Background: How AMP-activated protein kinase (AMPK) influences IL-4-induced human macrophage polarization is not completely understood.
Results: AMPK prevents arachidonate 15-lipoxygenase induction by IL-4 and abolishes the formation of 15-lipoxygenase arachidonic acid metabolites.
Conclusion: AMPK activation promotes an anti-inflammatory phenotype in IL-4-stimulated macrophages by reducing arachidonate 15-lipoxygenase expression.
Significance: This study supports an anti-inflammatory effect of AMPK activation.
Keywords: AMP-activated kinase (AMPK), arachidonic acid (AA), gene expression, inflammation, transcription
Abstract
Macrophages respond to the Th2 cytokine IL-4 with elevated expression of arachidonate 15-lipoxygenase (ALOX15). Although IL-4 signaling elicits anti-inflammatory responses, 15-lipoxygenase may either support or inhibit inflammatory processes in a context-dependent manner. AMP-activated protein kinase (AMPK) is a metabolic sensor/regulator that supports an anti-inflammatory macrophage phenotype. How AMPK activation is linked to IL-4-elicited gene signatures remains unexplored. Using primary human macrophages stimulated with IL-4, we observed elevated ALOX15 mRNA and protein expression, which was attenuated by AMPK activation. AMPK activators, e.g. phenformin and aminoimidazole-4-carboxamide 1-β-d-ribofuranoside inhibited IL-4-evoked activation of STAT3 while leaving activation of STAT6 and induction of typical IL-4-responsive genes intact. In addition, phenformin prevented IL-4-induced association of STAT6 and Lys-9 acetylation of histone H3 at the ALOX15 promoter. Activating AMPK abolished cellular production of 15-lipoxygenase arachidonic acid metabolites in IL-4-stimulated macrophages, which was mimicked by ALOX15 knockdown. Finally, pretreatment of macrophages with IL-4 for 48 h increased the mRNA expression of the proinflammatory cytokines IL-6, IL-12, CXCL9, and CXCL10 induced by subsequent stimulation with lipopolysaccharide. This response was attenuated by inhibition of ALOX15 or activation of AMPK during incubation with IL-4. In conclusion, limiting ALOX15 expression by AMPK may promote an anti-inflammatory phenotype of IL-4-stimulated human macrophages.
Introduction
Macrophages are highly plastic cells that respond to intra- and extracellular stimuli, generating diverse molecular and functional phenotypes (1). Upon exposure to the Th2 cytokines IL-4 or IL-13, macrophages are M2 or alternatively polarized (2, 3). Initially described in the context of helminth infection or allergic reactions, IL-4/IL-13 activation is now linked to a wide range of homeostatic functions, including maintenance of insulin sensitivity or regulation of thermogenesis (2). IL-4/IL-13 are anti-inflammatory cytokines and, indeed, they dampen monocyte/macrophage inflammatory responses toward LPS (4). However, macrophages pretreated with IL-4 enhance inflammatory responses to subsequent bacterial (5) or LPS stimulation (6). Neither molecular details how IL-4 elicits pro- versus anti-inflammatory signals nor the role of IL-4/IL-13-induced genes in controlling inflammation are fully understood.
Arachidonate 15-lipoxygenase (ALOX152 in the human nomenclature, alternatively called 15-LOX-1) has been described as a highly inducible IL-4/IL-13 target gene in mice (7) and humans (8, 9). ALOX15 belongs to a family of dioxygenases that convert unsaturated fatty acids, preferably arachidonic acid, to mono-oxygenated derivatives, such as 15(S)-HETE (10, 11). ALOX15 and its enzymatic products are implicated in many homeostatic as well as pathological processes, with roles both in the propagation and resolution of inflammation (12). The importance of ALOX15 for IL-4/IL-13-induced macrophage polarization has been indicated in mouse studies showing that the mouse ortholog 12/15-lipoxygenase produced endogenous ligands for the transcription factor PPARγ (7), thereby attenuating inflammatory responses. Whether ALOX15 exerts additional roles in IL-4/IL-13 macrophage polarization is unknown. Human macrophages also constitutively express ALOX15B, an isoform of arachidonate 15-lipoxygenase (13). This isoform is induced by hypoxia (14) and produces T cell-attractive chemokines in macrophages (15). However, the distinct roles of ALOX15 versus ALOX15B in human IL-4/IL-13-polarized macrophages are unclear.
Metabolism is recognized as an important determinant of macrophages activation. AMP-activated protein kinase (AMPK) is a major cellular metabolic sensor, responding to a decreased energy charge and adjusting cellular metabolism to prevent energy loss (16). AMPK is also involved in regulating inflammatory responses (17) and promotes an anti-inflammatory macrophage polarization (18, 19). This is facilitated by inhibiting NF-κB and stimulating Akt. Of note, the impact of AMPK on IL-4/IL-13-induced macrophage polarization has not yet been studied. Here we provide evidence that AMPK activation profoundly attenuated ALOX15 induction by reducing STAT3 activation by IL-4. ALOX15 generated 12(S)-HETE and 15(S)-HETE and potentiated inflammatory responses to LPS, which was prevented by AMPK activation.
Experimental Procedures
Cell Isolation and Culture
Human monocytes were isolated from buffy coats of anonymous donors (Deutsches Rotes Kreuz-Blutspendedienst Baden-Württemberg-Hessen, Institut für Transfusionsmedizin und Immunhämatologie, Frankfurt, Germany) using Ficoll density centrifugation, followed by magnetic separation with positive selection (CD14 MicroBeads, Miltenyi Biotec). Monocytes were differentiated into macrophages by culture in macrophage serum-free medium (Invitrogen) containing 50 ng/ml human recombinant macrophage colony-stimulating factor (Immunotools) for 7 days. Cells were treated (where indicated) with the following reagents: 20 ng/ml human recombinant IL-4 or IL-13 (Immunotools); 500μm 5-aminoimidazole-4-carboxamide 1-β-d-ribofuranoside (AICAR, EMD Biosciences); 500 μm A769662 (Tocris); 1 μm R419 (Rigel Pharmaceuticals); 100 nm 12(S)-HETE and 100 nm 15(S)-HETE (Cayman Chemical); and 100 μm phenformin, 20 μm nordihydroguaiaretic acid, 10 μm PD146716, 1 μm rosiglitazone, or 100 pg/ml LPS (Sigma-Aldrich). This investigation conforms to the principles outlined in the Declaration of Helsinki and was approved by the university ethics committee.
siRNA Transfection and Adenoviral Transduction of Macrophages
Silencing of AMPKα1, STAT3, and ALOX15 in human primary macrophages was achieved using corresponding siGENOME SMARTpools (Thermo Fisher Scientific) at 50 nm and Hyperfect transfection reagent (Qiagen). Cells were treated 72 h post-transfection, except for ALOX15 siRNA (24 h post-transfection). For adenoviral transduction, macrophages were incubated with a control adenovirus (Ad-Track-GFP, Addgene) or with an adenovirus coding for the AMPKγ1 regulatory subunit carrying an activating R70Q substitution (provided by Dr. Jason Dyck, Cardiovascular Research Centre, University of Alberta, Canada) for 48 h prior to treatments.
Quantitative PCR
Total RNA from human primary macrophages was isolated using the PeqGold RNAPure kit (PeqLab) and transcribed using a cDNA synthesis kit (Fermentas). Quantitative PCR was performed with iQ SYBR Green Supermix (Bio-Rad) using the CFX96 system (Bio-Rad). Primer sequences for quantitative PCR are available upon request. Expression was normalized to β-microglobulin.
Immunoprecipitation
Cells were lysed in HEPES-based buffer (20 mm HEPES (pH 7.4), 150 mm NaCl, 10% glycerol, and 1% Triton X-100) containing phosphatase and protease inhibitors and incubated overnight at 4 °C with Jak1 antibody (catalog no. 3332, Cell Signaling Technology) at 1:100 dilution, followed by 2 h of incubation with protein A/G-agarose beads (catalog no. sc-2003, Santa Cruz Biotechnology). After three washes with the same buffer, beads were eluted by heating at 95 °C into 2× electrophoresis sample buffer, separated on polyacrylamide gels, and analyzed by Western blotting using STAT3 and Jak1 antibodies.
Western Blot Analysis
Protein lysates were separated on polyacrylamide gels, followed by transfer onto nitrocellulose membranes. Membranes were incubated with antibodies against phospho-ACC (Ser-79, catalog no. 3661), phospho-AMPK (Thr-172, catalog no. 2531), phospho-STAT6 (Tyr-641, catalog no. 9361), phospho-STAT3 (Tyr-705, catalog no. 9145), phospho-Jak1 (Tyr-1022/1023, catalog no. 3331), ACC (catalog no. 3662), AMPK (catalog no. 2532), STAT6 (catalog no. 9362), STAT3 (catalog no. 9139), Jak1 (catalog no. 3332, Cell Signaling Technology), ALOX15 (catalog no. ab118774, Abcam), ALOX15B (catalog no. 10004454, Cayman Chemical), ALOX5 (catalog no. 610693, BD Biosciences), actin (catalog no. A2066, Sigma-Aldrich), or nucleolin (catalog no. sc-13057, Santa Cruz Biotechnology), followed by IRDye 800-coupled secondary antibodies (Licor Biosciences). Blots were visualized and quantified using the Odyssey imaging system (Licor Biosciences).
Chromatin Immunoprecipitation
10 × 106 macrophages were cross-linked for 10 min with 1% formaldehyde, and extracts were sonicated until the DNA fragments were 300–800 bp in average size. Cross-linked chromatin was immunoprecipitated with 2.5 μg of STAT6 antibody (catalog no. sc-981X, Santa Cruz Biotechnology), Lys-9-acetylated histone 3 antibody (catalog no. 04-1003, Millipore), or rabbit IgG overnight at 4 °C, followed by incubation with protein A/G-agarose beads (catalog no. sc-2003, Santa Cruz Biotechnology) for an additional 2 h. Immunoprecipitated DNA was recovered using a PCR purification kit (Qiagen) and analyzed using quantitative PCR. Primers corresponding to the human ALOX15 promoter (852 bp upstream of the transcription start site) or the CCL18 and Spint2 promoters (−5900 bp and −650 bp, respectively) were used for analysis. Data are shown as percentage of input DNA precipitated by a corresponding antibody.
Lipid Analysis
Leukotriene B4, 5(S)-HETE, 12(S)-HETE, 15(R)-HETE, and 15(S)-HETE in the extracted samples were analyzed employing LC-MS/MS. The LC/MS-MS system comprised an 5500 QTrap mass spectrometer (AB Sciex), an Agilent 1200 binary HPLC pump (Agilent), and an HTC Pal autosampler (Chromtech). Eicosanoid standards were obtained from Cayman Chemical. Sample extraction was performed with liquid-liquid extraction using ethyl acetate. The organic phase was removed under a stream of nitrogen, and the residues were reconstituted in methanol/water prior to injection into the LC-MS/MS system. For chromatographic separation, a Gemini NX C18 column and precolumn were used (Phenomenex). A linear gradient was employed at a flow rate of 0.5 ml/min with a total run time of 17.5 min. The mobile phase was water/ammonia (A, 100:0.05, v/v) and acetonitril/ammonia (B, 100:0.05, v/v). The gradient moved from 85% A to 10% within 12 min. The retention times of Leukotriene B4, 5(S)-HETE, 12(S)-HETE, and 15(S)-HETE were 7.94, 10.01, 9.62, and 9.92 min, respectively. Peak quantification was performed with Analyst software version 1.5 (Applied Biosystems) employing the internal standard method (isotope dilution mass spectrometry). The ratios of analyte peak area and internal standard area (y axis) were plotted against concentration (x axis), and calibration curves were calculated by least square regression.
Statistical Analysis
Data are presented as mean ± S.E. of at least three independent experiments. Data were analyzed by one-way analysis of variance with Bonferroni post hoc means comparison using OriginPro 8.5G software (OriginLab, Northampton, MA). Differences were considered statistically significant at p < 0.05.
Results
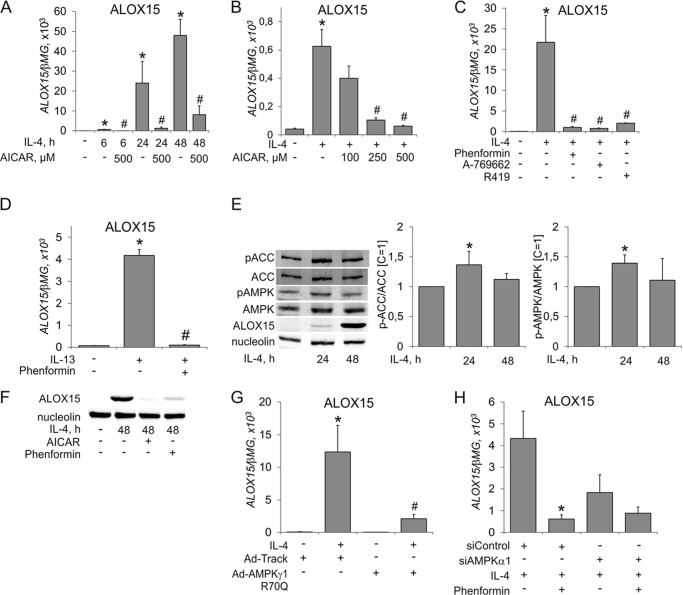
Initial experiments exploring the influence of AMPK activation on IL-4-induced gene expression in human macrophages revealed a striking inhibitory effect on mRNA induction of ALOX15. As shown in Fig. 1A, IL-4-induced up-regulation of ALOX15 mRNA started at 6 h and continued up to 48 h. This induction was suppressed in a concentration-dependent fashion at 6 h already by the AMP-mimetic AMPK activator AICAR (Fig. 1B) and at later time points (Fig. 1A). ALOX15 induction by IL-4 was abolished in the presence of pharmacological AMPK activators of different classes, including the allosteric AMPK activator A-769662, the biguanidine phenformin, a compound decreasing the cellular energy charge, and R419, a novel mitochondrial complex I inhibitor (20) (Fig. 1C). ALOX15 mRNA induction by IL-13, another Th2 cytokine, was inhibited by AMPK activation as well (Fig. 1D). We went on to analyze ALOX15 protein expression after IL-4 treatment in the absence or presence of AMPK activators. ALOX15 protein was induced prominently after 48 h of incubation with IL-4 (Fig. 1E). We questioned whether IL-4 treatment influenced the activity of AMPK by measuring the phosphorylation of AMPK and acetyl-CoA carboxylase (ACC). We noticed that IL-4 induced modest increases of ACC and AMPK phosphorylation at 24 h (1.4-fold) but not at 48 h (Fig. 1E). Incubating macrophages for 48 h with IL-4 in the presence of phenformin or AICAR fully antagonized ALOX15 protein induction (Fig. 1F). To validate the data obtained with pharmacological AMPK activators, we transduced macrophages with an adenoviral construct coding for a regulatory AMPK γ1 subunit with an R70Q substitution, which was previously shown to be an activating AMPK mutation, increasing the activity of AMPK heterotrimers (21). Fig. 1G shows that macrophages transduced with the AMPKγ1 R70Q adenovirus displayed reduced ALOX15 mRNA induction compared with control adenovirus transduced cells. Finally, we silenced the AMPK α1 catalytic subunit in macrophages, which do not express measurable amounts of AMPKα2 protein (18), by siRNA, followed by their exposure to IL-4 in the presence or absence of phenformin (Fig. 1H). Although phenformin inhibited IL-4-induced ALOX15 mRNA expression in siControl-transfected cells, this effect was lost in AMPKα1-silenced macrophages. Collectively, these data suggest an inhibitory effect of AMPK on IL-4-induced ALOX15 expression.
FIGURE 1.
Activated AMPK suppresses IL-4-induced ALOX15 gene expression. A and B, mRNA expression of ALOX15 in macrophages treated for the indicated times with 20 ng/ml IL-4 and 500 μm AICAR (A) or for 6 h with IL-4 and the indicated concentrations of AICAR (B). C, mRNA expression of ALOX15 in macrophages treated for 24 h with IL-4 and phenformin, A-769662, or R419. *, p < 0.05 versus untreated cells; #, p < 0.05 versus IL-4. D, mRNA expression of ALOX15 in macrophages treated for 24 h with IL-13 and phenformin. *, p < 0.05 versus untreated cells; #, p < 0.05 versus IL-13. E and F, Western blot analysis of ALOX15 expression and ACC and AMPK phosphorylation in macrophages treated for the indicated times with IL-4, AICAR, or phenformin. *, p < 0.05 versus untreated cells. G and H, mRNA expression of ALOX15 in macrophages infected with Ad-Track or Ad-AMPKγ1R70Q prior to treatment with IL-4 for 24 h (G) or transfected with control or AMPKα1 siRNA prior to treatment with IL-4 and phenformin for 24 h (H). *, p < 0.05 versus untreated cells; #, p < 0.05 versus Ad-Track+IL-4 (G). *, p < 0.05 versus siControl + IL-4 (H). Data represent mean ± S.E. of at least three independent experiments.
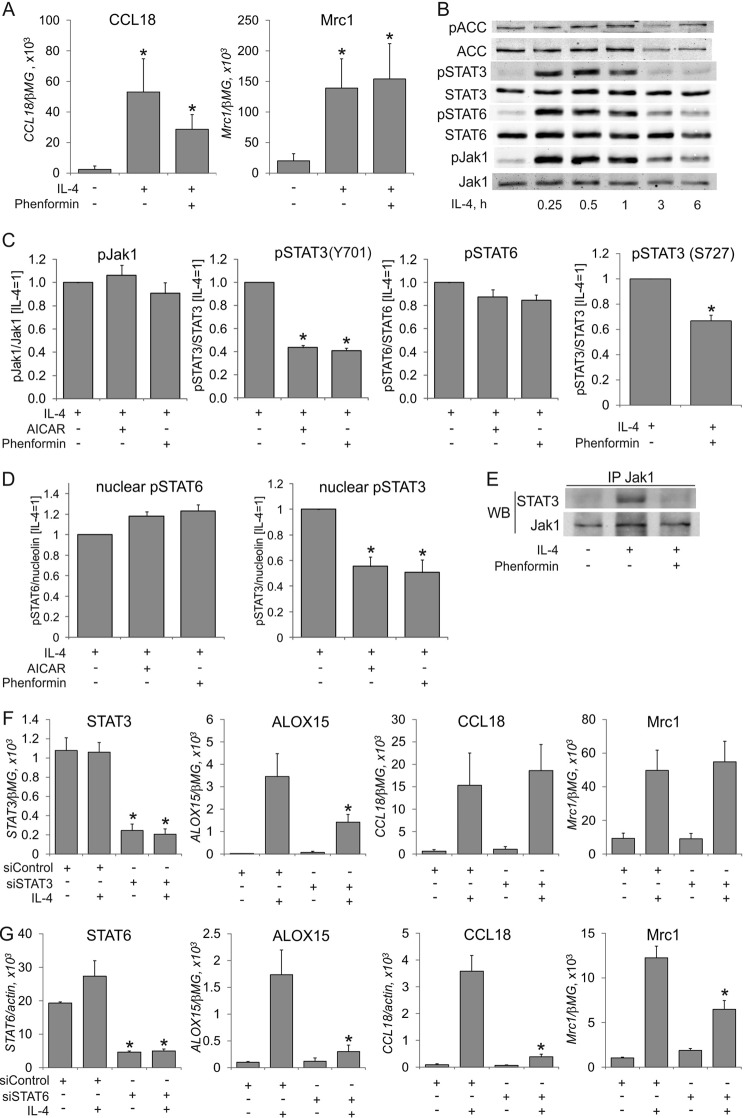
Next, we explored the mechanistic details of ALOX15 suppression by AMPK. First, we questioned whether AMPK activation generally affects IL-4-induced gene expression. As seen in Fig. 2A, this is not the case. The expression of typical IL-4 target genes in macrophages, such as chemokine (C-C motif) ligand 18 (CCL18) or mannose receptor C, type 1 (Mrc1) remained unaffected by phenformin, indicating that a typical transcriptional response to IL-4 stimulation remains unaffected by AMPK.
FIGURE 2.
AMPK activation inhibits STAT3 activation by IL-4. A, mRNA expression of CCL18 and Mrc1 in macrophages treated for 24 h with IL-4 and phenformin. *, p < 0.05 versus untreated cells. B–D, Western analysis of Jak1, STAT6, and STAT3 (Tyr-705 and Ser-727) phosphorylation in whole macrophage lysates (B and C) or nuclear extracts (D) of cells treated with IL-4, phenformin, or AICAR for the indicated times (B), 30 min (C), or 1 h (D). *, p < 0.05 versus IL-4. E, analysis of coimmunoprecipitation of Jak1 and STAT3 in macrophages treated for 10 min with IL-4 and phenformin. WB, Western blot. F and G, mRNA expression of STAT3, STAT6, ALOX15, CCL18, and Mrc1 in macrophages transfected with control siRNA, STAT3 (F), or STAT6 (G) siRNA for 72 h prior to treatment with IL-4 for 24 h. *, p < 0.05 versus siControl. Data represent mean ± S.E. of at least three independent experiments.
IL-4 activates genes predominantly via the STAT6 pathway (2). However, the STAT3 transcription factor is engaged in IL-4-induced ALOX15 expression in human monocytes (22). To question the involvement of STATs in AMPK-dependent ALOX15 suppression, we analyzed tyrosine phosphorylation and nuclear translocation of STAT3 and STAT6 as signs of their activation in IL-4-treated cells in the presence of AICAR or phenformin. Furthermore, we analyzed phosphorylation of Jak1 kinase, which was reported to phosphorylate STAT3 and STAT6 in IL-4-stimulated monocytes (22). IL-4 elicited robust phosphorylations of Jak1, STAT3, and STAT6 within 15 min, which returned to baseline after 3 h (Fig. 2B). Levels of ACC phosphorylation did not change at early time points of IL-4 stimulation. As seen in Fig. 2C, phosphorylation of STAT6 and Jak1 in whole-cell lysates was either not affected or only reduced slightly by AMPK activators. In contrast, both AICAR and phenformin lowered phosphorylation of STAT3 in IL-4-stimulated macrophages. Phenformin also inhibited phosphorylation of STAT3 at Ser-727, which is necessary for full transcriptional activity of STAT3. Apparently, the inhibitory effects of AMPK correlate only with attenuated STAT3 activation. Accordingly, the levels of phosphorylated STAT3 in nuclear extracts of IL-4-treated macrophages were reduced upon preincubation with AICAR or phenformin, whereas nuclear phospho-STAT6 remained unaffected (Fig. 2D). Association of STAT3 with its upstream kinase Jak1, an early step of IL4-induced STAT3 activation, was inhibited by phenformin as well (Fig. 2E). To demonstrate the role of STAT3 in IL-4-induced ALOX15 mRNA expression, we used RNA interference. STAT3-specific siRNA decreased its mRNA expression by 80% and reduced IL-4-induced ALOX15 mRNA expression by more than 50% (Fig. 2F) while leaving IL-4-induced CCL18 and Mrc1 mRNA expression intact. As a control, knockdown of STAT6 inhibited IL-4-induced mRNA expression of all target genes (Fig. 2G), confirming a central role of STAT6 in IL-4-elicited transcriptional responses. Taken together, these data suggest that AMPK prevents IL-4-induced ALOX15 expression by inhibiting STAT3.
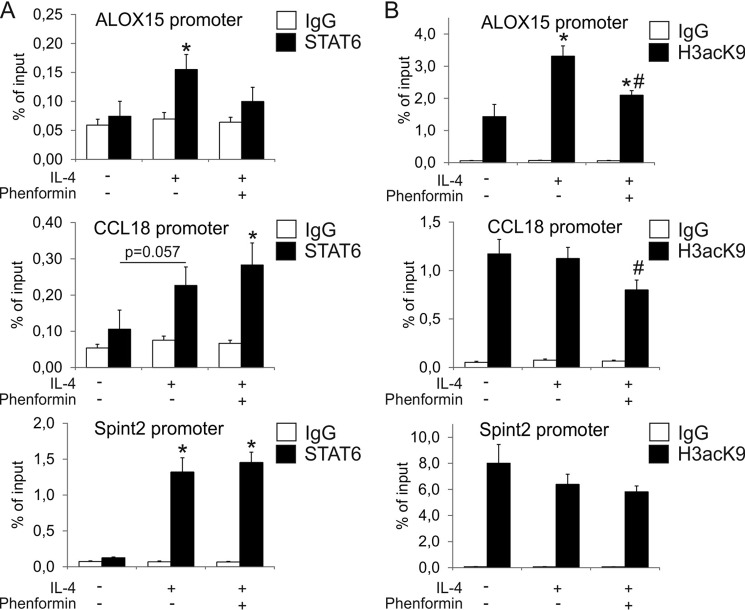
Tyrosine phosphorylation and nuclear translocation of STAT3 as well as STAT6 are early steps in an IL-4 signaling cascade leading to ALOX15 gene transcription. However, there is evidence that epigenetic modifications, such as histone acetylation, peak at later time points and are associated with delayed STAT6 association with the ALOX15 promoter (23). Although we failed to detect STAT6 binding to the ALOX15 promoter in chromatin IP experiments 1 h after IL-4 stimulation (data not shown), STAT6 binding significantly increased at 12 h. In the presence of phenformin, no significant increase of STAT6 binding was observed compared with untreated cells (Fig. 3A). Furthermore, Lys-9 acetylation of histone H3 at the ALOX15 promoter was elevated 12 h after IL-4 treatment, and this was attenuated significantly by phenformin (Fig. 3B). Increased histone acetylation after IL-4 treatment and its inhibition by phenformin was specific for the ALOX15 promoter because IL-4 failed to increase levels of Lys-9-acetylated histone H3 at the CCL18 and Spint2 promoters (Fig. 3B). In contrast, binding of STAT6 to the CCL18 and Spint2 promoters was still increased by IL-4 in the presence of phenformin (Fig. 3A), correlating with an inability of phenformin to affect CCL18 mRNA expression (Fig. 2A).
FIGURE 3.
AMPK activation prevents histone acetylation of the ALOX15 promoter. A and B, chromatin immunoprecipitation analysis of STAT6 (A) and Lys-9-acetlyated histone 3 (H3acK9) (B) binding to ALOX15, CCL18, and Spint2 promoters in macrophages treated for 24 h with IL-4 and phenformin. *, p < 0.05 versus untreated cells; #, p < 0.05 versus IL-4. Data represent mean ± S.E. of at least three independent experiments.
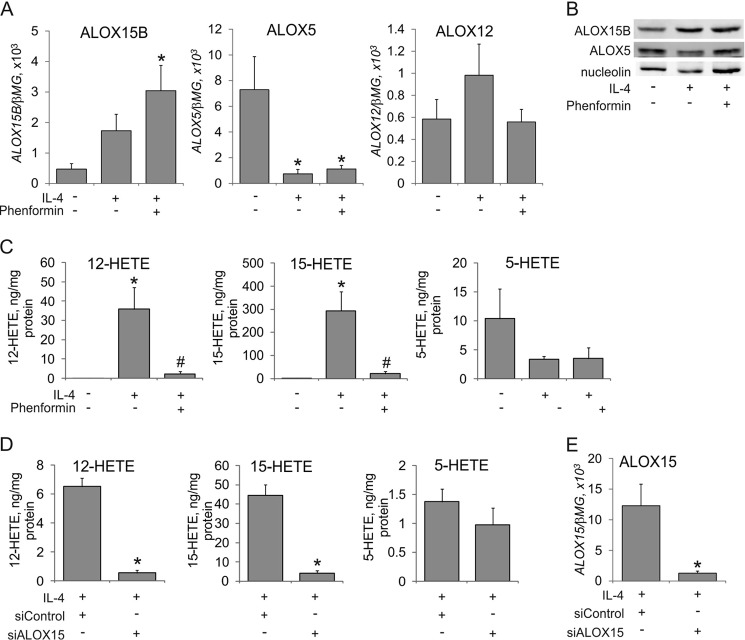
We then addressed the impact of IL-4 and AMPK on arachidonic acid lipoxygenase metabolism. As a first experiment, we explored whether the expression of other lipoxygenases involved in arachidonic acid metabolism is affected by IL-4 and AMPK. As depicted in Fig. 4A, the mRNA expression of the second human 15-lipoxygenase enzyme, ALOX15B, increased after IL-4-treatment both in the absence and presence of phenformin. ALOX5 mRNA was reduced by more than 80% after IL-4 treatment, an effect being not affected by phenformin. Platelet-specific ALOX12 mRNA was not significantly affected by IL-4 or phenformin. Western blot analysis confirmed the induction of ALOX15B and suppression of ALOX5 by IL-4, whereas these responses were unaffected by phenformin (Fig. 4B). Therefore, AMPK activation selectively affects ALOX15 expression in IL-4-treated macrophages without major effects on the expression of other lipoxygenases. As a second experiment, we analyzed lipoxygenase metabolites by LC-MS/MS. We noticed a massive increase of cellular 12(S)-HETE and 15(S)-HETE in macrophages exposed to IL-4 for 48 h, which was blocked by phenformin (Fig. 4C), whereas the level of 5(S)-HETE tended to decrease upon IL-4-treatment without reaching a statistical significance. Chromatographic separation of R- versus S-enantiomers of 15-HETE revealed that >95% of 15-HETE was in S-form, confirming the enzymatic character of 15-HETE formation. We did not observe measurable 12(S)-HETE and 15(S)-HETE levels in cell culture supernatants under these conditions and failed to detect leukotriene B4 in cell pellets and supernatants (data not shown). To determine the involvement of ALOX15 in accumulating cell-associated 12(S)-HETE and 15(S)-HETE after IL-4 treatment, we used an ALOX15 siRNA approach. Silencing ALOX15 reduced the production of cell-associated 12(S)-HETE and 15(S)-HETE by more than 90% (Fig. 4D). Quantitative PCR analysis confirmed the efficiency of ALOX15 knockdown (Fig. 4E). This confirms a predominant role of ALOX15 in generating 12(S)-HETE and 15(S)-HETE in IL-4-polarized macrophages, whereas 5(S)-HETE levels were not affected by ALOX15 silencing.
FIGURE 4.
AMPK activation blocks IL-4-induced 15-lipoxygenase arachidonate metabolites. A, mRNA expression of ALOX15B, ALOX5, and ALOX12 in cells treated with IL-4 and phenformin for 24 h. *, p < 0.05 versus untreated cells. B, Western blot analysis of ALOX15B and ALOX5 in macrophages treated for 48 h with IL-4 and phenformin. C and D, LC-MS/MS analysis of arachidonate lipoxygenase metabolites in cells treated for 48 h with IL-4 and phenformin (C) or in cells transfected with control or ALOX15 siRNAs for 24 h prior to treatment with IL-4 for 48 h (D). *, p < 0.05 versus untreated cells; #, p < 0.05 versus IL-4. E, mRNA expression of ALOX15 in macrophages transfected with control siRNA or ALOX15 siRNA for 24 h prior to treatments with IL-4 for 24 h. *, p < 0.05 versus siControl. Data represent mean ± S.E. of at least three independent experiments.
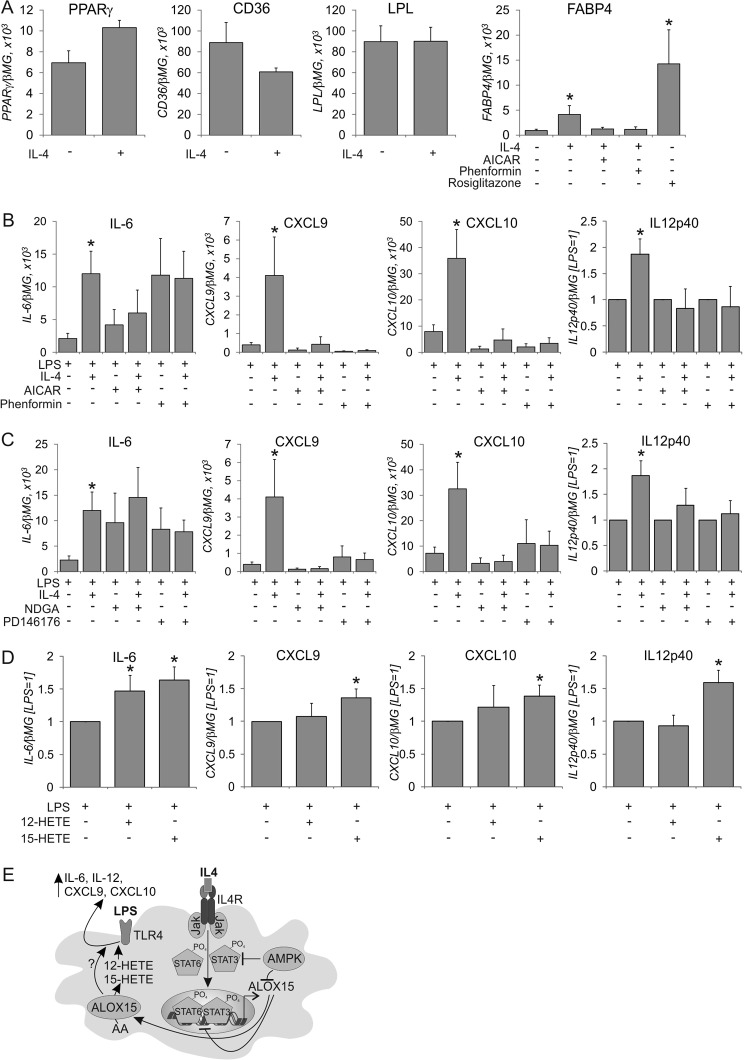
With the following experiments, we analyzed the impact of ALOX15 on the phenotype of IL-4-stimulated macrophages. ALOX15 is known to produce endogenous ligands for the transcription factor PPARγ (7), which regulates myeloid gene expression, e.g., to support macrophage M2 polarization (24), although this has not been confirmed in all studies (25). In IL-4-treated macrophages, we analyzed the expression of several PPARγ target genes, such as fatty acid binding protein 4 (FABP4), lipoprotein lipase, and CD36. As shown previously, these genes are induced PPARγ-dependently during monocyte-to-macrophage differentiation (26). IL-4 did not increase the mRNA expression of the PPARγ targets CD36 and lipoprotein lipase in fully differentiated macrophages, whereas PPARγ mRNA tended to increase, but this did not reach statistical significance (Fig. 5A). Only FABP4 mRNA expression was increased by IL-4, although the magnitude of its expression was only 30% compared with the synthetic PPARγ agonist rosiglitazone (Fig. 5A). IL-4 failed to increase FABP4 in the presence of phenformin or AICAR. Conclusively, IL-4 and AMPK have only a minor effect on PPARγ activity in fully matured macrophages.
FIGURE 5.
Role of 15-lipoxygenase in IL-4-induced priming of LPS responses. A, mRNA expression of PPARγ, CD36, lipoprotein lipase, and FABP4 in cells treated with IL-4, AICAR, rosiglitazone, and phenformin for 48 h. *, p < 0.05 versus untreated cells. B and C, mRNA expression of IL-6, CXCL9, CXCL10, and IL-12p40 in cells treated with IL-4, AICAR, and phenformin (B) for 48 h, followed by medium change and 3-h treatment with LPS. C, cells were treated with nordihydroguaiaretic acid or PD146176 during the last 24 h of IL-4 treatment and LPS stimulation. D, mRNA expression of IL-6, CXCL9, CXCL10, and IL-12p40 in cells treated with LPS in the presence of 12(S)-HETE or 15(S)-HETE. *, p < 0.05 versus LPS. Data represent mean ± S.E. of at least three independent experiments. E, schematic of AMPK and ALOX15 effects on IL-4-induced macrophage priming. AA, arachidonic acid.
Although IL-4 is known to inhibit the activation of inflammatory genes when added together with LPS, macrophages pretreated with IL-4 show enhanced proinflammatory responses to a subsequent LPS stimulus, an effect known as IL-4 priming (5, 6). Because 12/15-lipoxygenase or 12(S)/15(S)-HETEs have been reported to enhance proinflammatory responses in some systems (27, 28), we designed experiments to explore the role of ALOX15 and its regulation by AMPK in the priming effect of IL-4. Therefore, we pretreated macrophages with IL-4 for 48 h followed, by a 3-h treatment with LPS and subsequent analysis of proinflammatory mRNA expression. As summarized in Fig. 5B, IL-4 pretreatment enhanced LPS-induced mRNA expression of proinflammatory cytokines such as IL-6, chemokine (C-X-C motif) ligand 9 (CXCL9), CXCL10, and IL-12p40. The induction of other proinflammatory mediators, i.e. IL-1β, IL-8, CCL2, or CCL5, was not affected by IL-4 pretreatment (data not shown). When macrophages were pretreated with IL-4 and the AMPK activators AICAR or phenformin, the priming effect was lost. Similarly, priming was absent in the presence of the 15-lipoxygenase inhibitors nordihydroguaiaretic acid or PD146176 (Fig. 5C). Attempts to verify the specific role of ALOX15 by siRNA technology revealed that the transfection procedure using control siRNA greatly reduced the levels of arachidonic acid lipoxygenase metabolites in IL-4-treated cells (compare Fig. 4, C and D) and abolished the priming effect of IL-4 (data not shown). To investigate whether 15-lipoxygenase metabolites directly increase inflammatory LPS responses, we treated macrophages with LPS in the presence of 12(S)-HETE or 15(S)-HETE. As depicted in Fig. 5D, macrophages incubated in the presence of 15(S)-HETE showed small but significant increases in LPS-induced gene expression. In contrast, 12(S)-HETE potentiated only LPS-induced IL-6 mRNA expression. Therefore, 15-lipoxygenase metabolites of arachidonic acid may, at least in part, account for increased LPS-induced cytokine mRNA expression in IL-4-polarized macrophages. Collectively, these data imply that ALOX15 induction may play an important role in IL-4-induced macrophage priming, an effect that is under the control of AMPK.
Discussion
Our work aimed at understanding how AMPK activation impacts IL-4-induced gene expression in human macrophages. We obtained novel information showing that AMPK blocks IL-4-induced ALOX15 expression without generally suppressing IL-4 target genes. Moreover, ALOX15 alters inflammatory responses of IL-4-polarized macrophages toward TLR4 activation, potentiating the expression of a subset of LPS-targeted genes. AMPK, suppressing ALOX15 induction by IL-4,supports an anti-inflammatory phenotype of IL-4-polarized human macrophages.
A number of previous studies have reported the anti-inflammatory phenotype of AMPK-activated macrophages (18, 29, 30) with the notion that IL-4-induced polarization is attenuated in AMPKα1-deficient murine macrophages (19). In human macrophages, AMPK activation does not generally seem to affect IL-4-induced gene expression. We also noticed that, in contrast to the rapid activation of AMPK in response to IL-10 and TGFβ reported previously (18), acute IL-4 stimulation does not affect AMPK activity. The absence of a general effect of AMPK on IL-4-induced polarization is in line with the observation that the dominant IL-4-induced signaling pathway culminating in the activation of the transcription factor STAT6 remains intact in AMPK-stimulated macrophages. Interestingly, only a subset of IL-4-stimulated genes, most prominently ALOX15, is inhibited upon AMPK activation. It is known that, in addition to STAT6, STAT3 is involved in the regulation of ALOX15 in IL-4-treated human monocytes (22). Our data corroborate a role of STAT3 in regulating ALOX15 in response to IL-4, and we suggest that AMPK attenuates STAT3-dependent gene expression by preventing STAT3 association with upstream Jak1 tyrosine kinase, its tyrosine and serine phosphorylation, and nuclear translocation upon IL-4-treatment. These results concur with previous reports showing AMPK-dependent inhibition of STAT3 activation in hepatocytes (31), reduced STAT3 signaling in AMPK-activated astrocytes (32), and inhibition of STAT3 by metformin during monocyte-to-macrophage differentiation of THP-1 cells (33). At the moment, it is unclear how AMPK inhibits STAT3 activation. Because Jak1 and STAT6 phosphorylation was intact after AMPK activation, we suggest that AMPK specifically targets STAT3. This is also consistent with our observations that AMPK activators inhibit STAT3 phosphorylation induced by IL-6 or IL-10 in human macrophages (data not shown). However, our attempts to reveal possible posttranslational modifications of STAT3 after AMPK activation by mass spectrometry were unsuccessful. Therefore, the effect of AMPK on STAT3 may be indirect. One possibility is phosphorylation by AMPK of some unknown STAT3-interacting protein, the result of which could be sequestering STAT3 and preventing it from interacting with the IL-4 receptor. Exploring this possibility warrants further mechanistic investigation.
Interestingly, our data reveal a second mechanism of how AMPK may interfere with ALOX15 expression after IL-4 stimulation; that is, its preventing STAT6 binding and histone acetylation at the ALOX15 promoter. Furthermore, our data suggest that increased histone acetylation after IL-4 treatment specifically occurs at ALOX15 and not at promoters of other IL-4 target genes. Remarkably, STAT6 binding to the ALOX15 promoter is temporarily dissociated from the maximum of STAT6 nuclear translocation (23). The delayed recruitment of STAT6 to the ALOX15 promoter may be associated with histone and STAT6 acetylation by p300. Interestingly, AMPK has been reported to inhibit p300 activity (34). Although we failed to detect STAT6 acetylation in our system (data not shown), reduced histone acetylation at the ALOX15 promoter conformed with this mechanism. However, STAT3 did not bind to the STAT6-binding site of the ALOX15 promoter, and we could not identify any STAT3-binding site up to 50 kb upstream of ALOX15 transcriptional start site using chromatin immunoprecipitation (data not shown). The nature of STAT3-dependent gene regulatory elements on ALOX15 remains to be elucidated.
Although several high-throughput studies analyzed the transcriptome of human monocyte/macrophages polarized in response to IL-4 (8, 35–37), they did not specifically question the role of STAT3 in these responses. Therefore, it will be interesting to elucidate the full scope of STAT3-dependent gene expression changes in IL-4-polarized human macrophages, which are also likely to be affected by AMPK activation.
IL-4 stimulation greatly affects the way how macrophages handle arachidonate metabolism via lipoxygenases. Although ALOX5 expression is suppressed in IL-4-stimulated cells, both ALOX15 and ALOX15B are induced. Although ALOX15B is detectable at the protein level in resting macrophages, which corroborates an earlier report (13), only stimulation with IL-4 initiates the production of substantial amounts of the 15-lipoxygenase metabolites 12(S)-HETE and 15(S)-HETE, which is antagonized either by silencing ALOX15 or activating AMPK. Therefore, ALOX15 is the predominant lipoxygenase isoform producing 12(S)-HETE and 15(S)-HETE in IL-4-polarized macrophages. The ratio of 12(S)-HETE to 15(S)-HETE generated in IL-4-treated macrophages roughly corresponds to that reported previously for purified ALOX15 (38). It is surprising that IL-4-treated macrophages accumulate intracellular 12(S)-HETE and 15(S)-HETE without the addition of exogenous arachidonic acid. The mechanism of how free arachidonate, the substrate for 15-lipoxygenase, is generated under these conditions requires further investigation. 15-lipoxgenases are also implicated in the generation of proresolving lipid mediators form arachidonic, eicosapentaenoic, or docosahexaenoic acid (39). However, this mostly requires transcellular lipid modification and is unlikely to play a major role in IL-4-stimulated macrophages when ALOX5 and ALOX15 are regulated opposed to each other.
The physiological significance of 15-lipoxygenase in macrophages is not completely understood. ALOX15 has been proposed to generate ligands for the nuclear receptor PPARγ (7). Although PPARγ gene expression is induced by IL-4, which has also been reported previously, we observed only up-regulation of FABP4. FABP4 is a potently induced PPARγ target gene in different cell types, but its response to IL-4 stimulation was considerably lower compared with rosiglitazone. Because other PPARγ target genes, such as CD36 or lipoprotein lipase, were unaffected by IL-4, we suggest that the magnitude of PPARγ activation via the IL-4/ALOX15 axis may be quite small in fully matured human macrophages. 15-HETE has also been reported to be a ligand for the PPARδ transcription factor (40), but we did not observe any changes in the expression of the typical PPARδ target genes perilipin 2 and pyruvate dehydrogenase kinase 4 in IL-4-treated cells (data not shown).
The role of 15-lipoxygenase and the products of its enzymatic activity during inflammation is controversial (12). Although 12/15-lipoxygenase has been reported to be anti-inflammatory in mouse knockout models of rheumatoid arthritis or peritonitis (41, 42), other studies have suggested proinflammatory roles of 12/15-lipoxygenase and 12(S)-HETES or 15(S)-HETE in obese adipose tissue (43) or vascular endothelial cells (27, 44). Studies in 12/15-lipoxygenase-deficient murine peritoneal macrophages showed 12/15-lipoxygenase to augment toll-like receptor-induced expression of IL-12 (28), potentially involving interferon regulatory factor 8. Interestingly because the authors found no involvement of lipoxygenase-derived lipid mediators, they proposed the effect being mediated by lipoxygenase-derived reactive oxygen species. This concurs with reports showing 12/15-lipoxygenase-catalzed lipid peroxidation to modulate intracellular signaling, i.e. tyrosine phosphatase activity (45). Our data point to 15(S)-HETE being involved in proinflammatory macrophage activation, although a much lower efficacy was observed compared with IL-4 pretreatment. Therefore, we do not exclude lipid peroxidation-derived reactive oxygen species as codrivers of ALOX15-dependent proinflammatory skewing of human macrophages. Alternatively, the intracellular location of 12(S)-HETE and 15(S)-HETE may be important for their effects on inflammation, and, therefore, adding these lipids extracellularly may not deliver them to the appropriate sites.
Overexpression of ALOX15B in human THP-1 macrophages promoted CXCL10 secretion (15). Interestingly, the expression of several interferon-related genes, including CXCL9 and CXCL10, is characteristic for tumor-associated macrophages (46). Although the contribution of 12/15-lipoxygenase in shaping the tumor-associated macrophage phenotype is unknown, we recently found increased ALOX15 expression in macrophages phagocytosing dying tumor cells.3 Details of how 15-lipoxygenase contributes to the tumor-associated macrophage phenotype and the question of whether AMPK affects tumor-associated macrophages through inhibition of ALOX15 warrants further studies.
In summary, we provide evidence that AMPK interferes with IL-4-induced human macrophage polarization by attenuating STAT3 activation, which diminishes ALOX15 expression. In the absence of ALOX15, the corresponding arachidonic acid metabolites 12-HETE and 15-HETE are not produced. This, as well as other, still unknown factor(s), lowers the proinflammatory priming potency of IL-4 in macrophages (Fig. 5E), adding to the established anti-inflammatory role of AMPK in phagocytes.
Author Contributions
D. N. designed, performed, and analyzed the experiments and wrote the paper. R. G. S. and N. D. assisted with some of the experiments. C. A. and G. G. contributed to arachidonic acid metabolite analysis. N. G. provided critical reagents. B. B. supervised the study and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Yasumichi Hitoshi (Rigel Pharmaceuticals) for R419, Dr. Jason Dyck and Dr. Amy Barr (University of Alberta) for the AMPKγ1 R70Q adenovirus, and Isabelle Elschner for technical assistance.
This study was supported by Deutsche Forschungsgemeinschaft Grants NA429/2-1 and SFB 1039 Teilprojekt A05, B04, and Z01 and by the Else Kröner Fresenius Foundation (TRIP). The authors declare that they have no conflicts of interest with the contents of this article.
N. Grossmann, unpublished observations.
- ALOX15
- arachidonate 15-lipoxygenase
- AMPK
- AMP-activated protein kinase
- AICAR
- aminoimidazole-4-carboxamide 1-β-d-ribofuranoside
- ACC
- acetyl-CoA carboxylase
- 15(S)-HETE
- 15(S)-hydroxyeicosatetraenoic acid
- 12(S)-HETE
- 12(S)-hydroxyeicosatetraenoic acid
- 15(R)-HETE
- 15(R)-hydroxyeicosatetraenoic acid
- PPARγ
- peroxisome proliferator-activated receptor.
References
- 1. Mosser D. M., Edwards J. P. (2008) Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8, 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Van Dyken S. J., Locksley R. M. (2013) Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu. Rev. Immunol. 31, 317–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gordon S., Martinez F. O. (2010) Alternative activation of macrophages: mechanism and functions. Immunity 32, 593–604 [DOI] [PubMed] [Google Scholar]
- 4. Hart P. H., Vitti G. F., Burgess D. R., Whitty G. A., Piccoli D. S., Hamilton J. A. (1989) Potential antiinflammatory effects of interleukin 4: suppression of human monocyte tumor necrosis factor α, interleukin 1, and prostaglandin E2. Proc. Natl. Acad. Sci. U.S.A. 86, 3803–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Varin A., Mukhopadhyay S., Herbein G., Gordon S. (2010) Alternative activation of macrophages by IL-4 impairs phagocytosis of pathogens but potentiates microbial-induced signalling and cytokine secretion. Blood 115, 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. El Chartouni C., Rehli M. (2010) Comprehensive analysis of TLR4-induced transcriptional responses in interleukin 4-primed mouse macrophages. Immunobiology 215, 780–787 [DOI] [PubMed] [Google Scholar]
- 7. Huang J. T., Welch J. S., Ricote M., Binder C. J., Willson T. M., Kelly C., Witztum J. L., Funk C. D., Conrad D., Glass C. K. (1999) Interleukin-4-dependent production of PPAR-γ ligands in macrophages by 12/15-lipoxygenase. Nature 400, 378–382 [DOI] [PubMed] [Google Scholar]
- 8. Martinez F. O., Gordon S., Locati M., Mantovani A. (2006) Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 177, 7303–7311 [DOI] [PubMed] [Google Scholar]
- 9. Conrad D. J., Kuhn H., Mulkins M., Highland E., Sigal E. (1992) Specific inflammatory cytokines regulate the expression of human monocyte 15-lipoxygenase. Proc. Natl. Acad. Sci. U.S.A. 89, 217–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ivanov I., Heydeck D., Hofheinz K., Roffeis J., O'Donnell V. B., Kuhn H., Walther M. (2010) Molecular enzymology of lipoxygenases. Arch. Biochem. Biophys. 503, 161–174 [DOI] [PubMed] [Google Scholar]
- 11. Kuhn H., Banthiya S., van Leyen K. (2015) Mammalian lipoxygenases and their biological relevance. Biochim. Biophys. Acta 1851, 308–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uderhardt S., Krönke G. (2012) 12/15-lipoxygenase during the regulation of inflammation, immunity, and self-tolerance. J. Mol. Med. 90, 1247–1256 [DOI] [PubMed] [Google Scholar]
- 13. Wuest S. J., Crucet M., Gemperle C., Loretz C., Hersberger M. (2012) Expression and regulation of 12/15-lipoxygenases in human primary macrophages. Atherosclerosis 225, 121–127 [DOI] [PubMed] [Google Scholar]
- 14. Rydberg E. K., Krettek A., Ullström C., Ekström K., Svensson P. A., Carlsson L. M., Jönsson-Rylander A. C., Hansson G. I., McPheat W., Wiklund O., Ohlsson B. G., Hultén L. M. (2004) Hypoxia increases LDL oxidation and expression of 15-lipoxygenase-2 in human macrophages. Arterioscler. Thromb. Vasc. Biol. 24, 2040–2045 [DOI] [PubMed] [Google Scholar]
- 15. Danielsson K. N., Rydberg E. K., Ingelsten M., Akyürek L. M., Jirholt P., Ullström C., Forsberg G. B., Borén J., Wiklund O., Hultén L. M. (2008) 15-Lipoxygenase-2 expression in human macrophages induces chemokine secretion and T cell migration. Atherosclerosis 199, 34–40 [DOI] [PubMed] [Google Scholar]
- 16. Hardie D. G. (2011) AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 25, 1895–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O'Neill L. A., Hardie D. G. (2013) Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature 493, 346–355 [DOI] [PubMed] [Google Scholar]
- 18. Sag D., Carling D., Stout R. D., Suttles J. (2008) Adenosine 5′-monophosphate-activated protein kinase promotes macrophage polarization to an anti-inflammatory functional phenotype. J. Immunol. 181, 8633–8641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mounier R., Théret M., Arnold L., Cuvellier S., Bultot L., Göransson O., Sanz N., Ferry A., Sakamoto K., Foretz M., Viollet B., Chazaud B. (2013) AMPKα1 regulates macrophage skewing at the time of resolution of inflammation during skeletal muscle regeneration. Cell Metab. 18, 251–264 [DOI] [PubMed] [Google Scholar]
- 20. Jenkins Y., Sun T. Q., Markovtsov V., Foretz M., Li W., Nguyen H., Li Y., Pan A., Uy G., Gross L., Baltgalvis K., Yung S. L., Gururaja T., Kinoshita T., Owyang A., Smith I. J., McCaughey K., White K., Godinez G., Alcantara R., Choy C., Ren H., Basile R., Sweeny D. J., Xu X., Issakani S. D., Carroll D. C., Goff D. A., Shaw S. J., Singh R., Boros L. G., Laplante M. A., Marcotte B., Kohen R., Viollet B., Marette A., Payan D. G., Kinsella T. M., Hitoshi Y. (2013) AMPK activation through mitochondrial regulation results in increased substrate oxidation and improved metabolic parameters in models of diabetes. PLoS ONE 8, e81870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hamilton S. R., Stapleton D., O'Donnell J. B. Jr., Kung J. T., Dalal S. R., Kemp B. E., Witters L. A. (2001) An activating mutation in the γ1 subunit of the AMP-activated protein kinase. FEBS Lett. 500, 163–168 [DOI] [PubMed] [Google Scholar]
- 22. Bhattacharjee A., Shukla M., Yakubenko V. P., Mulya A., Kundu S., Cathcart M. K. (2013) IL-4 and IL-13 employ discrete signaling pathways for target gene expression in alternatively activated monocytes/macrophages. Free Radic. Biol. Med. 54, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shankaranarayanan P., Chaitidis P., Kühn H., Nigam S. (2001) Acetylation by histone acetyltransferase CREB-binding protein/p300 of STAT6 is required for transcriptional activation of the 15-lipoxygenase-1 gene. J. Biol. Chem. 276, 42753–42760 [DOI] [PubMed] [Google Scholar]
- 24. Odegaard J. I., Ricardo-Gonzalez R. R., Goforth M. H., Morel C. R., Subramanian V., Mukundan L., Red Eagle A., Vats D., Brombacher F., Ferrante A. W., Chawla A. (2007) Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature 447, 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Szanto A., Balint B. L., Nagy Z. S., Barta E., Dezso B., Pap A., Szeles L., Poliska S., Oros M., Evans R. M., Barak Y., Schwabe J., Nagy L. (2010) STAT6 transcription factor is a facilitator of the nuclear receptor PPARγ-regulated gene expression in macrophages and dendritic cells. Immunity 33, 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Namgaladze D., Kemmerer M., von Knethen A., Brüne B. (2013) AICAR inhibits PPARγ during monocyte differentiation to attenuate inflammatory responses to atherogenic lipids. Cardiovasc. Res. 98, 479–487 [DOI] [PubMed] [Google Scholar]
- 27. Li J., Rao J., Liu Y., Cao Y., Zhang Y., Zhang Q., Zhu D. (2013) 15-Lipoxygenase promotes chronic hypoxia-induced pulmonary artery inflammation via positive interaction with nuclear factor-κB. Arterioscler. Thromb. Vasc. Biol. 33, 971–979 [DOI] [PubMed] [Google Scholar]
- 28. Middleton M. K., Rubinstein T., Puré E. (2006) Cellular and molecular mechanisms of the selective regulation of IL-12 production by 12/15-lipoxygenase. J. Immunol. 176, 265–274 [DOI] [PubMed] [Google Scholar]
- 29. Galic S., Fullerton M. D., Schertzer J. D., Sikkema S., Marcinko K., Walkley C. R., Izon D., Honeyman J., Chen Z. P., van Denderen B. J., Kemp B. E., Steinberg G. R. (2011) Hematopoietic AMPK β1 reduces mouse adipose tissue macrophage inflammation and insulin resistance in obesity. J. Clin. Invest. 121, 4903–4915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang Z., Kahn B. B., Shi H., Xue B. Z. (2010) Macrophage α1 AMP-activated protein kinase (α1AMPK) antagonizes fatty acid-induced inflammation through SIRT1. J. Biol. Chem. 285, 19051–19059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nerstedt A., Johansson A., Andersson C. X., Cansby E., Smith U., Mahlapuu M. (2010) AMP-activated protein kinase inhibits IL-6-stimulated inflammatory response in human liver cells by suppressing phosphorylation of signal transducer and activator of transcription 3 (STAT3). Diabetologia 53, 2406–2416 [DOI] [PubMed] [Google Scholar]
- 32. Meares G. P., Qin H., Liu Y., Holdbrooks A. T., Benveniste E. N. (2013) AMP-activated protein kinase restricts IFN-γ signaling. J. Immunol. 190, 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vasamsetti S. B., Karnewar S., Kanugula A. K., Thatipalli A. R., Kumar J. M., Kotamraju S. (2015) Metformin inhibits monocyte-to-macrophage differentiation via AMPK-mediated inhibition of STAT3 activation: potential role in atherosclerosis. Diabetes 64, 2028–2041 [DOI] [PubMed] [Google Scholar]
- 34. Zhang Y., Qiu J., Wang X., Zhang Y., Xia M. (2011) AMP-activated protein kinase suppresses endothelial cell inflammation through phosphorylation of transcriptional coactivator p300. Arterioscler. Thromb. Vasc. Biol. 31, 2897–2908 [DOI] [PubMed] [Google Scholar]
- 35. Chaitidis P., O'Donnell V., Kuban R. J., Bermudez-Fajardo A., Ungethuem U., Kühn H. (2005) Gene expression alterations of human peripheral blood monocytes induced by medium-term treatment with the TH2-cytokines interleukin-4 and -13. Cytokine 30, 366–377 [DOI] [PubMed] [Google Scholar]
- 36. Martinez F. O., Helming L., Milde R., Varin A., Melgert B. N., Draijer C., Thomas B., Fabbri M., Crawshaw A., Ho L. P., Ten Hacken N. H., Cobos Jiménez V., Kootstra N. A., Hamann J., Greaves D. R., Locati M., Mantovani A., Gordon S. (2013) Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood 121, e57–69 [DOI] [PubMed] [Google Scholar]
- 37. Xue J., Schmidt S. V., Sander J., Draffehn A., Krebs W., Quester I., De Nardo D., Gohel T. D., Emde M., Schmidleithner L., Ganesan H., Nino-Castro A., Mallmann M. R., Labzin L., Theis H., Kraut M., Beyer M., Latz E., Freeman T. C., Ulas T., Schultze J. L. (2014) Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40, 274–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sloane D. L., Leung R., Craik C. S., Sigal E. (1991) A primary determinant for lipoxygenase positional specificity. Nature 354, 149–152 [DOI] [PubMed] [Google Scholar]
- 39. Buckley C. D., Gilroy D. W., Serhan C. N. (2014) Proresolving lipid mediators and mechanisms in the resolution of acute inflammation. Immunity 40, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Naruhn S., Meissner W., Adhikary T., Kaddatz K., Klein T., Watzer B., Müller-Brüsselbach S., Müller R. (2010) 15-hydroxyeicosatetraenoic acid is a preferential peroxisome proliferator-activated receptor β/δ agonist. Mol. Pharmacol. 77, 171–184 [DOI] [PubMed] [Google Scholar]
- 41. Krönke G., Katzenbeisser J., Uderhardt S., Zaiss M. M., Scholtysek C., Schabbauer G., Zarbock A., Koenders M. I., Axmann R., Zwerina J., Baenckler H. W., van den Berg W., Voll R. E., Kühn H., Joosten L. A., Schett G. (2009) 12/15-lipoxygenase counteracts inflammation and tissue damage in arthritis. J. Immunol. 183, 3383–3389 [DOI] [PubMed] [Google Scholar]
- 42. Dioszeghy V., Rosas M., Maskrey B. H., Colmont C., Topley N., Chaitidis P., Kühn H., Jones S. A., Taylor P. R., O'Donnell V. B. (2008) 12/15-Lipoxygenase regulates the inflammatory response to bacterial products in vivo. J. Immunol. 181, 6514–6524 [DOI] [PubMed] [Google Scholar]
- 43. Cole B. K., Lieb D. C., Dobrian A. D., Nadler J. L. (2013) 12- and 15-lipoxygenases in adipose tissue inflammation. Prostaglandins Other Lipid Mediat. 104–105, 84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kundumani-Sridharan V., Dyukova E., Hansen D. E. 3rd, Rao G. N. (2013) 12/15-Lipoxygenase mediates high-fat diet-induced endothelial tight junction disruption and monocyte transmigration: a new role for 15(S)-hydroxyeicosatetraenoic acid in endothelial cell dysfunction. J. Biol. Chem. 288, 15830–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Conrad M., Sandin A., Förster H., Seiler A., Frijhoff J., Dagnell M., Bornkamm G. W., Rådmark O., Hooft van Huijsduijnen R., Aspenström P., Böhmer F., Ostman A. (2010) 12/15-lipoxygenase-derived lipid peroxides control receptor tyrosine kinase signaling through oxidation of protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U.S.A. 107, 15774–15779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Biswas S. K., Gangi L., Paul S., Schioppa T., Saccani A., Sironi M., Bottazzi B., Doni A., Vincenzo B., Pasqualini F., Vago L., Nebuloni M., Mantovani A., Sica A. (2006) A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-κB and enhanced IRF-3/STAT1 activation). Blood 107, 2112–2122 [DOI] [PubMed] [Google Scholar]