δ mutant phenotype with full-length proteins from other species. In the case of another member of the Schizosaccharomyces genus, S. octosporus, full functional complementation was achieved with the full-length protein replacement despite only an overall 51% amino acid identity between the two species. This finding may be useful in future studies of protein-protein interaction domains in these proteins as well as analyses of sites of posttranslational modifications. Underway are studies with an even more distant relative from this genus, Schizosaccharomyces japonicus, which exhibits only 30% sequence identity to the S. pombe protein, mainly in their TZF domains (4). However, both full-length C. guilliermondii Zfs1 and M. musculus TTP were less than fully effective at complementing the null phenotype, although both seemed to exhibit some activity in terms of target transcript decrease compared with the zfs1Δ mutant strain and partial repair of the flocculation defect. This is not surprising in the case of the mouse protein because TTP family members from vertebrates, insects, nematodes, and even some plants, but not S. pombe and most other fungi, contain a conserved NOT1 binding domain in their extreme C termini (2, 26). Binding of NOT1 with its associated deadenylases has been proposed to be critical for TTP-mediated RNA decay (27). There are undoubtedly other species-specific protein-interacting domains in TTP family proteins that are required for in vivo activity. Interpretation of the mouse data were further complicated by the fact the construct was much more highly expressed than the others at both the mRNA and protein levels, the mechanism of which is not known. It seems possible that these very disparate proteins exert some “Zfs1-like” activity in these cells, suggesting at least the possibility of interactions between endogenous S. pombe proteins and the non-TZF domain regions of the foreign proteins.
In conclusion, we found that the RNA binding TZF domains from disparate species of plants, insects, and mammals could fully complement the zfs1Δ mutant phenotype in S. pombe both in terms of the molecular phenotype of altered gene expression and the sensitivity to calcium-induced flocculation. These findings suggest that this domain has remained functionally conserved in many species after nearly 2 billion years of evolution from its invention in a common ancestor. The many amino acid changes that have occurred during this time have produced many “experiments of nature” in terms of amino acid alterations and their presumed compensating changes in some cases, suggesting that maintaining this RNA binding capability was a high priority in the evolution of species expressing proteins of this family.
Author Contributions
M. L. W. and P. J. B. conceived and designed the study and wrote the manuscript. M. L. W. performed and analyzed the experiments shown in Figs. 1–4 and 6–8. S. N. H. performed and analyzed the experiments shown in Fig. 5. L. P. performed and analyzed the modeling shown in Fig. 9. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Drs. Jessica Williams and Robert Petrovich for critical comments on the manuscript and the Molecular Genomics Core for performing the NanoString analyses.
This research was supported, in whole or in part, by the intramural research program of the NIEHS/National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1–S4.
- TTP
- tristetraprolin
- TZF
- tandem zinc finger
- ARE
- AU-rich element
- MBP
- maltose binding protein.
References
- 1. Brooks S. A., Blackshear P. J. (2013) Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim. Biophys. Acta 1829, 666–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blackshear P. J., Perera L. (2014) Phylogenetic distribution and evolution of the linked RNA-binding and NOT1-binding domains in the tristetraprolin family of tandem CCCH zinc finger proteins. J. Interferon Cytokine Res. 34, 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ciais D., Cherradi N., Feige J. J. (2013) Multiple functions of tristetraprolin/TIS11 RNA-binding proteins in the regulation of mRNA biogenesis and degradation. Cell. Mol. Life Sci. 70, 2031–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wells M. L., Huang W., Li L., Gerrish K. E., Fargo D. C., Ozsolak F., Blackshear P. J. (2012) Posttranscriptional regulation of cell-cell interaction protein-encoding transcripts by Zfs1p in Schizosaccharomyces pombe. Mol. Cell Biol. 32, 4206–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi Y. J., Lai W. S., Fedic R., Stumpo D. J., Huang W., Li L., Perera L., Brewer B. Y., Wilson G. M., Mason J. M., Blackshear P. J. (2014) The Drosophila Tis11 protein and its effects on mRNA expression in flies. J. Biol. Chem. 289, 35042–35060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wells M. L., Washington O. L., Hicks S. N., Nobile C. J., Hartooni N., Wilson G. M., Zucconi B. E., Huang W., Li L., Fargo D. C., Blackshear P. J. (2015) Post-transcriptional regulation of transcript abundance by a conserved member of the tristetraprolin family in Candida albicans. Mol. Microbiol. 95, 1036–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cuthbertson B. J., Liao Y., Birnbaumer L., Blackshear P. J. (2008) Characterization of zfs1 as an mRNA-binding and -destabilizing protein in Schizosaccharomyces pombe. J. Biol. Chem. 283, 2586–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Puig S., Askeland E., Thiele D. J. (2005) Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120, 99–110 [DOI] [PubMed] [Google Scholar]
- 9. Blackshear P. J., Lai W. S., Kennington E. A., Brewer G., Wilson G. M., Guan X., Zhou P. (2003) Characteristics of the interaction of a synthetic human tristetraprolin tandem zinc finger peptide with AU-rich element-containing RNA substrates. J. Biol. Chem. 278, 19947–19955 [DOI] [PubMed] [Google Scholar]
- 10. Lai W. S., Carrick D. M., Blackshear P. J. (2005) Influence of nonameric AU-rich tristetraprolin-binding sites on mRNA deadenylation and turnover. J. Biol. Chem. 280, 34365–34377 [DOI] [PubMed] [Google Scholar]
- 11. Brewer B. Y., Malicka J., Blackshear P. J., Wilson G. M. (2004) RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of AU-rich MRNA-destabilizing motifs. J. Biol. Chem. 279, 27870–27877 [DOI] [PubMed] [Google Scholar]
- 12. Hudson B. P., Martinez-Yamout M. A., Dyson H. J., Wright P. E. (2004) Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 11, 257–264 [DOI] [PubMed] [Google Scholar]
- 13. Lai W. S., Perera L., Hicks S. N., Blackshear P. J. (2014) Mutational and structural analysis of the tandem zinc finger domain of tristetraprolin. J. Biol. Chem. 289, 565–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lai W. S., Kennington E. A., Blackshear P. J. (2002) Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J. Biol. Chem. 277, 9606–9613 [DOI] [PubMed] [Google Scholar]
- 15. Lai W. S., Carballo E., Thorn J. M., Kennington E. A., Blackshear P. J. (2000) Interactions of CCCH zinc finger proteins with mRNA: binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J. Biol. Chem. 275, 17827–17837 [DOI] [PubMed] [Google Scholar]
- 16. Lai W. S., Carballo E., Strum J. R., Kennington E. A., Phillips R. S., Blackshear P. J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell Biol. 19, 4311–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kedar V. P., Zucconi B. E., Wilson G. M., Blackshear P. J. (2012) Direct binding of specific AUF1 isoforms to tandem zinc finger domains of tristetraprolin (TTP) family proteins. J. Biol. Chem. 287, 5459–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Funakoshi M., Hochstrasser M. (2009) Small epitope-linker modules for PCR-based C-terminal tagging in Saccharomyces cerevisiae. Yeast 26, 185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Geiss G. K., Bumgarner R. E., Birditt B., Dahl T., Dowidar N., Dunaway D. L., Fell H. P., Ferree S., George R. D., Grogan T., James J. J., Maysuria M., Mitton J. D., Oliveri P., Osborn J. L., Peng T., Ratcliffe A. L., Webster P. J., Davidson E. H., Hood L., Dimitrov K. (2008) Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26, 317–325 [DOI] [PubMed] [Google Scholar]
- 20. Moon A. F., Mueller G. A., Zhong X., Pedersen L. C. (2010) A synergistic approach to protein crystallization: combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 19, 901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Case D. A., Babin V., Berryman J. T., Betz R. M., Cai Q., Cerutti D. S., Cheatham T. E. III, Darden T. A., Duke R. E., Gohlke H., Goetz A. W., Gusarov S., Homeyer N., Janowski P., Kaus J., Kolossváry I., Kovalenko A., Lee T. S., LeGrand S., Luchko T., Luo R., Madej B., Merz K. M., Paesani F., Roe D. R., Roitberg A., Sagui C., Salomon-Ferrer R., Seabra G., Simmerling C. L., Smith W., Swails J., Walker R. C., Wang J., Wolf R. M., Wu X., Kollman P. A. (2014) Amber 14. University of California, San Francisco [Google Scholar]
- 23. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 24. Kanoh J., Sugimoto A., Yamamoto M. (1995) Schizosaccharomyces pombe zfs1+ encoding a zinc-finger protein functions in the mating pheromone recognition pathway. Mol. Biol. Cell 6, 1185–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Taylor G. A., Lai W. S., Oakey R. J., Seldin M. F., Shows T. B., Eddy R. L. Jr., Blackshear P. J. (1991) The human TTP protein: sequence, alignment with related proteins, and chromosomal localization of the mouse and human genes. Nucleic Acids Res. 19, 3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fabian M. R., Frank F., Rouya C., Siddiqui N., Lai W. S., Karetnikov A., Blackshear P. J., Nagar B., Sonenberg N. (2013) Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat. Struct. Mol. Biol. 20, 735–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sandler H., Kreth J., Timmers H. T., Stoecklin G. (2011) Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 39, 4373–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.