Abstract
Despite decades of research, cancer metastasis remains an incompletely understood process that is as complex as it is devastating. In recent years, there has been an increasing push to investigate the biomechanical aspects of tumorigenesis, complementing the research on genetic and biochemical changes. In contrast to the high genetic variability encountered in cancer cells, almost all metastatic cells are subject to the same physical constraints as they leave the primary tumor, invade surrounding tissues, transit through the circulatory system, and finally infiltrate new tissues. Advances in live cell imaging and other biophysical techniques, including measurements of subcellular mechanics, have yielded stunning new insights into the physics of cancer cells. While much of this research has been focused on the mechanics of the cytoskeleton and the cellular microenvironment, it is now emerging that the mechanical properties of the cell nucleus and its connection to the cytoskeleton may play a major role in cancer metastasis, as deformation of the large and stiff nucleus presents a substantial obstacle during the passage through the dense interstitial space and narrow capillaries. Here, we present an overview of the molecular components that govern the mechanical properties of the nucleus and we discuss how changes in nuclear structure and composition observed in many cancers can modulate nuclear mechanics and promote metastatic processes. Improved insights into this interplay between nuclear mechanics and metastatic progression may have powerful implications in cancer diagnostics and therapy and may reveal novel therapeutic targets for pharmacological inhibition of cancer cell invasion.
Introduction
The cell nucleus was the first organelle discovered in the 17th century. In the oldest preserved depictions of the nucleus, Antonie van Leeuwenhoek described a central “clear area” in salmon blood cells that is now commonly acknowledged as the nucleus [1]. A more detailed description of the nucleus was subsequently provided by the botanist Robert Brown, who first articulated the concept of the nucleated cell as a structural unit in plants [1]. Today, the nucleus is recognized as the site of numerous essential functions in eukaryotes, including storage and organization of the genetic material, DNA synthesis, DNA transcription, transcriptional regulation, and RNA processing. In cancer biology, much of the research has traditionally been focused on this “DNA-centric view”, starting with the identification of oncogenes and tumor-suppressor genes to the establishment of the multiple “hits” (i.e., mutations) concept now commonly accepted as a requirement for cancer initiation and progression [2]. Recently, however, it has become apparent that in addition to these genetic components, it is necessary to take the physical, i.e., biomechanical, factors of tumor cells and their microenvironment into consideration. Research conducted within the last 10 years has revealed that cancer cells have reduced stiffness [3–7], generate increased contractile forces [8], and are strongly influenced by their biomechanical environment [9,10]. Furthermore, not only can cancer cells be mechanically distinguished from non-tumorigenic cells, but physical measurements also allow telling apart highly invasive cells from less invasive cells, for example, by their increased cell deformability [4] and increased traction forces [8], yielding the promise of future diagnostic and prognostic applications. Here, we focus on a particular aspect of cellular mechanics that has traditionally received less attention in cancer cell biology: the role of nuclear structure and mechanics in cancer progression.
Despite many advances in understanding the biology of cancer and its associated molecular changes, the most common and reliable diagnosis of cancer cells in tissue biopsies by pathologists still relies on the presence of morphological changes in nuclear structure, i.e., increased size, irregular shape and organization [11]. Nonetheless, the functional consequences of these characteristic changes have yet to be determined; thus, it remains unclear whether the observed morphological changes merely correlate with other, more difficult to observe cellular defects, or whether they can directly contribute to the disease progression.
In recent years, a growing number of studies have reported altered nuclear envelope composition in various cancers [12,13]. The structure and composition of the nucleus, particularly the nuclear envelope, play an important role in cellular mechanics and function, ranging from determining nuclear deformability and fragility [14–17] to participating in mechanotransduction signaling, i.e., the sensing of biomechanical factors and the corresponding signaling response [18,15]. One potential mechanism by which changes in nuclear envelope composition could contribute to cancer progression is that softer and more lobulated nuclei facilitate cancer cell invasion through dense tissues, where cells often have to pass through constrictions smaller than the nuclear diameter [19,20]. Furthermore, the physical coupling between the nucleus and the cytoskeleton is critical for cytoskeletal organization and cell polarization [21–24], which could further affect cancer cell migration. In the following, we provide a brief review of normal nuclear structure and mechanics, highlight changes that occur during oncogenic transformation, and discuss recent findings suggesting an important role of nuclear mechanics and nucleo-cytoskeletal coupling in cancer progression.
Normal nuclear compartmentalization and structure
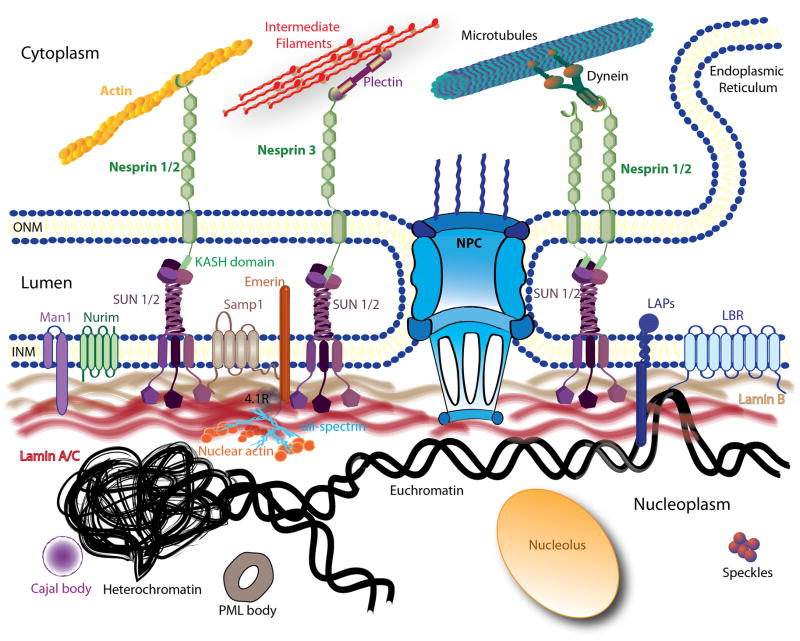
The nucleus is a highly compartmentalized organelle that can be roughly subdivided into the nuclear envelope and the nuclear interior (Fig. 1), the latter representing most of the chromatin in diverse states of organization [25], the nucleolus, and diverse smaller subnuclear structures such as Cajal bodies and nuclear speckles [26–28]. In addition, the nuclear interior contains a still incompletely defined structural network (i.e., the nucleoskeleton or nuclear matrix), which may provide additional mechanical support and also act as scaffold for transcriptional complexes and other nuclear processes. The nuclear envelope forms the physical barrier between the nucleus and the cytoplasm. It consists of two phospholipid bilayers, the inner and the outer membranes, and the underlying nuclear lamina, a dense protein meshwork mostly comprised of lamins. The inner and outer nuclear membranes are connected at the sites of nuclear pore complexes (NPCs) and encapsulate the perinuclear space or lumen.
Figure 1. Schematic overview of the nuclear structure and the LINC complex.
The nuclear envelope is composed of the inner nuclear membrane (INM) and the outer nuclear membrane (ONM) punctuated by nuclear pore complexes (NPC). The ONM is continuous with the endoplasmic reticulum (ER). Several structures of the nuclear interior are depicted here, including the nucleolus, Cajal bodies, promyelocytic leukemia bodies (PML) and speckles. Chromatin is shown in its two states, very condensed (heterochromatin) and loosely organized (euchromatin). Only a subset of nuclear membrane proteins are portrayed in this picture: lamin B receptor (LBR), emerin, MAN1, and nurim. The schematic also illustrates some of the interactions between these proteins with the lamina meshwork (lamins B and A/C). The LINC complex is represented by nesprins, Sad1p/UNC-84 (SUN) proteins and Samp1. On the outer membrane, nesprin-1 and -2 can directly bind to actin filaments or indirectly interact with microtubules through motors proteins (dyneins or kinesin). Nesprin-3 is shown interacting with intermediate filaments via plectin.
The outer nuclear membrane
The outer nuclear membrane is continuous with the endoplasmic reticulum (ER); like the ER, its surface is scattered with ribosomes. The outer nuclear membrane exhibits a high degree of similarity to the ER membrane in terms of protein, enzyme and lipid composition [29]. Nonetheless, recent studies have suggested that the outer nuclear membrane displays a certain degree of specialization [30] and participates in protein synthesis and processing [31]. The specialized protein composition of the outer nuclear membrane likely results from retention of specific proteins by direct interaction with inner nuclear membrane proteins across the lumen, thereby enriching them compared to the ER fraction [32,33]. In mammals, one particularly important family of outer nuclear membrane proteins are the nesprins [34], which play a central role in connecting the nucleus to the cytoskeleton [35–39].
The nuclear lumen and nuclear pore complexes
The nuclear lumen, also commonly termed the perinuclear space, is a 30 to 50 nm wide aqueous space separating the inner from the outer nuclear membrane that is continuous with the ER lumen [40]. It accommodates the luminal domains of integral nuclear membrane proteins [41]. The inner and outer nuclear membranes come together at sites of NPC insertion [42]. NPCs act as the main gateway for molecules between the cytoplasm and the nuclear interior (and also proteins of the inner nuclear membrane). Small molecules can diffuse freely through the NPC, while the exchange of macromolecules larger than ~40 kDa is mediated by a tightly controlled import and export mechanism requiring nuclear import and export signals and interaction with specific transport molecules [43–45].
The inner nuclear membrane
The inner nuclear membrane contains at least 70 to 100 unique membrane-associated and integral membrane proteins that are retained at the inner nuclear membrane through interaction with nucleoplasmic proteins (e.g., lamins) and chromatin [13]. Most of these proteins have only been identified in recent proteomic studies [46–50], and the function of several of the nuclear envelope transmembrane proteins remains unclear. Some well-characterized inner nuclear membrane proteins include lamin B receptor (LBR), lamina-associated polypeptides (LAPs) [30], emerin, MAN1, nurim, nesprins and Sad1p/UNC-84 (SUN) proteins [13]. Mislocalization or loss of these proteins due to mutations in nuclear envelope proteins causes a spectrum of diseases collectively known as laminopathies that include certain types of muscular dystrophies (e.g., Emery-Dreifuss muscular dystrophy and limb-girdle muscular dystrophy), dilated cardiomyopathy, and the premature aging disease Hutchinson-Gilford progeria syndrome [51].
The nuclear lamina
The lamina corresponds to a dense meshwork of proteins mainly composed of lamins underlying the inner nuclear membrane [52]. Lamins are type V intermediate filaments [53,54] and display the characteristic tripartite molecular organization of all intermediate filaments, which consists of a central α-helical rod domain flanked by a short non-helical N-terminal ‘head’ and a C-terminal ‘tail’ domain that includes an Ig-like fold [55].
In vertebrates, lamins are classified into two major classes, A-and B-type lamins, depending on their sequence, expression pattern, and biochemical properties [56,57]. A-type lamins, including lamins A, C, AΔ10 and C2, result from alternative splicing of the LMNA gene on chromosome 1. These proteins are expressed in a tissue-specific manner later in differentiation [58,59], have neutral isoelectric points, and are dispersed upon phosphorylation of lamins during mitosis [60]. Lamin A and C can be distinguished by their unique C-terminal tail and processing: the C-terminus of prelamin A contains a CaaX motif, which is subject to a series of post-translational modifications, including isoprenylation and proteolytic cleavage, to give rise to mature lamin A [61,62]. In contrast, the shorter lamin C has a unique C-terminus that lacks the CaaX motif and does not require post-translational processing. In addition to their localization at the nuclear lamina, A-type lamins are also present in the nuclear interior, where they form stable structures [63].
Unlike A-type lamins, B-type lamins are encoded by two separate genes: LMNB1 for lamin B1 [64,65] and LMNB2 for lamin B2 and B3 [66,67]. Only lamins B1 and B2 are found in somatic cells; expression of lamin B3 is restricted to germ cells. Unlike A-type lamins, at least one B-type lamin is expressed in all cells, including embryonic stem cells; B-type lamins are acidic and remain associated with membranes during mitosis [68]. The C-terminus of B-type lamins is also isoprenylated but, unlike prelamin A, does not undergo proteolytic cleavage. Consequently, B-type lamins remain permanently farnesylated, facilitating their attachment to the inner nuclear membrane.
The nuclear interior
In addition to DNA and histones, the nucleoplasm contains distinct structural and functional elements such as nucleoli [69], Cajal bodies [70], the Gemini of coiled bodies or gems [71], promyelocytic leukemia (PML) bodies [72], and splicing speckles [73]. The growing interest to decipher the detailed structure and composition of the nuclear interior has led to the recent discoveries that the nuclear interior contains actin [74,75], myosin [76,77], spectrin [78] and even titin [79]. It is now well established that actin oligomers or short polymers are present in the nucleus [80–82] and that all isoforms of actin contain nuclear export sequences [83], which may help prevent spontaneous assembly of actin filaments inside the nucleus. To date, many aspects of nuclear actin remain incompletely understood, including its precise structural organization [84]. Nonetheless, nuclear actin has been implicated in a number of functions highly relevant to tumorigenesis, including DNA organization, stabilization, and orientation during replication, determination of nuclear morphology, organization of gene regulatory complexes, and RNA synthesis [85]. The existence and function of the “nuclear matrix” or nucleoskeleton, typically defined as the insoluble structure remaining after nuclease, detergent and high salt treatment of isolated nuclei [86], remains a matter of lively debate, but given the plethora of structural proteins present in the nucleus and their often low diffusional mobility, it is likely that some (possibly local) structural frameworks exist in the nuclear interior.
Nuclear mechanics and mechanotransduction
In recent years, it has emerged that physical factors, such as the biomechanical properties of the microenvironment and the mechanical forces acting between cells and their environment, play an important role in cellular function [87]. With regards to cancer cells, modulation of cytoskeletal tension by Rho inhibition alone can be sufficient to phenotypically revert epithelial morphogenesis of malignant cells [10]. Rho proteins belong to the family of small signaling G-proteins (GTPases) that can act as “molecular switches” in regulating actin cytoskeleton dynamics, while also playing important roles in cell polarity, migration vesicle trafficking, mitosis, proliferation and apoptosis [88]. Furthermore, recent studies found that aggressive cancer cells can be distinguished from less invasive and non-tumorigenic cancer cells based on their cytoskeletal stiffness [3] and their contractile force generation [8]. What is now becoming apparent is that in addition to cytoskeletal stiffness and force generation, nuclear deformability, as well as the physical coupling between the nucleus and the cytoskeleton, play a critical role in cell motility in three-dimensional (3-D) environments [20,19]. In this section, we discuss the molecular players governing normal nuclear mechanics, i.e., nuclear deformability and nucleo-cytoskeletal coupling, as well as their potential contribution to cellular mechanosensing. Their involvement in cancer progression is then described in the subsequent section.
Nuclear deformability and stability
Over the years, a variety of experimental techniques has been developed to probe the mechanical properties of the nucleus, particularly its deformability under applied forces. These approaches include micropipette aspiration [89–93], atomic force microscopy [91,94–96], cell stretching [14,97–99], tracking of particles within the nucleoplasm [100], and, most recently, optical stretching [101] and measuring transit times through microfluidic constriction channels [102,103]. These experiments have revealed that the nucleus exhibits both elastic (the nuclear lamina) and viscoelastic (the nuclear interior) behavior and is typically ~2–10 times stiffer than the surrounding cytoplasm [99,104,93,105]. The precise measurements for the apparent Young’s modulus, a measure of material elasticity, range from ~0.1 to 10 kPa, depending on the experimental conditions and technique. This broad range of stiffness measures likely reflects a large degree of cell-to-cell variability, as well as different domains and mechanical behavior probed by the diverse experimental methods. For example, tracking of small particles within the nucleoplasm is sensitive to entanglement of the tracked particle within the nucleoskeleton/chromatin; in addition, the resulting measurements exclude contributions to nuclear stiffness from the nuclear envelope [90,91]. In contrast, cell stretch experiments and other techniques that result in large nuclear deformations will yield “bulk” measurements that combine contributions from the nuclear interior and the nuclear envelope, but may also depend on the mechanical properties of the cytoskeleton and its connection to the nucleus [17].
Micropipette aspiration experiments [90–92] and computational modeling [105] indicate that the mechanical deformability of the nucleus is mainly governed by the nuclear lamina and the nuclear interior; the relative contribution of each component depends on diverse factors such as mechanical load (e.g., applied tension vs. compression), the specific cell type, differentiation state, and chromatin configuration. The contribution of the inner and outer nuclear membranes to the deformability of the nucleus is largely negligible [106], as lipid membranes exhibiting relatively low bending stiffness and a 2-dimensional (2-D) liquid-like behavior, i.e., they can flow in response to applied shear stress, with connections to a large membrane reservoir in the form of the ER [106,16].
The importance of the nuclear lamina in providing structural support to the nucleus and controlling nuclear size is now well established [17,12], with the nuclear lamina acting as a load-bearing, elastic shell surrounding a viscoelastic nuclear interior [90,91,107]. Experiments on cells from gene-modified mice lacking specific lamin isoforms [98] and Xenopus oocytes ectopically expressing human lamins [95] suggest that lamins A and C are the main contributors to nuclear stiffness, with loss of lamin A or C resulting in softer, more deformable nuclei, while increased expression of lamin A results in stiffer, less deformable nuclei.. Given the structural similarities between A-type and B-type lamins, it may be somewhat surprising that these proteins have distinct roles in affecting nuclear deformability. However, recent findings suggest that A- and B-type lamins—and even lamins A and C—may form distinct but overlapping networks [108,109], and that A-type lamins may form a thicker protein network at the nuclear envelope [110]; however, as imaging the nuclear lamina in intact somatic cells with sufficiently high resolution remains technically extremely challenging, the exact structure and organization of the lamina and the different lamin isoforms at the nuclear envelope remains unclear. Interaction of specific lamin isoforms with other nuclear (envelope) proteins may serve as additional explanation for the distinct roles of the diverse lamins in nuclear mechanics. For example, loss of the inner nuclear membrane protein emerin, which directly interacts with lamins A/C, results in more deformable nuclei, although to a lesser degree than functional loss of lamins A/C [92,97]. In addition, functional loss of lamins due to mutations or (partial) deletion cam also affects chromatin organization [111–114], which could affect nuclear deformability.
Further illustrating the importance of A-type lamins in nuclear mechanics, lamin A/C-deficient cells have more deformable nuclei that are more susceptible to rupture under mechanical stress [115,14]. Of note, mutations in A-type lamins, as well as emerin, cause a spectrum of human diseases (laminopathies) that include Emery-Dreifuss muscular dystrophy, limb-girdle muscular dystrophy, dilated cardiomyopathy, Dunnigan-type familial partial lipodystrophy, and Hutchinson-Gilford Progeria syndrome [51]. In many cases, cells from affected patients show characteristic features such as misshapen nuclei, increased nuclear fragility, and herniations [16]; furthermore, LMNA mutations resulting in disease affecting cardiac and skeletal muscle often cause defects in nuclear mechanics [116], providing a potential disease mechanism for the muscular laminopathies.
Importantly, lamins also interact with other inner nuclear membrane proteins (e.g., emerin, LAPs and LBR), nuclear pore components, DNA, chromatin and transcription factors (e.g. retinoblastoma protein [Rb], SREBPs, GCL and MOK2), and structural proteins such as nuclear actin and titin [117]. These interactions could further modulate nuclear stiffness by forming nucleoskeletal structures or affecting chromatin organization and transcriptional regulation. For example, nuclear abnormalities have been observed in cells depleted of large repeat-domain proteins such as titin and αII-spectrin [118,119]. On the other hand, the role of nuclear actin in providing structural support to the nucleus remains unclear [84]. Through their interaction with SUN proteins, nesprins, and Samp1, lamins also play an important role in connecting the nucleus to the surrounding cytoskeleton [120], as discussed in more detail below.
Besides the nuclear lamina, chromatin is an important contributor to nuclear stiffness. Unlike the mostly elastic nuclear lamina, chromatin exhibits more viscoelastic material behavior, i.e., it flows when subjected to forces (Fig. 2) and undergoes plastic deformations [107,106]. Chromatin decondensation during initial lineage commitment of embryonic stem cells is associated with a significant softening of the nucleus [101]. Subsequently, the viscoelastic deformability of the cell nucleus in human embryonic stem cells changes during further cellular differentiation [107], becoming 6-times stiffer and also less fluid-like during terminal differentiation. It remains unclear, however, to what extent this behavior is caused by changes in chromatin organization, e.g., switching from loose euchromatin to more compacted heterochromatin, or results from the increased expression of A-type lamins in differentiated cells.
Figure 2. Invasive cancer cell MDA-MB-231 squeezing into 8 μm width constriction.
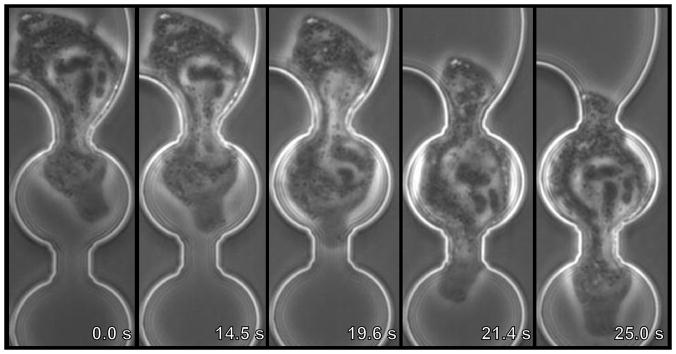
Image sequences of a cancer cell being perfused through 8 μm-wide constriction at a pressure (ΔP) of 10 psi. The viscoelastic deformation as the nucleus flows through the constriction is clearly visible.
Nucleo-cytoskeletal coupling
Over the last 10 years, it has become well established that the nucleus is physically coupled to the surrounding cytoskeleton [120]. Many of the molecular components are highly preserved throughout evolution, being present in unicellular organisms such as yeast all the way to mice and humans [121]. Building on work in yeast and drosophila, several of the molecular details of nucleo-cytoskeletal coupling were first unraveled in C. elegans, where UNC84 and ANC-1, in conjunction with Ce-lamin, participate in the actin-dependent anchorage and positioning of the nucleus [122–124,32,125]. Subsequent studies have confirmed that closely related proteins are also responsible for nucleo-cytoskeletal coupling in mammalian cells; this physical connection is now commonly referred to as the Linker of Nucleoskeleton and Cytoskeleton (LINC) complex [126]. In the strictest definition, the LINC complex contains two essential parts: (i) a member of the trimeric inner nuclear membrane SUN- [127] domain protein family, which engages with nucleoplasmic proteins such as lamins [128,129,121]; (ii), KASH- (Klarsicht, ANC-1, Syne Homology) domain containing nesprins located on the outer nuclear membrane that bind across the perinuclear space to the SUN domain of Sun1/Sun2 trimers [130]. The cytoplasmic ends of nesprins interact directly or indirectly with various components of the cytoskeleton, including actin, intermediate filaments (via plectin)[131], and microtubules (via microtubule-binding motors such as dynein and kinesin), thereby completing the physical connection across the nuclear envelope [121]. In many cases, lamins are considered an extended part of the LINC complex, as they bind to SUN proteins and inner membrane variants of nesprins and help tether these proteins to the nuclear interior [132]. Since the cytoskeleton also connects to focal adhesion and cell-cell junctions, cells contain a continuous mechanical network linking the nuclear interior and the extracellular matrix and neighboring cells, thereby allowing forces exerted from the cellular environment or the cytoskeleton to be transmitted directly to the nuclear interior [133,120,134,39].
A. SUN domain proteins
The characteristic feature of SUN domain family proteins is a 115–175 amino acid domain that shares homology with the Sad1 protein from S. pombe [135] and the UNC84 protein from C. elegans [122]. Mammalian cells have five SUN domain proteins, with two of these proteins (SUN1 and SUN2) present on the nuclear envelope in somatic cells (SUN3–5 are testis specific) [136]. SUN1 and SUN2 proteins consist of a helical N-terminal domain that can bind to lamins [137] and nuclear pore complex proteins [138,139], a single pass transmembrane domain to anchor the protein in the inner nuclear membrane [140], a luminal helical domain required for trimerization of SUN proteins [130], and the C-terminal SUN domain, which interacts with the KASH domain of nesprins [126].
B. Nesprins and other KASH domain proteins
Mammals have four nesprins (genes SYNE 1–4), with nesprins 1–3 having multiple isoforms resulting from alternative splicing, initiation, and termination [121,120,34]. Expression of various nesprin isoforms can be highly tissue-specific [34]. In skeletal muscle, levels of nesprin-1 (first described as Syne-1 for synaptic nuclear envelope protein-1) are highest in synaptic nuclei, suggesting that it might participate in the migration and anchoring of these specialized muscle nuclei [141]. Common to all nesprins is a central region containing multiple spectrin domains, whose number can greatly vary between isoforms [142]; all nesprins (but not all isoforms) contain a ~60 amino acid-long C-terminal KASH domain, consisting of a transmembrane domain and a short, highly conserved luminal domain, which is essential for anchoring nesprins to the nuclear envelop [59,142]. The N-terminal domain of nesprins typically contains specific motifs to interact with different cytoskeletal proteins. For instance, the nesprin-1 and -2 “giant” isoforms (1000 and 800 kDa in size, respectively) contain an actin-binding domain (ABD) composed of two calponin homology domains [143,37,35]; additionally, nesprins-1 and -2 can interact with the microtubule-associated motors dynein/dynactin and kinesin [120]. Nesprin-3 can connect to intermediate filaments via plectin [36]. Nesprin-4 binds the microtubule-associated motor kinesin, and ectopic expression of nesprin-4 induces dramatic changes in centrosome positioning in cells [144]. While localization of larger nesprin isoforms is restricted to the outer nuclear membrane, shorter isoforms can also be present at the inner nuclear membrane, where they can interact with lamins and emerin [145–147,38]. Nesprin isoforms lacking the KASH domain may also be found in other cellular structures. In addition to nesprins 1–4, mammals express at least one additional KASH-domain protein, aptly named KASH5, which is found exclusively in spermatocytes and oocytes, where it plays a critical role in meiosis [148].
C. Other molecules involved in nucleo-cytoskeletal coupling
With the growing interest in understanding the mechanics of the nucleus and its connection to the cytoskeleton, several recent studies have focused on identifying additional molecular players involved in nucleo-cytoskeletal coupling. Based on experimental findings in emerin-deficient cells, one study has proposed that emerin binds to microtubules and that a subset of emerin located on the outer nuclear membrane is involved in coupling the centrosome to the nuclear envelope [149], but it remains unclear whether the emerin-microtubule interaction is direct or mediated through other proteins such as nesprins.
A more recent candidate to be involved in nucleo-cytoskeletal coupling is the inner nuclear membrane protein Samp1 [150], which associates with lamin A/C, emerin, Sun1, and Sun2 [150–152]. During mitosis, Samp1 is associated with the mitotic spindle [150]; during interphase, however, Samp1 is an important component of transmembrane actin-associated nuclear (TAN) lines [152], which promote rearward nuclear movement in polarizing fibroblasts by connecting the nucleus to retrograde actin flow via nesprin-2giant and SUN2 [153]. The involvement of lamins A/C in nucleo-cytoskeletal coupling is further illustrated by the finding that lamin mutants associated with muscular dystrophies can disrupt this retrograde nuclear movement [132] and that lamin A/C is required for retaining Samp1 at the nuclear envelope [152]. Another potential mediator of nucleo-cytoskeletal coupling is the luminal protein torsinA, part of the AAA+ ATPase superfamily. TorsinA interacts with the KASH domains of nesprins 1–3, and loss of torsinA results in mislocalization of nesprin-3 from the nuclear envelope and impaired cell polarization and migration [131]. Given the promiscuous interaction of SUN domain proteins and nesprins [154], it is likely that tissue-specific expression of their isoforms, as well as potential interaction with other nuclear envelope proteins such as Samp1, play an important role in the spatial and temporal control of nucleo-cytoskeletal coupling.
D. Nucleo-cytoskeletal coupling is critical for many cell functions
Studies investigating molecules involved in connecting chromatin and cytoskeletal structures have often focused on processes during mitosis and meiosis. For instance, analysis of chromosome condensation during yeast prophase has unraveled a direct interaction between Sad1 (a Sun homologue protein) and meiotic-specific bouquet (Bqt) proteins [155]. Sad1 has also been linked to Kms1 protein [156] and this interaction is known to couple telomeres to microtubules and cytoplasmic dynein [157,158]. Similar results were obtained in C. elegans, where selective inactivation of Sun1 protein or Kdp-1 (KASH domain protein-1) protein delays cell cycle progression [159,160]. In mammalian cells, lamins, SUN proteins, KASH5 and Samp1 have all been implicated in specific roles during mitosis and/or meiosis, [161,148], and loss of A-type lamins causes telomere shortening defects and overall genomic instability [162].
In recent years, research has increasingly focused on the role of LINC complex proteins in interphase cells and consequences of LINC complex disruption. In C. elegans, deletion of the nesprin and SUN1 orthologues ANC-1 and UNC-84 result in impaired nuclear positioning and anchoring in muscle cells [122,32]. In mammalian cells, LINC complex disruption causes defects in nuclear positioning, cell polarization, and migration [133] by impairing force transmission between the nucleus and cytoskeleton [24,153]. LINC complex proteins are particularly important during cell migration in 3-D environments, for example, inside collagen matrices or tissues. In particular, lamins A/C, nesprin-2giant, and nesprin3 modulate perinuclear actin organization and actin protrusions; consequently, deletion of lamins A/C or LINC complex disruption results in significantly impaired migration of cells in 3-D collagen matrices [163]. The implications of impaired nucleo-cytoskeletal coupling in cancer progression are discussed in more detail below.
Nuclear mechanics stiffness and nucleo-cytoskeletal coupling in mechanotransduction
As described above, the cytoskeleton physically connects the nucleus to the cellular microenvironment. Consequently, pulling on integrins on the surface of intact endothelial cells results not only in reorientation of cytoskeletal filaments, but also in distortion of the nucleus and spatial redistribution of subnuclear structures [134]. Similar results, including force-induced dissociation of nuclear protein complexes, were recently obtained in HeLa cells subjected to forces applied via magnetic tweezers [164] and in human umbilical vein endothelial and osteosarcoma cells exposed to fluid shear stress [165]. It has long been speculated that such mechanically induced changes in nuclear structure and chromatin configuration could directly activate specific mechanosensitive genes, for example, by changing accessibility to transcription factors [166,18]. This idea is further supported by studies that have found interactions between applied forces, Rho signaling, cell shape, and histone acetylation [167–169]. Nonetheless, direct evidence for such nuclear mechanosensing remains scarce, and the majority of data are rather correlative, making it difficult to discern whether mechanical forces acting on the nucleus are sufficient to directly induce changes in gene regulation, or whether the observed activation of mechanosensitive genes is the downstream result of signaling cascades originating in the cytoskeleton or the plasma membrane [15]. A recent study [24] addressing this question found that LINC complex disruption had no discernible effect on the mechanically-induced expression of the mechanosensitive genes Iex-1 and Egr-1, whose activation is impaired in lamin A/C-deficient cells [14,170], even though LINC complex disruption resulted in substantially reduced nuclear deformation when the fibroblasts were subjected to substrate strain [24].
At the same time, changes in nuclear envelope composition undoubtedly affect cellular structure and function. For example, LINC complex disruption alters the mechanically induced proliferation of C2C12 myoblasts [171]; LINC complex depletion also causes impaired propagation of intracellular forces and disturbed organization of the perinuclear actin and intermediate filament networks, leading to defects in nuclear positioning and cell orientation [24,171,22]. In the case of impaired expression of mechanosensitive genes in lamin A/C- and emerin-deficient cells, it remains unclear whether this effect is due to direct mechanical defects or a consequence of altered interaction of lamins with specific transcriptional factors. An additional mechanism by which lamins and emerin can affect mechanotransduction signaling was recently identified, revealing that the actin polymerization-promoting activity of emerin at the nuclear envelope can influence nuclear and cytoskeletal actin dynamics, thereby modulating localization and activity of the mechanosensitive transcription factor MKL1 (also known as MRTF-A or MAL), whose localization is dependent on interaction with monomeric G-actin [172].
Relevance of nuclear mechanics and mechanotransduction in cancer progression
With growing advances in the understanding of the physics of cell motility, the mechanical properties of cancer cells have become an increasing area of interest [3]. As the nucleus is typically the largest and stiffest organelle, often occupying a large fraction of the cell’s volume, the properties of the nucleus can dominate the overall cellular mechanical response when cells are subjected to large deformations [17]. Several lines of evidence suggest that the ability of the nucleus to deform can impose a rate-limiting step in non-proteolytic cell migration in 3-D environments, when cells attempt to squeeze through narrow constrictions imposed by extracellular matrix fibers and other cells (Fig. 3) [20,19]. In this section, we summarize changes in nuclear structure and morphology observed in various cancers and describe the role of nuclear deformability in cell motility. In addition, we discuss the intricate feedback between the mechanics of the cellular microenvironment and intracellular organization and function.
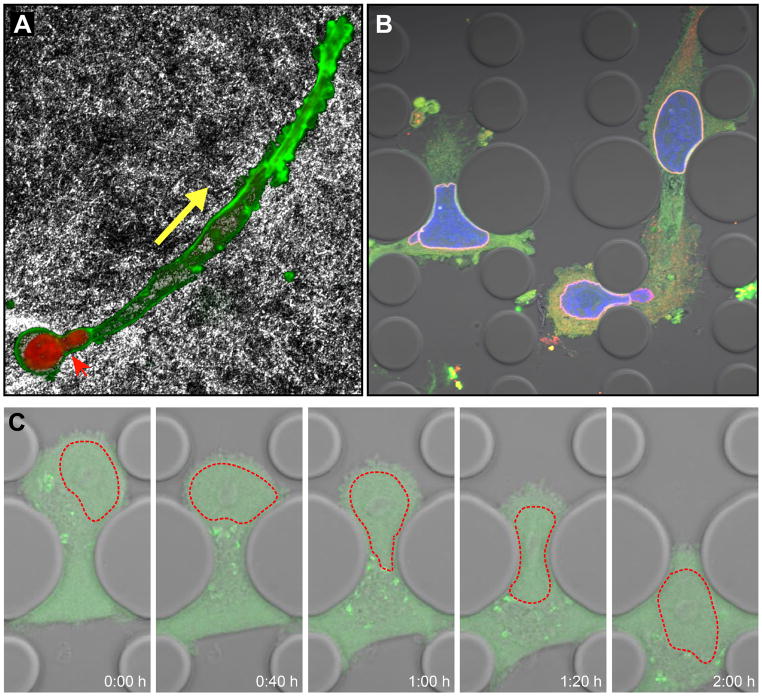
Figure 3. Migration of cancer cell in a constrained environment.
(A) Fibrosarcoma cell (HT1080 cell line) migrating through a dense collagen fiber matrix. The rat tail collagen matrix was imaged by reflection microscopy; the nucleus is visible in red (DAPI), F-actin in green (phalloidin). The cell body has already advanced in the direction of migration (yellow arrow), while the nucleus is still in the process of squeezing through constrictions in the collagen matrix (red arrow head). Image courtesy of Katarina Wolf, University of Nijmegen. (B) Fibrosarcoma cells (HT1080) migrating through 2 μm × 5 μm and 5 μm × 5 μm constrictions in a microfluidic channel. The cytoplasm is visible in green, the nucleus in blue, and the nuclear lamina (lamin B2) in red. (C) Time-lapse series of MDA-MB-231 breast cancer cell expressing a green fluorescent protein migrating through a 5 μm-wide constriction in a microfluidic channel. The nucleus is outlined in red (dashed line).
Altered nuclear structure and morphology in cancer cells
With few exceptions, the nuclei of normal cells have an ellipsoid shape with smooth outlines; in contrast, many cancer cells are easily identifiable by increased nuclear size, irregular nuclear contours, and disturbed chromatin distribution, making nuclear morphology one of the oldest and most commonly used cancer markers [11]. The irregular nuclear outline in cancer cells is mainly the result of grooving, convolutions and invaginations of the nuclear envelope [173]. While the characteristic changes in nuclear morphology in cancer cells are well documented, their cause and consequence remain unclear. Interestingly, the irregular nuclear morphology of cancer cells often bears striking resemblance to the abnormal nuclear shapes observed in cells lacking or expressing mutant nuclear envelope proteins such as lamins A/C, lamin B1/B2, or LBR [174,175], suggesting a possible involvement of dysregulated nuclear envelope proteins [173,176].
This idea is supported by a growing number of publications that report altered expression of lamins in a variety of human tumors, often associated with particularly malignant phenotypes (Table 1). Interestingly, while some cancers frequently show downregulation of lamin A/C [177–179], other cancers have upregulated levels of lamins A/C [177,180,181], and for some cancers, such as colon cancer, both increased [182] and decreased [183] levels of lamin A/C have been reported. Furthermore, even within single tumors and individual cancer cell lines [184], highly heterogeneous expression levels of lamin A/C can be found [185]. Similarly, both high and low levels of lamins A/C have been considered poor prognostic markers for cancer patients, depending on the specific study and cancer subtype. For example, reduced lamin A/C expression is a sign of poor prognosis for patients with gastric carcinoma [186], and patients with stage II and III colon cancer have a significantly increased risk of cancer recurrence when their tumors are marked by loss of lamin A/C expression [183]. At the same time, another study found that patients with increased expression of lamins A/C in colorectal cancer tumors were almost twice as likely to die of the disease than patients with tumors negative for lamin A/C [187], possibly by lamin A/C promoting cell motility [188]. These apparently inconsistent findings point at the multiple roles lamin can play in cancer progression, which will be discussed in more detail below.
Table 1.
Altered expression (and mutations) of nuclear envelope proteins in cancers.
| Protein | Cancer/tumor type | Reported change | Prognostic value | Reference |
|---|---|---|---|---|
| Emerin | Ovarian cancer | Loss of emerin | [222] | |
|
Lamin A Lamin C |
Lung cancer | Absence or very reduced expression in small cell lung carcinoma | [177] | |
| Colonic and gastric
adenocarcinomas Other cancers: Oesophagus cancer, cervical and uterine cancer, breast cancer. |
Reduced levels and mislocalization (aberrant cytoplasmic immunolabelling) | [178] | ||
| Basal cell skin carcinoma | Low levels or absence of lamin A | Increased proliferation | [179] | |
| Basal cell skin carcinoma | Low levels of lamin C | Low proliferation | [179] | |
| Skin cancer | High levels of lamin A and C in the basal cell layer of the epidermis overlying basal cell carcinomas, squamous cell carcinomas and actinic keratosis (AK) | Proliferative capacity | [180] | |
| Leukemia and lymphomas | Loss of gene expression by epigenetic silencing in nodal diffuse large B-cell lymphomas and acute lymphoblastic leukemias. | Poor outcome/overall survival | [223] | |
| Colorectal cancer | Increased expression (mainly lamin A) | Promote tumor invasiveness Poor prognosis (risk indicator of tumor related mortality) | [187] | |
| Ovarian serous cancer | High levels in all stages of ovarian serous carcinomas; increased immunoreactivity in the higher stage of tumor | Correlates with advanced stage | [224] | |
| Primary gastric carcinoma | Low levels | Poor histological differentiation; poor prognosis | [186] | |
| Prostate cancer | Low expression in lower grade; increased levels in higher grade | Correlates with advanced stage | [225] | |
| Colon cancer | Low expression in stage II and III patients | Correlates with increased relapse | [183] | |
| Ovarian cancer | Heterogeneous lamin A/C protein expression pattern or absence of lamin A/C and aneuploidy | [185] | ||
| Breast Cancer | Mutated | [190] | ||
| Lamin B | Colon cancer | Reduced expression | [178] | |
| Colorectal carcinoma | Increased levels | [226] | ||
| Ovarian cancer | Increased levels of lamin B1 and B2 in malignant cell compared to benign | [227] | ||
| Hepatocellular carcinoma | Increased levels of Lamin B1 in cirrhotic tissue | [228] | ||
| Liver cancer | Increased levels of lamin B1 in every stage (cirrhosis, early stage, late stage); presence of soluble lamin B1 in the circulation | Potential biomarker Correlate with the tumor development | [229] | |
| Prostate cancer | Increased levels of lamin B | Correlate with the tumor development | [230] | |
| Pancreatic cancer | Increased levels of lamin B1 | Correlate with decreased levels of tumor differentiation, high metastatic potential and poor overall survival | [231] | |
| LAP2 | Malignant lymphocytes | Increased levels | LAP2β correlates with highly proliferative malignant cells | [232] |
| Nesprins | Ovarian cancer | Nesprin 1 polymorphism. Down-regulation of a transcript (shorter isoform) | Associated with invasive ovarian cancer risk | [191] |
| Colorectal cancer | Nesprin 1 is candidate cancer gene (mutated in cancer) | [189] | ||
| Breast cancer | Nesprin 1 (mutations) | [190] | ||
| Breast cancer | Nesprin 2 (mutations) | [189,190] | ||
| NUP 88 | Ovarian cancer Different type of cancers: sarcomas, lymphomas, mesotheliomas and breast cancer | Increased levels Increased levels | Correlates with high-grade malignancies | [233] [234–237] |
| Colorectal cancer and hepatocellular carcinoma | Increased levels | Correlates with poor differentiation | [238,239] | |
| NUP 98 | Leukemia | Increased levels; may act as component of a chromosomal translocation | [240] | |
| NUP 214 | Uterine, stomach and rectal tumors, leukemias, breast cancer | Increased levels; may act as a multi –functional oncogene and as a component of a chromosomal translocation | [241,240, 242,189] |
In addition to lamins, other nuclear (envelope) proteins have recently been implicated in a variety of cancers. A genome-wide scan in several patients with either breast, colorectal or ovarian cancer revealed genetic alterations in nesprin-1 [189], and another genome-wide study identified mutations in nesprin-1, -2 and lamin A/C in a panel of 100 breast cancer patients [190]. Furthermore, downregulation and mutations in nesprin have been associated with an increased risk of invasive ovarian cancer [191]. Lastly, several “nuclear matrix” or nucleoskeletal-associated proteins such as NuMA or nucleoporin proteins (NUP 88, NUP 98) have been correlated with aggressive tumor phenotypes [192] and used as prognostic markers of disease [193].
Implications of altered nuclear envelope composition in cancer
What is the impact of altered nuclear envelope composition on nuclear mechanics? As lamin expression and chromatin organization determine nuclear deformability, it is expected that changes in nuclear architecture will alter the rigidity of the nucleus. In cancer, increased nuclear deformability may benefit metastatic cells that need to pass through narrow interstitial spaces or small capillaries, while defects in nucleo-cytoskeletal coupling may impair migration in 3-D tissues [20]. In addition to these mechanical functions, the nuclear envelope and nuclear interior play important roles in the processing of genetic information [194–196]. Thus, changes in nuclear organization could have consequences on gene expression or DNA stability with important implications in cancer progression.
A. Nuclear deformability and cell motility
The abnormal nuclear shapes observed in cancer cells and their resemblance to lamin-deficient or mutant cells, combined with the increasing reports of altered expression of nuclear envelope proteins in various cancers (Table 1), suggests that cancer cells may have altered nuclear mechanics. While direct measurements of nuclear deformability in cancer cells have not yet been reported, studies that measure whole-cell deformability consistently find that cancer cells, particularly highly invasive ones, have increased cellular deformability [4,3,7]. Why should (nuclear) deformability matter in cancer progression? During the metastatic process, cancer cells must undergo modifications and large elastic deformations to invade the tissue surrounding the primary tumor, intravasate blood vessels, survive the physical stresses during circulation in the blood stream, extravasate at new sites in the body, and eventually proliferate in a nutrient-deprived microenvironment [197]. Particularly during invasion and intra- and extravasation, cells penetrate through interstitial spaces and openings ranging in size from 2 to 30 μm [198,199]. Cytoskeletal shape is highly adaptive, owing to the rapid cytoskeletal remodeling and plasma membrane flexibility; consequently, cytoskeletal protrusions can invade spaces of less than 1 μm2 in cross-section [200,201]. In contrast, the ability of the nucleus to pass through narrow constrictions is more limited due to its size and stiffness. Transient nuclear deformations, resulting in hourglass- and cigar-shaped nuclei, as well as nuclear protrusions indicative of attempts to pass through narrow constrictions, can be observed (at least transiently) during cancer cell migration in vivo [20]. Importantly, a recent report by Friedl, Wolf and colleagues [19] found that deformation of the nucleus poses a rate-limiting step during proteolysis-independent cell migration. They found that in the absence of proteolysis, e.g., during matrix metalloprotease (MMP) inhibition or knockdown, migration of cancer cells through 3-D collagen matrices and polycarbonate filters is limited by the available pore size: cell migration speed and migration efficiency gradually drops with decreasing cross-sectional areas of the constrictions until cell body movement is completely stalled [19]. A similar size-dependent effect was observed by Tong and colleagues [202] when studying cell migration in microchannels with varying width. Indeed, decreasing channel width below 20 μm (at a fixed channel height of 10 μm) resulted in increasing reduction in migration speed. At the extreme, cells in 3 μm-wide channels had a 70% reduction in migration speed compared to 50 and 20 μm-wide channels. Interestingly, the minimum size requirement for (non-proteolytic) migration through 3-D environments was found to be independent of the shape of the constriction and only depends on the available cross-sectional area [19].
While these studies illustrate the importance of nuclear deformability in cell migration in confined environments, the role of the nuclear lamina and nuclear stiffness in this process remains to be explored [20]. At least in neutrophil-like cells, which normally have extremely low levels of lamins A/C and which can migrate through constrictions only a few micrometers in diameter, overexpression of lamin A results in less deformable nuclei that have reduced efficiency at crossing narrow constrictions and that take significantly longer to transit narrow microfluidic channels mimicking capillaries [103]. Similarly, fibroblasts expressing a mutant form of lamin A (progerin) that is responsible for Hutchinson-Gilford progeria syndrome have difficulties migrating through an array of microfabricated pillars spaced 6 μm apart [203], likely due to the increased nuclear stiffness caused by progerin [204,205], as migration on non-constricted surfaces was comparable to cells from healthy controls [203]. Although these findings suggest an important role of lamins A/C in moderating the ability of cells to pass through narrow constrictions, Wolf and colleagues [19] found that the maximal deformation the nucleus could achieve during passage through narrow constrictions, indicated as the ratio of the nuclear cross-section in the constriction to the undeformed nuclear cross-section, was consistently around 1:10, regardless of the cell type studied. These findings suggest that the size limit for nuclear passage through small constriction may be governed by the maximal compressibility of the nucleus. The theoretically maximal compression depends on the solid fraction of the nucleus, as the chromatin (and other nucleoplasmic proteins) can be no further compressed once all void spaces have been eliminated. This idea is consistent with the observed reduction in nuclear volume by up to 60% during migration of skin fibroblasts through microfabricated constrictions [203] and with micropipette aspiration experiments that revealed that the nuclear volume can be compacted to about 20 to 40% of its original size before reaching a state that resists further compression [92,106].
But what about cancers in which increased, rather than decreased, levels of lamin A/C have been reported, which is expected to result in reduced nuclear deformability [98]? Cancer cells are highly plastic and heterogeneous in their gene expression, so it is likely that different subpopulations of cells with distinct roles in cancer progression exist. Increased lamin levels could help protect cells from mechanical stress caused by the high hydrostatic pressure inside solid tumors. At the same time, lamins are also involved in multiple signaling pathways [117,51], which could modulate functions relevant to cancer progression. For example, increased levels of lamin A/C in prostate cancer cause changes in the PI3K/AKT/PTEN pathway [206], and upregulation of lamin A/C in colorectal cancer induces changes in cytoskeletal organization that promote cell motility [188]. As such, it is likely that different cells and tumors have found different approaches to find the best compromise between increasing nuclear deformability and activation of signaling pathways to increase cell motility and invasiveness.
B. Nuclear rupture of cancer cells
As described earlier, the nuclear envelope forms a well-defined compartment that acts as a protective shield for the genetic material. In normal cells, nuclear envelope breakdown and reassembly is limited to mitosis and precisely regulated [207]. Recently, Vargas et al. [208] reported that in many cancer cells, the nuclear envelope transiently ruptures and then reseals during interphase, resulting in temporary exchange between the nucleus and cytoplasm and the occasional entrapment of cytoplasmic organelles inside the nucleus. Nuclear envelope rupture was associated with the formation of micronuclei, portions of chromatin exiting the nuclear interior, and mislocalization of nucleoplasmic/cytoplasmic proteins. Importantly, the frequency of nuclear rupture events was increased in cells with small defects in the nuclear lamina [208]. These results are consistent with previous reports of increased nuclear fragility in lamin A/C-deficient mouse embryonic fibroblasts [14] and spontaneous (transient) nuclear rupture in these cells [209]. In our laboratory, we have frequently observed that cancer cells undergo transient nuclear rupture while migrating through narrow (~2 μm × 5 μm) microfluidic constrictions, with lamin-deficient cells displaying significantly increased rates of nuclear rupture (unpublished observations). Breakdown of the nuclear compartment during repetitive nuclear rupture could potentially result in increased genomic instability and chromatin rearrangements, which could further contribute to cancer progression, but this idea has not yet been experimentally tested.
C. Changes in chromatin organization in cancer cells
Epigenetic changes in chromatin configuration can directly impact nuclear stiffness. Therefore, the chromatin modifications frequently observed in cancer cells, including disturbed heterochromatin organization [11], could be associated with altered nuclear deformability and thereby affect 3-D cell migration, in addition to their role in transcriptional activity. Importantly, there is a strong interplay between nuclear envelope proteins and chromatin organization. Lamin A regulates dynamics of heterochromatin proteins in early embryonic stem cells [25]; lamins A/C-deficiency and mutations in the LMNA gene result in loss of heterochromatin [111,210]. Furthermore, lamins and lamin B receptor (LBR) play an important role in tethering specific chromatin regions to the nuclear periphery [211,212], which typically serves as a transcriptionally repressive environment [195]. LBR also interacts with heterochromatin protein 1 [213] and histones H3/H4 [213]. Lamin-associated polypeptide-2β (LAP2β) can modulate gene expression by regulating higher order chromatin structure or binding the transcriptional repressors germ cell less (GCL) [214] and histone deacetylase 3 [215], resulting in deacetylation of histone H4 [215]. Emerin can directly associate with chromatin modifiers and transcriptional repressors such as the death promoting factor Btf [216], the splicing associated factor YT521-B [217] and the transcriptional repressor GCL [218]. Given these findings, it is tempting to speculate that the altered expression of nuclear envelope proteins found in various cancers (Table 1) can directly affect chromatin organization and gene expression. Of course, the observed changes in expression of nuclear envelope proteins could also be the consequence, rather than the cause of altered chromatin organization. In this case, the changes in nuclear envelope composition could still result in further modifications of nuclear structure and organization while also directly altering nuclear mechanics.
Conclusion and future perspectives
The field of cancer cell biology has dramatically changed since 1943, when George Papanicolaou published his book Diagnosis of Uterine Cancer by the Vaginal Smear, which laid the basis for the now abundant “pap smear” to detect early signs of cervical cancer. Since then, researchers and clinicians have learned not only to identify and assess cancer cells based on characteristic morphological changes, but also to peek inside the inner life of cancer cells, including their genetic changes, biochemical composition, and metabolic state. In recent years, these approaches have been complemented by a new research direction, focused on the biophysical changes in cancer cells and their microenvironment. This research has already led to striking discoveries, including the role of the extracellular matrix stiffness, composition and topology in cancer progression [219] and the characteristic difference in cell deformability of cancer cells, which may lead to new diagnostic and prognostic applications [3]. Motivated by research in other diseases (laminopathies), it is now emerging that the mechanical properties of the cell nucleus, particularly its deformability and connection to the cytoskeleton, may play a similarly important role in cancer metastasis. The idea that deformation of the large and stiff nucleus presents a rate-limiting factor during the passage of metastatic cancer cells through tight interstitial spaces or narrow capillaries has recently found increasing experimental support [19,103,165]. Given the increasing reports of altered expression and mutations in nuclear envelope proteins responsible for determining nuclear stiffness, it is intriguing to speculate that (a subset of) cancer cells may have acquired specific adaptations in their nuclear structure and mechanics to promote metastatic spreading. Nonetheless, experimental verification of this idea is still lacking. Additional experiments, using sophisticated combinations of live cell imaging and measurements of subcellular mechanics, including primary tumor (and metastatic) cells from cancer patients and complemented by in vivo studies in mouse models, will be required to firmly establish this hypothesis. These experiments will also have to address why some cancers frequently have increased lamin levels while others have decreased or unchanged levels, and whether such changes in nuclear envelope composition can serve as reliable prognostic markers. Given the diverse functions of lamins, it is likely that (varying) combinations of altered cellular mechanics, cell signaling, and stem cell differentiation contribute to the increasingly emerging role of lamins in cancer progression. Done correctly, such experiments have the potential to not only address these key questions but to also produce novel insights into the dynamic nature of cancer cells, which may switch between different morphological and mechanical modes depending on their current role in cancer progression. Novel technology developments to probe single cell mechanics at substantial higher throughput than traditional methods [102,5,220,221] will enable detection of rare cell subpopulations, which could play a crucial role in cancer progression. Identifying key (mechanical) parameters that govern cancer cell metastasis may reveal novel therapeutic targets for pharmacological inhibition.
These clinical translation-driven experiments should be complemented by research to address some of the more fundamental questions in cancer cell biology, including the molecular mechanisms by which cells manage to squeeze the nucleus through constrictions only one tenth the diameter of the nucleus in size, and whether induced nuclear deformations can directly contribute to cellular mechanosensing. We are only at the beginning of a long road ahead, the destination a complete understanding of the physics of cancer progression und the underlying biology, but it will be exciting to see what is awaiting us around the next corner.
Acknowledgments
We apologize to all authors whose work could not be cited due to space constraints. This work was supported by National Institutes of Health awards [R01 NS059348 and R01 HL082792], a National Science Foundation CAREER award to J.L. [CBET-1254846], the Department of Defense Breast Cancer Idea Award [BC102152], and an award from the Progeria Research Foundation [PRF2011-0035].
References
- 1.Fawcett DW. An Atlas of fine structure (The Cell) 2. W. B. Saunders Company; 1966. [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Suresh S. Biomechanics and biophysics of cancer cells. Acta Biomater. 2007;3(4):413–438. doi: 10.1016/j.actbio.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guck J, Schinkinger S, Lincoln B, Wottawah F, Ebert S, Romeyke M, Lenz D, Erickson HM, Ananthakrishnan R, Mitchell D, Käs J, Ulvick S, Bilby C. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys J. 2005;88(5):3689–3698. doi: 10.1529/biophysj.104.045476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remmerbach TW, Wottawah F, Dietrich J, Lincoln B, Wittekind C, Guck J. Oral cancer diagnosis by mechanical phenotyping. Cancer Res. 2009;69(5):1728–1732. doi: 10.1158/0008-5472.can-08-4073. [DOI] [PubMed] [Google Scholar]
- 6.Byun S, Son S, Amodei D, Cermak N, Shaw J, Kang JH, Hecht VC, Winslow MM, Jacks T, Mallick P, Manalis SR. Characterizing deformability and surface friction of cancer cells. Proc Natl Acad Sci U S A. 2013;110(19):7580–7585. doi: 10.1073/pnas.1218806110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross SE, Jin YS, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol. 2007;2(12):780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 8.Kraning-Rush CM, Califano JP, Reinhart-King CA. Cellular traction stresses increase with increasing metastatic potential. PLoS One. 2012;7(2):e32572. doi: 10.1371/journal.pone.0032572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker EL, Lu J, Yu D, Bonnecaze RT, Zaman MH. Cancer cell stiffness: integrated roles of three-dimensional matrix stiffness and transforming potential. Biophys J. 2010;99(7):2048–2057. doi: 10.1016/j.bpj.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8(3):241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4(9):677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 12.Ho CY, Lammerding J. Lamins at a glance. J Cell Sci. 2012;125(Pt 9):2087–2093. doi: 10.1242/jcs.087288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Las Heras JI, Batrakou DG, Schirmer EC. Cancer biology and the nuclear envelope: a convoluted relationship. Semin Cancer Biol. 2013;23(2):125–137. doi: 10.1016/j.semcancer.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113(3):370–378. doi: 10.1172/jci19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahl KN, Ribeiro AJ, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circ Res. 2008;102(11):1307–1318. doi: 10.1161/circresaha.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zwerger M, Ho CY, Lammerding J. Nuclear mechanics in disease. Annu Rev Biomed Eng. 2011;13:397–428. doi: 10.1146/annurev-bioeng-071910-124736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lammerding J. Mechanics of the nucleus. Compr Physiol. 2011;1(2):783–807. doi: 10.1002/cphy.c100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10(1):75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 19.Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201(7):1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr Opin Cell Biol. 2011;23(1):55–64. doi: 10.1016/j.ceb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hale CM, Shrestha AL, Khatau SB, Stewart-Hutchinson PJ, Hernandez L, Stewart CL, Hodzic D, Wirtz D. Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys J. 2008;95(11):5462–5475. doi: 10.1529/biophysj.108.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chancellor TJ, Lee J, Thodeti CK, Lele T. Actomyosin tension exerted on the nucleus through nesprin-1 connections influences endothelial cell adhesion, migration, and cyclic strain-induced reorientation. Biophys J. 2010;99(1):115–123. doi: 10.1016/j.bpj.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider M, Lu W, Neumann S, Brachner A, Gotzmann J, Noegel AA, Karakesisoglou I. Molecular mechanisms of centrosome and cytoskeleton anchorage at the nuclear envelope. Cell Mol Life Sci. 2011;68(9):1593–1610. doi: 10.1007/s00018-010-0535-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem. 2011;286(30):26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melcer S, Hezroni H, Rand E, Nissim-Rafinia M, Skoultchi A, Stewart CL, Bustin M, Meshorer E. Histone modifications and lamin A regulate chromatin protein dynamics in early embryonic stem cell differentiation. Nat Commun. 2012;3:910. doi: 10.1038/ncomms1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Handwerger KE, Gall JG. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16(1):19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19(2):172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2(12):a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franke WW, Scheer U, Krohne G, Jarasch ED. The nuclear envelope and the architecture of the nuclear periphery. J Cell Biol. 1981;91(3 Pt 2):39s–50s. doi: 10.1083/jcb.91.3.39s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schirmer EC, Foisner R. Proteins that associate with lamins: many faces, many functions. Exp Cell Res. 2007;313(10):2167–2179. doi: 10.1016/j.yexcr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 31.Puddington L, Lively MO, Lyles DS. Role of the nuclear envelope in synthesis, processing, and transport of membrane glycoproteins. J Biol Chem. 1985;260(9):5641–5647. [PubMed] [Google Scholar]
- 32.Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298(5592):406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- 33.Fridkin A, Mills E, Margalit A, Neufeld E, Lee KK, Feinstein N, Cohen M, Wilson KL, Gruenbaum Y. Matefin, a Caenorhabditis elegans germ line-specific SUN-domain nuclear membrane protein, is essential for early embryonic and germ cell development. Proc Natl Acad Sci U S A. 2004;101(18):6987–6992. doi: 10.1073/pnas.0307880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajgor D, Mellad JA, Autore F, Zhang Q, Shanahan CM. Multiple novel nesprin-1 and nesprin-2 variants act as versatile tissue-specific intracellular scaffolds. PLoS One. 2012;7(7):e40098. doi: 10.1371/journal.pone.0040098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Padmakumar VC, Abraham S, Braune S, Noegel AA, Tunggal B, Karakesisoglou I, Korenbaum E. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp Cell Res. 2004;295(2):330–339. doi: 10.1016/j.yexcr.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, Raymond K, Sonnenberg A. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171(5):799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q, Skepper JN, Yang F, Davies JD, Hegyi L, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J Cell Sci. 2001;114(Pt 24):4485–4498. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Q, Ragnauth CD, Skepper JN, Worth NF, Warren DT, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J Cell Sci. 2005;118(Pt 4):673–687. doi: 10.1242/jcs.01642. [DOI] [PubMed] [Google Scholar]
- 39.Chen D, Zhao M, Mundy G. Bone morphogenetic proteins. Growth Factors. 2004;22(4):233–241. doi: 10.1080/08977190412331279890. HHV6108EX6P056CA [pii] [DOI] [PubMed] [Google Scholar]
- 40.WISCHNITZER S. An electron microscope study of the nuclear envelope of amphibian oocytes. J Ultrastruct Res. 1958;1(3):201–222. doi: 10.1016/s0022-5320(58)80001-5. [DOI] [PubMed] [Google Scholar]
- 41.Gerace L, Foisner R. Integral membrane proteins and dynamic organization of the nuclear envelope. Trends Cell Biol. 1994;4(4):127–131. doi: 10.1016/0962-8924(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 42.Grossman E, Medalia O, Zwerger M. Functional architecture of the nuclear pore complex. Annu Rev Biophys. 2012;41:557–584. doi: 10.1146/annurev-biophys-050511-102328. [DOI] [PubMed] [Google Scholar]
- 43.Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99(7):677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 44.D’Angelo MA, Anderson DJ, Richard E, Hetzer MW. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312(5772):440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- 45.D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18(10):456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dreger M, Bengtsson L, Schöneberg T, Otto H, Hucho F. Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc Natl Acad Sci U S A. 2001;98(21):11943–11948. doi: 10.1073/pnas.211201898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korfali N, Wilkie GS, Swanson SK, Srsen V, Batrakou DG, Fairley EA, Malik P, Zuleger N, Goncharevich A, de Las Heras J, Kelly DA, Kerr AR, Florens L, Schirmer EC. The leukocyte nuclear envelope proteome varies with cell activation and contains novel transmembrane proteins that affect genome architecture. Mol Cell Proteomics. 2010;9(12):2571–2585. doi: 10.1074/mcp.M110.002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schirmer EC, Florens L, Guan T, Yates JR, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301(5638):1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- 49.Wilkie GS, Korfali N, Swanson SK, Malik P, Srsen V, Batrakou DG, de las Heras J, Zuleger N, Kerr AR, Florens L, Schirmer EC. Several novel nuclear envelope transmembrane proteins identified in skeletal muscle have cytoskeletal associations. Mol Cell Proteomics. 2011;10(1):M110003129. doi: 10.1074/mcp.M110.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Korfali N, Wilkie GS, Swanson SK, Srsen V, de Las Heras J, Batrakou DG, Malik P, Zuleger N, Kerr AR, Florens L, Schirmer EC. The nuclear envelope proteome differs notably between tissues. Nucleus. 2012;3(6):552–564. doi: 10.4161/nucl.22257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schreiber KH, Kennedy BK. When lamins go bad: nuclear structure and disease. Cell. 2013;152(6):1365–1375. doi: 10.1016/j.cell.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paddy MR, Belmont AS, Saumweber H, Agard DA, Sedat JW. Interphase nuclear envelope lamins form a discontinuous network that interacts with only a fraction of the chromatin in the nuclear periphery. Cell. 1990;62(1):89–106. doi: 10.1016/0092-8674(90)90243-8. [DOI] [PubMed] [Google Scholar]
- 53.Strelkov S, Herrmann H, Aebi U. Molecular architecture of intermediate filaments. Bioessays. 2003;25(3):243–251. doi: 10.1002/bies.10246. [DOI] [PubMed] [Google Scholar]
- 54.Aebi U, Cohn J, Buhle L, Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323(6088):560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- 55.Herrmann H, Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu Rev Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- 56.Peter A, Reimer S. Evolution of the lamin protein family: what introns can tell. Nucleus. 2012;3(1):44–59. doi: 10.4161/nucl.18927. [DOI] [PubMed] [Google Scholar]
- 57.Batsios P, Peter T, Baumann O, Stick R, Meyer I, Gräf R. A lamin in lower eukaryotes? Nucleus. 2012;3(3):237–243. doi: 10.4161/nucl.20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly, and interactions. J Struct Biol. 1998;122(1–2):42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- 59.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6(1):21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 60.Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 2002;16(5):533–547. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- 61.Kitten GT, Nigg EA. The CaaX motif is required for isoprenylation, carboxyl methylation, and nuclear membrane association of lamin B2. J Cell Biol. 1991;113(1):13–23. doi: 10.1083/jcb.113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zastrow MS, Vlcek S, Wilson KL. Proteins that bind A-type lamins: integrating isolated clues. J Cell Sci. 2004;117(Pt 7):979–987. doi: 10.1242/jcs.01102. [DOI] [PubMed] [Google Scholar]
- 63.Dechat T, Gesson K, Foisner R. Lamina-independent lamins in the nuclear interior serve important functions. Cold Spring Harb Symp Quant Biol. 2010;75:533–543. doi: 10.1101/sqb.2010.75.018. [DOI] [PubMed] [Google Scholar]
- 64.Höger TH, Krohne G, Franke WW. Amino acid sequence and molecular characterization of murine lamin B as deduced from cDNA clones. Eur J Cell Biol. 1988;47(2):283–290. [PubMed] [Google Scholar]
- 65.Höger TH, Zatloukal K, Waizenegger I, Krohne G. Characterization of a second highly conserved B-type lamin present in cells previously thought to contain only a single B-type lamin. Chromosoma. 1990;99(6):379–390. doi: 10.1007/BF01726689. [DOI] [PubMed] [Google Scholar]
- 66.Lin F, Worman HJ. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J Biol Chem. 1993;268(22):16321–16326. [PubMed] [Google Scholar]
- 67.Lin F, Worman HJ. Structural organization of the human gene (LMNB1) encoding nuclear lamin B1. Genomics. 1995;27(2):230–236. doi: 10.1006/geno.1995.1036. [DOI] [PubMed] [Google Scholar]
- 68.Moir RD, Spann TP, Lopez-Soler RI, Yoon M, Goldman AE, Khuon S, Goldman RD. Review: the dynamics of the nuclear lamins during the cell cycle-- relationship between structure and function. J Struct Biol. 2000;129(2–3):324–334. doi: 10.1006/jsbi.2000.4251. [DOI] [PubMed] [Google Scholar]
- 69.Raska I, Koberna K, Malínský J, Fidlerová H, Masata M. The nucleolus and transcription of ribosomal genes. Biol Cell. 2004;96(8):579–594. doi: 10.1016/j.biolcel.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 70.Matera AG. Cajal bodies. Curr Biol. 2003;13(13):R503. doi: 10.1016/s0960-9822(03)00438-x. [DOI] [PubMed] [Google Scholar]
- 71.Matera AG, Frey MR. Coiled bodies and gems: Janus or gemini? Am J Hum Genet. 1998;63(2):317–321. doi: 10.1086/301992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dellaire G, Bazett-Jones DP. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays. 2004;26(9):963–977. doi: 10.1002/bies.20089. [DOI] [PubMed] [Google Scholar]
- 73.Nyman U, Hallman H, Hadlaczky G, Pettersson I, Sharp G, Ringertz NR. Intranuclear localization of snRNP antigens. J Cell Biol. 1986;102(1):137–144. doi: 10.1083/jcb.102.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pederson T, Aebi U. Actin in the nucleus: what form and what for? J Struct Biol. 2002;140(1–3):3–9. doi: 10.1016/s1047-8477(02)00528-2. [DOI] [PubMed] [Google Scholar]
- 75.Hofmann WA. Cell and molecular biology of nuclear actin. Int Rev Cell Mol Biol. 2009;273:219–263. doi: 10.1016/s1937-6448(08)01806-6. [DOI] [PubMed] [Google Scholar]
- 76.Nowak G, Pestic-Dragovich L, Hozák P, Philimonenko A, Simerly C, Schatten G, de Lanerolle P. Evidence for the presence of myosin I in the nucleus. J Biol Chem. 1997;272(27):17176–17181. doi: 10.1074/jbc.272.27.17176. [DOI] [PubMed] [Google Scholar]
- 77.de Lanerolle P, Serebryannyy L. Nuclear actin and myosins: life without filaments. Nat Cell Biol. 2011;13(11):1282–1288. doi: 10.1038/ncb2364. [DOI] [PubMed] [Google Scholar]
- 78.Young KG, Kothary R. Spectrin repeat proteins in the nucleus. Bioessays. 2005;27(2):144–152. doi: 10.1002/bies.20177. [DOI] [PubMed] [Google Scholar]
- 79.Zastrow MS, Flaherty DB, Benian GM, Wilson KL. Nuclear titin interacts with A- and B-type lamins in vitro and in vivo. J Cell Sci. 2006;119(Pt 2):239–249. doi: 10.1242/jcs.02728. [DOI] [PubMed] [Google Scholar]
- 80.Rando OJ, Zhao K, Crabtree GR. Searching for a function for nuclear actin. Trends Cell Biol. 2000;10(3):92–97. doi: 10.1016/s0962-8924(99)01713-4. [DOI] [PubMed] [Google Scholar]
- 81.McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol. 2006;172(4):541–552. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gieni RS, Hendzel MJ. Actin dynamics and functions in the interphase nucleus: moving toward an understanding of nuclear polymeric actin. Biochem Cell Biol. 2009;87(1):283–306. doi: 10.1139/o08-133. [DOI] [PubMed] [Google Scholar]
- 83.Wada A, Fukuda M, Mishima M, Nishida E. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. Embo J. 1998;17(6):1635–1641. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Belin BJ, Cimini BA, Blackburn EH, Mullins RD. Visualization of actin filaments and monomers in somatic cell nuclei. Mol Biol Cell. 2013;24(7):982–994. doi: 10.1091/mbc.E12-09-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Visa N, Percipalle P. Nuclear functions of actin. Cold Spring Harb Perspect Biol. 2010;2(4):a000620. doi: 10.1101/cshperspect.a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Berezney R, Coffey DS. Nuclear matrix. Isolation and characterization of a framework structure from rat liver nuclei. J Cell Biol. 1977;73(3):616–637. doi: 10.1083/jcb.73.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jaalouk DE, Lammerding J. Mechanotransduction gone awry. Nat Rev Mol Cell Biol. 2009;10(1):63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9(9):690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 89.Discher DE, Boal DH, Boey SK. Simulations of the erythrocyte cytoskeleton at large deformation. II. Micropipette aspiration. Biophys J. 1998;75(3):1584–1597. doi: 10.1016/s0006-3495(98)74076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dahl KN, Kahn SM, Wilson KL, Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci. 2004;117(Pt 20):4779–4786. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- 91.Dahl KN, Engler AJ, Pajerowski JD, Discher DE. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys J. 2005;89(4):2855–2864. doi: 10.1529/biophysj.105.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rowat AC, Lammerding J, Ipsen JH. Mechanical properties of the cell nucleus and the effect of emerin deficiency. Biophys J. 2006;91(12):4649–4664. doi: 10.1529/biophysj.106.086454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guilak F, Tedrow JR, Burgkart R. Viscoelastic properties of the cell nucleus. Biochem Biophys Res Commun. 2000;269(3):781–786. doi: 10.1006/bbrc.2000.2360. [DOI] [PubMed] [Google Scholar]
- 94.Jiménez-García LF, Fragoso-Soriano R. Atomic force microscopy of the cell nucleus. J Struct Biol. 2000;129(2–3):218–222. doi: 10.1006/jsbi.2000.4233. [DOI] [PubMed] [Google Scholar]
- 95.Schäpe J, Prausse S, Radmacher M, Stick R. Influence of lamin A on the mechanical properties of amphibian oocyte nuclei measured by atomic force microscopy. Biophys J. 2009;96(10):4319–4325. doi: 10.1016/j.bpj.2009.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kaufmann A, Heinemann F, Radmacher M, Stick R. Amphibian oocyte nuclei expressing lamin A with the progeria mutation E145K exhibit an increased elastic modulus. Nucleus. 2011;2(4):310–319. doi: 10.4161/nucl.2.4.16119. [DOI] [PubMed] [Google Scholar]
- 97.Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J Cell Biol. 2005;170(5):781–791. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem. 2006;281(35):25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- 99.Caille N, Thoumine O, Tardy Y, Meister JJ. Contribution of the nucleus to the mechanical properties of endothelial cells. J Biomech. 2002;35(2):177–187. doi: 10.1016/s0021-9290(01)00201-9. [DOI] [PubMed] [Google Scholar]
- 100.Tseng Y, Lee JS, Kole TP, Jiang I, Wirtz D. Micro-organization and visco-elasticity of the interphase nucleus revealed by particle nanotracking. J Cell Sci. 2004;117(Pt 10):2159–2167. doi: 10.1242/jcs.01073. [DOI] [PubMed] [Google Scholar]
- 101.Chalut KJ, Höpfler M, Lautenschläger F, Boyde L, Chan CJ, Ekpenyong A, Martinez-Arias A, Guck J. Chromatin decondensation and nuclear softening accompany Nanog downregulation in embryonic stem cells. Biophys J. 2012;103(10):2060–2070. doi: 10.1016/j.bpj.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Isermann P, Davidson PM, Sliz JD, Lammerding J. Assays to measure nuclear mechanics in interphase cells. Curr Protoc Cell Biol. 2012;Chapter 22(Unit 22.16) doi: 10.1002/0471143030.cb2216s56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rowat AC, Jaalouk DE, Zwerger M, Ung WL, Eydelnant IA, Olins DE, Olins AL, Herrmann H, Weitz DA, Lammerding J. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J Biol Chem. 2013;288(12):8610–8618. doi: 10.1074/jbc.M112.441535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kha HN, Chen BK, Clark GM, Jones R. Stiffness properties for Nucleus standard straight and contour electrode arrays. Med Eng Phys. 2004;26(8):677–685. doi: 10.1016/j.medengphy.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 105.Vaziri A, Mofrad MR. Mechanics and deformation of the nucleus in micropipette aspiration experiment. J Biomech. 2007;40(9):2053–2062. doi: 10.1016/j.jbiomech.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 106.Rowat AC, Lammerding J, Herrmann H, Aebi U. Towards an integrated understanding of the structure and mechanics of the cell nucleus. Bioessays. 2008;30(3):226–236. doi: 10.1002/bies.20720. [DOI] [PubMed] [Google Scholar]
- 107.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci U S A. 2007;104(40):15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, Goldman RD. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22(24):3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kolb T, Maass K, Hergt M, Aebi U, Herrmann H. Lamin A and lamin C form homodimers and coexist in higher complex forms both in the nucleoplasmic fraction and in the lamina of cultured human cells. Nucleus. 2011;2(5):425–433. doi: 10.4161/nucl.2.5.17765. [DOI] [PubMed] [Google Scholar]
- 110.Goldberg MW, Huttenlauch I, Hutchison CJ, Stick R. Filaments made from A- and B-type lamins differ in structure and organization. J Cell Sci. 2008;121(Pt 2):215–225. doi: 10.1242/jcs.022020. [DOI] [PubMed] [Google Scholar]
- 111.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147(5):913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Puckelwartz MJ, Depreux FF, McNally EM. Gene expression, chromosome position and lamin A/C mutations. Nucleus. 2011;2(3):162–167. doi: 10.1083/jcb.20110104610.4161/nucl.2.3.16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mewborn SK, Puckelwartz MJ, Abuisneineh F, Fahrenbach JP, Zhang Y, MacLeod H, Dellefave L, Pytel P, Selig S, Labno CM, Reddy K, Singh H, McNally E. Altered chromosomal positioning, compaction, and gene expression with a lamin A/C gene mutation. PLoS One. 2010;5(12):e14342. doi: 10.1371/journal.pone.0014342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bridger JM, Foeger N, Kill IR, Herrmann H. The nuclear lamina. Both a structural framework and a platform for genome organization. The FEBS journal. 2007;274(6):1354–1361. doi: 10.1111/j.1742-4658.2007.05694.x. [DOI] [PubMed] [Google Scholar]
- 115.Broers JL, Ramaekers FC. Dynamics of nuclear lamina assembly and disassembly. Symp Soc Exp Biol. 2004;(56):177–192. [PubMed] [Google Scholar]
- 116.Zwerger M, Jaalouk DE, Lombardi ML, Isermann P, Mauermann M, Dialynas G, Herrmann H, Wallrath LL, Lammerding J. Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo-cytoskeletal coupling. Hum Mol Genet. 2013;22(12):2335–2349. doi: 10.1093/hmg/ddt079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Simon DN, Wilson KL. Partners and post-translational modifications of nuclear lamins. Chromosoma. 2013;122(1–2):13–31. doi: 10.1007/s00412-013-0399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhong Z, Wilson KL, Dahl KN. Beyond lamins other structural components of the nucleoskeleton. Methods Cell Biol. 2010;98:97–119. doi: 10.1016/s0091-679x(10)98005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dahl KN, Kalinowski A. Nucleoskeleton mechanics at a glance. J Cell Sci. 2011;124(Pt 5):675–678. doi: 10.1242/jcs.069096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152(6):1376–1389. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rothballer A, Kutay U. The diverse functional LINCs of the nuclear envelope to the cytoskeleton and chromatin. Chromosoma. 2013 doi: 10.1007/s00412-013-0417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126(14):3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- 123.Lee KK, Starr D, Cohen M, Liu J, Han M, Wilson KL, Gruenbaum Y. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol Biol Cell. 2002;13(3):892–901. doi: 10.1091/mbc.01-06-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115(7):825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- 125.Starr DA, Han M. ANChors away: an actin based mechanism of nuclear positioning. J Cell Sci. 2003;116(Pt 2):211–216. doi: 10.1242/jcs.00248. [DOI] [PubMed] [Google Scholar]
- 126.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172(1):41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wagner N, Krohne G. LEM-Domain proteins: new insights into lamin-interacting proteins. Int Rev Cytol. 2007;261:1–46. doi: 10.1016/s0074-7696(07)61001-8. [DOI] [PubMed] [Google Scholar]
- 129.Méjat A, Misteli T. LINC complexes in health and disease. Nucleus. 2010;1(1):40–52. doi: 10.4161/nucl.1.1.10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149(5):1035–1047. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nery FC, Zeng J, Niland BP, Hewett J, Farley J, Irimia D, Li Y, Wiche G, Sonnenberg A, Breakefield XO. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J Cell Sci. 2008;121(Pt 20):3476–3486. doi: 10.1242/jcs.029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Folker ES, Ostlund C, Luxton GW, Worman HJ, Gundersen GG. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc Natl Acad Sci U S A. 2011;108(1):131–136. doi: 10.1073/pnas.1000824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lombardi ML, Lammerding J. Keeping the LINC: the importance of nucleocytoskeletal coupling in intracellular force transmission and cellular function. Biochem Soc Trans. 2011;39(6):1729–1734. doi: 10.1042/bst20110686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci U S A. 1997;94(3):849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129(4):1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sosa BA, Kutay U, Schwartz TU. Structural insights into LINC complexes. Curr Opin Struct Biol. 2013;23(2):285–291. doi: 10.1016/j.sbi.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26(10):3738–3751. doi: 10.1128/mcb.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu Q, Pante N, Misteli T, Elsagga M, Crisp M, Hodzic D, Burke B, Roux KJ. Functional association of Sun1 with nuclear pore complexes. J Cell Biol. 2007;178(5):785–798. doi: 10.1083/jcb.200704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lussi YC, Hügi I, Laurell E, Kutay U, Fahrenkrog B. The nucleoporin Nup88 is interacting with nuclear lamin A. Mol Biol Cell. 2011;22(7):1080–1090. doi: 10.1091/mbc.E10-05-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Razafsky D, Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186(4):461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Apel ED, Lewis RM, Grady RM, Sanes JR. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J Biol Chem. 2000;275(41):31986–31995. doi: 10.1074/jbc.M004775200. [DOI] [PubMed] [Google Scholar]
- 142.Mellad JA, Warren DT, Shanahan CM. Nesprins LINC the nucleus and cytoskeleton. Curr Opin Cell Biol. 2011;23(1):47–54. doi: 10.1016/j.ceb.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 143.Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J Cell Sci. 2002;115(Pt 15):3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- 144.Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci U S A. 2009;106(7):2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Libotte T, Zaim H, Abraham S, Padmakumar VC, Schneider M, Lu W, Munck M, Hutchison C, Wehnert M, Fahrenkrog B, Sauder U, Aebi U, Noegel AA, Karakesisoglou I. Lamin A/C-dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol Biol Cell. 2005;16(7):3411–3424. doi: 10.1091/mbc.E04-11-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mislow JM, Kim MS, Davis DB, McNally EM. Myne-1, a spectrin repeat transmembrane protein of the myocyte inner nuclear membrane, interacts with lamin A/C. J Cell Sci. 2002;115(Pt 1):61–70. doi: 10.1242/jcs.115.1.61. [DOI] [PubMed] [Google Scholar]
- 147.Mislow JM, Holaska JM, Kim MS, Lee KK, Segura-Totten M, Wilson KL, McNally EM. Nesprin-1alpha self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett. 2002;525(1–3):135–140. doi: 10.1016/s0014-5793(02)03105-8. [DOI] [PubMed] [Google Scholar]
- 148.Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K, Han M, Watanabe Y. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J Cell Biol. 2012;198(2):165–172. doi: 10.1083/jcb.201204085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Salpingidou G, Smertenko A, Hausmanowa-Petrucewicz I, Hussey PJ, Hutchison CJ. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J Cell Biol. 2007;178(6):897–904. doi: 10.1083/jcb.200702026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Buch C, Lindberg R, Figueroa R, Gudise S, Onischenko E, Hallberg E. An integral protein of the inner nuclear membrane localizes to the mitotic spindle in mammalian cells. J Cell Sci. 2009;122(Pt 12):2100–2107. doi: 10.1242/jcs.047373. [DOI] [PubMed] [Google Scholar]
- 151.Gudise S, Figueroa RA, Lindberg R, Larsson V, Hallberg E. Samp1 is functionally associated with the LINC complex and A-type lamina networks. J Cell Sci. 2011;124(Pt 12):2077–2085. doi: 10.1242/jcs.078923. [DOI] [PubMed] [Google Scholar]
- 152.Borrego-Pinto J, Jegou T, Osorio DS, Auradé F, Gorjánácz M, Koch B, Mattaj IW, Gomes ER. Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci. 2012;125(Pt 5):1099–1105. doi: 10.1242/jcs.087049. [DOI] [PubMed] [Google Scholar]
- 153.Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329(5994):956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314(8):1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Scherthan H. A bouquet makes ends meet. Nat Rev Mol Cell Biol. 2001;2(8):621–627. doi: 10.1038/35085086. [DOI] [PubMed] [Google Scholar]
- 156.Miki F, Kurabayashi A, Tange Y, Okazaki K, Shimanuki M, Niwa O. Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol Genet Genomics. 2004;270(6):449–461. doi: 10.1007/s00438-003-0938-8. [DOI] [PubMed] [Google Scholar]
- 157.Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125(1):59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 158.Shimanuki M, Miki F, Ding DQ, Chikashige Y, Hiraoka Y, Horio T, Niwa O. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol Gen Genet. 1997;254(3):238–249. doi: 10.1007/s004380050412. [DOI] [PubMed] [Google Scholar]
- 159.Penkner A, Tang L, Novatchkova M, Ladurner M, Fridkin A, Gruenbaum Y, Schweizer D, Loidl J, Jantsch V. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev Cell. 2007;12(6):873–885. doi: 10.1016/j.devcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 160.McGee MD, Stagljar I, Starr DA. KDP-1 is a nuclear envelope KASH protein required for cell-cycle progression. J Cell Sci. 2009;122(Pt 16):2895–2905. doi: 10.1242/jcs.051607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ottaviani A, Schluth-Bolard C, Rival-Gervier S, Boussouar A, Rondier D, Foerster AM, Morere J, Bauwens S, Gazzo S, Callet-Bauchu E, Gilson E, Magdinier F. Identification of a perinuclear positioning element in human subtelomeres that requires A-type lamins and CTCF. Embo J. 2009;28(16):2428–2436. doi: 10.1038/emboj.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gonzalez-Suarez I, Redwood AB, Gonzalo S. Loss of A-type lamins and genomic instability. Cell Cycle. 2009;8(23):3860–3865. doi: 10.4161/cc.8.23.10092. [DOI] [PubMed] [Google Scholar]
- 163.Khatau SB, Bloom RJ, Bajpai S, Razafsky D, Zang S, Giri A, Wu PH, Marchand J, Celedon A, Hale CM, Sun SX, Hodzic D, Wirtz D. The distinct roles of the nucleus and nucleus-cytoskeleton connections in three-dimensional cell migration. Sci Rep. 2012;2:488. doi: 10.1038/srep00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Poh YC, Shevtsov SP, Chowdhury F, Wu DC, Na S, Dundr M, Wang N. Dynamic force-induced direct dissociation of protein complexes in a nuclear body in living cells. Nat Commun. 2012;3:866. doi: 10.1038/ncomms1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Booth-Gauthier EA, Alcoser TA, Yang G, Dahl KN. Force-induced changes in subnuclear movement and rheology. Biophys J. 2012;103(12):2423–2431. doi: 10.1016/j.bpj.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Maniotis AJ, Bojanowski K, Ingber DE. Mechanical continuity and reversible chromosome disassembly within intact genomes removed from living cells. J Cell Biochem. 1997;65(1):114–130. [PubMed] [Google Scholar]
- 167.Ingber DE. Mechanobiology and diseases of mechanotransduction. Ann Med. 2003;35(8):564–577. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- 168.Ingber DE. Cellular mechanotransduction: putting all the pieces together again. FASEB J. 2006;20(7):811–827. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- 169.Moore KA, Polte T, Huang S, Shi B, Alsberg E, Sunday ME, Ingber DE. Control of basement membrane remodeling and epithelial branching morphogenesis in embryonic lung by Rho and cytoskeletal tension. Dev Dyn. 2005;232(2):268–281. doi: 10.1002/dvdy.20237. [DOI] [PubMed] [Google Scholar]
- 170.Lammerding J, Lee RT. The nuclear membrane and mechanotransduction: impaired nuclear mechanics and mechanotransduction in lamin A/C deficient cells. Novartis Found Symp. 2005;264:264–273. discussion 273-268. [PubMed] [Google Scholar]
- 171.Brosig M, Ferralli J, Gelman L, Chiquet M, Chiquet-Ehrismann R. Interfering with the connection between the nucleus and the cytoskeleton affects nuclear rotation, mechanotransduction and myogenesis. Int J Biochem Cell Biol. 2010;42(10):1717–1728. doi: 10.1016/j.biocel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 172.Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature. 2013;497(7450):507–511. doi: 10.1038/nature12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dey P. Nuclear margin irregularity and cancer: a review. Anal Quant Cytol Histol. 2009;31(5):345–352. [PubMed] [Google Scholar]
- 174.Vigouroux C, Auclair M, Dubosclard E, Pouchelet M, Capeau J, Courvalin JC, Buendia B. Nuclear envelope disorganization in fibroblasts from lipodystrophic patients with heterozygous R482Q/W mutations in the lamin A/C gene. J Cell Sci. 2001;114(Pt 24):4459–4468. doi: 10.1242/jcs.114.24.4459. [DOI] [PubMed] [Google Scholar]
- 175.Muchir A, Medioni J, Laluc M, Massart C, Arimura T, van der Kooi AJ, Desguerre I, Mayer M, Ferrer X, Briault S, Hirano M, Worman HJ, Mallet A, Wehnert M, Schwartz K, Bonne G. Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve. 2004;30(4):444–450. doi: 10.1002/mus.20122. [DOI] [PubMed] [Google Scholar]
- 176.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147(5):992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 177.Broers JL, Raymond Y, Rot MK, Kuijpers H, Wagenaar SS, Ramaekers FC. Nuclear A-type lamins are differentially expressed in human lung cancer subtypes. Am J Pathol. 1993;143(1):211–220. [PMC free article] [PubMed] [Google Scholar]
- 178.Moss SF, Krivosheyev V, de Souza A, Chin K, Gaetz HP, Chaudhary N, Worman HJ, Holt PR. Decreased and aberrant nuclear lamin expression in gastrointestinal tract neoplasms. Gut. 1999;45(5):723–729. doi: 10.1136/gut.45.5.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Venables RS, McLean S, Luny D, Moteleb E, Morley S, Quinlan RA, Lane EB, Hutchison CJ. Expression of individual lamins in basal cell carcinomas of the skin. Br J Cancer. 2001;84(4):512–519. doi: 10.1054/bjoc.2000.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tilli CM, Ramaekers FC, Broers JL, Hutchison CJ, Neumann HA. Lamin expression in normal human skin, actinic keratosis, squamous cell carcinoma and basal cell carcinoma. Br J Dermatol. 2003;148(1):102–109. doi: 10.1046/j.1365-2133.2003.05026.x. [DOI] [PubMed] [Google Scholar]
- 181.Hudson ME, Pozdnyakova I, Haines K, Mor G, Snyder M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc Natl Acad Sci U S A. 2007;104(44):17494–17499. doi: 10.1073/pnas.0708572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Willis ND, Wilson RG, Hutchison CJ. Lamin A: a putative colonic epithelial stem cell biomarker which identifies colorectal tumours with a more aggressive phenotype. Biochem Soc Trans. 2008;36(Pt 6):1350–1353. doi: 10.1042/bst0361350. [DOI] [PubMed] [Google Scholar]
- 183.Belt EJ, Fijneman RJ, van den Berg EG, Bril H, Delis-van Diemen PM, Tijssen M, van Essen HF, de Lange-de Klerk ES, Beliën JA, Stockmann HB, Meijer S, Meijer GA. Loss of lamin A/C expression in stage II and III colon cancer is associated with disease recurrence. Eur J Cancer. 2011;47(12):1837–1845. doi: 10.1016/j.ejca.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 184.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9(8):893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 185.Capo-chichi CD, Cai KQ, Simpkins F, Ganjei-Azar P, Godwin AK, Xu XX. Nuclear envelope structural defects cause chromosomal numerical instability and aneuploidy in ovarian cancer. BMC Med. 2011;9:28. doi: 10.1186/1741-7015-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wu Z, Wu L, Weng D, Xu D, Geng J, Zhao F. Reduced expression of lamin A/C correlates with poor histological differentiation and prognosis in primary gastric carcinoma. J Exp Clin Cancer Res. 2009;28:8. doi: 10.1186/1756-9966-28-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Willis ND, Cox TR, Rahman-Casañs SF, Smits K, Przyborski SA, van den Brandt P, van Engeland M, Weijenberg M, Wilson RG, de Bruïne A, Hutchison CJ. Lamin A/C is a risk biomarker in colorectal cancer. PLoS One. 2008;3(8):e2988. doi: 10.1371/journal.pone.0002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Foster CR, Robson JL, Simon WJ, Twigg J, Cruikshank D, Wilson RG, Hutchison CJ. The role of Lamin A in cytoskeleton organization in colorectal cancer cells: a proteomic investigation. Nucleus. 2011;2(5):434–443. doi: 10.4161/nucl.2.5.17775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 190.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerød A, Lee MT, Shen CY, Tee BT, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin SF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van ‘t Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SA, Salomon AV, Børresen-Dale AL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR OBCC. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Doherty JA, Rossing MA, Cushing-Haugen KL, Chen C, Van Den Berg DJ, Wu AH, Pike MC, Ness RB, Moysich K, Chenevix-Trench G, Beesley J, Webb PM, Chang-Claude J, Wang-Gohrke S, Goodman MT, Lurie G, Thompson PJ, Carney ME, Hogdall E, Kjaer SK, Hogdall C, Goode EL, Cunningham JM, Fridley BL, Vierkant RA, Berchuck A, Moorman PG, Schildkraut JM, Palmieri RT, Cramer DW, Terry KL, Yang HP, Garcia-Closas M, Chanock S, Lissowska J, Song H, Pharoah PD, Shah M, Perkins B, McGuire V, Whittemore AS, Di Cioccio RA, Gentry-Maharaj A, Menon U, Gayther SA, Ramus SJ, Ziogas A, Brewster W, Anton-Culver H, Pearce CL Group AOCSM, Cancer) ACS OCAC . ESR1/SYNE1 polymorphism and invasive epithelial ovarian cancer risk: an Ovarian Cancer Association Consortium study. Cancer Epidemiol Biomarkers Prev. 2010;19(1):245–250. doi: 10.1158/1055-9965.epi-09-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Barboro P, Repaci E, D’Arrigo C, Balbi C. The role of nuclear matrix proteins binding to matrix attachment regions (Mars) in prostate cancer cell differentiation. PLoS One. 2012;7(7):e40617. doi: 10.1371/journal.pone.0040617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Malonia SK, Sinha S, Lakshminarasimhan P, Singh K, Jalota-Badhwar A, Rampalli S, Kaul-Ghanekar R, Chattopadhyay S. Gene regulation by SMAR1: Role in cellular homeostasis and cancer. Biochim Biophys Acta. 2011;1815(1):1–12. doi: 10.1016/j.bbcan.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 194.Stein GS, Lian JB, van Wijnen AJ, Stein JL, Javed A, Montecino M, Zaidi SK, Young D, Choi JY, Gutierrez S, Pockwinse S. Nuclear microenvironments support assembly and organization of the transcriptional regulatory machinery for cell proliferation and differentiation. J Cell Biochem. 2004;91(2):287–302. doi: 10.1002/jcb.10777. [DOI] [PubMed] [Google Scholar]
- 195.Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152(6):1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 196.Burke B, Stewart CL. The nuclear lamins: flexibility in function. Nat Rev Mol Cell Biol. 2013;14(1):13–24. doi: 10.1038/nrm3488. [DOI] [PubMed] [Google Scholar]
- 197.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Stoitzner P, Pfaller K, Stössel H, Romani N. A close-up view of migrating Langerhans cells in the skin. J Invest Dermatol. 2002;118(1):117–125. doi: 10.1046/j.0022-202x.2001.01631.x. [DOI] [PubMed] [Google Scholar]
- 199.Weigelin B, Bakker G-J, Friedl P. Intravital third harmonic generation microscopy of collective melanoma cell invasion. Principles of interface guidance and microvesicle dynamics. IntraVital. 2012;1(1):32–43. doi: 10.4161/intv.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Shankar J, Messenberg A, Chan J, Underhill TM, Foster LJ, Nabi IR. Pseudopodial actin dynamics control epithelial-mesenchymal transition in metastatic cancer cells. Cancer Res. 2010;70(9):3780–3790. doi: 10.1158/0008-5472.can-09-4439. [DOI] [PubMed] [Google Scholar]
- 201.Schoumacher M, Goldman RD, Louvard D, Vignjevic DM. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J Cell Biol. 2010;189(3):541–556. doi: 10.1083/jcb.200909113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tong Z, Balzer EM, Dallas MR, Hung WC, Stebe KJ, Konstantopoulos K. Chemotaxis of cell populations through confined spaces at single-cell resolution. PLoS One. 2012;7(1):e29211. doi: 10.1371/journal.pone.0029211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Booth-Gauthier EA, Du V, Ghibaudo M, Rape AD, Dahl KN, Ladoux B. Hutchinson-Gilford progeria syndrome alters nuclear shape and reduces cell motility in three dimensional model substrates. Integr Biol (Camb) 2013;5(3):569–577. doi: 10.1039/c3ib20231c. [DOI] [PubMed] [Google Scholar]
- 204.Verstraeten VL, Ji JY, Cummings KS, Lee RT, Lammerding J. Increased mechanosensitivity and nuclear stiffness in Hutchinson-Gilford progeria cells: effects of farnesyltransferase inhibitors. Aging Cell. 2008;7(3):383–393. doi: 10.1111/j.1474-9726.2008.00382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dahl KN, Scaffidi P, Islam MF, Yodh AG, Wilson KL, Misteli T. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2006;103(27):10271–10276. doi: 10.1073/pnas.0601058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kong L, Schäfer G, Bu H, Zhang Y, Klocker H. Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis. 2012;33(4):751–759. doi: 10.1093/carcin/bgs022. [DOI] [PubMed] [Google Scholar]
- 207.Wandke C, Kutay U. Enclosing chromatin: reassembly of the nucleus after open mitosis. Cell. 2013;152(6):1222–1225. doi: 10.1016/j.cell.2013.02.046. [DOI] [PubMed] [Google Scholar]
- 208.Vargas JD, Hatch EM, Anderson DJ, Hetzer MW. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus. 2012;3(1):88–100. doi: 10.4161/nucl.18954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, Manders EM, Verstraeten VL, van Steensel MA, Marcelis CL, van den Wijngaard A, Vaux DJ, Ramaekers FC, Broers JL. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum Mol Genet. 2011;20(21):4175–4186. doi: 10.1093/hmg/ddr344. [DOI] [PubMed] [Google Scholar]
- 210.McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 2013;23(2):260–269. doi: 10.1101/gr.138032.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, Herrmann H, Blum H, Engelkamp D, Stewart CL, Leonhardt H, Joffe B. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152(3):584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 212.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer. 2013;13(7):497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Polioudaki H, Kourmouli N, Drosou V, Bakou A, Theodoropoulos PA, Singh PB, Giannakouros T, Georgatos SD. Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1. EMBO Rep. 2001;2(10):920–925. doi: 10.1093/embo-reports/kve199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Nili E, Cojocaru GS, Kalma Y, Ginsberg D, Copeland NG, Gilbert DJ, Jenkins NA, Berger R, Shaklai S, Amariglio N, Brok-Simoni F, Simon AJ, Rechavi G. Nuclear membrane protein LAP2beta mediates transcriptional repression alone and together with its binding partner GCL (germ-cell-less) J Cell Sci. 2001;114(Pt 18):3297–3307. doi: 10.1242/jcs.114.18.3297. [DOI] [PubMed] [Google Scholar]
- 215.Somech R, Shaklai S, Geller O, Amariglio N, Simon AJ, Rechavi G, Gal-Yam EN. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J Cell Sci. 2005;118(Pt 17):4017–4025. doi: 10.1242/jcs.02521. [DOI] [PubMed] [Google Scholar]
- 216.Haraguchi T, Holaska JM, Yamane M, Koujin T, Hashiguchi N, Mori C, Wilson KL, Hiraoka Y. Emerin binding to Btf, a death-promoting transcriptional repressor, is disrupted by a missense mutation that causes Emery-Dreifuss muscular dystrophy. Eur J Biochem. 2004;271(5):1035–1045. doi: 10.1111/j.1432-1033.2004.04007.x. [DOI] [PubMed] [Google Scholar]
- 217.Wilkinson FL, Holaska JM, Zhang Z, Sharma A, Manilal S, Holt I, Stamm S, Wilson KL, Morris GE. Emerin interacts in vitro with the splicing-associated factor, YT521-B. Eur J Biochem. 2003;270(11):2459–2466. doi: 10.1046/j.1432-1033.2003.03617.x. [DOI] [PubMed] [Google Scholar]
- 218.Holaska JM, Lee KK, Kowalski AK, Wilson KL. Transcriptional repressor germ cell-less (GCL) and barrier to autointegration factor (BAF) compete for binding to emerin in vitro. J Biol Chem. 2003;278(9):6969–6975. doi: 10.1074/jbc.M208811200. [DOI] [PubMed] [Google Scholar]
- 219.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol. 2011;3(12) doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Di Carlo D, Tse HT, Gossett DR. Introduction: why analyze single cells? Methods Mol Biol. 2012;853:1–10. doi: 10.1007/978-1-61779-567-1_1. [DOI] [PubMed] [Google Scholar]
- 221.Zhang W, Kai K, Choi DS, Iwamoto T, Nguyen YH, Wong H, Landis MD, Ueno NT, Chang J, Qin L. Microfluidics separation reveals the stem-cell-like deformability of tumor-initiating cells. Proc Natl Acad Sci U S A. 2012;109(46):18707–18712. doi: 10.1073/pnas.1209893109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Capo-chichi CD, Cai KQ, Testa JR, Godwin AK, Xu XX. Loss of GATA6 leads to nuclear deformation and aneuploidy in ovarian cancer. Mol Cell Biol. 2009;29(17):4766–4777. doi: 10.1128/mcb.00087-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Agrelo R, Setien F, Espada J, Artiga MJ, Rodriguez M, Pérez-Rosado A, Sanchez-Aguilera A, Fraga MF, Piris MA, Esteller M. Inactivation of the lamin A/C gene by CpG island promoter hypermethylation in hematologic malignancies, and its association with poor survival in nodal diffuse large B-cell lymphoma. J Clin Oncol. 2005;23(17):3940–3947. doi: 10.1200/jco.2005.11.650. [DOI] [PubMed] [Google Scholar]
- 224.Wang Y, Wu R, Cho KR, Thomas DG, Gossner G, Liu JR, Giordano TJ, Shedden KA, Misek DE, Lubman DM. Differential protein mapping of ovarian serous adenocarcinomas: identification of potential markers for distinct tumor stage. J Proteome Res. 2009;8(3):1452–1463. doi: 10.1021/pr800820z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Skvortsov S, Schäfer G, Stasyk T, Fuchsberger C, Bonn GK, Bartsch G, Klocker H, Huber LA. Proteomics profiling of microdissected low- and high-grade prostate tumors identifies Lamin A as a discriminatory biomarker. J Proteome Res. 2011;10(1):259–268. doi: 10.1021/pr100921j. [DOI] [PubMed] [Google Scholar]
- 226.Alfonso P, Cañamero M, Fernández-Carbonié F, Núñez A, Casal JI. Proteome analysis of membrane fractions in colorectal carcinomas by using 2D-DIGE saturation labeling. J Proteome Res. 2008;7(10):4247–4255. doi: 10.1021/pr800152u. [DOI] [PubMed] [Google Scholar]
- 227.Bengtsson S, Krogh M, Szigyarto CA, Uhlen M, Schedvins K, Silfverswärd C, Linder S, Auer G, Alaiya A, James P. Large-scale proteomics analysis of human ovarian cancer for biomarkers. J Proteome Res. 2007;6(4):1440–1450. doi: 10.1021/pr060593y. [DOI] [PubMed] [Google Scholar]
- 228.Lim SO, Park SJ, Kim W, Park SG, Kim HJ, Kim YI, Sohn TS, Noh JH, Jung G. Proteome analysis of hepatocellular carcinoma. Biochem Biophys Res Commun. 2002;291(4):1031–1037. doi: 10.1006/bbrc.2002.6547. [DOI] [PubMed] [Google Scholar]
- 229.Sun S, Xu MZ, Poon RT, Day PJ, Luk JM. Circulating Lamin B1 (LMNB1) biomarker detects early stages of liver cancer in patients. J Proteome Res. 2010;9(1):70–78. doi: 10.1021/pr9002118. [DOI] [PubMed] [Google Scholar]
- 230.Coradeghini R, Barboro P, Rubagotti A, Boccardo F, Parodi S, Carmignani G, D’Arrigo C, Patrone E, Balbi C. Differential expression of nuclear lamins in normal and cancerous prostate tissues. Oncol Rep. 2006;15(3):609–613. [PubMed] [Google Scholar]
- 231.Li L, Du Y, Kong X, Li Z, Jia Z, Cui J, Gao J, Wang L, Xie K. Lamin B1 Is a Novel Therapeutic Target of Betulinic Acid in Pancreatic Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013 doi: 10.1158/1078-0432.ccr-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Somech R, Gal-Yam EN, Shaklai S, Geller O, Amariglio N, Rechavi G, Simon AJ. Enhanced expression of the nuclear envelope LAP2 transcriptional repressors in normal and malignant activated lymphocytes. Ann Hematol. 2007;86(6):393–401. doi: 10.1007/s00277-007-0275-9. [DOI] [PubMed] [Google Scholar]
- 233.Martínez N, Alonso A, Moragues MD, Pontón J, Schneider J. The nuclear pore complex protein Nup88 is overexpressed in tumor cells. Cancer Res. 1999;59(21):5408–5411. [PubMed] [Google Scholar]
- 234.Agudo D, Gómez-Esquer F, Martínez-Arribas F, Núñez-Villar MJ, Pollán M, Schneider J. Nup88 mRNA overexpression is associated with high aggressiveness of breast cancer. Int J Cancer. 2004;109(5):717–720. doi: 10.1002/ijc.20034. [DOI] [PubMed] [Google Scholar]
- 235.Brustmann H, Hager M. Nucleoporin 88 expression in normal and neoplastic squamous epithelia of the uterine cervix. Ann Diagn Pathol. 2009;13(5):303–307. doi: 10.1016/j.anndiagpath.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 236.Emterling A, Skoglund J, Arbman G, Schneider J, Evertsson S, Carstensen J, Zhang H, Sun XF. Clinicopathological significance of Nup88 expression in patients with colorectal cancer. Oncology. 2003;64(4):361–369. doi: 10.1159/000070294. 70294. [DOI] [PubMed] [Google Scholar]
- 237.Gould VE, Orucevic A, Zentgraf H, Gattuso P, Martinez N, Alonso A. Nup88 (karyoporin) in human malignant neoplasms and dysplasias: correlations of immunostaining of tissue sections, cytologic smears, and immunoblot analysis. Human pathology. 2002;33(5):536–544. doi: 10.1053/hupa.2002.124785. [DOI] [PubMed] [Google Scholar]
- 238.Knoess M, Kurz AK, Goreva O, Bektas N, Breuhahn K, Odenthal M, Schirmacher P, Dienes HP, Bock CT, Zentgraf H, zur Hausen A. Nucleoporin 88 expression in hepatitis B and C virus-related liver diseases. World J Gastroenterol. 2006;12(36):5870–5874. doi: 10.3748/wjg.v12.i36.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhang ZY, Zhao ZR, Jiang L, Li JC, Gao YM, Cui DS, Wang CJ, Schneider J, Wang MW, Sun XF. Nup88 expression in normal mucosa, adenoma, primary adenocarcinoma and lymph node metastasis in the colorectum. Tumour Biol. 2007;28(2):93–99. doi: 10.1159/000099154. [DOI] [PubMed] [Google Scholar]
- 240.Xu S, Powers MA. Nuclear pore proteins and cancer. Semin Cell Dev Biol. 2009;20(5):620–630. doi: 10.1016/j.semcdb.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Cervoni N, Detich N, Seo SB, Chakravarti D, Szyf M. The oncoprotein Set/TAF-1beta, an inhibitor of histone acetyltransferase, inhibits active demethylation of DNA, integrating DNA methylation and transcriptional silencing. J Biol Chem. 2002;277(28):25026–25031. doi: 10.1074/jbc.M202256200. [DOI] [PubMed] [Google Scholar]
- 242.Hernández P, Solé X, Valls J, Moreno V, Capellá G, Urruticoechea A, Pujana MA. Integrative analysis of a cancer somatic mutome. Mol Cancer. 2007;6:13. doi: 10.1186/1476-4598-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]