Background: The yeast anaphase-promoting complex uses two E2 ubiquitin-conjugating enzymes, Ubc4 and Ubc1, to initiate and elongate polyubiquitin chains.
Results: Ubc1 carries a C-terminal UBA domain that binds the APC/C, enhancing competition with Ubc4.
Conclusion: The UBA domain balances Ubc1 affinity with that of Ubc4.
Significance: The UBA domain of Ubc1 enables efficient polyubiquitination of APC/C substrates.
Keywords: cell cycle, E3 ubiquitin ligase, ubiquitin, ubiquitin-conjugating enzyme (E2 enzyme), yeast
Abstract
The anaphase-promoting complex/cyclosome (APC/C) is a member of the RING family of E3 ubiquitin ligases, which promote ubiquitin transfer from an E2 ubiquitin-conjugating enzyme to a substrate. In budding yeast, the APC/C collaborates with two E2s, Ubc4 and Ubc1, to promote the initiation and elongation, respectively, of polyubiquitin chains on the substrate. Ubc4 and Ubc1 are thought to compete for the same site on the APC/C, but it is not clear how their affinities are balanced. Here, we demonstrate that a C-terminal ubiquitin-associated (UBA) domain enhances the affinity of Ubc1 for the APC/C. Deletion of the UBA domain reduced apparent APC/C affinity for Ubc1 and decreased polyubiquitin chain length. Surprisingly, the positive effect of the UBA domain was not due to an interaction with the acceptor ubiquitin attached to the APC/C substrate or the donor ubiquitin attached to Ubc1 itself. Instead, our evidence suggests that the UBA domain binds to a site on the APC/C core, thereby increasing Ubc1 affinity and enhancing its ability to compete with Ubc4. The UBA domain is required for normal Ubc1 function and E2 competition in vivo. Thus, the UBA domain of Ubc1 ensures efficient polyubiquitination of substrate by balancing Ubc1 affinity with that of Ubc4.
Introduction
The anaphase-promoting complex/cyclosome (APC/C)2 is a large, multisubunit E3 ubiquitin ligase that governs key mitotic events in eukaryotes (1, 2). Like other members of the RING family of ubiquitin ligases, the APC/C catalyzes the transfer of ubiquitin directly from an E2 ubiquitin-conjugating enzyme to a lysine residue on a protein substrate. Subsequent ubiquitin attachment to lysines on ubiquitin itself then leads to the assembly of polyubiquitin signals that mark substrates for destruction by the proteasome.
Polyubiquitin chain assembly by the APC/C depends on the sequential actions of two distinct E2s. In the budding yeast Saccharomyces cerevisiae, the APC/C collaborates with the E2s Ubc4 and Ubc1: first, APC/C interacts with Ubc4 to catalyze attachment of the initial ubiquitin to a lysine residue on the substrate, and then it interacts with Ubc1 to catalyze ubiquitin attachment to lysine 48 (Lys-48) of a preattached ubiquitin, thereby promoting Lys-48-linked polyubiquitin chain assembly (3, 4). In human cells, the APC/C collaborates with the E2s UBCH10 and UBE2S to initiate and elongate Lys-11-linked polyubiquitin chains, respectively (5–12).
Members of the RING family of ubiquitin ligases are generally thought to bind the E2 via a canonical interface between the RING subunit of the E3 and the conserved ubiquitin-conjugating (UBC) domain of the E2 (13). In many cases, this RING-E2 interaction enhances catalysis, primarily by promoting a productive “closed” orientation of the ubiquitin linked to the E2 active site cysteine (7, 14–19).
Interestingly, some E2s appear to interact with non-canonical sites on the E3. In the case of the human APC/C, the initiating E2, UBCH10, participates in canonical RING binding as well as binding the winged helix bundle of the cullin subunit Apc2 using the backside of the UBC domain (9). Both of these interaction surfaces are critical for APC/CUBCH10 activity (9).
The second E2 that operates with the human APC/C, UBE2S, appears to depend almost entirely on non-canonical interactions with the E3. UBE2S has a disordered C-terminal extension that binds a site on the APC2 subunit (5, 10–12, 20). Deletion of this C-terminal extension greatly reduces the binding of UBE2S to the APC/C in vitro (5, 8, 10). In contrast, mutations in the canonical E2-binding site of the RING subunit APC11 do not cause a defect in APC/CUBE2S activity (10). In addition, a distinct face of the RING subunit seems to interact with the ubiquitin that attacks the UBE2S-ubiquitin conjugate, suggesting that UBE2S is not activated by the canonical mechanism (10). Finally, there is recent evidence that the binding of the activator subunit CDH1 to the APC/C causes a conformational change that exposes the canonical E2-binding site of the RING subunit, thereby enhancing UBCH10 binding but having little effect on UBE2S binding (8, 10). These results suggest that UBCH10 and UBE2S bind to different sites on the human APC/C, raising the possibility that they can bind simultaneously to promote polyubiquitin chain assembly.
Unlike the E2s that operate with the human APC/C, the yeast E2s, Ubc4 and Ubc1, both seem to interact with the canonical RING-binding site. For example, Ubc1 inhibits the rapid substrate turnover catalyzed by Ubc4 in APC/C reactions in vitro, suggesting that the two E2s compete for the same binding site (4). Furthermore, addition of the activator Cdh1 to the APC/C promotes the binding of both Ubc4 and Ubc1, suggesting that both E2s utilize the canonical binding site on the RING subunit (21).
Although yeast Ubc1 seems to depend on a canonical RING interaction for its function, it also carries an additional feature that may modulate its interactions with the APC/C. The C terminus of Ubc1 is linked by a 22-residue flexible tether to a ubiquitin-associated (UBA) domain, a type of ubiquitin-binding domain characterized by a three-helix bundle of ∼50 residues (22). The UBA domain of Ubc1 has been shown to bind monoubiquitin with low affinity (KD ∼ 230 μm) (23) but has a ∼6-fold higher affinity (KD ∼ 37 nm) for Lys-48-linked tetraubiquitin (24).
Deletion of the UBA domain and flexible tether (Ubc1ΔUBA) results in a correctly folded and catalytically active UBC domain (25) that is charged normally with ubiquitin by E1 ubiquitin-activating enzyme and retains its catalytic specificity for Lys-48 of ubiquitin (4). Ubc1ΔUBA and wild-type Ubc1 exhibit similar APC/C-independent catalytic rates with ubiquitin as substrate (3), suggesting that the UBA domain is not required for catalytic activity.
Studies of the APC/C reaction in vitro indicate that deletion of the UBA domain reduces the length of Lys-48-linked polyubiquitin chains on APC/C substrates. In addition, the concentration of Ubc1ΔUBA required for half-maximal APC/C activity is increased 10-fold relative to wild type (4), suggesting that the UBA domain promotes binding to some site on the APC/C-substrate complex. However, previous studies indicate that the UBA domain does not bind the ubiquitin covalently linked to the UBC domain (the donor ubiquitin) (26) or the ubiquitin attacking the E2-ubiquitin conjugate (the acceptor ubiquitin) (3). UBA domains in other proteins have been shown to bind non-ubiquitin folds (27–29). It therefore remains unclear how a putative ubiquitin-binding domain promotes a productive interaction between Ubc1 and the APC/C.
Here, we set out to understand the mechanism by which the UBA domain exerts its effects on APC/C activity in vitro and in vivo. Our evidence suggests that the UBA domain binds not to ubiquitin but directly to the APC/C core, thereby boosting Ubc1 affinity and allowing it to compete effectively with Ubc4.
Experimental Procedures
Cloning, Expression, and Purification of Proteins
To make 32P-labeled K48R mutant ubiquitin for diubiquitin synthesis assays, GST-TEV-PKA-K48R-ubiquitin (a gift from Ray Deshaies (18)) was expressed in Escherichia coli, radioactively labeled, and purified as described (3). E2 constructs, including Ubc1-His6, Ubc1ΔUBA-His6, Ubc4-His6, and Ubc4-UBA-His6, were expressed in E. coli and purified as described (4). The Ubc4-UBA chimera was created by amplifying DNA encoding the flexible linker and UBA domain of Ubc1 (residues 151–215) from the Ubc1-His6 expression vector by PCR and ligating into the Ubc4-His6 vector.
The yeast E1 Uba1 was expressed in E. coli and purified as described (30). APC/C was purified from yeast cells using tandem affinity purification as described (30). Where indicated, APC/C carrying tandem affinity purification-tagged Cdc16 was immunoprecipitated from yeast cells using IgG-coupled Dynabeads (Invitrogen) and remained on beads for the duration of the experiment. Cdh1 was expressed in insect cells and purified as described (31). Sea urchin cyclin B N-terminal fragment (CycBN; residues 13–110), either wild-type CycBN or a version containing a single lysine (CycBN 1K; Lys-60 (32)), were expressed in E. coli, purified, and labeled with 125I (where indicated) as described previously (31). All APC/C substrates labeled with [35S]methionine were expressed and translated in rabbit reticulocyte lysates using the TnT Quick Coupled Transcription/Translation System (Promega, Madison, WI). Unlabeled Cdc20 and Cdh1 were also produced by this method where indicated. Proteins were purified from rabbit reticulocyte lysates by immunoprecipitation with IgG-coupled Dynabeads and cleavage from the beads with TEV protease.
Truncated APC11 (encoding residues 35–165) was amplified from yeast genomic DNA by PCR and ligated into a pGEX-4T1-derived expression vector containing an N-terminal GST. GST-Apc11ΔN was expressed in E. coli and purified with glutathione-Sepharose 4B (GE Healthcare).
DNA encoding the UBA domain of Ubc1 (residues 167–215) was amplified from the Ubc1-His6 expression vector by PCR and ligated into a pET28a-derived expression vector containing an N-terminal His61XGB1 tag followed by a recognition site for TEV protease. The UBA domain construct was expressed in E. coli and purified with nickel-nitrilotriacetic acid-agarose (Qiagen, Venlo, Holland). The His61XGB1 tag was cleaved by TEV protease during dialysis, and the tag and protease were removed by incubation with nickel-nitrilotriacetic acid-agarose prior to concentration. This leaves the N terminus of the protein with the sequence GGSGID, in which the last three residues, GID, are the final residues of the Ubc1 flexible linker.
Diubiquitin Synthesis Assays
All reactions were done in QAH buffer (50 mm Hepes, pH 7.4, 100 mm NaCl, 10% glycerol, 1 mm MgCl2) and stopped by addition of 6× non-reducing sample buffer (375 mm Tris-HCl, pH 6.8, 6% SDS, 30% glycerol, 0.03% bromphenol blue, 60 mm N-ethylmaleimide). Ubc1 was charged with 32P-radiolabeled K48R ubiquitin in the following manner. E1 (300 nm), ATP (1 mg/ml), 32P-labeled K48R ubiquitin (∼1 mg/ml), and Ubc1 (0.5 μm) were incubated at room temperature for 20 min. E1 and uncharged E2 were inactivated by incubation with N-ethylmaleimide (10 mm) and EDTA (50 mm) for 15 min at room temperature. Tubes were transferred to 4 °C and incubated with GST-Apc11ΔN (0–80 μm) for 3 min. Wild-type ubiquitin (Boston Biochem, Cambridge, MA) or CycBN (where indicated) was added to the reactions at the concentrations indicated to start the reactions. Proteins were separated by SDS-PAGE, and dried gels were exposed to a storage phosphor screen (GE Healthcare) overnight. Screens were scanned on a Typhoon phosphorimaging system (GE Healthcare), and autoradiographs were quantified using ImageQuant software (GE Healthcare). kobs was calculated by dividing the diubiquitin signal by the charged E2 signal and then dividing by the reaction time in seconds. Data were fit to the Michaelis-Menten equation in Prism software (GraphPad, La Jolla, CA).
APC/C Assays
E2s were charged in the following manner. E1 (300 nm), ATP (1 mg/ml), ubiquitin (100 μm), and E2 (0–40 μm) were incubated at room temperature for 20 min. APC/C, activator (either Cdh1 or Cdc20), and radiolabeled substrate were preincubated for 10 min, and reactions were started by mixing the E2 charging mixture with the APC/C mixture except where otherwise indicated. All reactions were carried out in QAH buffer, pH 7.4, for the amount of time indicated, and reaction products were separated by SDS-PAGE and visualized by autoradiography. APC/C activity was calculated by combining signals from all modified substrate bands and dividing by the reaction time in seconds. APC/C processivity was calculated by quantifying individual ubiquitinated products, multiplying the amount of product by the number of ubiquitins in the product, and dividing by the total amount of modified products.
For the assays in which substrate was fused to the APC/C, APC/C was immunoprecipitated from cdh1Δ doc1Δ cells using IgG-coupled Dynabeads (Invitrogen) and remained bead-bound throughout the course of the reactions. [35S]Methionine-labeled N-terminal securin fragment (residues 1–110) fused to the N terminus of Apc10 ([35S]SecurinN-Apc10) was incubated with the APC/C on beads, and unbound [35S]SecurinN-Apc10 was washed away. Ubc1 (either wild type or ΔUBA) charged with methylated ubiquitin (Boston Biochem) was added in increasing concentrations.
Yeast Strains and Analysis
All yeast strains were in the W303 background and are listed in Table 1. Strains were generated using standard yeast cloning techniques for transformation, mating, sporulation, and tetrad dissection. For yeast growth assays, strains were grown to midlog phase at 30 °C, diluted to an A600 of 0.1, and plated on the indicated medium. Plates were scanned, and images were prepared with Adobe Photoshop. For cell cycle analysis, asynchronous yeast cultures were grown to an A600 of 0.2 at 30 °C and then arrested in G1 by incubation with α factor (1 μg/ml) for at least 3 h. Cultures were released from G1 arrest by washing away α factor and resuspending in the indicated medium (zero time point). Cell samples were taken at the indicated times, lysed, and analyzed by Western blotting against the indicated proteins. Where shown, parallel samples were taken, and a budding index was counted by microscopy. For Western blot analysis, Securin-9XMyc and Ubc1–9XMyc were detected by monoclonal 9E10 anti-Myc antibody (Covance, Princeton, NJ; 1:1000), Cdk1 was detected by polyclonal sc-53 anti-Cdk1 antibody (Santa Cruz Biotechnology, Dallas, TX; 1:1000), and Ubc1–1XFLAGHis6 was detected by monoclonal M2 FLAG antibody (Sigma; 1:5000).
TABLE 1.
List of yeast strains
All strains listed are W303 and derived from AFS92 (from A. Straight).
| Strain | Genotype |
|---|---|
| DOM918 | MAT a; PDS1-13XMyc:HIS3 |
| yJG2 | MAT a; PDS1-13XMyc:HIS3; pGAL-3XHA-UBC1:TRP1; UBC1-9XMYC::LEU2 |
| yJG3 | MAT a; PDS1-13XMyc:HIS3; pGAL-3XHA-UBC1:TRP1; ubc1ΔUBA-9XMYC::LEU2 |
| yJG5 | MAT a; PDS1-13XMyc:HIS3; pGAL-3XHA-UBC1:TRP1; empty vector::LEU2 |
| yJG10 | MAT a; PDS1-13XMyc:HIS3; UBC1-His61XFLAG:URA3(Kluyveromyces lactis) |
| yJG11 | MAT a; PDS1-13XMyc:HIS3; ubc1ΔUBA-His61XFLAG:URA3(K. lactis) |
| yJG14 | MAT a; PDS1-13XMyc:HIS3; pGAL-3XHA-UBC1:TRP1; ubc4Δ:LEU2; UBC1-9XMYC::LEU2 |
| yJG15 | MAT a; PDS1-13XMyc:HIS3; pGAL-3XHA-UBC1:TRP1; ubc4Δ:LEU2; ubc1ΔUBA-9XMYC::LEU2 |
| yJG16 | MAT a; PDS1-13XMyc:HIS3; pGAL-3XHA-UBC1:TRP1; ubc4Δ:LEU2; empty vector::LEU2 |
| yJG81 | MAT α; PDS1-13XMyc:HIS3; UBC4-UBA-9XMyc:LEU2 |
| yJG82 | MAT a; PDS1-13XMyc:HIS3; UBC4-UBA-9XMyc:LEU2 |
| yJG85 | MAT a; PDS1-13XMyc:HIS3; pGAL-3XHA-UBC1:TRP1; UBC1-9XMYC::LEU2; UBC4-UBA-9XMyc:LEU2 |
| yJG86 | MAT α; PDS1-13XMyc:HIS3; pGAL-3XHA-UBC1:TRP1; UBC1-9XMYC::LEU2; UBC4-UBA-9XMyc:LEU2 |
| yJG87 | MAT a; PDS1-13XMyc:HIS3; pGAL-3XHA-UBC1:TRP1; ubc1ΔUBA-9XMYC::LEU2; UBC4-UBA-9XMyc:LEU2 |
| yJG88 | MAT α; PDS1-13XMyc:HIS3; pGAL-3XHA-UBC1:TRP1; ubc1ΔUBA-9XMYC::LEU2; UBC4-UBA-9XMyc:LEU2 |
Results
The UBA Domain Does Not Contribute to RING-mediated Stimulation of E2 Catalytic Activity
Previous evidence suggests that the UBA domain does not contribute to the APC/C-independent E2 catalytic rate or affinity for the acceptor ubiquitin (3). Here, we tested whether the UBA domain has an impact on these parameters when Ubc1 is bound to its RING E3 partner. In other systems, binding of an E2 to the RING orients the donor ubiquitin in a closed conformation that greatly enhances E2 catalytic function (7, 14–19). Moreover, the RING subunit of human APC/C promotes UBE2S activity through a unique interaction between the acceptor ubiquitin and a specific surface of the RING domain (10). It was therefore conceivable that the UBA domain could contribute to Ubc1 catalytic rate or acceptor ubiquitin affinity only in the presence of the RING, or that the UBA domain could bind directly to the RING. To explore these possibilities, we first set out to examine what role, if any, the RING subunit of APC/C plays in catalytic activation of Ubc1 or binding the acceptor ubiquitin.
We addressed this question with a diubiquitin synthesis assay in which Ubc1 is charged with radiolabeled K48R ubiquitin (donor) after which unlabeled wild-type ubiquitin (acceptor) is added at increasing concentrations, leading to the formation of radiolabeled Lys-48-linked diubiquitin, which cannot be elongated further. Ubc1 cannot be recharged by E1 in this assay due to inactivation of E1 and free E2, and thus the assay measures a single turnover of E2, allowing estimates of Ubc1 catalytic rate and affinity for acceptor ubiquitin (3, 18).
We carried out these studies with purified Apc11, the RING domain-containing subunit of APC/C. We found that Apc11 could be expressed recombinantly in E. coli after deletion of the N-terminal 34 residues, which contain the cullin-binding region and a flexible linker. The truncated Apc11 protein was expressed and purified as an N-terminal GST fusion (GST-Apc11ΔN) and added to diubiquitin synthesis assays.
GST-Apc11ΔN caused a massive stimulation of diubiquitin synthesis by Ubc1 (Fig. 1A) to the extent that reactions containing GST-Apc11ΔN were carried out on ice for 5 s to prevent depletion of the charged E2. Quantification of results from three separate experiments indicated that GST-Apc11ΔN stimulated the maximal catalytic rate of Ubc1 about 700-fold from 0.0002 to 0.14 s−1 (Fig. 1A). GST-Apc11ΔN caused a minor ∼2-fold reduction in Km for the acceptor ubiquitin from ∼840 to ∼435 μm (Fig. 1A), suggesting that Apc11 does not stimulate Ubc1 by interacting with the acceptor ubiquitin but rather by allosterically activating the E2 or by orienting the E2-donor ubiquitin conjugate for successful attack. This mechanism is distinct from the mechanism by which human APC/C stimulates the E2 UBE2S, which is primarily through decreasing the Km for acceptor ubiquitin by ∼40-fold via APC11 binding directly to the acceptor ubiquitin (10).
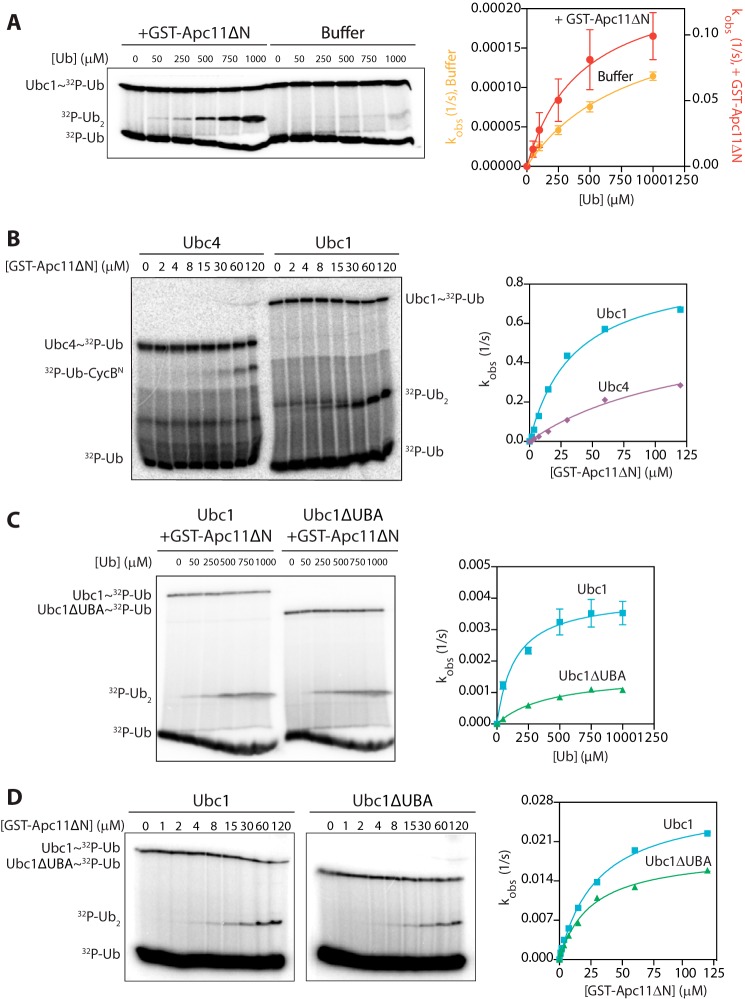
FIGURE 1.
The UBA domain does not contribute to acceptor ubiquitin binding or RING enhancement of E2 catalysis. A, Ubc1 (wild type; 0.5 μm) was charged with 32P-radiolabeled K48R ubiquitin and incubated with either GST-Apc11ΔN (80 μm) or buffer. Wild-type ubiquitin was added at the indicated concentrations, and reactions were carried out at 4 °C for 5 s (+GST-Apc11ΔN) or 10 min (buffer alone). Reaction products were analyzed by SDS-PAGE and autoradiography with a phosphorimaging system. The right panel displays the quantification of diubiquitin synthesis assays, showing the dependence of catalytic rate (kobs) on ubiquitin concentration. Autoradiographs were quantified using ImageQuant, and data were fit to the Michaelis-Menten equation in Prism software. The average of three experiments is shown. Error bars represent S.E. B, Ubc1 (wild type) and Ubc4 (each at 0.5 μm) were charged with 32P-radiolabeled K48R ubiquitin and incubated with buffer or increasing concentrations of GST-Apc11ΔN. Unlabeled CycBN (200 μm; Ubc4 reactions) or ubiquitin (100 μm; Ubc1 reactions) was added, and reactions were carried out at 4 °C for 5 s. Reactions were analyzed as in A. Results are representative of three independent experiments. C, Ubc1 (wild type or ΔUBA at 0.5 μm) reactions were carried out as in A except that a subsaturating concentration of GST-Apc11ΔN (6.5 μm) was used. Reactions were carried out at room temperature for 3 (wild-type Ubc1) or 5 min (Ubc1ΔUBA). The average of three experiments is shown. Error bars represent S.E. D, Ubc1 and Ubc1ΔUBA were charged as in A and incubated with buffer or increasing concentrations of GST-Apc11ΔN. Unlabeled wild-type ubiquitin (100 μm) was added, and reactions were carried out, visualized, and quantified as in A. Results are representative of three independent experiments. Ub, ubiquitin.
If Ubc1 and Ubc4 bind to the same site on Apc11, then one might expect that Apc11 can also stimulate Ubc4 catalytic activity. Because Ubc4 does not readily form diubiquitin in this assay, we used a modified assay in which unlabeled CycBN was used as an acceptor substrate instead of ubiquitin. Because of the low affinity of CycBN for Ubc4, it is difficult to saturate with substrate. Thus, instead of measuring activity across a range of substrate concentrations, we added increasing amounts of GST-Apc11ΔN to Ubc4 and Ubc1 assays with subsaturating substrate. The catalytic rate of both E2s increased significantly with the concentration of GST-Apc11ΔN (Fig. 1B). The concentration of GST-Apc11ΔN required for half-maximal activity was ∼4-fold lower for Ubc1 (35 μm) than it was for Ubc4 (135 μm), suggesting that Ubc1 might have a slightly higher affinity for the RING.
To measure the contribution of the UBA domain to stimulation by GST-Apc11ΔN, we compared Ubc1 and Ubc1ΔUBA in diubiquitin synthesis assays. We used a subsaturating concentration of GST-Apc11ΔN to prevent depletion of the charged E2 and reduce experimental variability; because GST-Apc11ΔN greatly stimulates Ubc1, RING-stimulated Ubc1 activity represents the majority of the activity in this assay. We found that deletion of the UBA domain had a minor effect on activation by Apc11; in multiple experiments, Ubc1ΔUBA exhibited a slightly lower catalytic rate than wild-type Ubc1 (0.0016 and 0.0040 s−1, respectively) and a slightly higher Km for the acceptor ubiquitin (425 and 145 μm, respectively) (Fig. 1C). In a GST-Apc11ΔN dose response, the UBA domain had no impact on the half-maximal concentration of GST-Apc11ΔN (25 μm for Ubc1ΔUBA and 30 μm for Ubc1), indicating that the UBA domain does not affect E2 affinity for Apc11 (Fig. 1D). Deletion of the UBA domain caused a minor decrease in maximal catalytic activity (0.03 s−1 for Ubc1 and 0.02 s−1 for Ubc1ΔUBA) (Fig. 1D). Our results suggest that the UBA domain does not contribute significantly to E2 affinity for the acceptor or donor ubiquitin, E2 affinity for the RING subunit, or RING-dependent Ubc1 activation.
The UBA Domain Does Not Bind Ubiquitin Attached to APC/C Substrate
Our results suggest that the UBA domain does not bind to the acceptor ubiquitin in assays where Ubc1 is bound to the RING subunit of the APC/C. It remained possible, however, that some other component of the APC/C orients the acceptor ubiquitin on an APC/C substrate, allowing the ubiquitin to bind the UBA domain. To rule out this possibility, we determined the effects of the UBA domain in APC/C reactions with a substrate that lacks preattached ubiquitin.
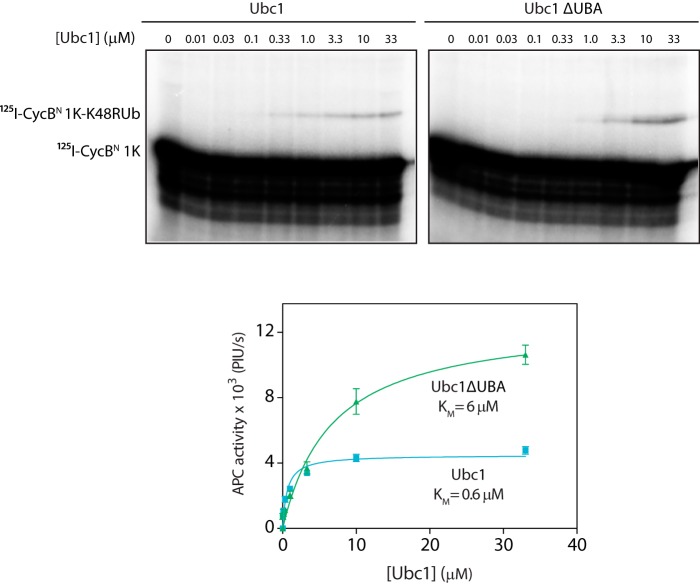
We used 125I-labeled single lysine CycBN as substrate and carried out APC/C reactions with K48R ubiquitin, ensuring that the substrate could be modified only once with a single ubiquitin. We found that the half-maximal concentration of Ubc1ΔUBA with this substrate was 10-fold higher than the half-maximal concentration of wild-type Ubc1 (0.6 μm for Ubc1 and 6 μm for Ubc1ΔUBA; Fig. 2). This is the same difference in apparent Ubc1 affinity that is seen with wild-type ubiquitin in reactions with conventional substrates (4), suggesting that Ubc1ΔUBA is defective in both initiation of a ubiquitin chain and subsequent polyubiquitination. Thus, the UBA domain promotes ubiquitination even when there is no ubiquitin on the substrate, further arguing that the effects of the UBA domain do not depend on its interaction with ubiquitin.
FIGURE 2.
The UBA domain is independent of ubiquitin on substrate. APC/C purified from yeast cells was mixed with Cdh1 and 125I-CycB 1K. Ubc1 (either wild type or ΔUBA) charged with K48R mutant ubiquitin was added at the indicated concentrations, and reactions were carried out at room temperature for 15 min. Reaction products were analyzed by SDS-PAGE and autoradiography with a phosphorimaging system. The bottom panel displays quantification of results, showing the dependence of APC/C activity on concentration of Ubc1 or Ubc1ΔUBA. Autoradiographs were quantified using ImageQuant, and data were fit to the Michaelis-Menten equation in Prism software. The average of four experiments is shown. Error bars represent S.E. PIU/s, phosphorimaging units/s.
The UBA Domain Does Not Bind to APC/C Activator
Because the UBA domain does not interact with ubiquitin attached to APC/C substrate, we asked whether the UBA domain binds other components of the APC/C. We first tested the possibility that the UBA domain interacts with the activator subunit, by analyzing the effect of the UBA domain in reactions where activator subunit is not present.
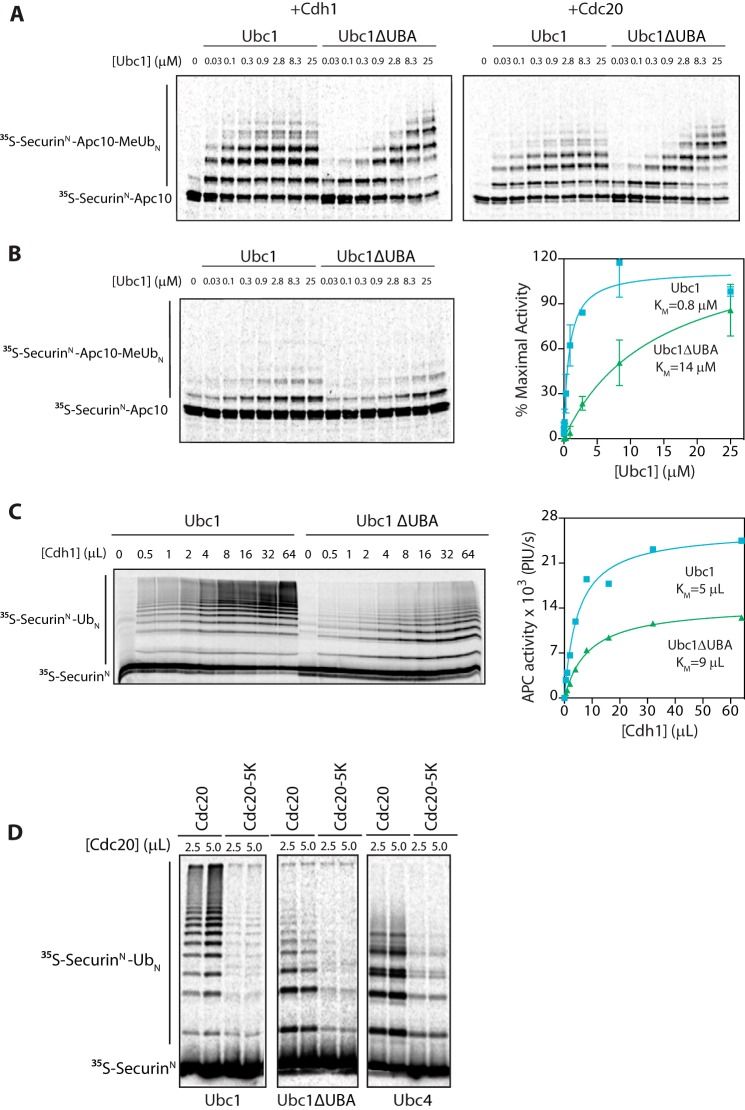
We used a recently devised APC/C assay in which it is possible to measure activity in the absence of activator (21). Although the activator subunit is normally required for substrate recruitment, this requirement can be bypassed by using a radiolabeled substrate (the N-terminal region of securin) directly fused to the Apc10 subunit of the APC/C (SecurinN-Apc10). Some ubiquitination of this substrate occurs in the absence of activator, but addition of activator enhances activity and E2 affinity due to an activator-induced conformational change (8, 21).
Deletion of the UBA domain increased the half-maximal E2 concentration in the presence of the APC/C activator Cdh1 or Cdc20 (Fig. 3A) as seen in previous studies with soluble substrate (4). Most importantly, deletion of the UBA domain also caused an ∼18-fold increase in the half-maximal E2 concentration in the absence of added activator from 0.8 to 14 μm (Fig. 3B). Thus, the enhanced binding provided by the UBA domain does not require activator or depend on the conformational change caused by activator. Notably, the maximal catalytic rate of Ubc1ΔUBA was comparable with that of wild-type Ubc1 (Fig. 3A; at maximal E2 concentrations), providing evidence that deletion of the UBA domain does not affect catalysis in the presence of the APC/C.
FIGURE 3.
UBA domain does not bind to APC/C activator. A, [35S]SecurinN-Apc10, Cdh1, and Cdc20 were generated by in vitro translation in rabbit reticulocyte lysate. [35S]SecurinN-Apc10 was bound to immunoprecipitated APC/C (apc10Δ cdh1Δ), and unbound substrate was washed away. Cdh1 and Cdc20 were purified from reticulocyte lysate and mixed with fusion substrate-bound APC/C. Ubc1 (either wild type or ΔUBA) charged with methylated ubiquitin (MeUb) was added in increasing concentrations, and reactions were carried out at room temperature for 15 min. Reaction products were analyzed by SDS-PAGE and autoradiography with a phosphorimaging system. Results are representative of three independent experiments. B, APC/C assays were performed as in A except that no activator subunit was added. The graph at right shows mean values ±S.E. normalized to maximal Ubc1 activity of three independent experiments. C, [35S]SecurinN and Cdh1 were generated by in vitro translation in rabbit reticulocyte lysate and purified. Immunoprecipitated APC/C was mixed with [35S]SecurinN and increasing amounts of Cdh1. Ubc1 or Ubc1ΔUBA (10 μm) was charged with wild-type ubiquitin (Ub), and reactions were carried out at room temperature for 15 min. Reaction products were analyzed as in A. The graph at right shows quantification of APC/C activity as a function of Cdh1 concentration for the experiment at left. Results are representative of three independent experiments. PIU/s, phosphorimaging units/s. D, [35S]SecurinN, Cdc20, and Cdc20-5K were generated as in C and mixed with purified APC/C. E2s (each at 5 μm) charged with wild-type ubiquitin were added, and reactions were carried out at room temperature for 30 min. Reaction products were analyzed as in A. Results are representative of three independent experiments. Error bars represent S.E.
To explore further whether the UBA domain interacts with the activator, we measured the concentration of Cdh1 needed for half-maximal APC/C activity, using soluble 35S-labeled SecurinN substrate and saturating amounts of either Ubc1 or Ubc1ΔUBA. Half-maximal Cdh1 concentrations were similar for mutant and wild-type Ubc1 (Fig. 3C), further suggesting that the UBA domain functions independently of activator.
The APC/C activator Cdc20 autoubiquitinates at multiple lysines during the course of the cell cycle (33). To test whether the UBA domain of Ubc1 binds to ubiquitin conjugated to Cdc20, we carried out APC/CUbc1 reactions with a mutant form of Cdc20, Cdc20-5K, that is poorly ubiquitinated because most of its ubiquitinated lysines are mutated to arginine (33). Although the activity of the Cdc20-5K mutant was low relative to wild-type Cdc20, the average chain length was the same, suggesting that there is no defect in Ubc1 binding (Fig. 3D). Similar results were obtained with APC/CUbc1ΔUBA, further indicating that the UBA domain does not bind ubiquitin conjugated to activator (Fig. 3D).
The UBA Domain Acts Independently to Promote APC/C Binding
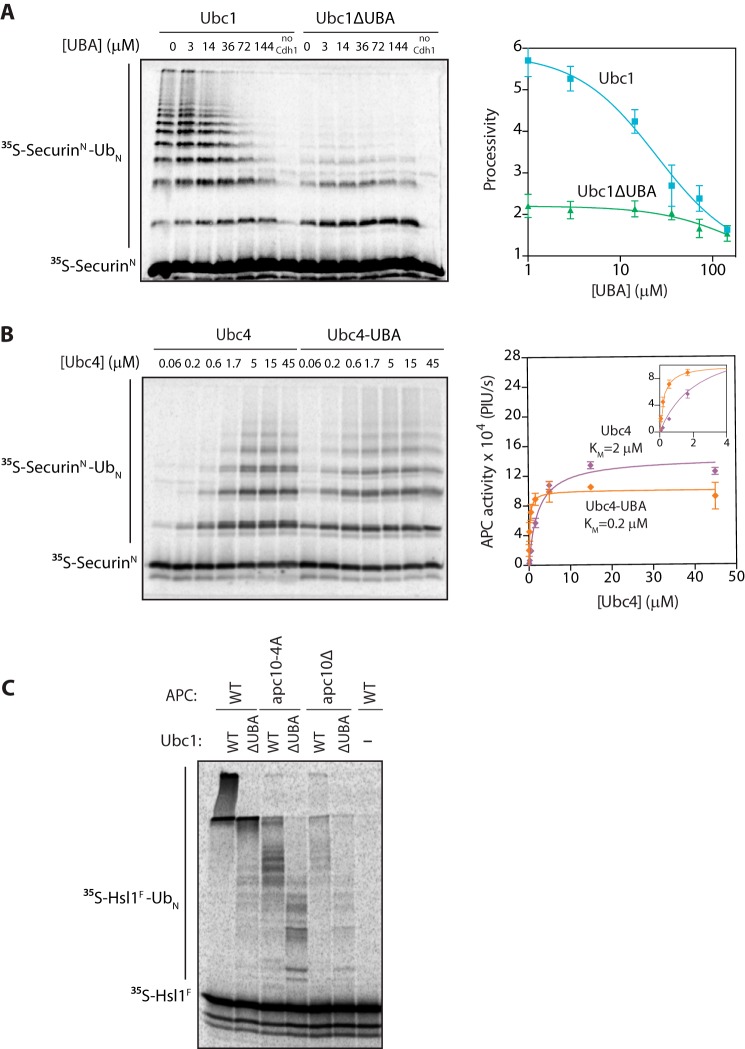
We next hypothesized that the UBA domain binds to the APC/C core. To explore this possibility, we tested whether adding the UBA domain alone to an APC/CUbc1 reaction inhibited processivity in trans. Recombinant UBA domain was prepared in E. coli, and high concentrations of the protein reduced chain length in APC/CUbc1 reactions to the length seen in reactions with Ubc1ΔUBA (Fig. 4A). UBA domain did not significantly affect APC/CUbc1ΔUBA reactions (Fig. 4A). The IC50 of the UBA domain in the wild-type Ubc1 reactions was ∼25 μm (Fig. 4A). These results suggest that the UBA domain can reduce reaction processivity independently of the UBC domain.
FIGURE 4.
The UBA domain promotes APC/C binding. A, [35S]SecurinN was translated in vitro in rabbit reticulocyte lysate, purified, and mixed with purified APC/C, Cdh1, and varying concentrations of recombinant UBA domain. Ubc1 (either wild type or ΔUBA; 10 μm) charged with wild-type ubiquitin (Ub) was added, and reactions were carried out at room temperature for 15 min. Reaction products were analyzed by SDS-PAGE and autoradiography with a phosphorimaging system. Autoradiographs were quantified using ImageQuant. The right panel displays quantification of processivity of APC/CUbc1 and APC/CUbc1ΔUBA as a function of the concentration of free UBA domain. Processivity was calculated by quantifying individual ubiquitinated products, multiplying the amount of product by the number of ubiquitins in the product, and dividing by the total amount of modified products. Data were fit to the log(inhibitor) versus response equation in Prism software. The average of three experiments is shown. Error bars represent S.E. B, purified [35S]SecurinN, APC/C, and Cdh1 were combined as in A. Ubc4 or Ubc4-UBA charged with wild-type ubiquitin was added at the indicated concentrations, and reactions were carried out at room temperature for 15 min. Reaction products were analyzed as in A. The right panel displays quantification of the dependence of APC/C activity on concentration of Ubc4 or Ubc4-UBA. Data were fit to the Michaelis-Menten equation in Prism software. The average of three experiments is shown. Error bars represent S.E. The inset shows a close-up of the graph at lower E2 concentrations. PIU/s, phosphorimaging units/s. C, [35S]Hsl1F was translated in vitro in rabbit reticulocyte lysate, purified, and mixed with Cdh1 and APC/C immunoprecipitated from wild-type, apc10Δ mutant, or apc10-4A mutant yeast. E2s (all at 5 μm final concentration) were charged with wild-type ubiquitin and added, and reactions were carried out at room temperature for 15 min. Reaction products were analyzed as in A.
We further explored the modularity of the UBA domain by creating a chimeric E2, Ubc4-UBA, in which the UBA domain and flexible tether of Ubc1 are fused to the C terminus of Ubc4. Adding the UBA domain to Ubc4 lowered its half-maximal concentration in a conventional APC/C reaction (Fig. 4B), suggesting that the UBA domain boosts Ubc4 affinity for the APC/C. The increase in apparent affinity was 10-fold, from 2 to 0.2 μm (Fig. 4B), identical to the loss of Ubc1 affinity upon deletion of the UBA domain.
Fusion of the UBA domain to Ubc4 did not significantly affect its maximal catalytic activity (Fig. 4B) or its specificity for ubiquitin chain initiation (see Fig. 5, A and B, first two lanes). Thus, the UBA domain can confer a boost in APC/C affinity to a distinct E2 with different lysine specificity, providing more evidence that the UBA domain does not interact with the UBC domain or Lys-48-linked polyubiquitin but is binding some site that is common to the functions of both E2s, such as the APC/C core.
FIGURE 5.
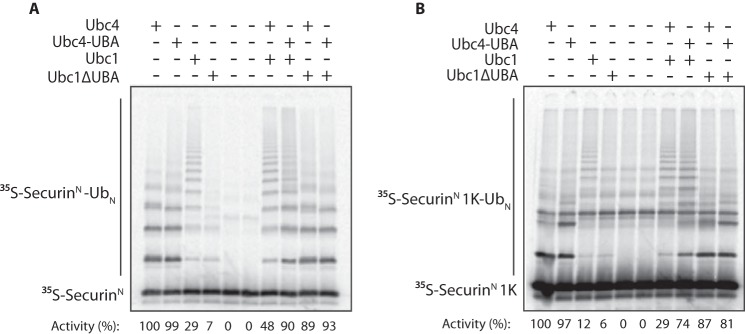
The UBA domain is important for E2 competition. A, [35S]SecurinN was translated in vitro in rabbit reticulocyte lysate, purified, and mixed with purified APC/C and Cdh1. The indicated E2s (each at 3 μm final concentration) were charged with wild-type ubiquitin (Ub) and added. Reactions were carried out at room temperature for 20 min. Reaction products were analyzed by SDS-PAGE and autoradiography with a phosphorimaging system. The numbers below show quantification of APC/C activity (i.e. total modified substrate) in each lane with Ubc4 activity normalized to 100. Results are representative of three independent experiments. B, APC/C assays were performed as in A except that [35S]SecurinN 1K was used as the substrate. Results are representative of three independent experiments.
The core subunit Apc10/Doc1 is a short distance from the E2-binding site of the APC/C. We tested its role in UBA domain binding by measuring Ubc1-dependent activity with APC/C lacking the Apc10 subunit. This subunit is involved in substrate binding, and deletion of Apc10 or mutation of key substrate-binding residues (the apc10-4A mutant) greatly reduces the processivity of ubiquitination (30, 34). For these experiments, we used a fragment of the APC/C substrate Hsl1, Hsl1F (residues 667–882), which binds extremely tightly to the APC/C and is modified with very high processivity. We found that apc10Δ APC/C and apc10-4A APC/C showed similar defects in polyubiquitin chain length with Ubc1, and deletion of the UBA domain from Ubc1 caused a major decrease in processivity regardless of the presence or absence of Apc10 (Fig. 4C). It is therefore unlikely that the UBA domain binds to the Apc10 subunit. Because our earlier work ruled out Apc11 as a binding site (Fig. 1B), it seems likely that the UBA domain binds some site on the nearby Apc1 or Apc2 subunits. We were unable to test this possibility because deletion of either of these subunits abolishes APC/C activity, and neither subunit can be expressed stably as a recombinant protein for binding experiments.
The UBA Domain Ensures E2 Competition
Ubc1 and Ubc4 likely compete for the same canonical binding site on the RING subunit Apc11. Our evidence suggests that the UBA domain of Ubc1 provides an extra affinity boost, and we hypothesized that deleting the UBA domain should decrease the ability of Ubc1 to compete with Ubc4. We assessed E2 competition by analyzing the products of APC/C reactions with each E2 alone or in combination. As seen in previous work (4), Ubc4 alone rapidly modified the substrate at multiple lysines to generate short monoubiquitinated products, whereas Ubc1 generated long polyubiquitin chains but turned over less substrate because it is less efficient than Ubc4 in attachment of the initial ubiquitin (Fig. 5, A and B). When the two E2s were mixed at equal concentrations, the high initiating activity of Ubc4 was reduced by competition with Ubc1, but the total amount of polyubiquitin chains increased slightly due to the increased number of initial ubiquitins relative to Ubc1 alone (see quantification of activity in Fig. 5, A and B). In addition, deletion of the UBA domain from Ubc1 decreased its ability to compete with Ubc4, resulting in higher substrate turnover, lower average polyubiquitin chain length, and a pattern of modification similar to that with Ubc4 alone (Fig. 5A). Similar results were obtained in reactions with a single lysine substrate, SecurinN 1K, which exhibits decreased substrate turnover (because Ubc4 can only modify the substrate with one or two ubiquitins) and very few ubiquitin chains (Fig. 5B).
Because attaching the UBA domain to Ubc4 increased its apparent affinity for APC/C, we hypothesized that Ubc4-UBA should compete more effectively with Ubc1 as compared with Ubc4. Indeed, addition of Ubc4-UBA to a Ubc1 reaction reduced average polyubiquitin chain length, resulting in a pattern of reaction products more closely resembling that seen in a reaction with Ubc4 alone (Fig. 5A). Also, APC/C activity was higher in the Ubc1 + Ubc4-UBA reaction relative to a Ubc1 + Ubc4 reaction, indicating that Ubc1 cannot compete as effectively with Ubc4-UBA as it can with Ubc4. The Ubc1 + Ubc4-UBA reaction exhibited a more heterogeneous banding pattern than the Ubc1 + Ubc4 reaction (Fig. 5A), likely because Ubc1 extended chains on substrates that had been monoubiquitinated at multiple lysines by Ubc4-UBA. The average ubiquitin chain length was also slightly shorter in a Ubc1 + Ubc4-UBA reaction with a single lysine substrate (Fig. 5B). Thus, we propose that the extra affinity provided by the UBA domain of Ubc1 ensures efficient polyubiquitination of substrate by balancing Ubc1 affinity with that of Ubc4, resulting in the optimal modification of APC/C substrates for proteasomal recognition.
The UBA Domain Is Important for APC/C Activity in Vivo
We assessed the importance of the UBA domain for Ubc1 function in vivo in S. cerevisiae. First, we deleted the UBA domain at the endogenous UBC1 locus and also introduced a C-terminal 1XFLAGHis6 tag. As a control, we introduced the same tag at the wild-type locus. Ubc1ΔUBA was expressed at a slightly lower level than Ubc1 (data not shown). Deletion of the UBA domain did not appear to have any effect on growth or the timing of destruction of the APC/C substrate securin (Fig. 6A).
FIGURE 6.
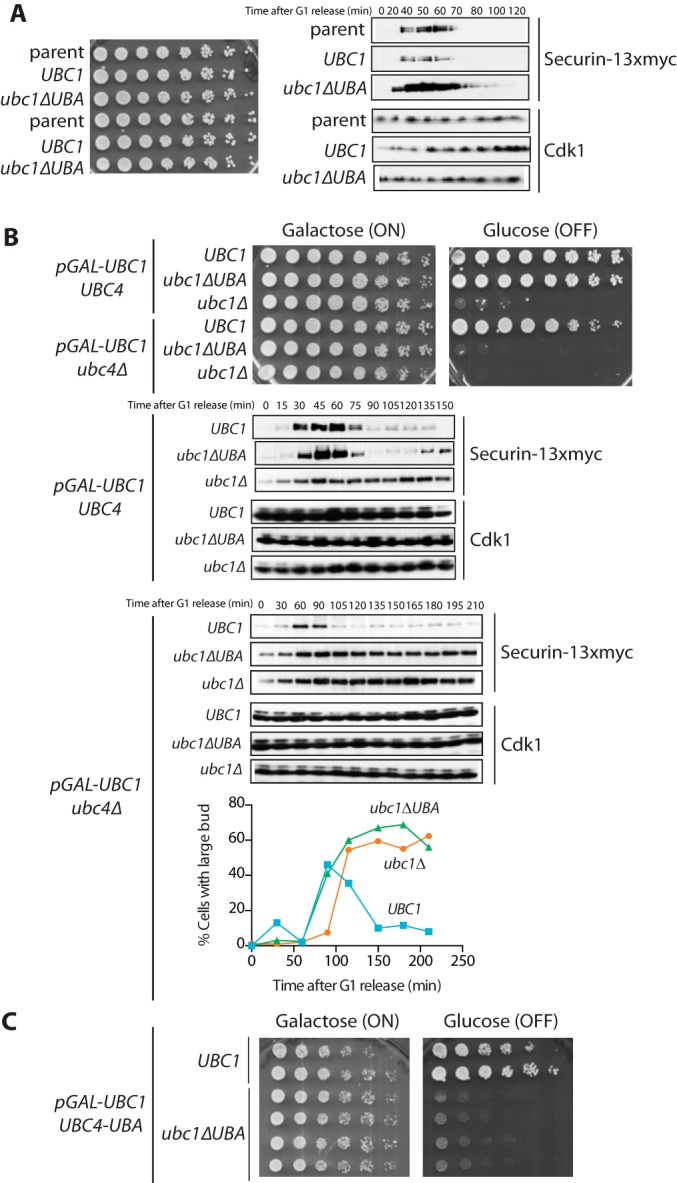
The UBA domain is important for Ubc1 function in vivo. A, left, strains were grown to midlog phase at 30 °C in medium containing 2% glucose, diluted to an A600 of 0.1, plated as serial dilutions on 2% glucose, and grown for 2 days at 30 °C. Right, asynchronous cultures (A600 = 0.2) were arrested in G1 with α factor (1 μg/ml) for 3 h and released from G1 arrest by washing away α factor (zero time point). Cell samples were taken at the indicated times, lysed, and analyzed by Western blotting against the indicated proteins. Results are representative of three independent experiments. B, top, strains were grown to midlog phase at 30 °C in medium containing 2% galactose and raffinose, diluted to an A600 of 0.1, plated as serial dilutions on 2% galactose and raffinose or 2% glucose, and grown for 2 days at 30 °C. Results are representative of three independent experiments. Bottom, asynchronous cultures were arrested in G1 with α factor (1 μg/ml) for 5 h. During the last 2 h of α factor treatment, cultures were incubated with 2% glucose. Cells were released from G1 by washing away α factor, and resuspended in medium containing 2% glucose (zero time point). Cell samples were taken at the indicated times, lysed, and analyzed by Western blotting against the indicated proteins. Parallel samples were taken, and a budding index was counted by microscopy. Results are representative of three independent experiments. C, the indicated strains were grown and plated as in B.
Tetrad analysis revealed that when we sensitized the system by deleting UBC4, the UBA domain became essential for yeast survival (data not shown). To further explore the phenotype of ubc4Δ ubc1ΔUBA strains, we created a conditional system in which we placed the endogenous copy of UBC1 under the control of the GAL promoter (with an N-terminal 3XHA tag) and introduced a second copy of UBC1 (either wild type, ubc1ΔUBA, or an empty vector) under the control of the endogenous promoter (tagged with 9XMYC) at the LEU2 locus. In this system, we could shut off expression of pGAL-UBC1 and observe the effects of the ubc1ΔUBA mutation in strains with either UBC4 or ubc4Δ. As in our earlier experiments, the ubc1ΔUBA mutant displayed no colony growth defect, but the ubc4Δ ubc1ΔUBA double mutant did not proliferate at all (Fig. 6B). Importantly, all the strains grew similarly when pGAL-UBC1 was expressed (Fig. 6B). The expression of Ubc1 and Ubc1ΔUBA were comparable in these strains (data not shown).
To determine whether ubc4Δ ubc1ΔUBA double mutants have a defect in cell cycle progression due to a loss of APC/C activity, we released these strains from a G1 arrest after shutting off pGAL-UBC1. ubc4Δ ubc1ΔUBA cells arrested with high levels of the APC/C substrate securin and large buds (Fig. 6B), consistent with a pre-anaphase arrest like that seen in apc mutants. We conclude that the UBA domain is particularly important for Ubc1 function in the absence of Ubc4. These results are consistent with our biochemical evidence that deletion of the UBA domain does not simply cause a defect in ubiquitin chain elongation by Ubc1 but also causes a defect in chain initiation (Figs. 2 and 5), and cell survival requires that Ubc1 must carry out this task efficiently in the absence of Ubc4.
We used a similar system to test the effect of attaching the Ubc1 UBA domain to Ubc4. We found that UBC4-UBA cells displayed wild-type viability (Fig. 6C). However, deletion of the UBA domain from Ubc1 together with attachment of the UBA domain to Ubc4 resulted in synthetic lethality (Fig. 6C). This effect was not due to low Ubc4-UBA expression (data not shown) or a lack of Ubc4-UBA activity, because our earlier results (Fig. 4B) indicate that this E2 is fully active in vitro. Instead, we believe that fusing the UBA domain to Ubc4 and deleting the UBA domain of Ubc1 increase the affinity of Ubc4 for the APC/C but decrease the affinity of Ubc1. This imbalance in E2 affinities cannot support proper APC/C activity, likely because Ubc1 cannot perform its essential function in chain elongation (Fig. 5A, last lane).
Discussion
We report that the UBA domain enhances Ubc1 affinity for the APC/C, ensuring that Ubc1 binds with sufficient affinity in the presence of the competing E2 Ubc4. The UBA domain is linked to the UBC domain of Ubc1 by a 22-residue flexible tether. In theory, a disordered linker of this size could reach up to ∼75 Å away from Ubc1, allowing it to interact with numerous sites on the 150-Å-wide APC/C (8, 12). However, assuming that the linker is not entirely unstructured, the UBA domain is most likely to interact with a site near the primary E2-binding site on the RING subunit Apc11. We ruled out an interaction with the Apc11 and Apc10 subunits, and so nearby regions of Apc2 or Apc1 represent the likeliest candidates.
It is unlikely that the flexible linker of Ubc1 contributes directly to APC/C binding, as the recombinant UBA domain alone (containing only the last 3 residues of the linker) inhibited APC/CUbc1 processivity in trans (Fig. 4A). Nevertheless, the length and flexibility of the tether are likely to be important for allowing the UBA domain to reach its binding site on the APC/C.
In the human APC/C, the chain-elongating E2, UBE2S, also uses a C-terminal extension to bind the APC/C at a site distinct from the canonical RING site (5, 10–12, 20). The C-terminal extension of UBE2S is required for its interaction with the APC/C (5, 8, 10), and deletion of the C-terminal extension decreases UBE2S processivity in vitro (6, 10). Thus, it appears that in both yeast and humans chain-elongating E2s have independently evolved extensions to enhance interactions with the APC/C, perhaps suggesting that this is a common feature of E3s that use sequential E2s for chain initiation and elongation.
In the case of the human APC/C, this strategy may allow both E2s to bind the APC/C simultaneously, perhaps enabling more efficient chain assembly. In the case of the yeast APC/C, however, the UBC domains of the two E2s are likely to interact with the same canonical binding site, resulting in competition and therefore requiring finely balanced affinities to allow the two E2s to alternate. The yeast strategy may be relevant to other E2-E3 modules, such as the human SCF complex, which uses the E2s UBCH5c and CDC34 to initiate and elongate Lys-48-linked ubiquitin chains, respectively (18, 35, 36). These E2s are thought to bind the canonical RING-binding site (35, 36). Here again, the chain-elongating E2, CDC34, has a C-terminal extension that binds to the cullin subunit of the SCF (37). This extension may tune CDC34 affinity for SCF in the face of competition from UBCH5c.
An intriguing possibility is that the UBC domain of Ubc1 can dissociate transiently while its UBA domain remains bound to the APC/C. This could be relevant for E2 competition: the same surface of the E2 UBC domain binds E3 and E1, and so the UBC domain must dissociate from the E3 to be recharged with ubiquitin by E1. However, if the UBA domain allows Ubc1 to remain bound to the APC/C while recharging, then it could perform multiple rounds of ubiquitin transfer in a single E3 binding event.
Ubiquitin chains containing four or more ubiquitins represent the canonical recognition motif for the proteasome (38), and several deubiquitinating enzymes in yeast exhibit preference for mono- and diubiquitinated substrates (32). Thus, the binding of the ubiquitin chain-elongating E2, Ubc1, is particularly important for the ability of the APC/C to effectively target its substrates for destruction, which is further demonstrated by the fact that Ubc1, but not Ubc4, is essential in vivo (4). The existence of a second APC/C-binding site for the chain-elongating E2 introduces the possibility that this site can be regulated; inhibition of this site, for example, could prevent the elongation of short ubiquitin chains that are spuriously initiated by APC/CUbc4, allowing their rapid removal by deubiquitinating enzymes. A similar sort of regulation has been demonstrated for the human APC/C inhibitor EMI1, which reduces chain-elongating APC/C activity by blocking binding of the UBE2S C-terminal tail to APC/C (20).
Given our result that the UBA domain of Ubc1 binds to the APC/C, it is surprising that this domain retains conserved ubiquitin-binding residues and has the ability to bind ubiquitin and tetraubiquitin with significant affinity. Previous evidence suggests that the UBA domain does not bind to the donor ubiquitin, and evidence presented here suggests that it does not bind to the acceptor ubiquitin or ubiquitin conjugated to the APC/C activator. To our knowledge, there is no experimental evidence to suggest that ubiquitin is present in significant quantities on any other APC/C subunit. There are also no obvious ubiquitin or ubiquitin-like sequences encoded by any APC/C subunit.
Nevertheless, it is possible that the UBA domain binds both the APC/C and ubiquitin under certain circumstances. Several previously characterized UBA domains can bind both ubiquitin and another partner, sometimes simultaneously (27–29). Because the affinity of the UBA domain for a single ubiquitin is low (∼230 μm) but is considerably higher for a ubiquitin chain (∼37 nm for Lys-48-linked tetraubiquitin), it is possible that as the ubiquitin chain on substrates grows longer it interacts with the UBA domain. It is unclear whether the UBA domain of Ubc1 could bind ubiquitin and the APC/C simultaneously or whether it switches from binding APC/C to binding the ubiquitin chain. This mechanism could allow Ubc1 to compete more effectively with Ubc4 when the substrate carries a polyubiquitin chain. Also, it may explain why mixing the two E2s at equal concentrations leads to a banding pattern that is identical to that of Ubc1 alone: Ubc1 binding may become dominant at longer chain lengths, thereby ensuring that the substrate has an adequate signal for recognition by the proteasome.
Author Contributions
J. R. G. and D. O. M. designed the study. J. L. T. designed, performed, and analyzed experiments in Fig. 1. J. R. G. designed, performed, and analyzed experiments in all figures. J. R. G. and D. O. M. wrote the paper.
Acknowledgments
We thank D. Lu, L. Rosen, M. Galli, T. Noriega, and A. Mizrak for advice on the manuscript and M. Rodrigo-Brenni and R. Deshaies for protocols and reagents.
This work was supported, in whole or in part, by National Institutes of Health Grants R37-GM053270 (to D. O. M.) and R25-GM056847 (to J. R. G.). This work was also supported by a fellowship from the National Science Foundation (to J. R. G.). The authors declare that they have no conflicts of interest with the contents of this article.
- APC/C
- anaphase-promoting complex/cyclosome
- UBA
- ubiquitin-associated
- UBC
- ubiquitin-conjugating
- RING
- really interesting new gene
- CycBN
- sea urchin cyclin B N-terminal fragment
- TEV
- tobacco etch virus
- SCF
- Skp cullin F-box.
References
- 1. Primorac I., Musacchio A. (2013) Panta rhei: the APC/C at steady state. J. Cell Biol. 201, 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chang L., Barford D. (2014) Insights into the anaphase-promoting complex: a molecular machine that regulates mitosis. Curr. Opin. Struct. Biol. 29, 1–9 [DOI] [PubMed] [Google Scholar]
- 3. Rodrigo-Brenni M. C., Foster S. A., Morgan D. O. (2010) Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol. Cell 39, 548–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rodrigo-Brenni M. C., Morgan D. O. (2007) Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell 130, 127–139 [DOI] [PubMed] [Google Scholar]
- 5. Williamson A., Wickliffe K. E., Mellone B. G., Song L., Karpen G. H., Rape M. (2009) Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl. Acad. Sci. U.S.A. 106, 18213–18218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu T., Merbl Y., Huo Y., Gallop J. L., Tzur A., Kirschner M. W. (2010) UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc. Natl. Acad. Sci. U.S.A. 107, 1355–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wickliffe K. E., Lorenz S., Wemmer D. E., Kuriyan J., Rape M. (2011) The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang L., Zhang Z., Yang J., McLaughlin S. H., Barford D. (2014) Molecular architecture and mechanism of the anaphase-promoting complex. Nature 513, 388–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown N. G., VanderLinden R., Watson E. R., Qiao R., Grace C. R., Yamaguchi M., Weissmann F., Frye J. J., Dube P., Ei Cho S., Actis M. L., Rodrigues P., Fujii N., Peters J. M., Stark H., Schulman B. A. (2015) RING E3 mechanism for ubiquitin ligation to a disordered substrate visualized for human anaphase-promoting complex. Proc. Natl. Acad. Sci. U.S.A. 112, 5272–5279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown N. G., Watson E. R., Weissmann F., Jarvis M. A., VanderLinden R., Grace C. R., Frye J. J., Qiao R., Dube P., Petzold G., Cho S. E., Alsharif O., Bao J., Davidson I. F., Zheng J. J., Nourse A., Kurinov I., Peters J. M., Stark H., Schulman B. A. (2014) Mechanism of polyubiquitination by human anaphase-promoting complex: RING repurposing for ubiquitin chain assembly. Mol. Cell 56, 246–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kelly A., Wickliffe K. E., Song L., Fedrigo I., Rape M. (2014) Ubiquitin chain elongation requires E3-dependent tracking of the emerging conjugate. Mol. Cell 56, 232–245 [DOI] [PubMed] [Google Scholar]
- 12. Chang L., Zhang Z., Yang J., McLaughlin S. H., Barford D. (2015) Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature 522, 450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 14. Dou H., Buetow L., Sibbet G. J., Cameron K., Huang D. T. (2012) BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 19, 876–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Plechanovová A., Jaffray E. G., Tatham M. H., Naismith J. H., Hay R. T. (2012) Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pruneda J. N., Littlefield P. J., Soss S. E., Nordquist K. A., Chazin W. J., Brzovic P. S., Klevit R. E. (2012) Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell 47, 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ozkan E., Yu H., Deisenhofer J. (2005) Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc. Natl. Acad. Sci. U.S.A. 102, 18890–18895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petroski M. D., Deshaies R. J. (2005) Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123, 1107–1120 [DOI] [PubMed] [Google Scholar]
- 19. Saha A., Lewis S., Kleiger G., Kuhlman B., Deshaies R. J. (2011) Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell 42, 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang W., Kirschner M. W. (2013) Emi1 preferentially inhibits ubiquitin chain elongation by the anaphase-promoting complex. Nat. Cell Biol. 15, 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Voorhis V. A., Morgan D. O. (2014) Activation of the APC/C ubiquitin ligase by enhanced E2 efficiency. Curr. Biol. 24, 1556–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hofmann K., Bucher P. (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci. 21, 172–173 [PubMed] [Google Scholar]
- 23. Merkley N., Shaw G. S. (2004) Solution structure of the flexible class II ubiquitin-conjugating enzyme Ubc1 provides insights for polyubiquitin chain assembly. J. Biol. Chem. 279, 47139–47147 [DOI] [PubMed] [Google Scholar]
- 24. Sims J. J., Haririnia A., Dickinson B. C., Fushman D., Cohen R. E. (2009) Avid interactions underlie the Lys63-linked polyubiquitin binding specificities observed for UBA domains. Nat. Struct. Mol. Biol. 16, 883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Merkley N., Shaw G. S. (2003) Interaction of the tail with the catalytic region of a class II E2 conjugating enzyme. J. Biomol. NMR 26, 147–155 [DOI] [PubMed] [Google Scholar]
- 26. Merkley N., Barber K. R., Shaw G. S. (2005) Ubiquitin manipulation by an E2 conjugating enzyme using a novel covalent intermediate. J. Biol. Chem. 280, 31732–31738 [DOI] [PubMed] [Google Scholar]
- 27. Hobeika M., Brockmann C., Gruessing F., Neuhaus D., Divita G., Stewart M., Dargemont C. (2009) Structural requirements for the ubiquitin-associated domain of the mRNA export factor Mex67 to bind its specific targets, the transcription elongation THO complex component Hpr1 and nucleoporin FXFG repeats. J. Biol. Chem. 284, 17575–17583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jung J., Byeon I. J., DeLucia M., Koharudin L. M., Ahn J., Gronenborn A. M. (2014) Binding of HIV-1 Vpr protein to the human homolog of the yeast DNA repair protein RAD23 (hHR23A) requires its xeroderma pigmentosum complementation group C binding (XPCB) domain as well as the ubiquitin-associated 2 (UBA2) domain. J. Biol. Chem. 289, 2577–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peschard P., Kozlov G., Lin T., Mirza I. A., Berghuis A. M., Lipkowitz S., Park M., Gehring K. (2007) Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol. Cell 27, 474–485 [DOI] [PubMed] [Google Scholar]
- 30. Carroll C. W., Morgan D. O. (2002) The Doc1 subunit is a processivity factor for the anaphase-promoting complex. Nat. Cell Biol. 4, 880–887 [DOI] [PubMed] [Google Scholar]
- 31. Carroll C. W., Morgan D. O. (2005) Enzymology of the anaphase-promoting complex. Methods Enzymol. 398, 219–230 [DOI] [PubMed] [Google Scholar]
- 32. Schaefer J. B., Morgan D. O. (2011) Protein-linked ubiquitin chain structure restricts activity of deubiquitinating enzymes. J. Biol. Chem. 286, 45186–45196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Foster S. A., Morgan D. O. (2012) The APC/C subunit Mnd2/Apc15 promotes Cdc20 autoubiquitination and spindle assembly checkpoint inactivation. Mol. Cell 47, 921–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carroll C. W., Enquist-Newman M., Morgan D. O. (2005) The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr. Biol. 15, 11–18 [DOI] [PubMed] [Google Scholar]
- 35. Saha A., Deshaies R. J. (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu K., Kovacev J., Pan Z. Q. (2010) Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol. Cell 37, 784–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kleiger G., Saha A., Lewis S., Kuhlman B., Deshaies R. J. (2009) Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139, 957–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]