Abstract
Ras genes are frequently activated in human cancers, but the mutant Ras proteins remain largely “undruggable” by the conventional small-molecule approach due to absence of any obvious binding pockets on their surfaces. By screening a combinatorial peptide library followed by structure-activity relationship analysis, we discovered a family of cyclic peptides possessing both Ras-binding and cell-penetrating properties. These cell-permeable cyclic peptides inhibited Ras signaling by binding to Ras-GTP and blocking its interaction with downstream proteins and induced apoptosis of cancer cells. Our results demonstrate the feasibility of developing cyclic peptides for inhibition of intracellular protein-protein interactions and direct Ras inhibitors as a novel class of anticancer agents.
Keywords: Cancer, cell-penetrating peptide, cyclic peptide, protein-protein interaction, Ras
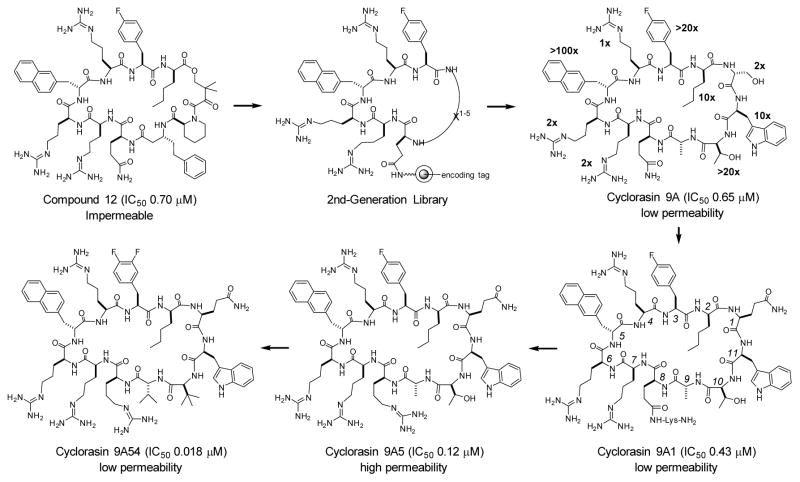
The monomeric GTPases K-Ras, H-Ras, and N-Ras play critical roles in many signaling pathways and regulate cell proliferation, differentiation, and survival.[1] Wild-type Ras oscillates between inactive GDP-bound (Ras-GDP) and active GTP-bound forms (Ras-GTP), with the latter interacting with and activating multiple effector proteins including Raf, PI3K, and Ral-GDS. Somatic mutations that cause constitutive activation of Ras are the most common activating lesions and found in ~30% human cancers.[2] These mutations impair GTP hydrolysis, thereby increasing the Ras-GTP population and causing uncontrolled cell growth. Genetic studies suggest that blocking the Ras-effector protein interaction should have therapeutic benefits in cancer patients;[3,4] however, doing so pharmacologically has been challenging, because the Ras protein surface has no obvious pockets for small-molecule drugs to bind.[5] Consequently, most of the drug discovery efforts have so far been focused on inhibiting the signaling molecules downstream of Ras,[6] the posttranslational processing/membrane anchoring of Ras,[6,7] or the nucleotide exchange activity of Ras.[8–14] Inhibitors that physically block the Ras-effector protein interactions have generally lacked potency, selectivity, and/or membrane permeability.[15,16] Here we report a family of cyclic peptides possessing both Ras-binding and cell-penetrating properties. These cell-permeable cyclic peptides bind potently to Ras-GTP near the effector-binding site and block its interaction with downstream proteins, resulting in growth inhibition and apoptosis of cancer cells. We previously reported a cyclic peptide inhibitor against K-Ras, compound 12 (Figure 1), which blocks the Ras-effector protein interaction in vitro, but lacks cellular activity due to poor membrane permeability.[16] Interestingly, compound 12 contains an amphipathic sequence motif, Arg-Arg-nal-Arg-Fpa (where Fpa is L-4-fluorophenylalanine and nal is D-β-naphthylalanine), which resembles a recently discovered cyclic cell-penetrating peptide (CPP).[17,18] To improve the potency and membrane permeability of compound 12, we designed a second-generation library in which the CPP-like motif was retained, while the remaining structure was replaced with a random peptide sequence of 0–5 amino acids (X1–5). The library (~1.3 × 106 compounds) was constructed with 28 different amino acids[19] at the X1-X5 positions on spatially segregated TentaGel beads,[20] with each bead displaying a unique cyclic peptide on its surface and a linear peptide of the same sequence in its interior as an encoding tag (Figure 1). Because the effector-binding site of Ras is highly negatively charged,[21] we anticipated that screening the library against K-Ras might select one or more additional arginine and/or aromatic hydrophobic residues at the random positions which, together with the Arg-Arg-nal-Arg-Fpa motif, might generate a functional CPP.[17.18]
Figure 1.
Flowchart showing the evolution of Ras inhibitors. The boldfaced numbers next to the structure of cyclorasin 9A indicate the fold of activity loss upon replacing each residue with alanine (or D-alanine). Residue numbering shown in the structure of 9A1 is adopted for compounds 9A1 to 9A54.
Screening of the peptide library against K-Ras(G12V) identified 13 hits (Table S1 in Supporting Information). When assayed in solution by fluorescence anisotropy (FA), hits 4A, 5A, 7A, 9A (Figure 1), 12A, and 13A showed strong binding to K-Ras (Figure S1). In a homogeneous time-resolved fluorescence (HTRF) assay, hits 9A and 12A inhibited the Ras-Raf interaction with IC50 values of 0.65 and 1.0 μM, respectively (Figure 2a). Gratifyingly, hits 9A and 12A contained additional Trp and/or Arg residues in the X2-X5 region, were cell permeable, and exhibited weak anti-proliferative activity against lung cancer cells (Figure S2). These two peptides were named as cyclorasin (for cyclic Ras inhibitor) 9A and 12A, respectively.
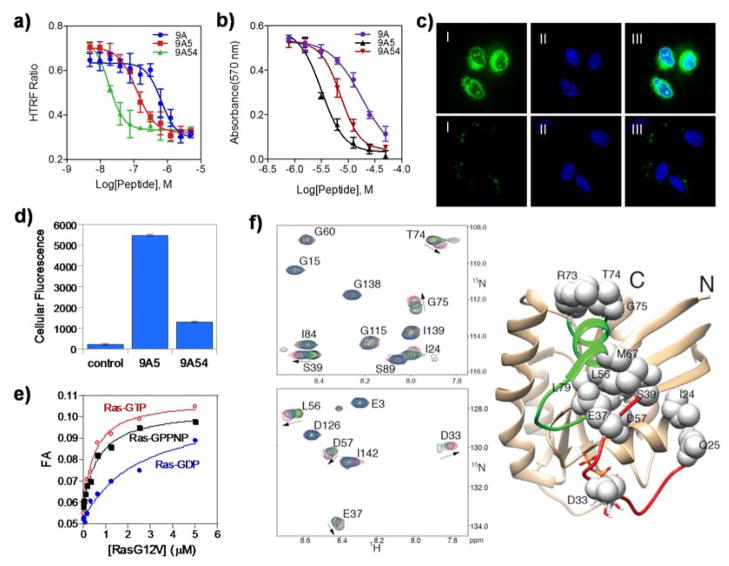
Figure 2.
K-Ras binding, cellular uptake, and anti-proliferative activity of cyclic peptides. a) HTRF assay of inhibition of Ras-Raf interaction by 9A, 9A5 and 9A54. b) Effect of 9A, 9A5 and 9A54 on H1299 cell viability. c) Live-cell confocal microscopic images of A549 cells treated with 5 μM FITC-9A5 (top panel) or FITC-9A54 (bottom panel) for 30 min. I, FITC fluorescence; II, nuclear stain with DRAQ5; and III, merge of I and II. d) Total fluorescence of cells untreated (control) or treated with FITC-9A5 or FITC-9A54 (from C) as determined by flow cytometry. e) Binding of FITC-9A5 to Ras-GTP, Ras-GDP, and Ras-GPPNP as monitored by FA. f) Overlay of 1H–15N HSQC spectra of K-Ras(G12V)-GDP in the presence of 0 (black), 0.5 eq. (green), 0.75 eq. (blue), and excess (red) of 9A5 at 298 K. K-Ras residues perturbed by 9A5 are mapped to the structure of wild-type K-Ras-GDP (4LPK) and shown as spheres. The switch I and II regions are colored red and green, respectively, and GDP is shown as sticks.
Because of its relatively high potency, cyclorasin 9A was chosen for optimization. Alanine scan analysis revealed that nal, Fpa, Thr, norleucine (nle), and Trp are critical for K-Ras binding, as replacement of each of these residues with L- or D-alanine reduced the Ras inhibitory activity by ≥10-fold. Substitution of alanine for the remaining residues had relatively minor effects (≤2-fold). Since the D-serine residue was not critical for Ras binding, it was replaced by an L-Gln to provide an alternative site for attachment to the solid support and designated as position 1 (Figure 1). The resulting peptides (cyclorasin 9A1-4), which contained different glutamine derivatives at position 8, had similar inhibitory activity against the Ras-Raf interaction (Table S2), suggesting that Gln-8 is not critical for Ras binding. Gln-8 was therefore replaced with L- or D-arginine, to potentially improve the membrane permeability of the peptide.[17,18] To our delight, the resulting peptides (9A5 and 9A6) showed both improved cell permeability and a ~4-fold higher affinity for K-Ras (IC50 = 0.12 and 0.17 μM, respectively) (Figure 2a). Further modifications of 9A5 and 9A6 produced mixed results, with a few resulting in significant improvements (Table S2). Replacement of the D-Ala at position 9 by D-valine (val-9), Thr-10 by L-tert-leucine (Tle), or Fpa-3 by 3,4-difluorophenylalanine (F2pa) increased the affinity by ~2-fold (9A14, 9A16, and 9A43, respectively). Finally, combinations of val-9, Tle-10, and F2pa-3 substitutions produced cyclorasin 9A51 and 9A54 as highly potent Ras inhibitors (IC50 = 0.014 and 0.018 μM, respectively) (Figure 2a).
Peptides with strong K-Ras binding (IC50 ≤0.2 μM) were tested for anti-proliferative activity against H1299 lung cancer cells (Figure 2b). Cyclorasin 9A5 was most potent (LD50 ~3 μM), whereas peptides of greater Ras-binding affinities (e.g., 9A54) were less active in the cellular assay. To test whether poor membrane permeability limited the cellular activity of the latter peptides, we treated A549 lung cancer cells with fluorescein isothiocyanate (FITC)-labeled 9A5 or 9A54 and examined the internalization of the peptides by confocal microscopy (Figure 2c). Cells treated with FITC-9A5 exhibited intense diffuse fluorescence throughout the cytoplasm, whereas cells treated with FITC-9A54 had much weaker and predominantly punctate fluorescence, which is indicative of endosomal entrapment. Flow cytometry analysis showed that FITC-9A5 entered the lung cancer cells ~5-fold more efficiently than FITC-9A54 (Figure 2d).
Cyclorasin 9A5 was selected to investigate the mechanism of the observed anti-proliferative activity. The ability of 9A5 to inhibit the Ras-Raf interaction suggests that it binds to Ras-GTP. To test whether it is selective for Ras-GTP, we prepared K-Ras G12V loaded with GTP, GDP, or GPPNP (a nonhydrolyzable GTP analogue) and tested them for binding to FITC-9A5 by FA. FITC-9A5 bound to the Ras-GTP, Ras-GPPNP, and Ras-GDP with KD values of 0.44, 0.64, and 2.5 μM, respectively (Note that labeling with FITC required replacing Arg-6 with a lysine, which reduced its affinity for K-Ras) (Figure 2e and Figure S3). FITC-9A5 also bound to wild-type K- and H-Ras (which were mixtures of GTP- and GDP-bound forms) with apparent KD values of 1.2 and 1.4 μM, respectively, but not to a panel of five arbitrarily selected control proteins (KD ≥ 30 μM; Figure S3). Cyclorasin 9A5 is therefore a selective ligand of Ras-GTP.
To map the binding site of 9A5, 15N-labeled K-Ras-GPPNP was mixed with increasing concentrations of 9A5 and analyzed by 1H-15N heteronuclear single quantum correlation (HSQC) NMR spectroscopy (Figure 2f). Unfortunately, most of the switch I and II residues were invisible in the HSQC spectra (Figure S4). A few K-Ras residues that exhibited detectable chemical shift perturbation upon binding to 9A5 were exemplified by Gln25, Thr74, and Gly75. We therefore repeated the HSQC experiment with K-Ras-GDP. Most of the K-Ras residues were unaffected by 9A5, indicating that binding of 9A5 does not induce global changes in the K-Ras structure (Figure S5). K-Ras residues that underwent detectable spectral shifts included Ile24, Gln25, Asp33, Glu37, and Ser39, which are within the switch I loop, the primary binding site of effector proteins[21] (Figure 2f). The other perturbed residues (Leu56, Asp57, Met67, Arg73, Thr74, Gly75, and Leu79) are clustered around a small pocket between the switch I and II loops, which had previously been found to bind to small-molecule ligands.[9,10] The overlapping binding sites of 9A5 and Ras effector proteins are consistent with the observed inhibition of Ras-Raf interaction by 9A5.
The Raf/MEK/ERK and PI3K/PDK1/Akt signaling pathways represent the signature events downstream of Ras.[1] To determine whether 9A5 inhibited the intracellular Ras activity, H358 lung cancer cells were treated with increasing concentrations of 9A5 and the Ras-Raf interaction was examined by precipitating the cell lysate with an anti-Ras antibody and immunoblotting with an anti-B-Raf antibody (Figure 3a). In the absence of 9A5, B-Raf co-precipitated with Ras. However, treatment of the cells with 9A5 dose-dependently inhibited the Ras-B-Raf interaction. Next, we examined Akt, MEK, and ERK1/2 phosphorylation in H1299 cells (Figure 3b). Cyclorasin 9A5 inhibited EGF-stimulated phosphorylation of Akt at Thr308 and MEK in dose-dependent manners (IC50 ~3 μM) and abolished the phosphorylation events at ≥12 μM, while the total Akt and MEK protein levels remained constant. 9A5 also decreased the phosphorylation of ERK1/2 and Akt at Ser473, but less effectively, as expected from the fact that ERK1/2 functions downstream of MEK and Akt Ser473 is phosphorylated by mTOR instead of PDK1.[22] A time-course experiment showed that the dephosphorylation of MEK and Akt was largely complete after 10 min of exposure to 9A5 whereas a comparable degree of dephosphorylation of ERK was reached after 20 min (Figure S6a). H358 and H1299 cells express mutant K-Ras G12C and mutant N-Ras G12V, respectively. Cyclorasin 9A5 also reduced the MEK and Akt phosphorylation in lung cancer cell lines H1975 and H1650, which express wild-type Ras proteins but mutant EGFR (Figure S6b).
Figure 3.
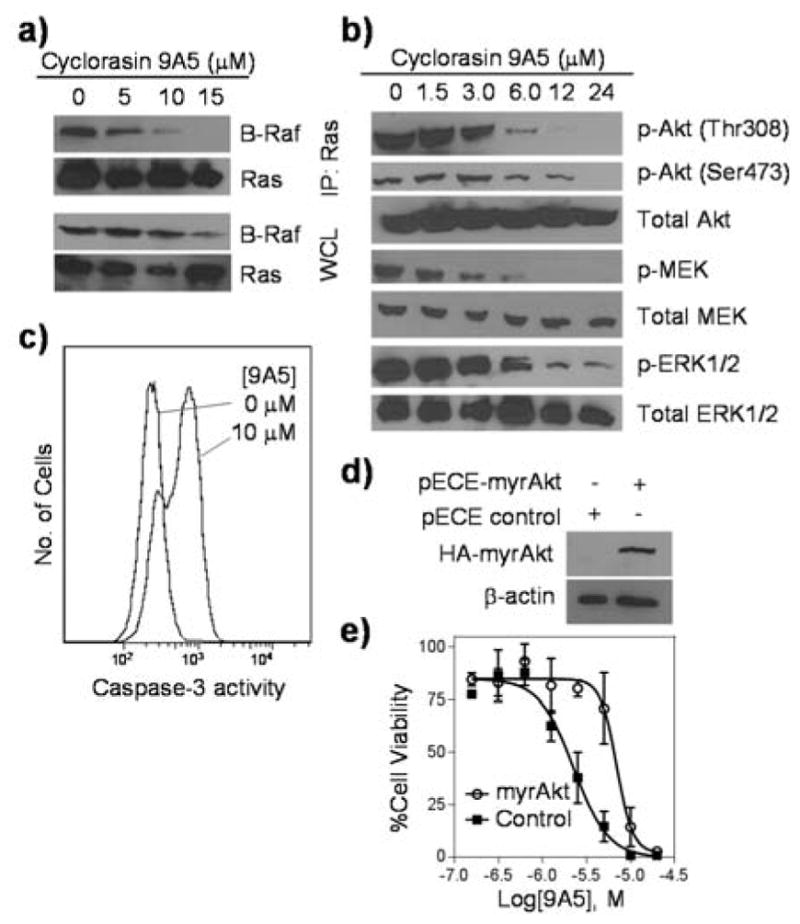
Inhibition of Ras signaling by 9A5. a) Inhibition of Ras-B-Raf interaction in H358 cells by 9A5. WCL, whole cell lysate. b) Dose-dependent inhibition of Akt, MEK, and ERK phosphorylation in H1299 cells by 9A5. Cells were treated with the indicated concentrations of 9A5 for 10 min and stimulated with EGF (50 ng/ml) for 5 min before lysis. c) Activation of caspase-3 activity in H1299 cells by 9A5 (10 μM treatment for 3 h) as monitored by anti-caspase-3 immunostaining and flow cytometry. d, e) Protection of H1299 cells against 9A5-induced apoptosis by ectopic expression of a constitutively active, HA-tagged, and myristoylated Akt (HA-myrAkt). The pECE-control plasmid contained a frame-shift mutation in the myrAkt gene and the myrAkt protein levels in cells were determined by immunoblotting with anti-HA antibody. Actin was used as loading control. Cell viability was determined by the MTT assay and relative to that of untreated cells.
Dual inhibition of MEK and PI3K signaling results in synergistic decrease in cell viability and increase in apoptosis of Ras mutant cancer cells.[23–25] Given its ability to inhibit both Raf/MEK/ERK and PI3K/PDK1/Akt pathways, 9A5 is expected to inhibit cell growth and induce apoptosis of cancer cells. Indeed, treatment of H1299 cells with 10 μM 9A5 for 3 h resulted in a 2.3-fold increase in caspase-3 activity (Figure 3c). Flow cytometry analysis of the treated cells showed positive annexin-FITC and propidium iodide staining (Figure S7). The treated cells underwent morphological changes including cell rounding and reduction in the cell size. Consistent with these morphological changes, LIM domain kinase and cofilin, which function downstream of Ras and regulate the assembly and disassembly of actin filaments, showed decreased phosphorylation upon treatment of 9A5. Finally, transfection of H1299 cells with a constitutively active form of Akt[26] rendered the cells substantially more resistant to 9A5 (LD50 ~8 μM), whereas transfection with a nonfunctional Akt gene had no effect (Figure 3d,e). Taken together, our data suggest that cyclorasin 9A5 was able to enter human cancer cells and bind directly to Ras proteins, thereby blocking the Ras-effector protein interactions and causing cell growth inhibition and apoptosis, although we cannot rule out potential off-target effects that may also contribute to the observed cell growth inhibition and apoptotic cell death.
In summary, we have developed a family of cell-permeable cyclic peptides as potent and selective inhibitors of Ras-GTP by screening a peptide library followed by preliminary SAR studies. These peptides bind directly to Ras-GTP, blocking its interaction with effector proteins and causing growth inhibition and apoptosis of cancer cells. Our compounds are among the first biologically active Ras inhibitors that act by physically blocking the Ras-effector protein interactions. With sizes similar to that of cyclosporine A (an orally available cyclic peptide drug), these cyclic peptides should serve as useful leads for further development into therapeutic agents. Our strategy of integrating target-binding and cell-penetrating motifs into a single cyclic peptide should be applicable to the development of biologically active inhibitors against other intracellular protein-protein interactions.
Footnotes
This work was supported by grants from the National Institutes of Health (GM062820, GM110208, and CA132855 to D.P.). We thank Prof. Andrea Doseff for helpful suggestions to some of the cellular tests.
Contributor Information
Dr. Punit Upadhyaya, Department of Chemistry and Biochemistry, The Ohio State University, 484 West 12th Avenue, Columbus, OH 43210 (USA)
Dr. Ziqing Qian, Department of Chemistry and Biochemistry, The Ohio State University, 484 West 12th Avenue, Columbus, OH 43210 (USA)
Dr. Nicholas G. Selner, Department of Chemistry and Biochemistry, The Ohio State University, 484 West 12th Avenue, Columbus, OH 43210 (USA)
Sarah R. Clippinger, Department of Chemistry and Biochemistry, The Ohio State University, 484 West 12th Avenue, Columbus, OH 43210 (USA)
Prof. Dr. Zhengrong Wu, Department of Chemistry and Biochemistry, The Ohio State University, 484 West 12th Avenue, Columbus, OH 43210 (USA)
Dr. Roger Briesewitz, Email: Roger.Briesewitz@osumc.edu, Department of Pharmacology, The Ohio State University, 5065 Graves Hall, 333 West 10th Avenue, Columbus, OH 43210 (USA)
Prof. Dr. Dehua Pei, Email: pei.3@osu.edu, Department of Chemistry and Biochemistry, The Ohio State University, 484 West 12th Avenue, Columbus, OH 43210 (USA)
References
- 1.a) Karnoub AE, Weinberg RA. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Young A, Lyons J, Miller AL, Phan VT, Alarcon IR, McCormick F. Adv Cancer Res. 2009;102:1–17. doi: 10.1016/S0065-230X(09)02001-6. [DOI] [PubMed] [Google Scholar]; c) Campbell PM, Der CJ. Seminars Cancer Biol. 2004;14:105–114. doi: 10.1016/j.semcancer.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Prior IA, Lewis PD, Mattos CA. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 4.Castellano E, Sheridan C, Thin MZ, Nye E, Spencer-Dene B, Diefenbacher ME, Moore C, Kumar MS, Murillo MM, Gronroos E, Lassailly F, Stamp G, Downward J. Cancer Cell. 2013;24:617–630. doi: 10.1016/j.ccr.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Wang W, Fang G, Rudolph J. Bioorg Med Chem Lett. 2012;22:5766–5776. doi: 10.1016/j.bmcl.2012.07.082. [DOI] [PubMed] [Google Scholar]; b) Spiegel J, Cromm PM, Zimmermann G, Grossmann TN, Waldmann H. Nat Chem Biol. 2014;10:613–622. doi: 10.1038/nchembio.1560. [DOI] [PubMed] [Google Scholar]; c) Stephen AG, Esposito D, Bagni RK, McCormick F. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 6.a) Gysin S, Salt M, Young A, McCormick F. Genes Cancer. 2011;2:359–372. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Baines AT, Xu D, Der CJ. Future Med Chem. 2011;3:1787–1808. doi: 10.4155/fmc.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PI, Waldmann H. Nature. 2013;497:638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 8.Taveras AG, Remiszewski SW, Doll RJ, Cesarz D, Huang EC, Kirschmeier P, Parmanik BN, Snow ME, Wang Y–S, del Rosario Jd, Vibulbhan B, Bauer BB, Brown JE, Carr D, Catino J, Evans CA, Girijavallabhan V, Heimark L, James L, Liberles S, Nash C, Perkins L, Senior MM, Tsarbopoulos A, Ganguly AK, Aust R, Brown E, Delisle D, Fuhrman S, Hendrickson T, Kissinger C, Love R, Sisson W, Villafranca E, Webber SE. Bioorg Med Chem. 1997;5:125–133. doi: 10.1016/s0968-0896(96)00202-7. [DOI] [PubMed] [Google Scholar]
- 9.Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, Ludlam MJ, Bowman KK, Wu J, Giannetti AM, Starovasnik MA, Mellman I, Jackson PK, Rudolph J, Wang W, Fang G. Proc Natl Acad Sci U S A. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, Lee T, Rossanese OW, Fesik SW. Angew Chem Int Ed. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patgiri A, Yadav KK, Arora PS, Bar-Sagi D. Nat Chem Biol. 2011;7:585–587. doi: 10.1038/nchembio.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim SM, Westover KD, Ficarro SB, Harrison RA, Choi HG, Pacold ME, Carrasco M, Hunter J, Kim ND, Xie T, Sim T, Jänne PA, Meyerson M, Marto JA, Engen JR, Gray NS. Angew Chem Int Ed. 2014;53:199–204. doi: 10.1002/anie.201307387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leshchiner ES, Parkhitko A, Bird GH, Luccarelli J, Bel-lairs JA, Escudero S, Opoki-Nsiah K, Godes M, Perrimon N, Walensky LD. Proc Natl Acad Sci U S A. 2015;112:1761–1766. doi: 10.1073/pnas.1413185112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Bernard D, Sun H, Baker L, Marshall MS. Biochem Biophys Commun. 1998;247:176–180. doi: 10.1006/bbrc.1998.8746. [DOI] [PubMed] [Google Scholar]; b) Kato-Stankiewicz J, Hakimi I, Zhi G, Serebriiskii I, Guo L, Edamatsu H, Koide H, Menon S, Ecki R, Sakamuri S, Liu Y, Chen QZ, Agarwal S, Baumbach WR, Golemis EA, Tamanoi F, Khazak V. Proc Natl Acad Sci U S A. 2002;99:14398–14403. doi: 10.1073/pnas.222222699. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) González-Pérez V, Reiner DJ, Alan JK, Mitchell C, Edwards LJ, Khazak V, Der CJ, Cox AD. J Mol Signal. 2010;5:2. doi: 10.1186/1750-2187-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Shima F, Shima F, Yoshikawa Y, Ye M, Araki M, Matsumoto S, Liao J, Hu L, Sugimoto T, Ijiri Y, Takeda A, Nishiyama Y, Sato C, Muraoka S, Tamura A, Osoda T, Tsuda K, Miyakawa T, Fukunishi H, Shimada J, Kumasaka T, Yamamoto M, Kataoka T. Proc Natl Acad Sci U S A. 2013;110:8182–8187. doi: 10.1073/pnas.1217730110. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Upadhyaya P, Qian Z, Habir NAA, Pei D. Tetrahedron. 2014;70:7714–7720. doi: 10.1016/j.tet.2014.05.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu X, Upadhyaya P, Villalona-Calero MA, Briesewitz R, Pei D. Med Chem Commun. 2013;4:378–382. doi: 10.1039/C2MD20329D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qian Z, Liu T, Liu YY, Briesewitz R, Barrios AM, Jhiang SM, Pei D. ACS Chem Biol. 2013;8:423–431. doi: 10.1021/cb3005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian Z, LaRochelle JR, Jiang B, Lian W, Hard RL, Selner NG, Luechapanichkul R, Barrios AM, Pei D. Biochemistry. 2014;53:4034–4046. doi: 10.1021/bi5004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The 28-amino acid set included 11 proteinogenic amino acids [Arg, Asp, Gln, Gly, His, Ile, Leu, Pro, Thr, Trp, and Tyr], 4 nonproteinogenic α-L-amino acids [L-4-fluorophenylalanine (Fpa), L-2-amino butyric acid (Abu), L-ornithine (Orn), and L-phenylglycine (Phg)], 9 α-D-amino acids [D-2-naphthylalanine (D-Nal), D-Ala, D-Asn, D-Glu, D-Lys, D-Nle, D-Phe, D-Ser and D-Val] and 4 L-Nα-methylated amino acids [sarcosine (Sar), MeAla, MeLeu and MePhe].
- 20.Joo SH, Xiao Q, Ling Y, Gopishetty B, Pei D. J Am Chem Soc. 2006;128:13000–13009. doi: 10.1021/ja063722k. [DOI] [PubMed] [Google Scholar]
- 21.Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, Hawkins PT, Stephens L, Eccleston JF, Williams RL. Cell. 2000;103:931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 22.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 23.Shelton JG, Steelman LS, White ER, McCubrey JA. Cell Cycle. 2004;3:372–379. [PubMed] [Google Scholar]
- 24.Sos ML, Fischer S, Ullrich R, Peifer M, Heuckmann JM, Koker M, Heynck S, Stuckrath I, Weiss J, Fisher F, Michel K, Goel A, Regales L, Politi KA, Perera S, Getlik M, Heukamp LC, Ansen S, Zander T, Beroukhim R, Kashkar H, Shokat KM, Sellers WR, Rauh D, Orr C, Hoeflich KP, Friedman L, Wong KK, Pao W, Thomas RK. Proc Natl Acad Sci U S A. 2009;106:18351–18356. doi: 10.1073/pnas.0907325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, Chirieac LR, Kaur R, Lightbown A, Simendinger J, Li T, Padera RF, Garcia-Echeverria C, Weissleder R, Mahmood U, Cantley LC, Wong KK. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohn AD, Takeuchi F, Roth RA. J Biol Chem. 1996;271:21920–21926. doi: 10.1074/jbc.271.36.21920. [DOI] [PubMed] [Google Scholar]