Background: The ABCG1 lipid transporter is regulated via protein ubiquitination.
Results: We identify two E3 ligases that regulate the protein stability and activity of ABCG1 and ABCG4.
Conclusion: The ligases, HUWE1 and NEDD4-1, are involved in the regulation of cholesterol export from cells.
Significance: Understanding the fine tuning of cholesterol homeostasis will help to understand how dysregulation can cause disease.
Keywords: ABC transporter, cholesterol regulation, E3 ubiquitin ligase, membrane lipid, post-transcriptional regulation
Abstract
The ATP-binding cassette transporter ABCG1 has an essential role in cellular cholesterol homeostasis, and dysregulation has been associated with a number of high burden diseases. Previous studies reported that ABCG1 is ubiquitinated and degraded via the ubiquitin proteasome system. However, so far the molecular mechanism, including the identity of any of the rate-limiting ubiquitination enzymes, or E3 ligases, is unknown. Using liquid chromatography mass spectrometry, we identified two HECT domain E3 ligases associated with ABCG1, named HUWE1 (HECT, UBA, and WWE domain containing 1, E3 ubiquitin protein ligase) and NEDD4-1 (Neural precursor cell-expressed developmentally down regulated gene 4), of which the latter is the founding member of the NEDD4 family of ubiquitin ligases. Silencing both HUWE1 and NEDD4-1 in cells overexpressing human ABCG1 significantly increased levels of the ABCG1 monomeric and dimeric protein forms, however ABCA1 protein expression was unaffected. In addition, ligase silencing increased ABCG1-mediated cholesterol export to HDL in cells overexpressing the transporter as well as in THP-1 macrophages. Reciprocally, overexpression of both ligases resulted in a significant reduction in protein levels of both the ABCG1 monomeric and dimeric forms. Like ABCG1, ABCG4 protein levels and cholesterol export activity were significantly increased after silencing both HUWE1 and NEDD4-1 in cells overexpressing this closely related ABC half-transporter. In summary, we have identified for the first time two E3 ligases that are fundamental enzymes in the post-translational regulation of ABCG1 and ABCG4 protein levels and cellular cholesterol export activity.
Introduction
The ATP-binding cassette (ABC)4 transporter, ABCG1, is involved in exporting lipids from cells to external acceptors such as high density lipoprotein (HDL) (1). ABCG1 belongs to the G-subfamily of ABC half-transporters (2), and is thought to function as a homodimer (3). The transporter is highly expressed in macrophages and neurons, where it is involved in the export of cholesterol, phospholipids as well as oxysterols (2, 4). Dysregulation of ABCG1 has been implicated in a number of diseases, such as atherosclerosis, lung disease, Type 2 diabetes, Alzheimer, and more recently, cancer and immune function (5, 6). Although its individual role in the development of atherosclerosis in mouse models has at times been confusing, its important role in conjunction with ABCA1 in maintaining macrophage cholesterol homeostasis has been more evident (7). In addition, ABCG1 (in conjunction with ABCA1) has been implicated in the regulation of β-cell function and insulin secretion (8), and more recently in the regulation of triglyceride storage in adipocytes (9).
ABCG4 is highly homologous to ABCG1, sharing 72% sequence identity in humans (10). However, its expression has traditionally been thought to be limited to cell types in the brain and eye (11), with co-expression of both ABCG1 and ABCG4 in neurons and astrocytes (12). Overexpression studies and analyses of tissues from knock-out mice have identified potential substrates for ABCG4, including cholesterol, oxysterols, and cholesterol intermediates (11–13). Interestingly, while the brains of single ABCG1 or ABCG4-null mice show limited accumulation of lipids, the double ABCG1/ABCG4 knock-out mouse has significant brain accumulation of a number of oxysterols (such as 24(S), 25- and 27-OH-cholesterol), cholesterol intermediates (such as lathosterol, lanosterol, and desmosterol), as well as cholesterol itself. This suggests a potential compensatory role for these two half-transporters in brain cells (11, 12). It is not yet fully determined whether these two half-transporters function on the plasma membrane or whether they also have a role intracellularly, potentially in proper trafficking of lipids (14, 15).
The transcriptional regulation of ABCG1 has been well established, with the liver X-receptor, or LXR, transcription factor identified as an important regulator of ABCG1 mRNA expression (16). The post-translational regulation of the transporter is less well established, but thought to be equally important in the fine-tuning of transporter activity. Previously, it has been demonstrated that ABCG1 is ubiquitinated and degraded via ubiquitin-mediated proteasomal degradation (17). In addition, Hsieh et al. showed that the cellular cholesterol status is important in the control of the proteasomal degradation of ABCG1 (18). Low cellular cholesterol levels increased the extent of ABCG1 ubiquitination and degradation, while this process was significantly repressed by addition of cholesterol (18). These findings highlight a pivotal role for ubiquitination in the regulation of ABCG1 protein expression and cholesterol export activity.
Ubiquitination involves the covalent attachment of one or more ubiquitin molecules to a lysine residue of proteins (19), which can target proteins for proteasome-mediated degradation (20). Apart from regulating protein turnover, this post-translational modification can also play a crucial role in other important cellular processes, including the regulation of protein trafficking and subcellular distribution, signal transduction, cell cycle, apoptosis, and DNA repair (21–23). Protein ubiquitination is carried out by a trio of enzymes, named ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin protein ligase (E3). The E3 ligases are thought to determine the substrate specificity of the ubiquitination reactions (19, 24). Humans express in excess of 600 E3 ligases, which are classified into two main subfamilies, depending on their enzymatic domain structure and mechanism of action. Approximately 90% of human E3 ligases belong to the Really Interesting New Gene (RING) domain ligases, while 28 members belong to the Homologous to E6Ap C terminus (HECT) domain ligases (25).
In the present study, using nano-liquid chromatography mass spectrometry (LC/MS), we identified several HECT domain E3s associated with ABCG1 and further investigated two of these, namely HUWE1 (HECT, UBA, and WWE domain containing 1, E3 ubiquitin protein ligase) and NEDD4-1 (Neural precursor cell-expressed developmentally down-regulated gene 4), which is the founding member of the NEDD4 family of ubiquitin protein ligases. The aim of this study was to establish the role of these two proteins on the stability and activity of ABCG1 as well as ABCG4.
Experimental Procedures
Reagents
Protease and phosphatase inhibitor cocktails, phorbol myristate acetate (PMA), BSA (essentially fatty acid free), IGEPAL, and scrambled control siRNA were purchased from Sigma-Aldrich. Zeocin, Lipofectamine 2000, Lipofectamine RNAiMAX, Opti-Mem, and all cell culture media were purchased from Life Technologies. BCA protein reagents and protein G Sepharose beads were purchased from Thermo Scientific. Anti-HUWE1 polyclonal antibody was purchased from Bethyl. Anti-ABCA1 monoclonal, anti-NEDD4 polyclonal and VeriBlot anti-rabbit secondary antibody for immunoprecipitation was from Abcam. Anti-ABCG4 polyclonal, anti-Myc polyclonal, secondary anti-mouse, and anti-rabbit antibodies, anti-α-tubulin monoclonal and anti-flag polyclonal antibodies were from Sigma-Aldrich. Anti-ABCG1 polyclonal antibody was purchased from Novus Biologicals. Anti-V5 monoclonal antibody was purchased from Novex. [1α,2α(n)-3H]cholesterol was from Perkin Elmer. Nitrocellulose membrane and ECL reagents were from Millipore and Amersham Biosciences. Reagents for hand-casting SDS-PAGE gels, including acrylamide, Tris-HCl, glycine, SDS, and TEMED were purchased from Amresco. Mini-PROTEAN precast gels were purchased from Bio-Rad. HDL2 was a generous gift from Professor Wendy Jessup of the Anzac Research Institute, Sydney, Australia.
Plasmid Constructs and siRNA Oligonucleotides
Plasmid DNA encoding human NEDD4-1 (cat. no. 27002) and HUWE1 (cat. no. 37431) were purchased from Addgene, and subcloned into pcDNA3.1/V5 using polymerase incomplete primer extension cloning (26). Two independent siRNA target sequences for both hamster HUWE1 and NEDD4-1 were custom designed and purchased from Sigma-Aldrich. Target sequences for NEDD4-1 were CTATGAATGGATTTGCTGA and GAGCCTGGCTGGGTTGTTT, and for HUWE1: GGAACAGTACAATTATAGT and GAGAAGATTCCATGAATAT. A standard scrambled control siRNA was also purchased from Sigma-Aldrich.
Generation of ABCG4-overexpressing Cells
Human ABCG4 cDNA, a generous gift from Professor Helen Hobbs, UT Southwestern, Dallas Texas, and described in Graf et al. (27), was subcloned into pcDNA4.0Myc/His using polymerase incomplete primer extension cloning (26). CHO-K1 cells were transfected and selected in zeocin (1 mg/ml), followed by single cell dilution. After screening for ABCG4 expression, positive clones were selected and expanded, followed by routine culture in zeocin at 200 μg/ml.
Cell Culture
CHO-K1 cells stably overexpressing c-terminally Myc-tagged human ABCG1 (described in Refs. 3 and 28) were maintained in Ham's F12 medium containing 10% (v/v) heat-inactivated FBS, and supplemented with l-glutamine (2 mm), penicillin (100 units/ml), streptomycin (100 μg/ml), and zeocin (200 μg/ml) at 37 °C in 5% CO2. Cells individually overexpressing two isoforms of ABCG1, named ABCG1(+12) or ABCG1(−12), as described in Gelissen et al. (3, 28), were utilized for the LC/MS analyses while all other experiments were carried out with cells expressing ABCG1(−12) only. ABCG1(−12) and ABCG1(+12) differ by a 12-amino acid peptide that is present between the ATP-cassette and the trans-membrane domains in ABCG1(+12) but absent from ABCG1(−12), attributed to alternative splicing of the ABCG1 gene. Both ABCG1 isoforms are expressed in human cells. However, ABCG1(+12) is not expressed in some rodents(3, 28).
CHO-K1 cells stably over expressing human ABCG4 were cultured under the same conditions as CHO-K1 cells overexpressing ABCG1. THP-1 monocytes (American Type Culture Collection) were routinely maintained at a density ranging from 2 × 105 to 1 × 106 cell/ml in RPMI 1640 medium containing 10% (v/v) FBS, l-glutamine (2 mm), penicillin (100 units/ml), and streptomycin (100 μg/ml). For experiments, the cells were seeded at a density 1 × 106 cell/ml and incubated with PMA (50 ng/ml) for 72 h to induce differentiation into macrophages. PMA was excluded during incubations with siRNA oligos (described below). After 48 h of siRNA transfection, macrophage cells were incubated with 5 μg/ml cholesterol/cyclodextrin (29) for 16 h to up-regulate ABCG1 expression.
LC/MS Identification of ABCG1-interacting Partners
CHO-K1 cells overexpressing Myc-tagged ABCG1 (ABCG1(+12) or ABCG1(−)) and CHO-K1 parental cells were harvested in RIPA buffer (20 mm Tris, 150 mm NaCl, 0.1% SDS, 1% IGEPAL, 0.5% deoxycholate; pH 7.5) containing protease inhibitors (5 μl/ml) and phosphatase inhibitor (5 μl/ml). ABCG1-Myc was immunoprecipitated using protein G-Sepharose beads and anti-Myc polyclonal antibody. Parental CHO-K1 cells were also subjected to immunoprecipitation (IP) as a control for nonspecific protein binding. Beads were washed five times with RIPA buffer and once with PBS, and proteins eluted by boiling in 1× SDS-PAGE loading dye at 95 °C for 10 min, with vortexing every 2 min. Proteins eluted were separated by SDS-PAGE using a precast “Any kDa” polyacrylamide gel to minimize keratin contamination. The gel was fixed in 50/5 % (v/v) methanol/acetic acid for 30 min followed by washing twice with 50% (v/v) methanol for 10 min, and rinsed twice in deionized water for 5 min. The gel was sensitized by incubation in 0.02% (w/v) sodium thiosulfate for 2 min, then rinsed three times with water, followed by incubation with 0.1% (w/v) silver nitrate for 25 min, rinsed three times in water, then developed in a solution of 2% (w/v) sodium carbonate, 0.04% (v/v) formaldehyde, and 0.0004% (w/v) sodium thiosulfate. The developing reaction was stopped with 37.6 mm EDTA (10 min) and the gel stored in water. Gel bands were excised as indicated in Fig. 1A. Bands were transferred to 1.5 ml tubes and rinsed in water. Proteins were reduced with 10 mm DTT in 50 mm ammonium bicarbonate at 37 °C for 30 min. DTT was removed, and proteins were alkylated with 25 mm iodoacetamide in 50 mm ammonium bicarbonate at 37 °C for 30 min. The solutions were removed, and the gel bands dehydrated with acetonitrile, followed by protein digestion with trypsin (2 ng/ml in 20 mm ammonium bicarbonate) at 37 °C for 17 h. The trypsin solution was removed, and peptides extracted from the gel with 1% (v/v) formic acid for 10 min. Gels were dehydrated with 25 μl of acetonitrile. The peptide extracts were transferred to new 1.5 ml tubes and evaporated to dryness under vacuum (SpeedVac, Savant). Peptides were re-suspended in 20 μl 0.05%/1% (v/v) heptafluorobutyric acid/formic acid and transferred to vials for LC/MS. LC/MS was performed using an LTQ Orbitrap Velos (Thermo Scientific) with electrospray ionization (ESI) injections, with injection volumes of 5 μl. Peptide hits were screened against the rodent database using the Mascot search engine (Matrix Science). We used the following criteria for inclusion of hits: 1) a required protein score of >30, 2) two or more peptide hits for at least one ABCG1 isoform, and 3) protein hits were absent from the CHO-K1 control IP.
FIGURE 1.
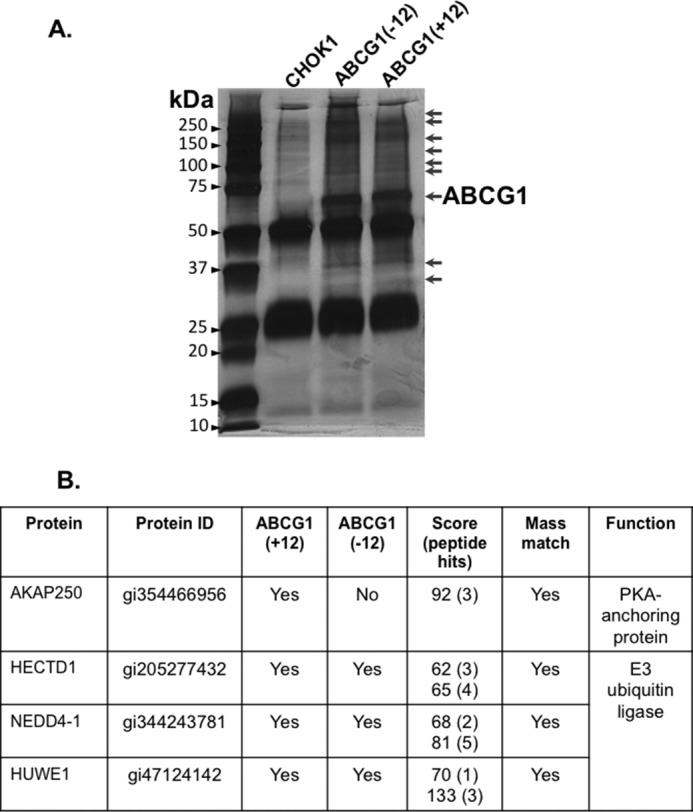
SDS-PAGE separation of IP products before preparation for LC/MS. A, cells overexpressing either ABCG1(−12) or ABCG1(+12), or CHO-K1 parental cells were lysed and subjected to IP using an anti-Myc antibody as described under “Experimental Procedures.” Proteins were separated by SDS-PAGE, then visualized via silver-staining. From left to right are the protein molecular mass marker (lane 1), CHO-K1 control (lane 2), ABCG1(-12) (lane 3), and ABCG1(+12) (lane 4). Arrows on the right indicate areas from which bands were excised to analyze via LC/MS. ABCG1 is labeled. B, table indicating some of the proteins identified (for full table see supplemental Table S1). Protein ID refers to the ID from the rodent database. Scores and peptide hits are indicated for ABCG1(+12) and ABCG1(−12), respectively.
SiRNA and Plasmid Transfections
Cells were transfected with siRNAs oligo's using Lipofectamine RNAiMAX at a ratio of 1 μl of siRNA/3 μl lipofectamine. Cells were incubated with a total siRNA concentration of 0.125 μm, which consisted of only negative control (scrambled) siRNA, a combination of 0.0625 μm control plus 0.0625 μm of either HUWE1 or NEDD4-1 siRNAs, or 0.0625 μm of HUWE1 plus NEDD4-1 siRNAs. After 24 h, cells were incubated with fresh Ham's F12 medium for a further 24 h for CHO-K1 cells (or 48 h for the data presented in Fig. 8) or with RPMI 1640 medium containing PMA (50 ng/ml) for a further 48 h for THP-1 cells.
FIGURE 8.
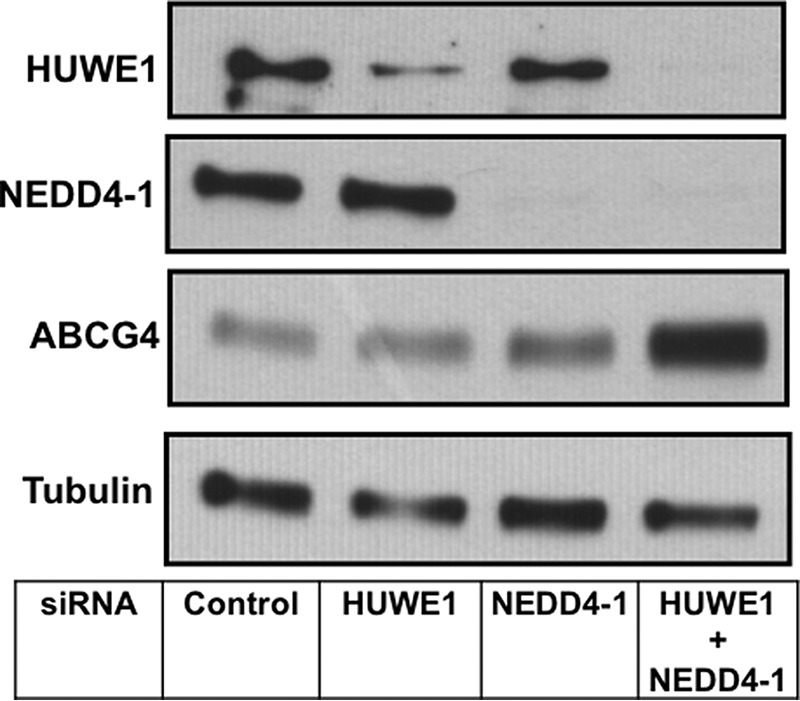
SiRNA silencing of HUWE1 and NEDD4-1 in CHO-K1 cells overexpressing ABCG4. CHO-K1 cells stably overexpressing ABCG4 were transfected with control siRNA, siRNAs for HUWE1, or NEDD4-1 individually, or both for a total of 72 h as described under “Experimental Procedures.” Cells were harvested and cell proteins separated via SDS-PAGE, followed by immunoblotting with the indicated antibodies.
CHO-K1 cells overexpressing ABCG1(−12) were transiently transfected with plasmid DNA expressing NEDD4-1/V5 or HUWE1/V5 using Lipofectamine 2000 at a ratio of 0.25 μg DNA/μl lipofectamine.
Cell Lysis and Western Blotting
Cells were washed with ice-cold PBS and lysed in cell lysis buffer (1% IGEPAL in 50 mm Tris-HCl, 150 mm NaCl, pH 7.8) with the addition of protease and phosphatase inhibitor cocktails (5 μl/ml). Cell protein levels were measured using the BCA assay, and equal amounts of cell protein per lane were separated using either 8% (v/v) SDS-PAGE or 4–15% (v/v) Mini-PROTEAN precast gels, and proteins were transferred onto nitrocellulose membranes. Gels to separate HUWE1 protein were transferred for 16 h due to its large size (∼480 kDa). Membranes were probed using the following antibody dilutions: anti-HUWE1 (1:2000), anti-NEDD4-1 (1:5000), anti-Myc (1:5000), anti-V5 (1:5000), anti-tubulin (1:3000), anti-ABCA1 (1:1000), anti-ABCG4 (1:5000), anti-ABCG1 (1:500). HRP-conjugated anti-rabbit or anti-mouse secondary antibodies were used at 1:10,000. Protein bands were visualized by chemiluminescence and quantified using Image J software (NIH).
Co-IP of ABCG1 and E3 Ligases
CHO-K1 cells stably overexpressing ABCG1-Myc were transiently transfected with plasmids encoding NEDD4-1/V5 or HUWE1/V5 for 24 h. Cells were harvested in RIPA buffer (20 mm Tris-HCl, 150 mm NaCl, 0.1% (w/v) SDS, 1% (v/v) IGEPAL, pH 7.5) supplemented with protease and phosphates inhibitors (5 μl/ml). Protein G Sepharose beads were incubated with anti-Myc antibody for 1 h at 25 °C to form antibody/bead complexes. Aliquots of cell lysates were pre-cleared with fresh beads, then added to the antibody/bead complexes and gently rotated overnight at 4 °C. The beads were washed extensively and boiled in 1× SDS-PAGE loading dye (30 mm Tris-base, 10 mm EDTA, 6% (v/v) glycerol, 2% (w/v) SDS, 0.005% (w/v) bromphenol blue, 10 mm DTT, pH 6.8) at 95 °C for 7 min, followed by SDS-PAGE and Western blot analysis.
Cholesterol Efflux Assay
Cholesterol efflux assays were conducted as previously described (28) with minor modifications as follows. Cells were incubated with siRNA oligos for 24 h for CHO-K1 cells overexpressing ABCG1 or ABCG4, or 48 h for THP-1 cells as described above, followed by radio-labeling overnight with [1α,2α(n)-3H]cholesterol at 1 μCi/ml in serum-containing medium. The labeling medium was removed, and the cells washed twice with PBS, then incubated for 30 min in serum-free medium containing 0.1% BSA to equilibrate. Cells were washed once with PBS and incubated in serum-free medium containing 0.1% BSA alone or with the addition of HDL2 at 10 μg/ml for 6 h for CHOK-1 cells and for 24 h for THP-1 macrophages. Radioactivity was counted in medium and cells (washed with PBS and lysed in 0.1% (v/v) Triton X-100 in PBS), and cholesterol efflux expressed as the percentage of radioactivity released into the media relative to the total radioactive pool. In the case of ABCG1- and ABCG4-overexpressing cells, background efflux to CHO-K1 parental cells was subtracted. Data are presented either as percentage efflux or for ease of presentation, as fold increase over control siRNA treatment, with the latter including subtraction of background efflux to BSA alone.
Statistical Analyses
Data are expressed as means ± S.E. and were analyzed for statistical differences using one way ANOVA and Student's t-tests using Prism version 6 (GraphPad Software). A p value of < 0.05 was considered significant.
Results
LC/MS Identification of ABCG1 Interacting Partners
Using immunoprecipitation followed by LC/MS, we identified a total of 88 proteins that were associated with ABCG1, with the full list of candidates presented in supplemental Table S1. Only 25 binding partners identified were exclusively associated with one isoform of ABCG1, with the remainder associated with both. Of specific interest was AKAP250, a protein kinaseA (PKA) anchoring protein, that was associated with ABCG1(+12) but not ABCG1(−12). We have previous published that ABCG1(+12) is phosphorylated by PKA at a serine residue near the 12 amino acid region that is specific for ABCG(+12) but absent from ABCG1(−12) (28). Finding that only ABCG1(+12) is associated with this specific PKA anchoring protein suggests the involvement of AKAP250 in this process. However, further work is required to confirm this observation.
Considering the findings that ABCG1 is ubiquitinated and degraded by the ubiquitin proteasomal system (17, 18), a process that involves the activity of E3 ligases, we specifically looked for the presence of these proteins among the ABCG1 binding partners. We identified three HECT-domain E3 ligases that were associated with both ABCG1 isoforms (Fig. 1B), namely HECTD1, HUWE1, and NEDD4-1. In humans and rodents, two isoforms of NEDD4 are expressed, with the peptide identified via LC/MS representing the equivalent of the human NEDD4-1 isoform. Pilot experiments, silencing all three E3 ligases listed in Fig. 1B, indicated that siRNA knockdown of HUWE1 and NEDD4-1, but not HECTD1, increased ABCG1 protein levels, hence only NEDD4-1 and HUWE1 were investigated further. Experiments were carried out with ABCG1(−12) only as this isoform is more ubiquitously expressed (30).
NEDD4-1 Interacts with ABCG1
Firstly, we aimed to confirm a direct interaction between ABCG1 and HUWE1 and/or NEDD4-1, using an alternate approach. CHO-K1 cells stably overexpressing ABCG1(−12)-Myc were transiently transfected with overexpression plasmids for either NEDD4-1/V5 or HUWE1/V5, followed by IP of ABCG1-Myc. We were able to co-IP NEDD4-1 with ABCG1 (Fig. 2A), confirming the direct interaction of these two proteins. However, we were unsuccessful in our attempts to co-IP HUWE1/V5 with ABCG1 (data not shown), most likely due to sensitivity issues. Fig. 2B indicates that, although both plasmids were expressing HUWE1/V5 and NEDD4-1/V5, respectively, expression of HUWE1/V5 was much less compared with NEDD4-1/V5 as can be seen from the V5 blot. This may have been caused by differences in the transfection efficiency as the HUWE1 gene and protein are significantly larger than NEDD4-1 (480 kDa versus 120 kDa). In summary, we were able to confirm a direct interaction between NEDD4-1 and ABCG1.
FIGURE 2.
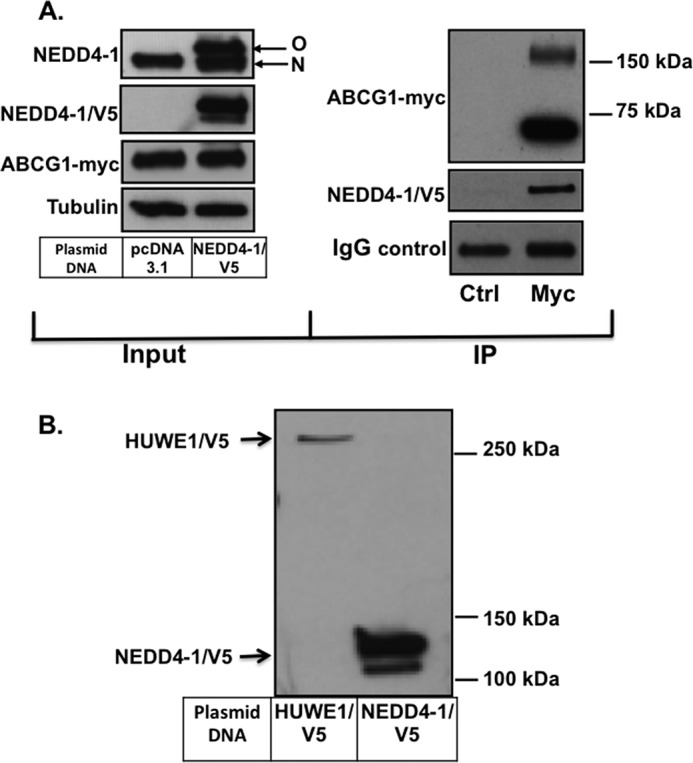
NEDD4-1/V5 and ABCG1-Myc co-IP. A, left: CHO-K1 cells stably overexpressing ABCG1 were transfected with plasmid DNA encoding either pcDNA3.1 as control or NEDD4-1/V5 as described under “Experimental Procedures.” Immunoblots show expression of NEDD4-1 (top; native (N) and overexpressed (O) NEDD4-1 protein), ABCG1-Myc, and tubulin, respectively. Right: IP of ABCG1-Myc-overexpressing cells, transfected with NEDD4-1/V5. ABCG1-Myc band is only detected when cell lysate was IPed with anti-Myc (Myc) antibodies and absent with anti-flag control antibody (Ctrl). Results are representative of two independent experiments. B, CHO-K1 cells overexpressing ABCG1 were transiently transfected with HUWE1/V5 or NEDD4-1/V5 plasmids as described under “Experimental Procedures.” Cells were harvested and subjected to SDS-PAGE and Western blotting, and membranes probed with V5 antibodies, detecting only the overexpressed proteins.
SiRNA Silencing of HUWE1 and NEDD4-1 Increases ABCG1 Protein Expression
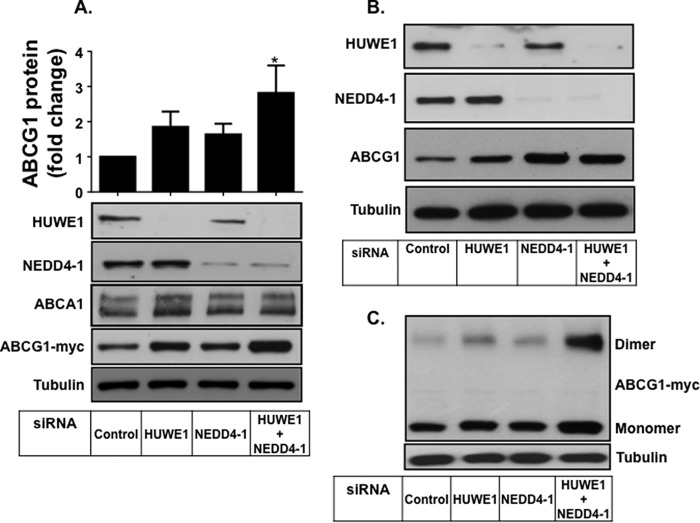
To investigate whether HUWE1 and NEDD4-1 are involved in ABCG1 degradation, we performed siRNA knockdown of both ligases individually and in combination in CHO-K1 cells stably overexpressing ABCG1(−12) (Fig. 3). Single knockdown of either HUWE1 or NEDD4-1, using two independent sets of siRNA primers (Fig. 3, A and B), modestly increased ABCG1 protein levels, with effects from HUWE1 silencing being greater than NEDD4-1 silencing. Importantly, double knockdown of both HUWE1 and NEDD4-1 significantly increased ABCG1 protein levels ∼3-fold and substantially more than the individual knockdowns (Fig. 3, A and C), suggesting that these two E3 ligases have an additive affect on ABCG1 protein expression. ABCA1 protein levels were also measured, but were shown to be unaffected by silencing of either of the two ligases (Fig. 3A).
FIGURE 3.
SiRNA silencing of HUWE1 and NEDD4-1 increases ABCG1 protein expression. A, CHO-K1 cells overexpressing ABCG1 were transfected with control siRNA, siRNAs for HUWE1 or NEDD4-1 individually, or both as described under “Experimental Procedures.” Cells were harvested and proteins separated via SDS-PAGE followed by immunoblotting with the indicated antibodies. Immunoblots are representative of four independent experiments with similar results. Bar graphs represent quantification of ABCG1 protein relative to tubulin, of four independent experiments (average ± S.E.), performed in duplicate cultures, with * indicating p < 0.05 compared with Control. B, CHO-K1 cells overexpressing ABCG1 were transfected with alternate primers as described under A. Results are representative of two independent experiments. C, ABCG1 monomer as well as dimer after siRNA treatment as described under A.
We have previously shown that ABCG1 homodimerizes to form an active transporter (3). Our SDS-PAGE shows a dimeric form of the ABCG1 protein at twice the molecular weight as the monomer (Fig. 3C). We determined the effect of HUWE1 and NEDD4-1 silencing on the presence of this ABCG1 dimeric protein (Fig. 3C). Consistent with effects on the monomeric form, silencing of HUWE1 and NEDD4-1 led to an increase in ABCG1 dimer levels.
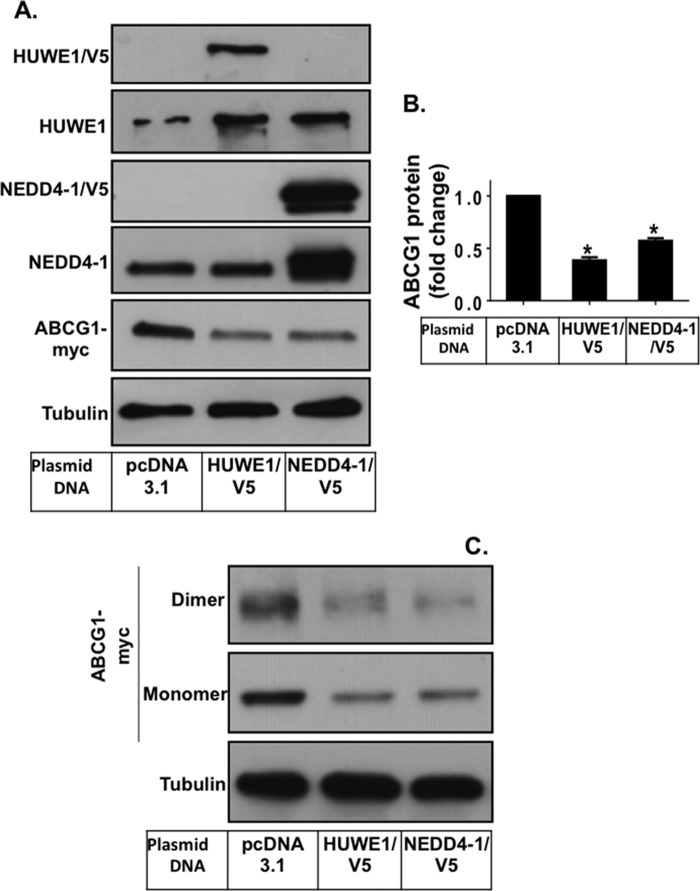
Overexpression of HUWE1 and NEDD4-1 Promotes ABCG1 Degradation
To confirm that HUWE1 and NEDD4-1 are indeed involved in the regulation of ABCG1 degradation, we overexpressed exogenous HUWE1 or NEDD4-1 together with ABCG1(−12), followed by measurement of ABCG1. Fig. 4A shows that overexpression of either HUWE1 or NEDD4-1 led to an approximate 2-fold increase in protein expression of the E3s compared with those expressed endogenously. In addition, overexpression of the ligases resulted in a significant reduction in protein levels of the ABCG1 monomeric form (Fig. 4, A and B), with effect of HUWE1 overexpression more profound than NEDD4-1. Furthermore, the ABCG1 dimeric form was also reduced by ligase overexpression (Fig. 4C).
FIGURE 4.
HUWE1 and NEDD4-1 overexpression reduces ABCG1 protein levels. A, CHO-K1 cells overexpressing ABCG1 were transiently transfected with either control (pcDNA3.1), HUWE1/V5 or NEDD4-1/V5 plasmids as described under “Experimental Procedures.” Cells were harvested and cell proteins separated via SDS-PAGE followed by immunoblotting with the indicated antibodies, detecting the native and overexpressed proteins. Immunoblots are representative of two independent experiments. B, quantification of ABCG1 protein levels relative to tubulin. Results presented are mean ± S.E. from two independent experiments, both performed in duplicate, with * indicating p < 0.05 compared with pcDNA3.1. C, immunoblots showing the ABCG1 monomer as well as the dimer.
SiRNA Silencing of HUWE1 and NEDD4-1 Enhances ABCG1-mediated Cholesterol Export to HDL2
Next, we examined the functional consequences of HUWE1 and NEDD4-1 silencing on ABCG1 transporter activity. HDL2 mediated cholesterol efflux was measured to evaluate efflux activity of human ABCG1 overexpressed in CHOK-1 cells. Basal cholesterol efflux to BSA was not affected after ligase silencing (Fig. 5A). However, HDL2 mediated cholesterol export was increased modestly with individual ligase silencing (Fig. 5, A and B). Knockdown of both ligases resulted in a significant increase in cholesterol export, with levels approximately double those observed with the control siRNA treatment (Fig. 5, A and B). This finding indicates that the increased ABCG1 protein levels after ligase silencing translates to up-regulation of transporter activity. Furthermore, it implies that these two E3s have a fundamental role in regulating ABCG1-mediated cellular cholesterol export activity.
FIGURE 5.
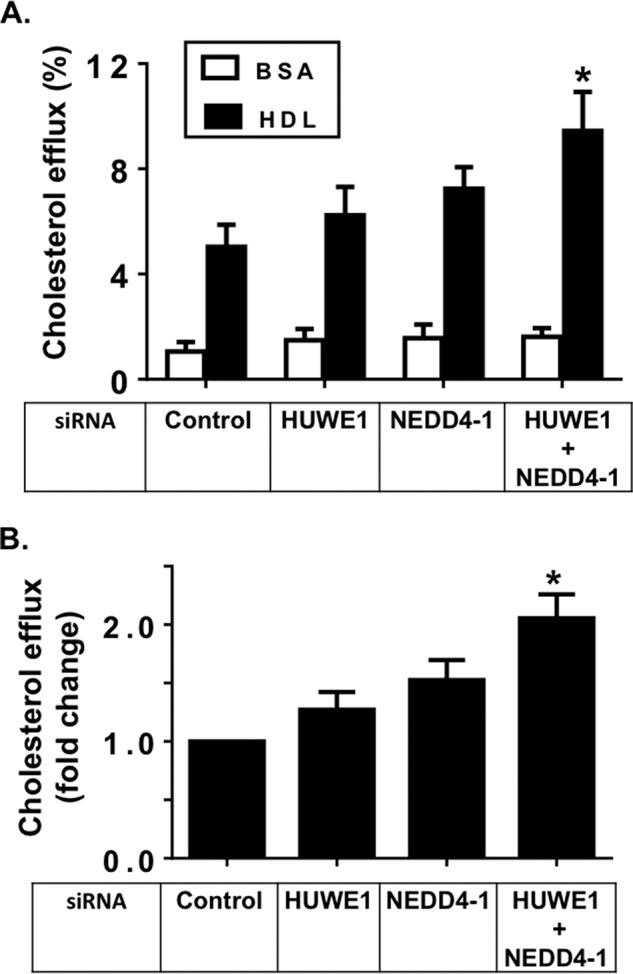
SiRNA silencing of HUWE1 and NEDD4-1 enhances ABCG1 efflux activity in CHO-K1 cells overexpressing ABCG1. A, ABCG1-mediated cholesterol efflux in CHO-K1 cells overexpressing ABCG1 after 6 h incubation with either 0.1% BSA alone or with addition of HDL2 (10 μg/ml) as described under “Experimental Procedures.” Results are mean ± S.E. of three independent experiments, performed in triplicate cultures, with * indicating p < 0.05 compared with Control siRNA. B, ABCG1-mediated cholesterol efflux to HDL2, expressed as fold change, relative to Control siRNA treatment. For each individual experiment, efflux to BSA was subtracted. Results are mean ± S.E. of three independent experiments, performed in triplicate cultures, with * indicating p < 0.05 compared with Control siRNA.
To assess whether cholesterol export was affected by ligase modulation in cells that express endogenous ABCG1, we performed siRNA silencing in THP-1 macrophages. SiRNA silencing of ligases was successfully achieved in these cells (Fig. 6A). Although no measurable increase in total cellular ABCG1 protein levels could be observed (Fig. 6A), HDL2-mediated cholesterol export was significantly increased after silencing of both ligases. Again, only modest increases were observed after individual silencing of HUWE1 or NEDD4-1 (Fig. 6B). ABCA1 proteins levels, as seen before in the CHOK1 cells, were unaffected by ligase silencing (Fig. 6A). Altogether, the significant increase in cholesterol export to HDL2 after ligase silencing may imply that there is only a small but active pool of ABCG1 that is affected by both of these ligases in macrophages.
FIGURE 6.
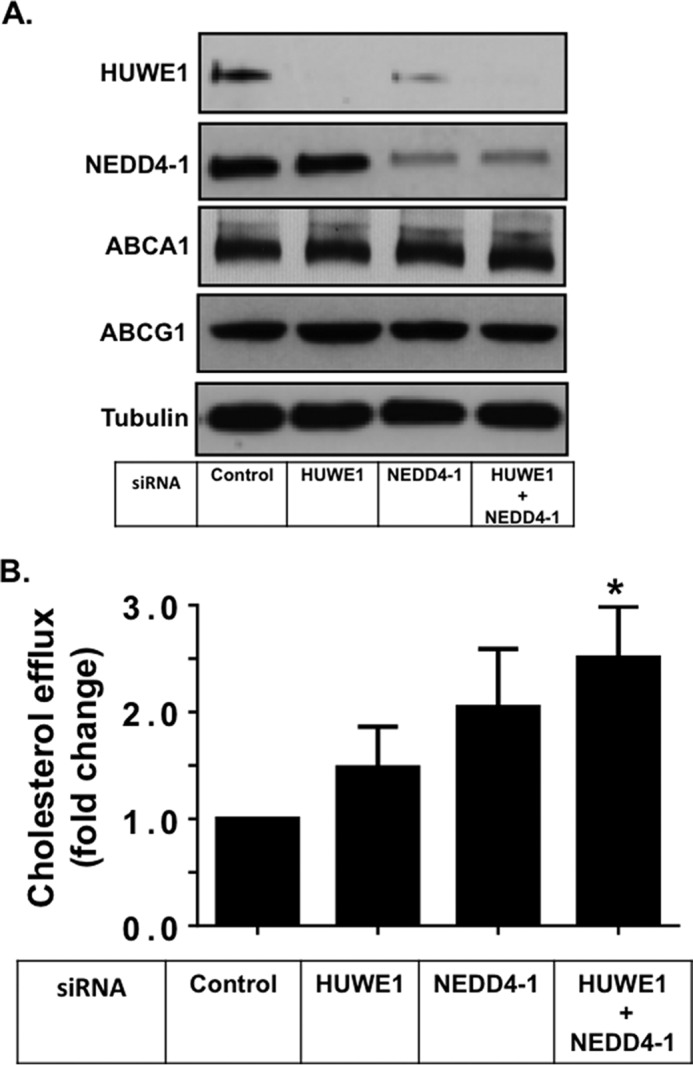
SiRNA silencing of HUWE1 and NEDD4-1 enhances ABCG1 efflux activity in THP-1 macrophages. A, THP-1 macrophages were transfected with control siRNA, siRNAs for HUWE1 or NEDD4-1 individually, or both as described under “Experimental Procedures.” Cells were harvested and cell proteins separated via SDS-PAGE followed by immunoblotting with the indicated antibodies, detecting the native proteins. Immunoblots are representative of 2 independent experiments. B, ABCG1-mediated cholesterol efflux to HDL2 (10 μg/ml) in THP-1 macrophages after 24 h as described under “Experimental Procedures.” Values are expressed as fold change relative to control siRNA and are mean ± S.E. of three independent experiments performed in triplicate cultures, with * indicating p < 0.05 compared with Control.
SiRNA Silencing of HUWE1 and NEDD4-1 Increases ABCG4 Protein Expression and Activity
CHO-K1 cells stably overexpressing ABCG4 were generated and treated with siRNAs targeting HUWE1 and NEDD4-1. As for ABCG1, ABCG4 protein levels were significantly increased after double knockdown of both HUWE1 and NEDD4-1, with modest but significant increases observed when individual ligases were silenced (Fig. 7A). Furthermore, cholesterol export from ABCG4 overexpressing cells to HDL2 was modestly but significantly increased after silencing of both ligases (Fig. 7B). The extent of protein stabilization and effects on cholesterol export were less pronounced than those seen for ABCG1. Extending the siRNA treatment time by another 24 h revealed a further stabilization of ABCG4 protein (Fig. 8), hence we hypothesize that this may be due to ABCG4 having a longer half-life than ABCG1. Collectively, these results indicate that HUWE1 and NEDD4-1 are also involved in the post-translational regulation of ABCG4 protein levels and activity.
FIGURE 7.
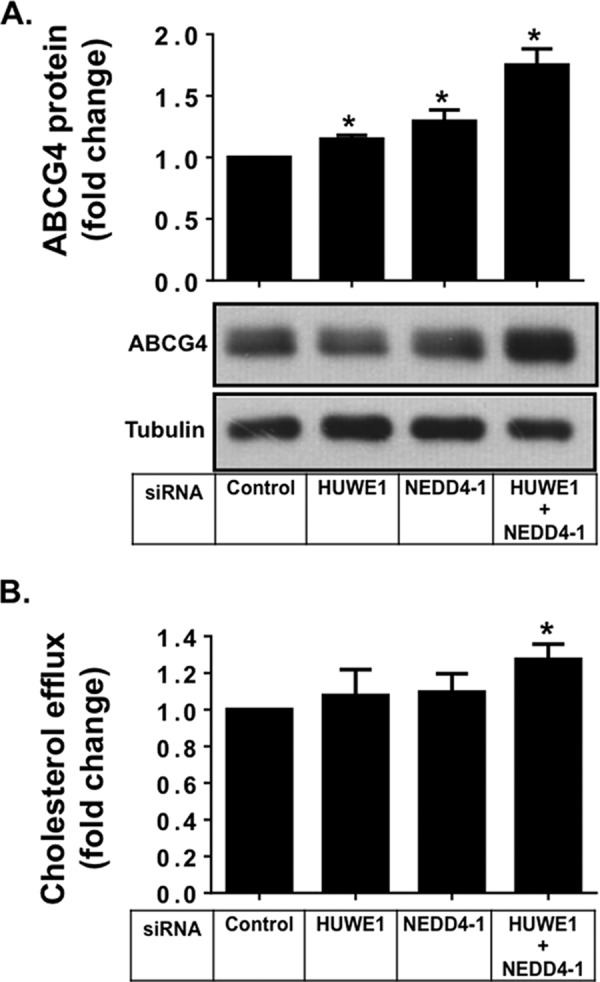
SiRNA silencing of HUWE1 and NEDD4-1 increases ABCG4 protein and activity. A, CHO-K1 cells stably overexpressing ABCG4 were transfected with control siRNA, siRNAs for HUWE1, or NEDD4-1 individually, or both for a total of 48 h as described under “Experimental Procedures.” Cells were harvested and cell proteins separated via SDS-PAGE followed by immunoblotting with the indicated antibodies. Immunoblots are representative of 4 independent experiments. Bar graphs represent quantification of ABCG4 protein relative to tubulin, of four independent experiments (average ± S.E.), performed in duplicate cultures, with * indicating p < 0.05 compared with Control. B, ABCG4-mediated cholesterol efflux in CHO-K1 cells overexpressing ABCG4 after 6 h of incubation with either 0.1% BSA alone or with addition of HDL2 (10 μg/ml) as described under “Experimental Procedures.” Results are mean ± S.E. of three independent experiments performed in triplicate cultures. Values are expressed as fold change relative to control siRNA.
Discussion
It is becoming increasingly clear that the ubiquitin proteasome system has a key role in the post-translational regulation of various integral proteins involved in cellular cholesterol homeostasis (reviewed in (31)). These include key enzymes involved in the transcriptional regulation, synthesis, uptake and ABC transporter-mediated export of cholesterol from cells (18, 31). However, the actual E3 ligases that control the last step in the ubiquitination cascade have only been identified and characterized for a limited number of these proteins. In the present study, we have uncovered HUWE1 and NEDD4-1 as important enzymes in the regulation of ABCG1 and ABCG4 mediated export of cholesterol from cells. Our results show that NEDD4-1 and HUWE1 modulate ABCG1 as well as ABCG4 protein levels and cholesterol export activity. These findings highlight a new molecular mechanism that regulates these ABCG half-transporters at the post-translational level, and identify ABCG1 and ABCG4 as novel candidate substrates for the E3 ligases HUWE1 and NEDD4-1. Although a significant number of others targets of both HUWE1 and NEDD4-1 have been identified, neither of these E3 ligases have been studied with regards to a role in ABC transporter mediated sterol export in humans. However, it is noteworthy that the yeast homologue of NEDD4 proteins, Rsp5p, has been assigned a function in lipid homeostasis in yeast, including the regulation of enzymes involved in ergosterol synthesis (32).
Considering the roles of ABCG1 and ABCG4 in cholesterol homeostasis in various tissues, it is of interest to note the overlap between these and functions ascribed to NEDD4-1, which has to date been characterized more extensively than HUWE1. Nedd4 (the murine homologue of NEDD4-1) knock-out mice display delayed embryonic development, greatly reduced growth and abnormalities in neurons and cardiovascular development (33). Hence, NEDD4-1 has been studied extensively with respect to a role in various stages of neuronal growth (reviewed in Ref. 34). More recently, the development of heterozygous Nedd4 knock-out models has allowed for better characterization of the role of Nedd4 in more mature animals. Camera et al. confirmed its importance in neuronal development by showing that Nedd4 heterozygous mice have abnormalities in gait, with greatest expression of Nedd4 in Purkinje neurons of the cerebellum (35). It would be interesting to measure whether Nedd4 ablation in neuronal cells in these animals coincides with overexpression of ABCG1 and ABCG4, which in turn may cause disturbances in lipid homeostasis. More recently, another study investigated insulin resistance and obesity-related parameters in a heterozygous Nedd4 knock-out mouse (36) and showed that glucose-stimulated insulin secretion in these mice was significantly increased (36). Conversely, in the ABCG1−/− mouse model, insulin secretion from ABCG1−/− islets was significantly reduced upon glucose challenge (8). One can speculate that the changes observed in the Nedd4 heterozygotes may be associated with increases in ABCG1 transporter levels and activity. These observations clearly warrant further investigation.
Apart from lipid transporters, the ABC transporter family contains a significant number of drug transporters, including its archetypical member ABCB1 (also known as p-glycoprotein). This transporter was recently shown to be a substrate for NEDD4-1 (37). The authors investigated ABCB1 in the context of its proposed role on the blood brain barrier, where it has been hypothesized to be involved in the efflux of the β-amyloid peptide in the context of Alzheimer disease (38). Overexpression of NEDD4-1 in cells expressing ABCB1 lead to reduced surface expression of the ABC transporter (37), suggesting that NEDD4-1 may be involved in the internalization and subsequent degradation of ABCB1. This may provide a mechanism by which NEDD4-1 can affect ABC transporter activity, which is currently being investigated in our laboratory with respect to ABCG1 and ABCG4.
HUWE1 (also known as UREB1, HECTH9, ARF-BP1, MULE, E3 Histone, and LASU1) is less well characterized than NEDD4-1. HUWE1 has been investigated previously with regards to its regulation of a wide array of proteins with important roles in cell stress response pathways and has been reported to be overexpressed in multiple human tumors (24, 39). Cell specific knockouts such as those in B-lymphocytes delineate an important role for this ligase in cell proliferation, apoptosis (by regulating p53 levels) and B-cell homeostasis (40). There are no other ABC transporter or lipid-associated targets identified for HUWE1 or its homologous to date. Hence its role in cholesterol homeostasis has thus far been unexplored.
Both HUWE1 and NEDD4-1 are members of the HECT-domain subfamily of E3 ligases that has only 28 known members, which are distinguished by the sequential manner in which they transfer ubiquitin molecules to their target protein. First, the ubiquitin-E2 conjugate binds to the HECT domain, followed by the catalytic cys-residue unloading the ubiquitin from the E2 by forming a ubiquitin-thioester intermediate. After that, the ubiquitin is transferred to a specific lysine residue of the target protein (19). To facilitate these steps, HECT domain E3 ligases have been shown to associate with their targets in two possible ways. Firstly, by binding of the so-called WW-domain(s) of the E3 to a conserved proline-rich PPXY or PY domain of the target protein. Second, as has been shown for NEDD4 proteins, targets that do not contain PPXY domains have been shown to require the recruitment of interacting or adaptor proteins that contain such domains (24, 41). Investigation of the protein sequence of ABCG1 and ABCG4 shows that neither of these ABC transporters contains proline-rich domains in cytosolic regions, hence potential adaptor proteins may be required for the E3 ligases to interact with ABCG1 and ABCG4. As a precedent, another key player in cholesterol homeostasis, namely HMG-CoA reductase, is also regulated via ubiquitination and was shown to make use of its regulatory proteins Insig-1 and -2 to recruit specific E3 ligases when cellular cholesterol levels are increased (42).
In conclusion, we have identified two HECT-domain E3 ligases that are involved in the regulation of protein levels and activity of two ABCG-subfamily members that are important players in the regulation of cholesterol homeostasis. Because of HECT-domain E3 ligases being studied as potential anticancer targets (39, 43), peptide and small molecular inhibitors for this subfamily of ligases are being developed (44). These could potentially be used to investigate whether up-regulation of ABCG-mediated lipid export can result in an increase in lipid removal from cells in vivo, and study potential effects on associated disease states such as atherosclerosis.
Author Contributions
S. M. A. acquired the majority of the data, helped with the data interpretation, and drafted the article. V. H. performed the mass spectrometry identification, helped in the conception of this project and interpretation of data, as well as critically revising the article. L. J. S. generated molecular tools, helped with the conception and interpretation, and critically revised the article. A. Y. contributed the ABCG4 data, including the generation of the stable cell line, and critically revised the article. G. R. helped with experimental procedures and critically revised the article. A. J. B. helped in the conception of the project, the interpretation of the data, and in the drafting and critical revision of the article. I. C. G. is responsible for the conception of the project, the development of the ideas, the interpretation of data, and the management of the overall project and revision of the manuscript. All authors have approved the final version of this article.
Supplementary Material
Acknowledgments
We thank Professor Wendy Jessup for her generous donation of HDL2, Professor Helen Hobbs for the ABCG4 cDNA, and Dr. Julian Stevenson for assistance with the cloning of the HUWE1 and NEDD4-1 constructs.
This project was funded by the National Health and Medical Research Council (NHMRC) of Australia (APP1004392). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
- ABC
- ATP-binding cassette
- HECT
- homologous to E6Ap C-terminus
- HECTD1
- HECT domain containing 1, E3 ubiquitin protein ligase
- HUWE1
- HECT, UBA, and WWE domain containing 1, E3 ubiquitin protein ligase
- HDL
- high density lipoprotein
- IP
- immunoprecipitation
- LC/MS
- liquid chromatography mass spectrometry
- NEDD4-1
- neural precursor cell-expressed developmentally down-regulated gene 4
- PMA
- phorbol myristate acetate
- RING
- really interesting new gene.
References
- 1. Klucken J., Büchler C., Orsó E., Kaminski W. E., Porsch-Ozcürümez M., Liebisch G., Kapinsky M., Diederich W., Drobnik W., Dean M., Allikmets R., Schmitz G. (2000) ABCG1 (ABC8), the human homolog of the Drosophila white gene, is a regulator of macrophage cholesterol and phospholipid transport. Proc. Natl. Acad. Sci. U.S.A. 97, 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kerr I. D., Haider A. J., Gelissen I. C. (2011) The ABCG family of membrane-associated transporters: you don't have to be big to be mighty. Br. J. Pharmacol. 164, 1767–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gelissen I. C., Harris M., Rye K. A., Quinn C., Brown A. J., Kockx M., Cartland S., Packianathan M., Kritharides L., Jessup W. (2006) ABCA1 and ABCG1 synergize to mediate cholesterol export to apoA-I. Arterioscler. Thromb. Vasc. Biol. 26, 534–540 [DOI] [PubMed] [Google Scholar]
- 4. Terasaka N., Wang N., Yvan-Charvet L., Tall A. R. (2007) High-density lipoprotein protects macrophages from oxidized low-density lipoprotein-induced apoptosis by promoting efflux of 7-ketocholesterol via ABCG1. Proc. Natl. Acad. Sci. U.S.A. 104, 15093–15098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tarr P. T., Tarling E. J., Bojanic D. D., Edwards P. A., Baldán A. (2009) Emerging new paradigms for ABCG transporters. Biochim. Biophys. Acta 1791, 584–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sag D., Cekic C., Wu R., Linden J., Hedrick C. C. (2015) The cholesterol transporter ABCG1 links cholesterol homeostasis and tumour immunity. Nat. Commun. 6, 6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Westerterp M., Bochem A. E., Yvan-Charvet L., Murphy A. J., Wang N., Tall A. R. (2014) ATP-Binding Cassette Transporters, Atherosclerosis, and Inflammation. Circ. Res. 114, 157–170 [DOI] [PubMed] [Google Scholar]
- 8. Kruit J. K., Wijesekara N., Westwell-Roper C., Vanmierlo T., de Haan W., Bhattacharjee A., Tang R., Wellington C. L., LütJohann D., Johnson J. D., Brunham L. R., Verchere C. B., Hayden M. R. (2012) Loss of both ABCA1 and ABCG1 results in increased disturbances in islet sterol homeostasis, inflammation, and impaired beta-cell function. Diabetes 61, 659–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frisdal E., Le Lay S., Hooton H., Poupel L., Olivier M., Alili R., Plengpanich W., Villard E. F., Gilibert S., Lhomme M., Superville A., Miftah-Alkhair L., Chapman M. J., Dallinga-Thie G. M., Venteclef N., Poitou C., Tordjman J., Lesnik P., Kontush A., Huby T., Dugail I., Clement K., Guerin M., Le Goff W. (2015) Adipocyte Atp-Binding Cassette G1 Promotes Triglyceride Storage, Fat Mass Growth And Human Obesity. Diabetes 64, 840–855 [DOI] [PubMed] [Google Scholar]
- 10. Engel T., Lorkowski S., Lueken A., Rust S., Schlüter B., Berger G., Cullen P., Assmann G. (2001) The human ABCG4 gene is regulated by oxysterols and retinoids in monocyte-derived macrophages. Biochem. Biophys. Res. Commun. 288, 483–488 [DOI] [PubMed] [Google Scholar]
- 11. Wang N., Yvan-Charvet L., Lütjohann D., Mulder M., Vanmierlo T., Kim T. W., Tall A. R. (2008) ATP-binding cassette transporters G1 and G4 mediate cholesterol and desmosterol efflux to HDL and regulate sterol accumulation in the brain. FASEB J. 22, 1073–1082 [DOI] [PubMed] [Google Scholar]
- 12. Tarr P. T., Edwards P. A. (2008) ABCG1 and ABCG4 are coexpressed in neurons and astrocytes of the CNS and regulate cholesterol homeostasis through SREBP-2. J. Lipid Res. 49, 169–182 [DOI] [PubMed] [Google Scholar]
- 13. Wang N., Lan D., Chen W., Matsuura F., Tall A. R. (2004) ATP-binding cassette transporters G1 and G4 mediate cellular cholesterol efflux to high-density lipoproteins. Proc. Natl. Acad. Sci. U.S.A. 101, 9774–9779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tarling E. J., Edwards P. A. (2011) ATP binding cassette transporter G1 (ABCG1) is an intracellular sterol transporter. Proc. Natl. Acad. Sci. U.S.A. 108, 19719–19724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sano O., Ito S., Kato R., Shimizu Y., Kobayashi A., Kimura Y., Kioka N., Hanada K., Ueda K., Matsuo M. (2014) ABCA1, ABCG1, and ABCG4 Are Distributed to Distinct Membrane Meso-Domains and Disturb Detergent-Resistant Domains on the Plasma Membrane. PLoS ONE 9, e109886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Venkateswaran A., Repa J. J., Lobaccaro J. M., Bronson A., Mangelsdorf D. J., Edwards P. A. (2000) Human white/murine ABC8 mRNA levels are highly induced in lipid-loaded macrophages. A transcriptional role for specific oxysterols. J. Biol. Chem. 275, 14700–14707 [DOI] [PubMed] [Google Scholar]
- 17. Ogura M., Ayaori M., Terao Y., Hisada T., Iizuka M., Takiguchi S., Uto-Kondo H., Yakushiji E., Nakaya K., Sasaki M., Komatsu T., Ozasa H., Ohsuzu F., Ikewaki K. (2011) Proteasomal inhibition promotes ATP-binding cassette transporter A1 (ABCA1) and ABCG1 expression and cholesterol efflux from macrophages in vitro and in vivo. Arterioscler. Thromb. Vasc. Biol. 31, 1980–1987 [DOI] [PubMed] [Google Scholar]
- 18. Hsieh V., Kim M. J., Gelissen I. C., Brown A. J., Sandoval C., Hallab J. C., Kockx M., Traini M., Jessup W., Kritharides L. (2014) Cellular cholesterol regulates ubiquitination and degradation of the cholesterol export proteins ABCA1 and ABCG1. J. Biol. Chem. 289, 7524–7536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rotin D., Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 20. Schrader E. K., Harstad K. G., Matouschek A. (2009) Targeting proteins for degradation. Nat. Chem. Biol. 5, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Metzger M. B., Hristova V. A., Weissman A. M. (2012) HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 125, 531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Teixeira L. K., Reed S. I. (2013) Ubiquitin ligases and cell cycle control. Annu. Rev. Biochem. 82, 387–414 [DOI] [PubMed] [Google Scholar]
- 23. Clague M. J., Liu H., Urbé S. (2012) Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev. Cell 23, 457–467 [DOI] [PubMed] [Google Scholar]
- 24. Scheffner M., Kumar S. (2014) Mammalian HECT ubiquitin-protein ligases: Biological and pathophysiological aspects. Biochim. Biophys. Acta 1843, 61–74 [DOI] [PubMed] [Google Scholar]
- 25. Li W., Bengtson M. H., Ulbrich A., Matsuda A., Reddy V. A., Orth A., Chanda S. K., Batalov S., Joazeiro C. A. (2008) Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE 3, e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klock H. E., Lesley S. A. (2009) The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol. Biol. 498, 91–103 [DOI] [PubMed] [Google Scholar]
- 27. Graf G. A., Yu L., Li W. P., Gerard R., Tuma P. L., Cohen J. C., Hobbs H. H. (2003) ABCG5 and ABCG8 are obligate heterodimers for protein trafficking and biliary cholesterol excretion. J. Biol. Chem. 278, 48275–48282 [DOI] [PubMed] [Google Scholar]
- 28. Gelissen I. C., Sharpe L. J., Sandoval C., Rao G., Kockx M., Kritharides L., Jessup W., Brown A. J. (2012) Protein kinase A modulates the activity of a major human isoform of ABCG1. J. Lipid Res. 53, 2133–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sharpe L. J., Rao G., Jones P. M., Glancey E., Aleidi S. M., George A. M., Brown A. J., Gelissen I. C. (2015) Cholesterol sensing by the ABCG1 lipid transporter: Requirement of a CRAC motif in the final transmembrane domain. Biochim. Biophys. Acta 1851, 956–964 [DOI] [PubMed] [Google Scholar]
- 30. Burns V., Sharpe L. J., Gelissen I. C., Brown A. J. (2013) Species variation in ABCG1 isoform expression: implications for the use of animal models in elucidating ABCG1 function. Atherosclerosis 226, 408–411 [DOI] [PubMed] [Google Scholar]
- 31. Sharpe L. J., Cook E. C., Zelcer N., Brown A. J. (2014) The UPS and downs of cholesterol homeostasis. Trends Biochem. Sci. 39; 527–535 [DOI] [PubMed] [Google Scholar]
- 32. Kaliszewski P., Szkopiñska A., Ferreira T., Swiezewska E., Berges T., Zoładek T. (2008) Rsp5p ubiquitin ligase and the transcriptional activators Spt23p and Mga2p are involved in co-regulation of biosynthesis of end products of the mevalonate pathway and triacylglycerol in yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1781, 627–634 [DOI] [PubMed] [Google Scholar]
- 33. Cao X. R., Lill N. L., Boase N., Shi P. P., Croucher D. R., Shan H., Qu J., Sweezer E. M., Place T., Kirby P. A., Daly R. J., Kumar S., Yang B. (2008) Nedd4 controls animal growth by regulating IGF-1 signaling. Sci. Signal. 1, ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Donovan P., Poronnik P. (2013) Nedd4 and Nedd4–2: ubiquitin ligases at work in the neuron. Int. J. Biochem. Cell Biol. 45, 706–710 [DOI] [PubMed] [Google Scholar]
- 35. Camera D., Boase N. A., Kumar S., Pow D. V., Poronnik P. (2014) Subtle gait abnormalities in Nedd4 heterozygous mice. Behav. Brain Res. 260, 15–24 [DOI] [PubMed] [Google Scholar]
- 36. Li J. J., Ferry R. J. Jr., Diao S., Xue B., Bahouth S. W., Liao F. F. (2015) Nedd4 haploinsufficient mice display moderate insulin resistance, enhanced lipolysis, and protection against high-fat diet-induced obesity. Endocrinology 156, 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Akkaya B. G., Zolnerciks J. K., Ritchie T. K., Bauer B., Hartz A. M., Sullivan J. A., Linton K. J. (2015) The multidrug resistance pump ABCB1 is a substrate for the ubiquitin ligase NEDD4-1. Mol. Membr. Biol. 32, 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cirrito J. R., Deane R., Fagan A. M., Spinner M. L., Parsadanian M., Finn M. B., Jiang H., Prior J. L., Sagare A., Bales K. R., Paul S. M., Zlokovic B. V., Piwnica-Worms D., Holtzman D. M. (2005) P-glycoprotein deficiency at the blood-brain barrier increases amyloid-beta deposition in an Alzheimer disease mouse model. J. Clin. Invest. 115, 3285–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bernassola F., Karin M., Ciechanover A., Melino G. (2008) The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14, 10–21 [DOI] [PubMed] [Google Scholar]
- 40. Hao Z., Duncan G. S., Su Y. W., Li W. Y., Silvester J., Hong C., You H., Brenner D., Gorrini C., Haight J., Wakeham A., You-Ten A., McCracken S., Elia A., Li Q., Detmar J., Jurisicova A., Hobeika E., Reth M., Sheng Y., Lang P. A., Ohashi P. S., Zhong Q., Wang X., Mak T. W. (2012) The E3 ubiquitin ligase Mule acts through the ATM-p53 axis to maintain B lymphocyte homeostasis. J. Exp. Med. 209, 173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shearwin-Whyatt L., Dalton H. E., Foot N., Kumar S. (2006) Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. Bioessays 28, 617–628 [DOI] [PubMed] [Google Scholar]
- 42. Jo Y., Lee P. C., Sguigna P. V., DeBose-Boyd R. A. (2011) Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. Proc. Natl. Acad. Sci. U.S.A. 108, 20503–20508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boase N. A., Kumar S. (2015) NEDD4: The founding member of a family of ubiquitin-protein ligases. Gene 557, 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mund T., Lewis M. J., Maslen S., Pelham H. R. (2014) Peptide and small molecule inhibitors of HECT-type ubiquitin ligases. Proc. Natl. Acad. Sci. U.S.A. 111, 16736–16741 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.