Background: A unique Glu(2.50) in the NK1 receptor interacts directly with Ser(3.39) and Asn(7.49).
Results: Mutational changes in this interface create receptors that selectively signal through Gq or β-arrestin versus Gs.
Conclusion: A focal point in differentiation between Gs, Gq, and β-arrestin signaling was identified.
Significance: This network constitutes an allosteric interface essential for 7TM receptor fine-tuning toward different signaling pathways.
Keywords: cell signaling, G protein-coupled receptor (GPCR), homology modeling, molecular pharmacology, receptor structure-function, functional selectivity, hydrogen bond network
Abstract
X-ray structures, molecular dynamics simulations, and mutational analysis have previously indicated that an extended water hydrogen bond network between trans-membranes I–III, VI, and VII constitutes an allosteric interface essential for stabilizing different active and inactive helical constellations during the seven-trans-membrane receptor activation. The neurokinin-1 receptor signals efficiently through Gq, Gs, and β-arrestin when stimulated by substance P, but it lacks any sign of constitutive activity. In the water hydrogen bond network the neurokinin-1 has a unique Glu residue instead of the highly conserved AspII:10 (2.50). Here, we find that this GluII:10 occupies the space of a putative allosteric modulating Na+ ion and makes direct inter-helical interactions in particular with SerIII:15 (3.39) and AsnVII:16 (7.49) of the NPXXY motif. Mutational changes in the interface between GluII:10 and AsnVII:16 created receptors that selectively signaled through the following: 1) Gq only; 2) β-arrestin only; and 3) Gq and β-arrestin but not through Gs. Interestingly, increased constitutive Gs but not Gq signaling was observed by Ala substitution of four out of the six core polar residues of the network, in particular SerIII:15. Three residues were essential for all three signaling pathways, i.e. the water-gating micro-switch residues TrpVI:13 (6.48) of the CWXP motif and TyrVII:20 (7.53) of the NPXXY motif plus the totally conserved AsnI:18 (1.50) stabilizing the kink in trans-membrane VII. It is concluded that the interface between position II:10 (2.50), III:15 (3.39), and VII:16 (7.49) in the center of the water hydrogen bond network constitutes a focal point for fine-tuning seven trans-membrane receptor conformations activating different signal transduction pathways.
Introduction
Within the last decade, high resolution x-ray structures of a large number of family A seven trans-membrane (7TM),4 G protein-coupled receptors have underlined the association between highly conserved residues in the trans-membrane segments and receptor function (1–4). These conserved residues are particularly located in the intracellular parts of the trans-membrane helices where they function either as micro-switches or are involved in the large water-mediated hydrogen bond network. This network constitutes an extended allosteric interface between the helices that perform the large scale movements when the receptor alternates between inactive and active conformations (2, 5). The water-mediated hydrogen bond network is limited at the extracellular side by TrpVI:13 (6.48)5 of the so-called CWXP motif in the middle of TM-VI, which forms the “gate” between the network and the extracellular facing ligand-binding pocket (2). At the cytoplasmic side, the network is limited by TyrVII:20 (7.53), which is part of the NPXXY motif at the intracellular end of TM-VII (Fig. 1A). Previously, by use of molecular dynamics simulations and molecular pharmacological means, we have presented evidence indicating that key polar residues of this interface are essential for the signal transduction process in the β2-adrenergic receptor (B2AR) and that TrpVI:13 (6.48) and TyrVII:20 (7.53) appear to function as gates for the movements of water molecules in and out of the network (5).
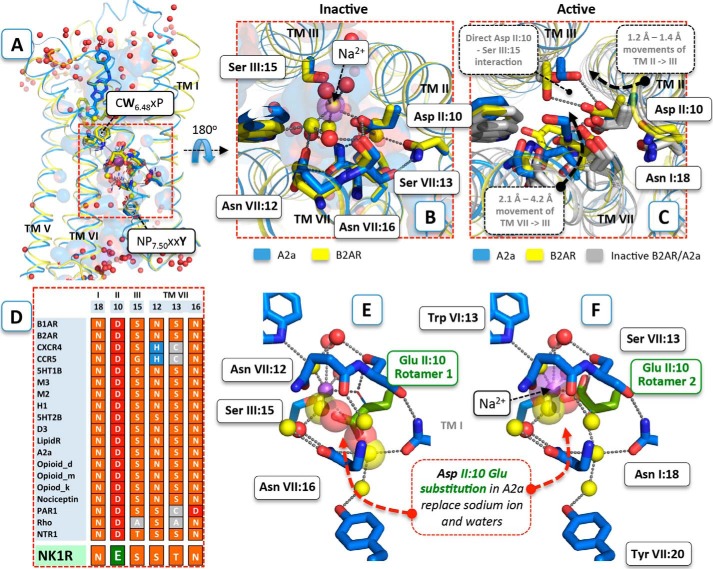
FIGURE 1.
Structural conservation of the polar interface between TM-I–III and -VII. A, structural distribution of conserved polar residues between the intracellular half of TM-I–III and -VII of 18 unique inactive GPCR structures. The residues AsnI:18 (1.50), AspII:10 (2.50), SerIII:15 (3.39), SerVII:12 (7.45), ThrVII:13 (7.46), and AsnVII:16 (7.49) of the conserved polar interface restricted by the functionally important and conserved TrpVI:13 (6.48) and TyrVII:20 (7.53) in the antagonist-bound structures of A2AAR (PDB code 4EIY solved at 1.8 Å resolution) (blue) and B2AR (PDB code 2RH1) (yellow) are shown as sticks. The corresponding residues in the other inactive structures are shown by gray lines. B, close-up view of the polar interface comprising a sodium ion (purple sphere) and water molecules (red spheres). Co-localized water molecules in the A2AAR and the B2AR structure are shown as yellow spheres. The salt bridge between sodium and AspII:10 (2.50) and water-mediated hydrogen bond interactions are shown as gray dotted lines. C, close-up view of the polar pocket in the active-like agonist-bound A2AAR/UK432,097 structure (PDB code 3QAK) (blue) and the active agonist-bound B2AR structure in complex with Gs (PDB code 3SN6). D, polar pocket alignment of the distinct inactive experimental structures compared with the conforming sequence in the NK1 receptor, which have a glutamic acid instead of the highly conserved AspII:10 (2.50). E and F, structural details of AspII:10 Glu substitution (green) in the inactive A2AAR structure (E, first rotamer conformation of AspII:10 Glu substitution; F, second rotamer conformation of AspII:10 Glu substitution). Water molecules that make steric clashes with the substituted glutamic acid are shown as big transparent spheres.
The polar residues of the water hydrogen bond network are not only highly conserved in the primary structure (Fig. 1D) but are also almost superimposable in all of the other 20 unique inactive x-ray structures (Fig. 1A) solved to date (6–26). Surprisingly, the structures revealed that in the inactive state almost no hydrogen bonds are observed between the polar residues themselves. Instead, they interact through a number of structural water molecules generating the extended hydrogen bond network as shown for the inactive adenosine A2A receptor (A2AAR) and B2AR structures in Fig. 1, A–C (10, 16). Interestingly, a sodium ion was identified as an important part of this network located almost identically in the A2AAR, the protease-activated receptor 1, and most recently in the 1.8-Å high resolution structure of the δ-opioid receptor (12, 26–28). In these structures, the sodium ion is coordinated by the highly conserved AspII:10 (2.50) in TM-II, SerIII:15 (3.39) in TM-III and nearby water molecules in a position previously shown to be taken up by either structural water molecules or left as a relatively large “empty space” in x-ray structures of inactive forms of other 7TM receptors (28). Importantly, in the active structure of the B2AR and the active-like structures of the A2AAR, this region has undergone major conformational changes, including the well recognized major tilts of the intracellular parts of TM-V (inward) and TM-VI (outward) combined with an inward tilt of TM-VII (more than 2 Å combined with rotation), and an axial shift of TM-III (29, 30). These movements eliminate much of the space occupied by water and the sodium ion in the inactive conformation so that in the active state the polar residues instead mainly make hydrogen bonds directly with each other (Fig. 1C). The notion that a sodium ion stabilizes the inactive conformation of 7TM receptors, and acts as negative allosteric modulators, was well established 1½ decades ago through studies of the α2A adrenergic receptor, the A2AAR, the μ- and δ-opioid receptors, and the D2 dopamine receptor, for example (31–37). However, we now have structural proof of this interaction in the A2AAR and the δ-opioid receptor, which provides a detailed picture of how other residues besides AspII:10 (2.50) (in particular SerIII:15 (3.39)) are also involved in the binding of the allosteric negative regulator Na+ (16, 27, 30).
In this study, we investigate the relative role of different residues in the conserved polar interface in the neurokinin-1 (NK1) receptor, which is activated by substance P and signals through both the Gq and the Gs pathway (38–41). A unique structural feature of the NK1 receptor is a glutamic acid residue in position II:10 (2.50), in contrast to aspartic acid found in more than 98% of other family A receptors (42). Interestingly, the unstimulated NK1 receptor is very “silent” with no evidence of constitutive activity resulting from spontaneous transition to the active state. For more than 2 decades we have performed structural-functional studies on the NK1 receptor and have never observed measurable constitutive activity, and in contrast to most other 7TM receptors, we have never identified an NK1 mutant showing ligand-independent signaling (38, 43–47). This is particularly surprising as it is generally known in the field that it is rather easy to find mutations in 7TM receptors that make them signal with high constitutive activity.
We now find that in molecular models of the NK1 receptor the γ-carboxylic acid group of the larger GluII:10 (2.50) side chain (compared with Asp) is located at or very close to the position where the negative allosteric modulating sodium ion is found in the A2AAR, the δ-opioid receptor, and the β1-adrenergic receptor, for example (27, 28, 32, 48), and that it makes direct hydrogen bonds to other members of the water-hydrogen bond network, SerIII:15 (3.39), and AsnVII:16 (7.49). Mutational analysis indicates that GluII:10 and its two hydrogen bond partners in different ways are involved in tuning Gs versus Gq and β-arrestin signaling and that negative modulatory Na+ effects could be induced in the NK1 receptor by “reintroducing” Asp in position II:10.
Experimental Procedures
Comparative Homology Modeling
The sequence of the human NK1 receptor was obtained at the Uniprot web page. The distinct x-ray structures (available at the time the modelings were conducted) of 7TM receptors in inactive-like states (adenosine A2A (PDB code 4EIY), β1-adrenergic receptor (PDB code 2VT4), β2-adrenergic receptor (PDB code 2RH1), chemokine CXCR4 (PDB code 3ODU), chemokine CCR5 (PDB code 4MBS), dopamine D3 (PDB code 3PBL), histamine H1 (PDB code 3RZE), muscarinic M2 (PDB code 3OUN), muscarinic M3 (PDB code 4DAJ), serotonin 5HT1B (PDB code 4IAR), serotonin 5HT2B (PDB code 4IB4), μ-opioid (PDB code 4DKL), δ-opioid (PDB code 4EJ4), κ-opioid (PDB code 4DJH), nociceptin (PDB code 4EA3), protease-activated receptor 1 (PDB code 3VW7), sphingosine S1P (PDB code 3V2Y), and rhodopsin (PDB code 1GZM)) as well as the structures in active-like states (adenosine A2A (PDB code 3QAK), β2-adrenergic receptor (PDB code 3SN6), muscarinic M2 (PDB code 4MQS), neurotensin 1 receptor (PDB code 4GRV)) were obtained from the PDB database. The structures were superimposed with respect to the intracellular half of TM-I–III and -VII using the CEAlign protocol in PyMOL. Pairwise sequence alignments and comparative homology models (excluding intra- and extracellular loops) of the NK1 receptor were produced in ICM 3.7b using the inactive- and active-like structures of A2AAR and B2AR as templates that devised reasonable high sequence identity to the intracellular half of TM-I–III and -VII. GluII:10 was assumed to be negatively charged as it is located in-between SerIII:15 and ThrVII:13 in an optimal position to be involved in hydrogen bond interactions, the hydroxyl side chain of SerIII:15, and the co-existence of “conserved” bulky water molecules observed in other receptors with similar environments. Initially, hydrogen atoms and side chains were optimized using the comparative modeling protocol in ICM. When modeling the inactive state of NK1R, the sodium ion and structural water molecules of the template structures were “copied” to the comparative NK1 receptor models. The location of the sodium ion and water molecules was initially accessed in the presence of a fixed NK1 receptor model. The sodium ion and a nearby water molecule, which are involved in hydrogen bond interactions with AspII:10 and SerIII:15 in the parent A2AAR and B2AR structures, were removed from the NK1 receptor models as they make steric clashes with the larger (compared with Asp) carboxyl acid side chain of GluII:10 in the NK1 receptor models. Local optimization of side chains within 8 Å of this co-localized sodium ion/water molecule was unable to release the strain when optimizing all atoms using the biased probability Monte Carlo optimization protocol (200 global moves and 100 local minimization calls). To ensure acceptance of the remaining water molecules, side chains and water molecules were globally optimized using the Monte Carlo conformational sampling procedure in the presence of a fixed main chain conformation in ICM 3.7b. Finally, the resulting NK1 models were minimized in 300 steps of steepest decent, using the MMFF force field, and all atoms were free to move. In this work we only present the NK1 receptor model based on A2AAR as the corresponding NK1 receptor model based on B2AR is highly similar.
Materials
Substance P and ghrelin were purchased from Bachem and 125I-labeled Lys3-SP from PerkinElmer Life Sciences. Pindolol was purchased from Sigma, and AR-231453 was a generous gift from Arena Pharmaceuticals (San Diego).
Molecular Biology
The NK1, B2AR, and GHSR cDNA were cloned into the eukaryotic expression vector pCMV-Tag(2B) (Stratagene), and GPR119 cDNA was cloned into the pcDNA3.1 vector (Invitrogen). Mutations were constructed by PCR using the QuikChange method. All PCR experiments were performed using Pfu polymerase (Stratagene) according to the instructions of the manufacturer. All mutations were verified by DNA sequence analysis by MWG (Ebersberg, Germany). For β-arrestin recruitment, cDNAs encoding native and mutant NK1 receptors were tagged at the C terminus with an enzyme donor fragment of β-galactosidase.
Transfections and Tissue Culture
COS-7 cells were grown in Dulbecco's modified Eagle's medium 1885 supplemented with 10% fetal calf serum, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were transfected with 20 μg/75 cm2 DNA using the calcium phosphate precipitation method with chloroquine addition as described previously (45).
CHO-K1 cells stably expressing β-arrestin2 tagged with the enzyme-accepting portion of a β-galactosidase enzyme were grown in Ham's F-12 media supplemented with 10% fetal calf serum, 2 mm glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 300 μg/ml hygromycin. Cells were transfected using 0.15 μl of FuGENE (Promega, Madison, WI)/well in opaque white 96-well plates using 50 ng of DNA/well according to the manufacturer's protocol.
Cell Surface Expression (ELISA)
Cells transfected and seeded for IP3 or cAMP were in parallel seeded for ELISA. The cells were washed twice with 200 μl of phosphate-buffered saline (PBS), fixed with 150 μl of paraformaldehyde for 10 min, and finally incubated in blocking solution (PBS plus 3% dry milk) for 30 min at room temperature. Subsequently, the cells were incubated for 1 h at room temperature with anti-FLAG (M2) (Sigma) antibody diluted 1:1000. The cells were washed three times with PBS and incubated for 1 h at room temperature with anti-mouse horseradish peroxidase-conjugated antibody (Sigma) diluted 1:1250. After three additional washing steps with PBS, immunoreactivity was discovered by addition of horseradish peroxidase. The absorbance was read by TopCount (PerkinElmer Life Sciences).
Competition Binding Assay
Transfected COS-7 cells were plated in poly-d-lysine-coated 96-well plates at a density of 500–20,000 cells/well aiming at 5–10% binding of the radioactive ligand. The following day the binding experiments were performed for 3 h at 4 °C using ∼25 pm 125I-labeled SP (PerkinElmer Life Sciences). Binding assays were performed in 0.1 ml of a HEPES buffer, pH 7.4, supplemented with 1 mm CaCl2, 5 mm MgCl2, 0.1% (w/v) bovine serum albumin, and 40 μg/ml bacitracin with or without increasing concentrations of NaCl. Nonspecific binding was determined as the binding in the presence of 1 μm unlabeled SP. After two washes in cold buffer, Lysis buffer (1% SDS, 200 mm NaOH) was added for 30 min, and radioactivity was counted.
Phosphatidylinositol Turnover Assay
One day after transfection, COS-7 cells were incubated for 24 h with 5 μCi of myo-[3H]inositol (catalogue no. PT-271, Amersham Biosciences) in 300 μl of medium supplemented with 10% fetal calf serum, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin per well. Cells were washed twice in 20 mm HEPES buffer (pH 7.4, supplemented with 140 mm NaCl, 5 mm KCl, 1 mm MgSO4, 1 mm CaCl2, 10 mm glucose, 0.05% (w/v) fetal bovine serum) and then incubated in 0.2 ml of HEPES buffer supplemented with 10 mm LiCl at 37 °C for 15 min. After stimulation with SP for 45 min at 37 °C, cells were extracted with 10 mm formic acid followed by incubation on ice for 30 min. The resulting supernatant was purified on AG 1-X8 anion-exchange resin (Bio-Rad) to isolate the negatively charged inositol phosphates. After application of the cell extract to the column, the columns were washed twice with GPI buffer (60 mm sodium formate and 100 mm formic acid) to remove glycerophosphoinositol. Inositol phosphates were eluded by the addition of elution buffer (1 mm ammonium formate, 100 mm formic acid). Determinations were made in duplicate. The columns containing AG 1-X8 anion-exchange resin were regenerated by the addition of 3 ml of regeneration buffer (3 m ammonium formate, 100 mm formic acid) and 10 ml of water.
cAMP Assay
One day after transfection, COS-7 cells were plated into white 96-well plates (20,000 cells/well). The next day, the cAMP assay was performed using DiscoveRx HitHunterTM cAMPxs+ kit (Fremont, CA) according to the manufacturer's protocol.
β-Arrestin2 Recruitment
The day after transfection, β-arrestin2 recruitment was determined using DiscoveRx PathHunter® β-arrestin GPCR assay (Fremont, CA) according to the manufacturers protocol.
Calculations
EC50 values were determined by nonlinear regression using the Prism 6.0 software (GraphPad Software, San Diego). Unpaired t test (p < 0.05) was performed using the Prism 6.0 software (GraphPad software, San Diego).
Results
Modeling of the Polar Network in the NK1 Receptor
Analysis of 20 unique inactive 7TM receptor structures in complexes with inverse agonists or antagonists demonstrated that polar, water-, and sodium ion-coordinating residues of the conserved interface between the intracellular segments of TM-I–III and -VII, can be almost perfectly superimposed (Fig. 1, A and B) (7–21, 23–26, 50). This is in contrast to residues in the extracellular ligand-binding pocket, which shows larger variations in sequence and structure. Although a sodium ion has only been directly identified in this polar interface of the A2AAR, protease-activated receptor 1, and the δ-opioid receptors, there is also clearly room for it between the highly conserved AspII:10 (2.50) and SerIII:15 (3.39) in all other inactive receptor conformations (37). In contrast, in the molecular model of the inactive conformation of the NK1 receptor, the γ-carboxyl side chain of GluII:10 replaces the sodium ion and/or one or more water molecules making direct rather than water-mediated interactions with not only SerIII:15 (3.39) but also with AsnVII:16 (7.49) (Figs. 1, E and F, and 2A). The helix organization and side chain packing of the energy-optimized NK1 receptor models are highly similar to the inactive experimental structures when superimposed with respect to conserved key residues in the intracellular half of TM-I–III and -VII. Either of the water molecules in the refined models deviate more than 0.75 Å from their original positions, and rotamer states of side chains involved in the water-mediated hydrogen bonds remain unchanged. In addition, GluII:10 (2.50) interacts with TyrVII:20 (7.53) through hydrogen bond interactions with a single water molecule, in contrast to the interaction of AspII:10 (2.50) with TyrVII:20 (7.53) in the inactive structures of A2AAR and B2AR, which is mediated through two water molecules (5, 10, 51).
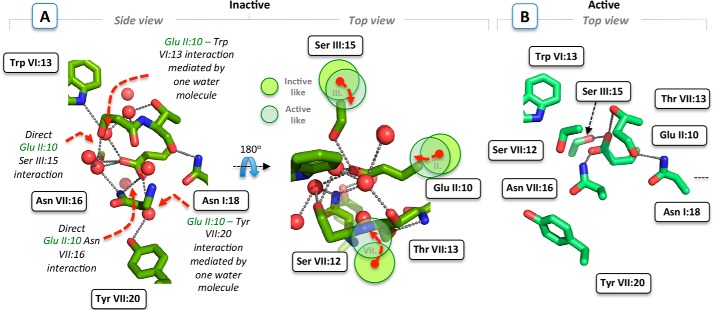
FIGURE 2.
NK1 receptor models. A, side and top close-up view of the conserved polar interface, including AsnI:18 (1. 50), GluII:10 (2.50), SerIII:15 (3.39), SerVII:12 (7.45), ThrVII:13 (7.46), and AsnVII:16 (7.49) in the inactive NK1 receptor model restricted by the functionally important and conserved residues TrpVI:13 (6.48) and TyrVII:20 (7.53). Water molecules are shown as red spheres and hydrogen bonds as gray dotted lines. In contrast to highly conserved AspII:10, the longer GluII:10 side chain in the NK1 model is predicted to make direct rather than water-mediated interactions with SerIII:15 and AsnVII:16. Dashed red arrows show the conformational rearrangements of TM-II, -III, and -VII (illustrated by light green and transparent circles) required to obtain an “active” NK1 receptor structure consistent with common conformational changes observed in distinct active structures. B, model of the conserved polar interface of the NK1 receptor in an active state, based on the agonist-bound A2AAR structure (PDB code 3QAK).
In the models of the active conformation of the NK1 receptor built over the active structures of the B2AR-Gs and A2AAR receptors (16, 29), the volume of the polar interface is greatly reduced due to the inward movement of TMVII toward TMIII by more than 2 Å, which effectively “squeezes” out the possible sodium ion and most of the structural water molecules (Fig. 2, A and B). In this model, the carboxylic acid side chain of GluII:10 (2.50) could either be making direct interactions with SerII:15 (3.39), ThrVII:13 (7.46), and AsnVII:16 (7.49) or direct interactions with SerIII:15 (3.39) and AsnVII:16 (7.49) depending on the detailed side chain packing (Fig. 2B). In conclusion, the γ-carboxyl side chain of GluII:10 (2.50) in the NK1 receptor appears to be substituting the Na+ in the polar interface making direct instead of water-mediated hydrogen bond interactions to other members of the hydrogen bond network (Fig. 2B).
Functional Effects of Ala Substitution of the Polar Residues in the NK1 Hydrogen Bond Network
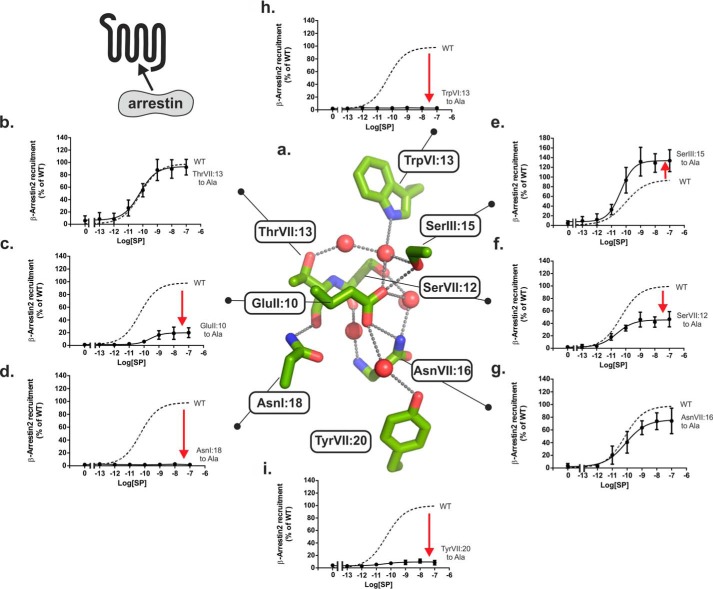
The functional importance of each residue in the water hydrogen bond network of the NK1 receptor was probed by Ala substitutions, and constitutive and SP-induced signaling were determined for the three different signal transduction pathways as follows: 1) Gq signaling monitored by IP3 accumulation assay (Fig. 3); 2) Gs signaling monitoring by cAMP production (Fig. 4); and 3) β-arrestin mobilization measured by an enzyme complementation assay (Fig. 5). The effect of the mutations on cell surface expression was determined for each of the mutants using cell surface ELISA (Fig. 3, insets).
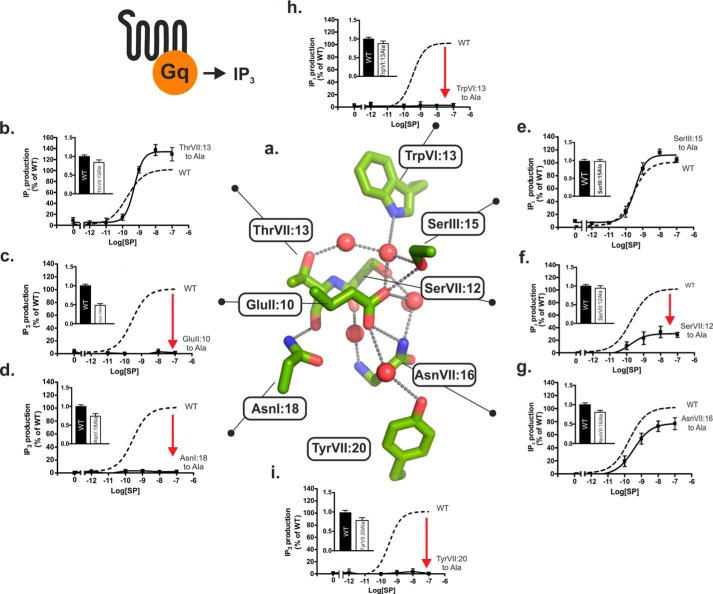
FIGURE 3.
Functional consequence of Ala substitution on Gq signaling of the conserved polar residues in the NK1 receptor. a, structure of the hydrogen bond network in the NK1 receptor model based on A2AAR (PDB code 4EIY) as a template structure. Key side chains are shown as sticks and water molecules as red spheres. b–h, agonist (SP)-induced IP3 production in COS-7 cells transiently transfected with either wild type NK1 (dotted lines) or mutant forms of ThrVII:13Ala (7.46) (b), GluII:10Ala (2.50) (c), AsnI:18Ala (1.50) (d), SerIII:15Ala (3.39) (e), SerVII:12Ala (7.45) (f), AsnVII:16Ala (7.49) (g), TrpVI:13Ala (6.48) (h), and TyrVII:20Ala (7.53) (i). Cell surface receptor expression measured by ELISA is shown in the column diagram insets in each panel.
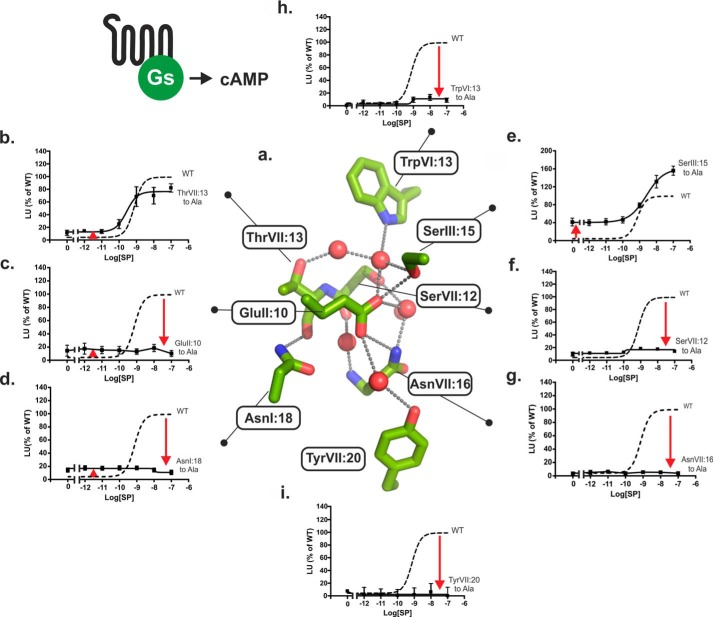
FIGURE 4.
Functional consequence of Ala substitution on Gs signaling of the conserved polar residues in the NK1 receptor. a, structure of the hydrogen bond network in the NK1 receptor model based on A2AAR (PDB code 4EIY) as a template structure. Key side chains are shown as sticks and water molecules as red spheres. b–h, agonist (SP)-induced cAMP production in COS-7 cells transiently transfected with either wild type NK1 (dotted lines) or mutant forms of ThrVII:13Ala (7.46) (b), GluII:10Ala (2.50) (c), AsnI:18Ala (1.50) (d), SerIII:15Ala (3.39) (e), SerVII:12Ala (7.45) (f), AsnVII:16Ala (7.49) (g), TrpVI:13Ala (6.48) (h), and TyrVII:20Ala (7.53) (i).
FIGURE 5.
Functional consequence of Ala substitution on β-arrestin2 mobilization of the conserved polar residues in the NK1 receptor. a, structure of the hydrogen bond network in the NK1 receptor model based on A2AAR (PDB code 4EIY) as a template structure. Key side chains are shown as sticks and water molecules as red spheres. b–h, agonist (SP)-induced β-arrestin2 mobilization in CHOK1 cells transiently transfected with either wild type NK1 (dotted lines) or mutant forms of ThrVII:13Ala (7.46) (b), GluII:10Ala (2.50) (c), AsnI:18Ala (1.50) (d), SerIII:15Ala (3.39) (e), SerVII:12Ala (7.45) (f), AsnVII:16Ala (7.49) (g), TrpVI:13Ala (6.48) (h), and TyrVII:20Ala (7.53) (i).
Effects of Ala Substitutions on Gq Signaling
Ala substitution of each of the two micro-switch residues TrpVI:13 (6.48) of the CWXP motif in TM-VI and TyrVII:20 (7.53) of the NPXXY motif in TM-VII, which place respective extracellular and intracellular limits on the hydrogen bond network, eliminated SP-induced signaling of the NK1 receptor through the Gq pathway as determined by IP3 accumulation (Fig. 3, h and i, and Table 1). Ala substitution of the almost 100% conserved AsnI:18 (1.50), which in the inactive receptor structures forms a direct hydrogen bond to the backbone carbonyl group of ThrVII:13 (7.46) that is exposed at the helical kink in TM-VII (2), also eliminated SP-induced Gq signaling (Fig. 3d). Similarly, Ala substitution of GluII:10 (2.50) totally eliminated SP-induced Gq signaling (Fig. 3c). There was no indication of increased constitutive Gq signaling in the GluII:10 to Ala mutant receptor (Fig. 3c), which otherwise could have been expected based on the potential function of GluII:10 as a tethered negative allosteric regulator as discussed in the modeling section above. In TM-VII, Ala substitution of SerVII:12 (7.45) impaired the SP-induced IP3 accumulation down to 35% of the Emax values observed in the wild type receptor (Fig. 3f and Table 1). In contrast, Ala substitution of AsnVII:16 (7.49) one helical turn below, which we previously found to be essential for Gs signaling in the B2AR, had surprisingly almost no effect on the SP-induced signaling through the Gq pathway in the NK1 receptor (Fig. 3g). Similarly, Ala substitution of ThrVII:13 (7.46) and SerIII:15 (3.39) in TM-III located at the top of the water hydrogen bond network had no effect or very little effect on the Gq signaling in the NK1 receptor (Fig. 3, b and e).
TABLE 1.
Gq, Gs, and β-arrestin signaling of the WT NK1 receptor and mutant forms with Ala substitution in positions AsnI:18 (1.50), GluII:10 (2.50), SerIII:15Ala (3.39), TrpVI:13 (6.48), SerVII:12 (7.45), ThrVII:13 (7.46), AsnVII:16 (7.49), and TyrVII:20 (7.53) and Asp substitution in position GluII:10 (2.50)
The constructs were expressed in transiently transfected COS-7 cells (Gq and Gs) and CHOK1 cells (β-arrestin). The Emax values are expressed as percentage of the ligand-induced signaling for the WT NK1 receptor. Values are shown ± S.E. Comparison is between constitutive activity of WT NK1 and the mutant receptors (p < 0.05). ns = not significant.
| Construct | IP3 |
cAMP |
β-Arrestin2 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Expression level | n | EC50 (nm) | Emax | n | Constitutive activity | EC50 (nm) | Emax | n | EC50 (nm) | Emax | n | |
| NK1 WT | 1 | 8 | 0.31 ± 0.11 | 100 | 7 | 1.7 ± 1.4 | 0.62 ± 0.18 | 100 | 5 | 0.06 ± 0.01 | 100 | 8 |
| AsnI:18Ala (1.50) | 0.81 ± 0.07 | 3 | 3 | 9.3 ± 6a | 4 | 4 | ||||||
| GluII:10Ala (2.50) | 0.56 ± 0.05 | 4 | 3 | 10.8 ± 6.7b | 4 | 0.29 ± 0.03 | 20 ± 3.4 | 6 | ||||
| GluII:10Gln | 1.31 ± 0.09 | 6 | 0.72 ± 0.25 | 38.6 ± 2.5 | 3 | 37.7 ± 4.6c | 3 | 0.46 ± 0.25 | 98.06 ± 11.2 | 4 | ||
| GluII:10Asp | 1.49 ± 0.09 | 4 | 0.35 ± 0.12 | 85.2 ± 6.3 | 4 | 12.0 ± 6.6ns | 20.3 ± 9.6 | 60.9 ± 6.5 | 4 | 3 | ||
| SerIII:15Ala (3.39) | 0.85 ± 0.05 | 3 | 0.42 ± 0.11 | 113 ± 5.3 | 6 | 37.4 ± 6.1c | 1.29 ± 0.35 | 156 ± 17.7 | 6 | 0.07 ± 0.02 | 157 ± 21.0 | 10 |
| TrpVI:13Ala (6.48) | 0.73 ± 0.05 | 3 | 3 | 0.95 ± 0.43 | 14.0 ± 4.9 | 4 | 4 | |||||
| SerVII:12Ala (7.45) | 0.93 ± 0.06 | 6 | 0.17 ± 0.06 | 35.0 ± 6.7 | 4 | 4.2 ± 2.1ns | 5 | 0.03 ± 0.007 | 45.8 ± 4.5 | 6 | ||
| ThrVII:13Ala (7.46) | 0.84 ± 0.07 | 4 | 0.58 ± 0.13 | 133.4 ± 9.4 | 4 | 13.9 ± 4.4d | 0.23 ± 0.08 | 85.5 ± 4.8 | 4 | 0.08 ± 0.04 | 95.2 ± 5.7 | 6 |
| AsnVII:16Ala (7.49) | 0.90 ± 0.06 | 5 | 0.35 ± 0.07 | 76.7 ± 8.1 | 5 | 4 | 0.23 ± 0.07 | 74.8 ± 8.9 | 6 | |||
| TyrVII:20Ala (7.53) | 0.78 ± 0.07 | 3 | 3 | 5 | 0.05 ± 0.02 | 9.3 ± 2.0 | 4 | |||||
a p = 0.0009.
b p = 0.0015.
c p < 0.0001.
d p = 0.0108.
Effects of Ala Substitutions on Gs Signaling
As observed for Gq signaling, Ala substitution of the two water-gating micro-switch residues TrpVI:13 (6.48) and TyrVII:20 (7.53) as well as Ala substitution of AsnI:18 (1.50) and GluII:10 (2.50) eliminated Gs signaling through the NK1 receptor as measured by SP-induced cAMP accumulation and showed no sign of constitutive Gs signaling either (Fig. 4, c, d, h, and i). In contrast, increased basal cAMP production in the absence of agonist was observed not only in GluII:10 but in all of the Ala mutations of the conserved polar residues, except for the mutant forms of SerVII:12 (7.45) and AsnVII:16 (7.49) (Fig. 4 and Table 1). In particular, the SerIII:15 (3.39) to Ala mutant showed a highly increased constitutive Gs signaling of 37 ± 6% of Emax of the wild type receptor corresponding to 24% of its own somewhat increased Emax value (Fig. 4). In the case of the AsnI:18 and GluII:10 substitutions, the SP-induced cAMP response was totally eliminated despite the slight but significantly increased basal Gs signaling of the AnsI:18 and GluII:10 mutants. Surprisingly, in view of the lack of effect on Gq signaling, removal of the polar side chain of AsnVII:16 (7.49) by Ala substitution totally eliminated Gs signaling (Figs. 3g and 4g), just as we had observed for Ala substitution of this residue in the B2AR (5). Furthermore, the AsnVII:16 to Ala substitution did not increase basal Gs signaling (Fig. 4).
Effect of Ala Substitutions on β-Arrestin Mobilization
As shown in Fig. 5, the effects of Ala substitution of the residues in the water hydrogen bond network upon β-arrestin mobilization were rather similar to the effects observed in Gq signaling (Fig. 3). Thus, as observed in the IP3 assay, Ala substitution of TrpVI:13 (6.48), TyrVII:20 (7.53), and AsnI:18 (1.50) in all cases eliminated SP-induced β-arrestin mobilization, and Ala substitution of ThrVII:13 (7.46) and SerIII:15 (3.39) located at the top of the hydrogen bond network had almost no effect or induced a slight increase in β-arrestin mobilization. As for IP3 accumulation assays, a partial inhibition of the β-arrestin response was observed following SerVII:12 (7.46) to Ala mutation (Fig. 5f). In the AsnVII:16 (7.49) to Ala mutant, which selectively inhibited Gs but not Gq, the β-arrestin response was almost similar to that observed for wild type NK1 receptor (Fig. 5g). Interestingly, in the case of GluII:10 (2.50), which was totally unresponsive in both Gq and Gs signaling, SP responses were evident from β-arrestin mobilization, albeit only ∼25% of the response observed in the wild type NK1 receptor (Fig. 5c).
Effect of Substituting GluII:10 with Gln on NK1 Signaling
To examine the effect of removing the potential negative charge of GluII:10 while retaining the ability to participate in hydrogen bond formation, we introduced Gln at this position. Cell surface expression of the Gln mutant was comparable with the WT receptor as shown in Table 1. Through the Gs pathway the Gln substitution of GluII:10 resulted in a very high constitutive signaling corresponding to almost 40% of Emax of the WT receptor, i.e. similar to what was observed with the SerIII:15 to Ala mutant (Table 1). The high constitutive Gs signaling could not be further increased by the agonist SP in the GluII:10 to Gln mutant (Table 1). In respect of Gq signaling, sub-nanomolar potency of SP was retained, but the Emax value was reduced to 40% of the WT receptor (Table 1), and in respect of β-arrestin mobilization, a 10-fold reduction in potency of SP was observed, although the maximum signaling efficacy resembled the WT receptor (Table 1). It is concluded that GluII:10 very likely in its charged form acts as a negative regulator particularly of Gs signaling because substitution with Gln results in very high constitutive signaling.
Effect of “Reintroduction” of Asp for GluII:10 on NK1 Signaling and Na+ Modulation
Mutating GluII:10 (2.50) into aspartic acid, as found in the vast majority of 7TM receptors, increased cell surface expression by ∼50% as compared with wild type receptor (Fig. 6a, inset). In contrast to the Ala substitution, the AspII:10 mutant signaled as wild type NK1 through Gq as determined by IP3 accumulation (Fig. 6a). However, in respect to Gs signaling, the AspII:10 mutant, in contrast to the Ala substitution, displayed no significant constitutive activity in the cAMP assay and had a seriously impaired SP-induced cAMP response with a reduction in potency of at least 2 orders of magnitude and not reaching an Emax with the highest concentration tested (Fig. 6b). In contrast to the GluII:10 to Ala mutant, which did show a clear but diminished β-arrestin response to SP, the GluII:10Asp mutant did not recruit β-arrestin at all upon stimulation with SP (Fig. 6c).
FIGURE 6.
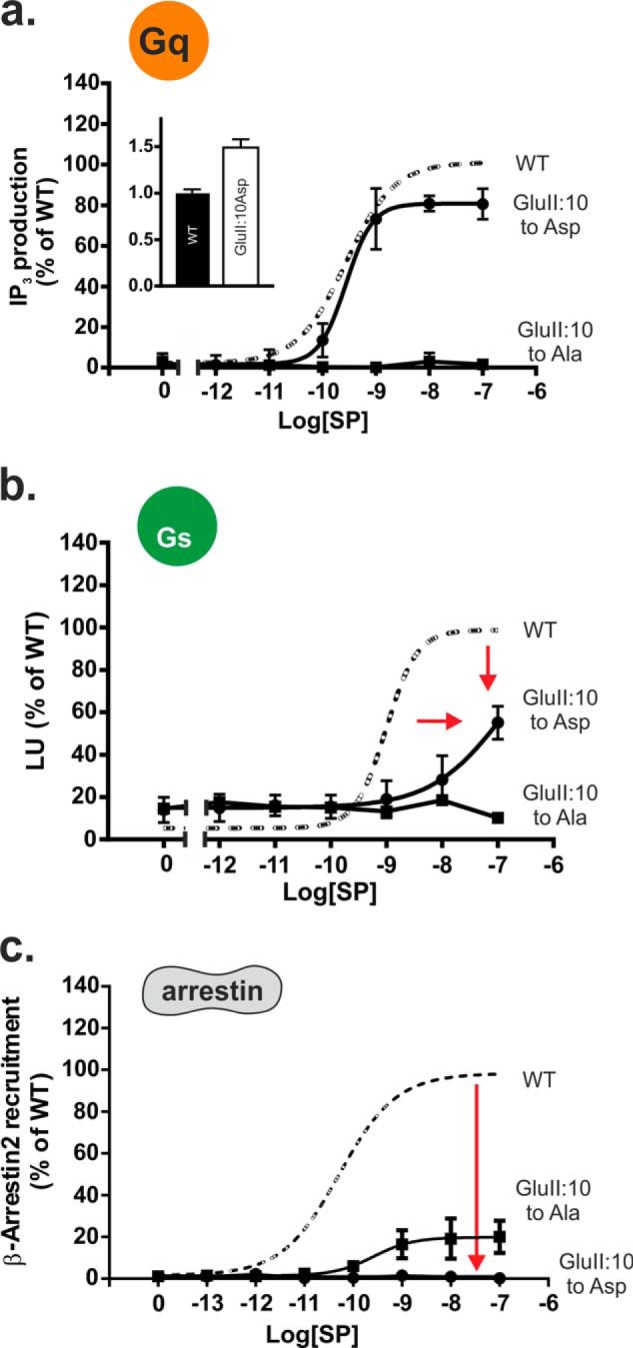
Functional consequence of Asp substitution of the GluII:10 (2.50) residue in the NK1 receptor. a–c, agonist (SP)-induced IP3 (a), cAMP (b), or β-Arrestin2 (c) mobilization in COS-7 cells (IP3 and cAMP) or CHOK1 cells (β-arrestin2) transiently transfected with either wild type NK1 (dotted lines) or mutant GluII:10Ala/Asp. Cell surface receptor expression measured by ELISA is shown in the column diagram inset in a.
The most clear allosteric modulatory effect of Na+ in receptors with an Asp in position II:10 is on ligand binding affinity as recently reported for the δ-opioid receptor (27). In the wild type NK1 receptor, no effect of Na+ on SP binding was observed in whole cell competition binding using radiolabeled SP in transfected COS-7 cells (Table 2). However, when Asp was reintroduced instead of Glu in position II:10, 300 mm Na+ impaired the SP affinity from 0.11 ± 0.05 nm in its absence to 1.2 ± 0.5 nm in its presence (Table 2), i.e. a negative modulating effect similar to what has been reported for the δ-opioid receptor (27). Ala substitution of SerIII:15 (3.39), another residue potentially involved in coordinating Na+ in the water hydrogen bond network, had no effect on the binding of SP in the NK1 receptor, and here no effect on Na+ was observed (Table 2).
TABLE 2.
Ligand binding properties of SP to WT NK1 receptor and substitutions in position GluII:10 (2.50) and SerIII:15 (3.39) in the NK1 receptor using 125I-labeled SP as a radioligand
The constructs were expressed in transiently transfected COS-7 cells. Values are shown ± S.E.
| Construct | Kd (nm) |
|||||
|---|---|---|---|---|---|---|
| n | 0 mm NaCl | 50 mm NaCl | 100 mm NaCl | 200 mm NaCl | 300 mm NaCl | |
| WT NK1 | 5 | 0.125 ± 0.06 | 0.116 ± 0.03 | 0.187 ± 0.09 | 0.314 ± 0.16 | 0.253 ± 0.08 |
| GluII:10A (2.50) | 5 | 0.657 ± 0.28 | 0.520 ± 0.13 | 0.778 ± 0.11 | 1.45 ± 0.43 | 3.18 ± 2.09 |
| GluII:10D | 5 | 0.114 ± 0.04 | 0.128 ± 0.02 | 0.204 ± 0.06 | 0.290 ± 0.07 | 1.20 ± 0.54 |
| SerIII:15A (3.39) | 4 | 0.172 ± 0.04 | 0.469 ± 0.32 | 0.284 ± 0.13 | 0.310 ± 0.10 | 0.446 ± 0.09 |
Effect of Introducing Glu at Position II:10 in GPR119, B2AR, and the Ghrelin Receptor
The B2AR, GPR119, and the Ghrelin receptors were chosen because they display variable degrees of constitutive activity, and all carry an Asp in position II:10. Introduction of a Glu residue at this position as in the NK1 receptor decreased the cell surface expression of all three receptors, although they were still expressed at more than 50% of the corresponding WT receptors (Table 3). In the case of GPR119, the effect of the mutation was difficult to interpret as both the constitutive activity and the Emax were reduced to ∼50% of WT, i.e. corresponding to the reduction in expression level. However, the potency of the agonist AR231453 was also reduced, i.e. EC50 increased from 2.2 to 17 nm (Table 3). In the case of B2AR, introduction of Glu at position II:10 reduced the Emax to 17% of WT and the constitutive activity from 9.9 to 2% (Table 3). However, the negative effect on signal transduction was most clearly observed in the ghrelin receptor, where substitution of AspII:10 with Glu totally eliminated both the constitutive signaling and the ghrelin-induced signaling (Table 3).
TABLE 3.
Gq or Gs signaling of the WT GHSR, B2AR, and GPPR119 and the AspII:10Glu mutant variant of these receptors
The constructs were expressed in transiently transfected COS-7 cells. The constitutive and ghrelin-induced Gq signaling was determined for the ghrelin receptor, whereas signaling through the Gs signaling pathway was determined for B2AR (pindolol) and GPR119 (AR-231453). The Emax values are expressed as percentage of the ligand-induced signaling for the WT receptors. Values are shown ± S.E. CA means constitutive activity.
| Construct | Expression level | n | CA | EC50(nm) (ligand) | Emax | n |
|---|---|---|---|---|---|---|
| GHSR 1a WT | 1 ± 0.02 | 3 | 43.4 ± 2.06 | 0,36 ± 0.27 (Ghrelin) | 100 | 3 |
| AspII:10Glu | 0.60 ± 0.06 | 3 | 3 | |||
| B2AR WT | 1 ± 0.03 | 5 | 9.9 ± 2.55 | 2.97 ± 1.09 (Pindolol) | 100 | 6 |
| AspII:10Glu | 0.68 ± 0.06 | 5 | 2 ± 1.92 | 5.68 ± 2.90 | 16.6 ± 5.35 | 5 |
| GPR119 WT | 1 ± 0.09 | 5 | 43.6 ± 3.43 | 2.22 ± 0.34 (AR-231453) | 100 | 5 |
| AspII:10Glu | 0.51 ± 0.11 | 5 | 27.5 ± 5.3 | 17.0 ± 5.01 | 56 ± 5.01 | 5 |
Discussion
In this study, we tested the unique role of GluII:10 (2.50) of the water hydrogen bond network in the NK1 receptor. Molecular modeling suggests that the side chain of GluII:10 replaces one or more water molecules that normally mediate interactions between the conserved polar residues in inactive conformations of family A GPCRs carrying an Asp residue in position II:10. Consequently, GluII:10 makes direct hydrogen bond interactions to SerIII:15 (3.39) in TM-III and AsnVII:16 (7.49) of the NPXXY motif in TM-VII and thereby conceivably mimics movable water molecules and/or a putative allosteric modulating Na+ ion. As observed previously in the B2AR (5), we find here that the water-gating residues of the hydrogen bond network are essential for signal transduction, whereas GluII:10 and its interaction partners are important for fine-tuning the signaling through Gs versus Gq and β-arrestin (Fig. 7).
FIGURE 7.
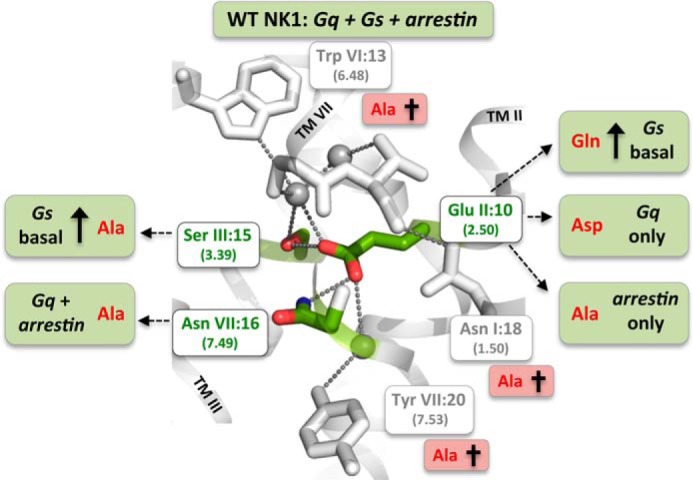
Residues of the water hydrogen bond network of the NK1 receptor identified to be either essential for overall signal transduction or for the fine-tuning and bias of signaling through the three different signaling pathways: Gs, Gq, and β-arrestin. Three residues were by Ala substitutions identified to be essential for all three signal transduction pathways as follows: the water-gating TrpVI:13 (6.48) of the CWXP motif and the TyrVII:20 (7.53) of the NPXXY motif as well as the ultra-highly conserved TM-VII kink-stabilizing AsnI:18 (1.50). Mutations of GluII:10 (2.50) and the two residues SerIII:15 (3.39) and AsnVII:16 (7.49), to which GluII:10 according to the molecular models makes direct hydrogen bond interactions, result in receptor mutants with biased signaling as follows: β-arrestin only (GluII:10 to Ala), Gq only (GluII:10 to Asp), high constitutive Gs signaling (GluII:10 to Gln), and Gq plus β-arrestin but not Gs (AsnVII:16 to Ala). Ala substitution of SerIII:15 selectively increases Gs signaling from zero to ∼40% of Emax.
Is GluII:10 a Tethered Negative Allosteric Regulator in the NK1 Receptor?
The initial hypothesis was that GluII:10 (2.50), by replacing the Na+ ion, would function as a tethered negative allosteric modulator and account for the unusual lack of constitutive signaling by the NK1 receptor. The very high maximal constitutive signaling through Gs, observed when GluII:10 was substituted with Gln, supports this notion (Table 1). These data also indicate that it is the deprotonated and charged form of the γ-carboxylic acid group of GluII:10 that acts as an allosteric negative modulator in the NK1 receptor. Introduction of Gln in this position, which has the same size and similar hydrogen bond potential as Glu, increased the constitutive Gs signaling.
Surprisingly, however, we only observed marginal constitutive activity through Gs and no constitutive signaling through Gq when GluII:10 was substituted with Ala (Figs. 3 and 4). One explanation could be that GluII:10 not only functions as an allosteric negative regulator (stabilizing an inactive conformation) but also is essential for not only the SP-induced Gq signaling, as it clearly is (Fig. 3), but is also essential for the ligand-independent constitutive signaling. In other words, GluII:10 is also involved in stabilizing active conformations of the receptor, where it can be replaced by Gln, for example, but not by Ala. In that case, we would not be able to observe any high constitutive signaling in the Ala -substituted receptor because the general importance of GluII:10 for receptor signaling would prevent that. Interestingly, although G-protein signaling was eliminated, SP-induced arrestin signaling was still observed in the GluII:10 to Ala mutant.
The notion that a Glu residue at position II:10 (2.50) can act as an allosteric negative regulator of receptor signaling was further supported by the deleterious effects observed when Glu was introduced instead of AspII:10 in the B2AR, GPR119, and in particular in the ghrelin receptor, where signaling was essentially eliminated. The cell surface expression was somewhat decreased in the AspII:10 to Glu mutant form of the ghrelin receptor; however, we have previously studied a number of ghrelin receptor mutants having similar or even lower expression levels, who nevertheless all signaled efficiently (see Table 2 in Ref. 52).
Differential Gs Versus Gq Signaling through the Water Hydrogen Bond Network
Although most 7TM receptors are known to signal mainly through one G protein pathway, they quite often are able to activate several G proteins (53–55). The NK1 receptor, for example, is best known as a Gq-coupled receptor activating IP3 and mobilizing calcium. However, substance P in fact activates both Gs and Gq pathways efficiently with similar nanomolar potency (38). Interestingly, the mutational substitutions of the water hydrogen bond network of the NK1 receptor in several aspects affected Gq and Gs signaling differently. Thus, removal of the polar side chain of four out of the six residues of the water hydrogen bond network individually increased the constitutive and ligand-independent Gs signaling without affecting the corresponding basal Gq signaling (Fig. 4). Previously, we have observed a similar phenomenon in the B2AR, where Ala substitution of AsnI:18, AspII:10, and SerVII:13, which jointly are coordinating the mobile water molecules in this receptor, also increased the basal, ligand-independent Gs signaling (5).
The agonist-induced Gs versus Gq signaling was also differentially affected by mutational substitutions of the hydrogen bond network. Most notably, Ala substitution of AsnVII:16 (7.49) of the NPXXY motif selectively eliminated Gs signaling with no effect on the Gq pathway. The SerVII:12 (7.45) to Ala mutant located one helical turn above in TM-VII had a similar phenotype but with a somewhat impaired Gq response (Figs. 3 and 4). Similarly, in the B2AR, Ala substitution of the corresponding two residues both abolished agonist-induced receptor activation through Gs (5). Interestingly, molecular modeling studies of the Gs-coupled thyrotropin receptor suggested that AsnVII:16 (7.49) acted as an on/off switch in receptor activation, which was confirmed in functional studies as the AsnVII:16 to Ala mutant form could not be activated by thyrotropin, although it bound the agonist with wild type affinity and displayed a normal high constitutive basal Gs signaling (56). In the cholecystokinin receptor, the AsnVII:16 (7.49) residue has been demonstrated to be essential for Gq signaling (57). Apparently, the interface between TM-II, TM-III, and TM-VII is particularly important for Gs versus Gq signaling. Previously, we identified another residue located at this interface but more toward the extracellular side between TM-II and TM-III, i.e. PheIII:07 (3.31), which upon substitution to Ser also selectively uncoupled the NK1 receptor from the Gs pathway while leaving Gq signaling intact (38).
β-Arrestin Versus G Protein Signaling through the Water Hydrogen Bond Network
Both ligands and mutations have been shown to be able to stabilize distinct conformations leading to biased receptor signaling within specific GPCRs (4, 38, 58–62). As shown in Fig. 7, we find that mutations of GluII:10 and AsnVII:16, which according to the molecular modeling in the NK1 receptor, are interacting directly with each other, result in biased signaling, i.e. where the three signaling pathways Gs, Gq, and β-arrestin mobilization are differentially affected. Thus, in the AsnVII:16 to Ala mutant, the Gs signaling was selectively eliminated, although both Gq signaling and β-arrestin mobilization were similar to wild type NK1. In contrast, in the GluII:10 (2.50) to Ala mutant, both SP-induced Gq and Gs signaling were totally eliminated, although the SP-induced recruitment of β-arrestin was rather well preserved in particular when taking into account the somewhat reduced cell surface expression (Fig. 5). Thus, removal of the acidic side chain of GluII:10 by Ala substitution converted the NK1 receptor into a clean β-arrestin-biased receptor. However, when GluII:10 instead was substituted with the shorter acidic Asp residue, as found in most other receptors, β-arrestin mobilization was totally eliminated and Gs signaling was seriously impaired, although Gq signaling was similar to wild type NK1 (Fig. 6). This underlines the crucial importance of the water hydrogen bond network for fine-tuning the signaling of the NK1 receptor through different signal transduction pathways (Fig. 7).
Interestingly, an interaction between AspII:10 (2.50) and AsnVII:16 (7.49) of the NPXXY motif in TM-VII was proposed early on in several receptors based on observations that paired swap of these two residues could restore receptor signaling, compared with the detrimental effects of each of the single mutations (50, 63–65). Furthermore, rearrangements in this region during activation have also been shown for rhodopsin using radiolytic protein footprinting (66).
Water-gating Residues of the Hydrogen Bond Network Are Essential for Signaling in General
Three residues appeared to be essential for all three signaling pathways, Gq, Gs, and β-arrestin mobilization; the two water-gating micro-switch residues, TrpVI:13 (6.48) and TyrVII:20 (7.53), and the totally conserved AsnI:18 (1.50) (Fig. 7).
Originally, we demonstrated by molecular dynamics simulations based on the high resolution structures of rhodopsin and B2AR that most of the water molecules of the network are highly mobile and that the entry and exit of these waters in and out of the network is gated by micro-switch residues, i.e. toward the ligand-binding pocket by TrpVI:13 and toward the intracellular cytosol by the rotary micro-switch TyrVII:20 (see central vignette in Figs. 3–5) (5). Structures of active receptor conformations subsequently demonstrated that the “water volume” of the hydrogen bond network changes considerably in the transition between inactive and active conformations in which most of the water molecules are “squeezed” out (16, 27). Thus, the movement of water molecules in and out of the network and the gating of these must be essential for receptor function (5). Accordingly, in this study of the NK1 receptor, we find that both of the “water-gating” residues, TrpVI:13 and TyrVII:20, are essential for all three signaling pathways tested, just as we found they were essential for the Gs signaling in the B2AR (5). TrpVI:13 has been extensively studied and determined to be essential for receptor activation in multiple 7TM receptors (46). The rotameric micro-switch, TyrVII:20, in its inward rotameric form stabilizes the outward tilt of TM-VI (2), which opens a pocket between the helices allowing for G protein binding (29, 67–70).
AsnI:18 (1.50), which in the inactive receptor conformation makes a helix kink-stabilizing hydrogen bond to the backbone carbonyl of the residue in position VII:13 (7.46) exposed by the highly conserved ProVII:17 (7.50) of the NPXXY motif, isthe only residue that is totally conserved throughout the entire large rhodopsin-like family A of 7TM receptors (42). In the NK1 receptor the AsnI:18 (1.50) to Ala mutant abolished all receptor signaling. The extraordinary conservation of this residue in 7TM receptors would suggests that it is essential in receptor signaling in general. However, in the B2AR we found that the corresponding AsnI:18 to Ala substitution surprisingly had no effect on the agonist-induced Gs signaling, and in fact it increased the constitutive activity (5).
Conclusion
Based on the x-ray structures, molecular dynamics simulations, and mutational analysis, we have previously proposed that the extended water hydrogen bond network between TM-I–III, -VI, and -VII constitutes an allosteric interface essential for stabilizing different active and inactive helical constellations during receptor activation (5). Here, we confirm that the water-gating micro-switch residues TrpVI:13 (6.48) of the CWXP motif and TyrVII:20 (7.53) of the NPXXY motif are essential for all signaling also in the NK1 receptor. Importantly, we identify the interaction between the unique Glu in position II:10 (2.50), SerIII:15 (3.39), and AsnVII:16 (7.49) of the NPXXY motif in TM-VII as a key point for determining Gs versus Gq signaling as well as β-arrestin mobilization. Specifically, the interface between TM-II, -III, and -VII appears to be particularly important for Gs signaling, which could potentially be used in future drug discovery efforts. For example, it may be possible to generate agonists with an appropriate signaling bias ensuring proper balance in efficacy through the Gs and the Gq pathways as recently described for the long chain fatty acid receptor GPR40 to obtain appropriate in vivo incretin-releasing efficacy (49).
Author Contributions
L. V. H., T. M. F., and T. W. S. conceived the study and wrote the manuscript. L. V. H., J. M., and N. D. H. performed and analyzed the experiments. All comparative homology modeling was conducted and analyzed by T. M. F. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
The Novo Nordisk Foundation Center for Basic Metabolic Research is based on an unconditional grant from the Novo Nordisk Foundation to the Faculty of Health Sciences at University of Copenhagen.
The authors declare that they have no conflicts of interest with the contents of this article.
The Schwartz/Baldwin generic numbering system for 7TM receptors, which is based on the actual location of the residues in each trans-membrane helix, is used throughout the article (41).
- 7TM
- 7-trans-membrane
- TM
- trans-membrane
- B2AR
- β2-adrenergic receptor
- A2AAR
- adenosine A2A receptor
- NK1
- neurokinin-1
- PDB
- Protein Data Bank
- IP3
- inositol 1,4,5-trisphosphate
- SP
- substance P.
References
- 1. Rosenbaum D. M., Rasmussen S. G., Kobilka B. K. (2009) The structure and function of G-protein-coupled receptors. Nature 459, 356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nygaard R., Frimurer T. M., Holst B., Rosenkilde M. M., Schwartz T. W. (2009) Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 30, 249–259 [DOI] [PubMed] [Google Scholar]
- 3. Katritch V., Cherezov V., Stevens R. C. (2013) Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 53, 531–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wisler J. W., Xiao K., Thomsen A. R., Lefkowitz R. J. (2014) Recent developments in biased agonism. Curr. Opin. Cell Biol. 27, 18–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nygaard R., Valentin-Hansen L., Mokrosinski J., Frimurer T. M., Schwartz T. W. (2010) Conserved water-mediated hydrogen bond network between TM-I, -II, -VI, and -VII in 7TM receptor activation. J. Biol. Chem. 285, 19625–19636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stevens R. C., Cherezov V., Katritch V., Abagyan R., Kuhn P., Rosen H., Wüthrich K. (2013) The GPCR network: a large-scale collaboration to determine human GPCR structure and function. Nat. Rev. Drug Discov. 12, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Srivastava A., Yano J., Hirozane Y., Kefala G., Gruswitz F., Snell G., Lane W., Ivetac A., Aertgeerts K., Nguyen J., Jennings A., Okada K. (2014) High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature 513, 124–127 [DOI] [PubMed] [Google Scholar]
- 8. Zhang K., Zhang J., Gao Z. G., Zhang D., Zhu L., Han G. W., Moss S. M., Paoletta S., Kiselev E., Lu W., Fenalti G., Zhang W., Müller C. E., Yang H., Jiang H., et al. (2014) Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature 509, 115–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tan Q., Zhu Y., Li J., Chen Z., Han G. W., Kufareva I., Li T., Ma L., Fenalti G., Li J., Zhang W., Xie X., Yang H., Jiang H., Cherezov V., et al. (2013) Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science 341, 1387–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chien E. Y., Liu W., Zhao Q., Katritch V., Han G. W., Hanson M. A., Shi L., Newman A. H., Javitch J. A., Cherezov V., Stevens R. C. (2010) Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science 330, 1091–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Granier S., Manglik A., Kruse A. C., Kobilka T. S., Thian F. S., Weis W. I., Kobilka B. K. (2012) Structure of the δ-opioid receptor bound to naltrindole. Nature 485, 400–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haga K., Kruse A. C., Asada H., Yurugi-Kobayashi T., Shiroishi M., Zhang C., Weis W. I., Okada T., Kobilka B. K., Haga T., Kobayashi T. (2012) Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature 482, 547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hanson M. A., Roth C. B., Jo E., Griffith M. T., Scott F. L., Reinhart G., Desale H., Clemons B., Cahalan S. M., Schuerer S. C., Sanna M. G., Han G. W., Kuhn P., Rosen H., Stevens R. C. (2012) Crystal structure of a lipid G protein-coupled receptor. Science 335, 851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kruse A. C., Hu J., Pan A. C., Arlow D. H., Rosenbaum D. M., Rosemond E., Green H. F., Liu T., Chae P. S., Dror R. O., Shaw D. E., Weis W. I., Wess J., Kobilka B. K. (2012) Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature 482, 552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu W., Chun E., Thompson A. A., Chubukov P., Xu F., Katritch V., Han G. W., Roth C. B., Heitman L. H., IJzerman A. P., Cherezov V., Stevens R. C. (2012) Structural basis for allosteric regulation of GPCRs by sodium ions. Science 337, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Manglik A., Kruse A. C., Kobilka T. S., Thian F. S., Mathiesen J. M., Sunahara R. K., Pardo L., Weis W. I., Kobilka B. K., Granier S. (2012) Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature 485, 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruprecht J. J., Mielke T., Vogel R., Villa C., Schertler G. F. (2004) Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 23, 3609–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shimamura T., Shiroishi M., Weyand S., Tsujimoto H., Winter G., Katritch V., Abagyan R., Cherezov V., Liu W., Han G. W., Kobayashi T., Stevens R. C., Iwata S. (2011) Structure of the human histamine H1 receptor complex with doxepin. Nature 475, 65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thompson A. A., Liu W., Chun E., Katritch V., Wu H., Vardy E., Huang X. P., Trapella C., Guerrini R., Calo G., Roth B. L., Cherezov V., Stevens R. C. (2012) Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature 485, 395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wacker D., Wang C., Katritch V., Han G. W., Huang X. P., Vardy E., McCorvy J. D., Jiang Y., Chu M., Siu F. Y., Liu W., Xu H. E., Cherezov V., Roth B. L., Stevens R. C. (2013) Structural features for functional selectivity at serotonin receptors. Science 340, 615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang C., Jiang Y., Ma J., Wu H., Wacker D., Katritch V., Han G. W., Liu W., Huang X. P., Vardy E., McCorvy J. D., Gao X., Zhou X. E., Melcher K., Zhang C., et al. (2013) Structural basis for molecular recognition at serotonin receptors. Science 340, 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Structure of a β1-adrenergic G-protein-coupled receptor. Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu B., Chien E. Y., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherezov V., Stevens R. C. (2010) Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu H., Wacker D., Mileni M., Katritch V., Han G. W., Vardy E., Liu W., Thompson A. A., Huang X. P., Carroll F. I., Mascarella S. W., Westkaemper R. B., Mosier P. D., Roth B. L., Cherezov V., Stevens R. C. (2012) Structure of the human κ-opioid receptor in complex with JDTic. Nature 485, 327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang C., Srinivasan Y., Arlow D. H., Fung J. J., Palmer D., Zheng Y., Green H. F., Pandey A., Dror R. O., Shaw D. E., Weis W. I., Coughlin S. R., Kobilka B. K. (2012) High-resolution crystal structure of human protease-activated receptor 1. Nature 492, 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fenalti G., Giguere P. M., Katritch V., Huang X. P., Thompson A. A., Cherezov V., Roth B. L., Stevens R. C. (2014) Molecular control of δ-opioid receptor signalling. Nature 506, 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gutiérrez-de-Terán H., Massink A., Rodríguez D., Liu W., Han G. W., Joseph J. S., Katritch I., Heitman L. H., Xia L., Ijzerman A. P., Cherezov V., Katritch V., Stevens R. C. (2013) The role of a sodium ion binding site in the allosteric modulation of the A2A adenosine G protein-coupled receptor. Structure 21, 2175–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rasmussen S. G., DeVree B. T., Zou Y., Kruse A. C., Chung K. Y., Kobilka T. S., Thian F. S., Chae P. S., Pardon E., Calinski D., Mathiesen J. M., Shah S. T., Lyons J. A., Caffrey M., Gellman S. H., et al. (2011) Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu F., Wu H., Katritch V., Han G. W., Jacobson K. A., Gao Z. G., Cherezov V., Stevens R. C. (2011) Structure of an agonist-bound human A2A adenosine receptor. Science 332, 322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Costa T., Lang J., Gless C., Herz A. (1990) Spontaneous association between opioid receptors and GTP-binding regulatory proteins in native membranes: specific regulation by antagonists and sodium ions. Mol. Pharmacol. 37, 383–394 [PubMed] [Google Scholar]
- 32. Gao Z. G., Ijzerman A. P. (2000) Allosteric modulation of A(2A) adenosine receptors by amiloride analogues and sodium ions. Biochem. Pharmacol. 60, 669–676 [DOI] [PubMed] [Google Scholar]
- 33. Horstman D. A., Brandon S., Wilson A. L., Guyer C. A., Cragoe E. J. Jr., Limbird L. E. (1990) An aspartate conserved among G-protein receptors confers allosteric regulation of α2-adrenergic receptors by sodium. J. Biol. Chem. 265, 21590–21595 [PubMed] [Google Scholar]
- 34. Selent J., Sanz F., Pastor M., De Fabritiis G. (2010) Induced effects of sodium ions on dopaminergic G-protein coupled receptors. PLoS Comput. Biol. 6, e1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilson M. H., Highfield H. A., Limbird L. E. (2001) The role of a conserved inter-trans-membrane domain interface in regulating α(2a)-adrenergic receptor conformational stability and cell-surface turnover. Mol. Pharmacol. 59, 929–938 [DOI] [PubMed] [Google Scholar]
- 36. Schwartz T. W., Holst B. (2007) Allosteric enhancers, allosteric agonists and ago-allosteric modulators: where do they bind and how do they act? Trends Pharmacol. Sci. 28, 366–373 [DOI] [PubMed] [Google Scholar]
- 37. Katritch V., Fenalti G., Abola E. E., Roth B. L., Cherezov V., Stevens R. C. (2014) Allosteric sodium in class A GPCR signaling. Trends Biochem. Sci. 39, 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Holst B., Hastrup H., Raffetseder U., Martini L., Schwartz T. W. (2001) Two active molecular phenotypes of the tachykinin NK1 receptor revealed by G-protein fusions and mutagenesis. J. Biol. Chem. 276, 19793–19799 [DOI] [PubMed] [Google Scholar]
- 39. Martini L., Hastrup H., Holst B., Fraile-Ramos A., Marsh M., Schwartz T. W. (2002) NK1 receptor fused to β-arrestin displays a single-component, high-affinity molecular phenotype. Mol. Pharmacol. 62, 30–37 [DOI] [PubMed] [Google Scholar]
- 40. Quartara L., Maggi C. A. (1997) The tachykinin NK1 receptor. Part I: ligands and mechanisms of cellular activation. Neuropeptides 31, 537–563 [DOI] [PubMed] [Google Scholar]
- 41. Schwartz T. W., Frimurer T. M., Holst B., Rosenkilde M. M., Elling C. E. (2006) Molecular mechanism of 7TM receptor activation–a global toggle switch model. Annu. Rev. Pharmacol. Toxicol. 46, 481–519 [DOI] [PubMed] [Google Scholar]
- 42. Mirzadegan T., Benkö G., Filipek S., Palczewski K. (2003) Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin. Biochemistry 42, 2759–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gether U., Johansen T. E., Snider R. M., Lowe J. A. 3rd, Nakanishi S., Schwartz T. W. (1993) Different binding epitopes on the NK1 receptor for substance P and non-peptide antagonist. Nature 362, 345–348 [DOI] [PubMed] [Google Scholar]
- 44. Gether U., Yokota Y., Emonds-Alt X., Brelière J. C., Lowe J. A. 3rd, Snider R. M., Nakanishi S., Schwartz T. W. (1993) Two nonpeptide tachykinin antagonists act through epitopes on corresponding segments of the NK1 and NK2 receptors. Proc. Natl. Acad. Sci. U.S.A. 90, 6194–6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holst B., Zoffmann S., Elling C. E., Hjorth S. A., Schwartz T. W. (1998) Steric hindrance mutagenesis versus alanine scan in mapping of ligand binding sites in the tachykinin NK1 receptor. Mol. Pharmacol. 53, 166–175 [DOI] [PubMed] [Google Scholar]
- 46. Holst B., Nygaard R., Valentin-Hansen L., Bach A., Engelstoft M. S., Petersen P. S., Frimurer T. M., Schwartz T. W. (2010) A conserved aromatic lock for the tryptophan rotameric switch in TM-VI of seven-trans-membrane receptors. J. Biol. Chem. 285, 3973–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nielsen S. M., Elling C. E., Schwartz T. W. (1998) Split-receptors in the tachykinin neurokinin-1 system–mutational analysis of intracellular loop 3. Eur. J. Biochem. 251, 217–226 [DOI] [PubMed] [Google Scholar]
- 48. Miller-Gallacher J. L., Nehmé R., Warne T., Edwards P. C., Schertler G. F., Leslie A. G., Tate C. G. (2014) The 2.1 A resolution structure of cyanopindolol-bound β1-adrenoceptor identifies an intramembrane Na+ ion that stabilises the ligand-free receptor 2. PLoS One 9, e92727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hauge M., Vestmar M. A., Husted A. S., Ekberg J. P., Wright M. J., Di Salvo J., Weinglass A. B., Engelstoft M. S., Madsen A. N., Lückmann M., Miller M. W., Trujillo M. E., Frimurer T. M., Holst B., Howard A. D., Schwartz T. W. (2015) GPR40 (FFAR1)–combined Gs and Gq signaling in vitro is associated with robust incretin secretagogue action ex vivo and in vivo. Mol. Metab. 4, 3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou W., Flanagan C., Ballesteros J. A., Konvicka K., Davidson J. S., Weinstein H., Millar R. P., Sealfon S. C. (1994) A reciprocal mutation supports helix 2 and helix 7 proximity in the gonadotropin-releasing hormone receptor. Mol. Pharmacol. 45, 165–170 [PubMed] [Google Scholar]
- 51. Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sivertsen B., Lang M., Frimurer T. M., Holliday N. D., Bach A., Els S., Engelstoft M. S., Petersen P. S., Madsen A. N., Schwartz T. W., Beck-Sickinger A. G., Holst B. (2011) Unique interaction pattern for a functionally biased ghrelin receptor agonist. J. Biol. Chem. 286, 20845–20860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jin L. Q., Wang H. Y., Friedman E. (2001) Stimulated D(1) dopamine receptors couple to multiple Gα proteins in different brain regions. J. Neurochem. 78, 981–990 [DOI] [PubMed] [Google Scholar]
- 54. Kilts J. D., Gerhardt M. A., Richardson M. D., Sreeram G., Mackensen G. B., Grocott H. P., White W. D., Davis R. D., Newman M. F., Reves J. G., Schwinn D. A., Kwatra M. M. (2000) β(2)-adrenergic and several other G protein-coupled receptors in human atrial membranes activate both G(s) and G(i). Circ. Res. 87, 705–709 [DOI] [PubMed] [Google Scholar]
- 55. Laugwitz K. L., Allgeier A., Offermanns S., Spicher K., Van Sande J., Dumont J. E., Schultz G. (1996) The human thyrotropin receptor: a heptahelical receptor capable of stimulating members of all four G protein families. Proc. Natl. Acad. Sci. U.S.A. 93, 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Govaerts C., Lefort A., Costagliola S., Wodak S. J., Ballesteros J. A., Van Sande J., Pardo L., Vassart G. (2001) A conserved Asn in trans-membrane helix 7 is an on/off switch in the activation of the thyrotropin receptor. J. Biol. Chem. 276, 22991–22999 [DOI] [PubMed] [Google Scholar]
- 57. Galés C., Kowalski-Chauvel A., Dufour M. N., Seva C., Moroder L., Pradayrol L., Vaysse N., Fourmy D., Silvente-Poirot S. (2000) Mutation of Asn-391 within the conserved NPXXY motif of the cholecystokinin B receptor abolishes Gq protein activation without affecting its association with the receptor. J. Biol. Chem. 275, 17321–17327 [DOI] [PubMed] [Google Scholar]
- 58. Webb D. R., Handel T. M., Kretz-Rommel A., Stevens R. C. (2013) Opportunities for functional selectivity in GPCR antibodies. Biochem. Pharmacol. 85, 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Luttrell L. M. (2014) Minireview: More than just a hammer: ligand “bias” and pharmaceutical discovery. Mol. Endocrinol. 28, 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shukla A. K., Violin J. D., Whalen E. J., Gesty-Palmer D., Shenoy S. K., Lefkowitz R. J. (2008) Distinct conformational changes in β-arrestin report biased agonism at seven-trans-membrane receptors. Proc. Natl. Acad. Sci. U.S.A. 105, 9988–9993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wei H., Ahn S., Shenoy S. K., Karnik S. S., Hunyady L., Luttrell L. M., Lefkowitz R. J. (2003) Independent β-arrestin 2 and G protein-mediated pathways for angiotensin II activation of extracellular signal-regulated kinases 1 and 2. Proc. Natl. Acad. Sci. U.S.A. 100, 10782–10787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Steen A., Thiele S., Guo D., Hansen L. S., Frimurer T. M., Rosenkilde M. M. (2013) Biased and constitutive signaling in the CC-chemokine receptor CCR5 by manipulating the interface between trans-membrane helices 6 and 7. J. Biol. Chem. 288, 12511–12521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Urizar E., Claeysen S., Deupí X., Govaerts C., Costagliola S., Vassart G., Pardo L. (2005) An activation switch in the rhodopsin family of G protein-coupled receptors: the thyrotropin receptor. J. Biol. Chem. 280, 17135–17141 [DOI] [PubMed] [Google Scholar]
- 64. Sealfon S. C., Chi L., Ebersole B. J., Rodic V., Zhang D., Ballesteros J. A., Weinstein H. (1995) Related contribution of specific helix 2 and 7 residues to conformational activation of the serotonin 5-HT2A receptor. J. Biol. Chem. 270, 16683–16688 [DOI] [PubMed] [Google Scholar]
- 65. Nikiforovich G. V., Zhang M., Yang Q., Jagadeesh G., Chen H. C., Hunyady L., Marshall G. R., Catt K. J. (2006) Interactions between conserved residues in trans-membrane helices 2 and 7 during angiotensin AT1 receptor activation. Chem. Biol. Drug Des. 68, 239–249 [DOI] [PubMed] [Google Scholar]
- 66. Angel T. E., Gupta S., Jastrzebska B., Palczewski K., Chance M. R. (2009) Structural waters define a functional channel mediating activation of the GPCR, rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 106, 14367–14372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Choe H. W., Kim Y. J., Park J. H., Morizumi T., Pai E. F., Krauss N., Hofmann K. P., Scheerer P., Ernst O. P. (2011) Crystal structure of metarhodopsin II. Nature 471, 651–655 [DOI] [PubMed] [Google Scholar]
- 68. Park J. H., Scheerer P., Hofmann K. P., Choe H. W., Ernst O. P. (2008) Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454, 183–187 [DOI] [PubMed] [Google Scholar]
- 69. Rasmussen S. G., Choi H. J., Fung J. J., Pardon E., Casarosa P., Chae P. S., Devree B. T., Rosenbaum D. M., Thian F. S., Kobilka T. S., Schnapp A., Konetzki I., Sunahara R. K., Gellman S. H., Pautsch A., et al. (2011) Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature 469, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., Ernst O. P. (2008) Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502 [DOI] [PubMed] [Google Scholar]