Abstract
A PP2C homolog gene was cloned from the drought-treated cDNA library of Populus euphratica. Multiple sequence alignment analysis suggested that the gene is a potential ortholog of HAB1. The expression of this HAB1 ortholog (PeHAB1) was markedly induced by drought and moderately induced by ABA. To characterize its function in ABA signaling, we generated transgenic Arabidopsis thaliana plants overexpressing this gene. Transgenic lines exhibited reduced responses to exogenous ABA and reduced tolerance to drought compared to wide-type lines. Yeast two-hybrid analyses indicated that PeHAB1 could interact with the ABA receptor PYL4 in an ABA-independent manner. Taken together; these results indicated that PeHAB1 is a new negative regulator of ABA responses in poplar.
Introduction
The phytohormone abscisic acid (ABA) plays a key role in different plant developmental processes as well as in the perception of abiotic stresses such as drought, salt and cold [1,2]. ABA is well established as a mediator of physiological responses to water deficit. Water stress can lead to an increase of ABA level in plants, resulting in various adaptive responses, including stomatal closure and gene expression [3,4]. The ABA signal transduction system involves a complex network of both positive and negative regulators. Previous reports reviewed Clade A PP2C/Protein Phosphatase 2C suppressed the release of signaling triggered by ABA [5,6]. In Arabidopsis, PP2CA, ABI1 and ABI2 were characterized as key negative regulators of ABA signaling [7]. Studies on loss-of-function alleles of ABI1 and ABI2 and the generation of double revealed that ABI1 and ABI2 function partially redundant as inhibitors of ABA signaling [8]. Experimental data also alternatively support that PP2C-like gene could act as a positive regulator rather than a negative regulator of ABA signaling [9], which indicated that PP2C play complicated roles in plants. ABA-related Clade A PP2Cs triggered the regulation of numerous processes by interaction with multiple proteins. Recently, crystal graphic studies revealed the complex structure of PP2C-ABA-PYR1/PYL/RCAR (Pyrabactin Resistance 1 /PYR1-Like /Regulatory component of ABA receptor), among which PYL was identified as an intracellular ABA receptor [10–16]. Ongoing researches also demonstrates that PYR/PYL/RCAR receptors have shown preferences in substrate specificity and selectively inhibit specific PP2Cs [17]. These studies triggered considerable interest in ABA signaling in different plant species.
As the second published genome of poplar species [18], Populus euphratica is a famous poplar species that is mainly distributed in deserts and noted for its high abiotic stress tolerance. However, to our knowledge, only one PP2C member has been characterized in poplar [19], and few studies have focused on interactions between type 2C protein phosphatases and putative ABA receptors in poplar. To test whether PP2C member may be crucial for ABA response in P. euphratica and whether it could function through PP2C-PYL interaction, we cloned a potential HAB1 ortholog gene from P. euphratica and generated Arabidopsis thaliana transgenic lines to characterize the function of this gene in ABA signaling. An attempt also has been made to identify the interacting protein of this HAB1 ortholog gene by screening a cDNA library to illustrate its potential function.
Results
PeHAB1 identification
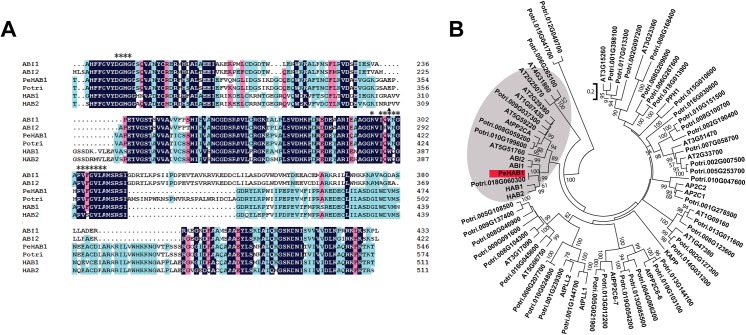
To study the response of drought, a cDNA library was constructed by using leaves of P. eupharatica as the source of mRNA. The titers of primary cDNA library and amplified library were 2.2×106 and 1.2×1010, respectively, and the recombinant was >87%. Over 1500 clones were randomly sequenced. The inserted fragments ranged from 500 bp to 3000 bp. Among the fragments, a putative PP2C member has been screened as full length for five times. According to the BLAST searching method, this sequence was supposed to encode a Clade A PP2C family member and was given a temporary name PeHAB1. Full-length PeHAB1 was then isolated from cDNAs of roots, leaves and stems, indicating that PeHAB1 could be expressed in all these organs. The isolated cDNA fragment contains 1641 bp and encodes a protein with 546 amino acids (Genebank ID: KP055179), with a calculated molecular mass of 58.79 kD and a predicted pI of 4.47. The overall sequence similarity between AtHAB1 and PeHAB1 is 45.08% (Fig 1A). This percentage is reasonable, considering that PP2C did not share similarities in the C-terminal part with their paralogs or orthologs [20]. Certain motifs, such as the PYL interaction site, are highly conserved in PeHAB1 (Fig 1A). We further compiled an alignment that included PeHAB1, 45 annotated PP2C candidates from poplar genome, and 27 representative Arabidopsis PP2Cs, and then constructed a phylogenetic tree. As expected, PeHAB1 was clustered in Clade A with a position close to HAB1 (Fig 1B). Therefore, the P. euphratica gene was designated as PeHAB1.
Fig 1. Sequence analysis of PeHAB1.
(A) Sequence alignment of the deduced amino acid sequence of PeHAB1 with four Clade A Arabidopsis PP2C members including AtABI1, AtABI2, AtHAB1, AtHAB2, and one Populus trichocarpa gene Potri.018G060300. Identical amino acid residues are highlighted in black, while conserved amino acid residues are indicated by colored shading. Asterisk codes indicated the amino acid residues involved in interaction with PYLs. Trp420 was marked with a small triangle. (B) Phylogenetic tree of PeHAB1, 27 representative PP2C members from Arabidopsis, and 45 PP2C candidates from P. trichocarpa. The ABA-related Clade A PP2Cs were indicated by the grey shade. The tree was constructed by Neighbor–Joining method based on multiple alignments of full-length amino acid sequences with bootstrap values of 1000 replicates.
Expression pattern of four annotated ABA-related PP2Cs from Populus tricocarpa
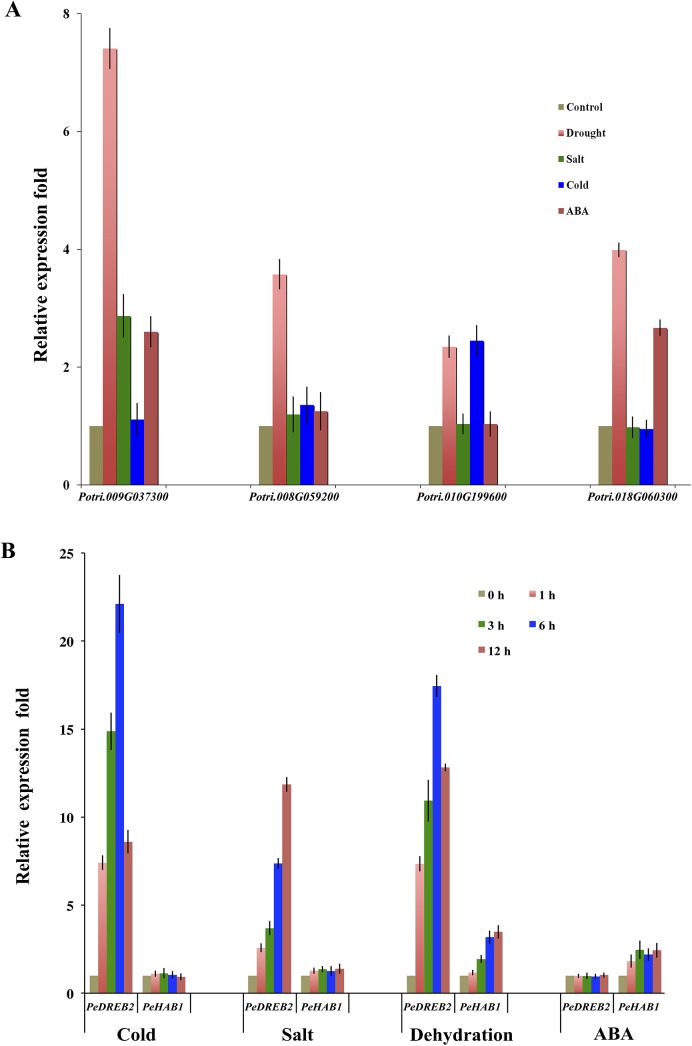
Our qPCR analysis indicated that all the Clade A PtPP2C candidates were differentially expressed in response to various abiotic stress treatments. In general, all four genes involved in analysis were induced by drought treatment, and the highest expression level was observed in Potri.009G037300. Besides drought treatment, Potri.009G037300 also showed a high induction under salt treatment, and Potri.010G199600 was seen to be accumulated under cold treatment. The accumulation of Potri.018G060300 increased after both drought and ABA treatments, whereas Potri.008G059200 was the member whose expression was alerted only after drought treatment (Fig 2A).
Fig 2. Real-time PCR analysis of several Clade A PtPP2Cs and PeHAB1 under ABA, NaCl, drought, and cold treatment.
(A) Relative gene expression analysis of four Clade A PtPP2Cs under various abiotic stress treatments. (B) Quantitative RT-PCR was performed with PeActin as an internal reference and PeDREB2 as an experimental control. The expression level in untreated leaves was assigned a value of 1. The capital characters represent 200 μM ABA (ABA), 300 mM NaCl (S), dehydration (D), and 4°C cold, respectively. Data are presented as means ± SE from three independent biological replicates.
PeHAB1 is induced by dehydration and ABA
The expression level of PeHAB1 under different abiotic stresses was determined by quantitative real-time PCR analyses, with PeActin as a housekeeping gene. A multiple-abiotic-stress- inducible gene, PeDREB2, was introduced in real-time PCR analysis as an experimental control [21]. The results indicated that PeHAB1 accumulates its transcripts after ABA and drought treatments, with a relative higher accumulation under drought treatment (Fig 2B). The expression of PeHAB1 was not significantly stimulated by high salt and cold stresses. Gene expression patterns often illustrate the gene function. Thus, our results suggested that PeHAB1 may participate in ABA or drought responses.
PeHAB1 interacts with abscisic acid receptor PePYL4
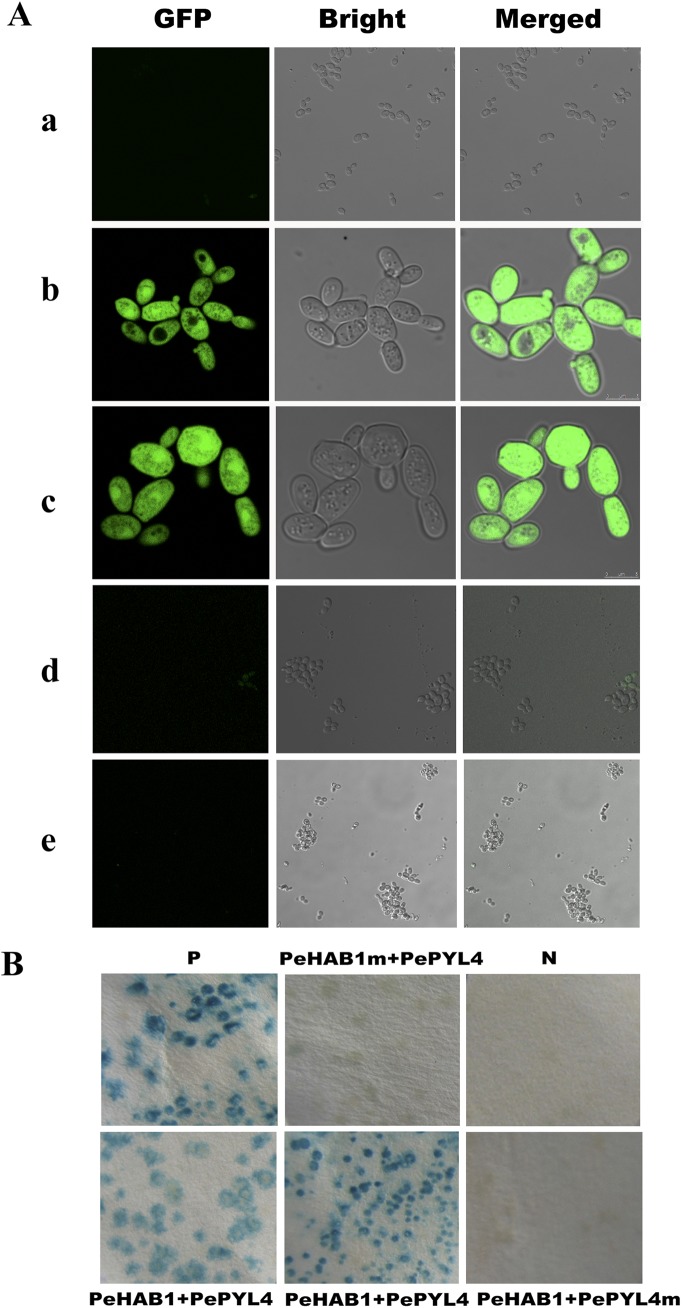
A new yeast-two-hybrid method, with PeHAB1 protein as bait, was applied to search the potential PeHAB1-interacting proteins in P. euphratica. After transformation with the prey cDNA library of P. euphratica, dozens of putative clones were screened from the total of 1 × 106 recombinant colonies. To discriminate true positive clones from spontaneous yeast mutations, we retransformed the yeast reporter strain with the cDNA of each potential candidate. Finally, five positives clones encoding the same gene were retrieved. The isolated cDNA encompassed an open reading frame (ORF) of 666 bp with a predicted protein of 221 amino acids (Genebank ID: KP064303, S1A Fig). A BLAST search against the NCBI database indicated that the cDNA belongs to the RCAR/PYR/PYL gene family. It shares 98% identity with Potri.006G104100 from P. trichocarpa, 82% identity of KDP22835.1 from Jatropha curcas, and 65% identity of AtPYL4 from Arabidopsis at the amino acid level. The gene contains conserved gate region and latch region, which suggested that the gate-latch-lock mechanism is employed in its hormone binding [22]. The phylogenic tree that was constructed by using all Arabidopsis PYLs indicates that its most likely orthologous gene is PYL4 (S1B Fig). Thus, this gene was hereby named PePYL4. We further confirmed the interaction between PeHAB1 and PePYL4 by both MoBiTec Grow’n’Glow and GAL4 Clontech systems, and the results showed that PeHAB1 can interact with PePYL4 without ABA supplementation (Fig 3A).
Fig 3. Yeast two-hybrid assay for PeHAB1- interacting proteins.
(A) Yeast two-hybrid interaction test of PeHAB1 with PePYL4. Interactions were detected by growth of yeast on selective medium and the detection of a GFP signal. (a-d) indicated negative control; yeast harbored PeHAB1 and PePYL4; Supplied interacting pairs that served as positive control; yeast harbored PePYL4 and PeHAB1 of substitution mutation W420A and yeast harbored PeHAB1 and PePYL4 of substitution mutation P124S. (B) β-galactosidase activity (blue) in streaks of yeast cells on a culture plate as readouts of protein-protein interactions by using the yeast two-hybrid system.
Certain PP2C members have been reported to interact with PYL through the region around its active site and a flap sub-domain [22,23]. The critical active site residues of Arg199, Glu203, Asp204, Asp243, Gly244, His245, Asp432, and Asp492, which were previously reported in Arabidopsis, were conserved in PeHAB1 [23,24], corresponding to the residues of Arg242, Glu246, Asp247, Asp288, Gly289, His290, Asp465, and Asp527, respectively (Fig 1). Previous reports have established a critical tryptophan residue that can be bound by PYLs and/or ABA by pointing into the hydrophobic pocket of PYLs [12,22]. Thus, we introduced a mutation to the corresponding residue of Try420 in PeHAB1 by substituting Try to Ala. As a result, W420A mutate showed a remarkable reduction of affinity to PePYL4 (Fig 3). We also introduced a P124S mutation to PeHAB1, which resulted in a disability of affinity. These results indicated that PeHAB1 may interact with PePYL4 using the gate-latch-lock mechanism.
Overexpression of PeHAB1 reduces ABA sensitivity
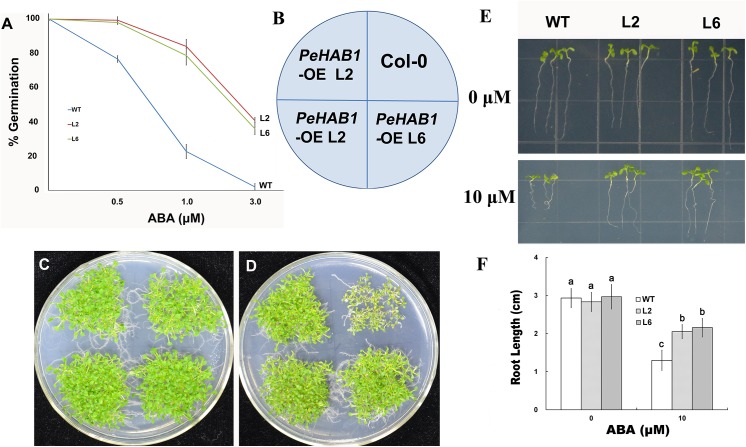
To investigate whether sustained transcriptional overexpression of PeHAB1 affects ABA sensitivity, we analyzed two homozygous lines with similar expression levels of PeHAB1 by performing assays on the germination and early seeding development of ABA treated seeds. For germination assay, seeds were planted in MS medium supplemented with different concentrations of exogenous ABA. As a result, 35S::PeHAB1 homozygous lines showed similar germination ratios to wild-type in the absence of ABA. However, the germination ratio of wide-type plants was strongly inhibited and only less inhibitory effect was found in transgenic lines after ABA was added (Fig 4A). We also tested the germination rate and development performance at an ABA concentration of 0.75 μM. As expected, PeHAB1 overexpressing lines showed a higher frequency of seedlings with green cotyledons and a faster growth rate (Fig 4B and 4D).
Fig 4. Constitutive expression of PeHAB1confers ABA insensitivity in transgenic plants during seed germination and root growth development.
(A) The emergence rate of green cotyledons from Col-0 and PeHAB1 transgenic seeds plated on MS medium supplemented with different concentration of ABA. Approximately 100 seeds were used in each experiment. Error bars represent the standard deviation. (B-D) Col-0 and two independent 35S::PeHAB1 lines were germinated and grown in medium (MS) supplemented with 0.75 μM ABA. Photographs were taken after 14 days. (E-F) Root length in MS medium supplemented with 10 μM ABA and its correspondingly statistical analysis. Data means ±SE from three independent experiments (n = 30 seeds).
Root growth is usually considered as a standard criterion in evaluating the ABA sensitivity of plants because of the inhibitory role of ABA plays on roots [25]. To test the sensitivity of root in response to exogenous ABA, we transferred 5-day-old seedlings to solid MS media supplemented with 10 μM ABA and then incubated plates in a vertical position in a growth chamber. As a result, a significant reduction of root growth in wild-type plants was observed. However, the phenotypes of transgenic lines did not considerably change with exogenous ABA (Fig 4E and 4F), indicating that PeHAB1 overexpression reduced the sensitivity of the root to ABA.
Overall, the overexpression of PeHAB1 resulted in an ABA-insensitive phenotype with ABA-mediated inhibition of seedling germination and root growth. This result indicated that PeHAB1 was a negative regulator of ABA signaling.
Overexpression of PeHAB1 enhances water loss of plants
Transpiration rate assays showed that detached leaves of PeHAB1-overexpressing plants lose water more quickly in comparison with those of wild-type plants (Fig 5A; Student’s t-test), indicating a reduced tolerance of transgenic plants to drought stress. To assess whether the drought tolerance reduction was related to an alteration in sensitivity to ABA, we measured the effect of ABA on stomatal aperture. The results showed a significant difference of stomatal aperture between PeHAB1 overexpressing lines and wild-type under 10 μM ABA (Fig 5B and 5C), indicating that ABA-triggered stomatal closure is blocked in transgenic lines, which may result in an enhanced transpiration rate afterwards in PeHAB1 overexpressing lines.
Fig 5. Phenotype tests for water loss and stomata opening under ABA treatment.
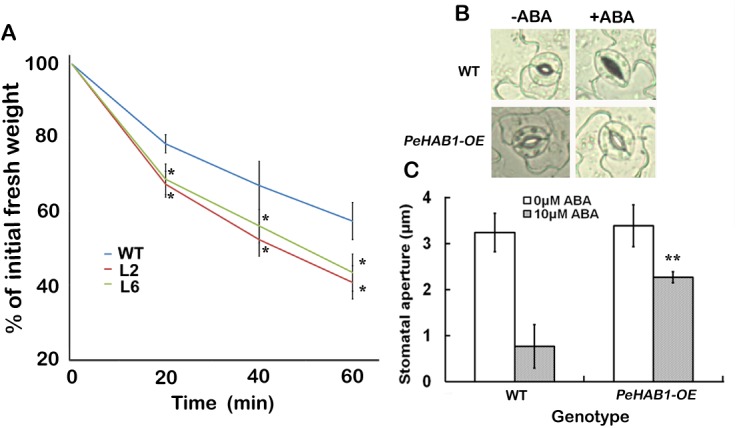
(A) Water loss analysis. Time courses for the percentage of initial fresh weight were recorded from the detached leaves of wild-type controls, L2, and L6. Data are averages ±SE from three independent experiments (n = 10 plants). Asterisks indicate significant difference between transgenic lines and wild-type at different time points (* P<0.05; Student’s t-test). (B) Stomatal photographs of 35S::PeHAB1 transgenic lines and wild-type plants in the presence or absence of ABA. (C) Average stomatal aperture of PeHAB1 overexpression transgenic lines and wild type plants in response to ABA. Data are averages ±SE from three independent experiments (n > 20 stomata). Asterisks indicate significant difference between transgenic lines and wild-type with or without ABA (** P<0.01; Student’s t-test).
Discussion
Notable progress has been recently made in ABA signaling by identifying the Arabidopsis PYR/PYL/RCAR gene family as ABA receptors that inhibit group-A PP2C activity in response to ABA. PYL-PP2C interaction has been identified in organisms, such as Artemisia annua, Oryza sativa, and soybean, by experimental analysis [25]. To explore ABA components in P. euphratica, we cloned a putative PP2C gene by cDNA library screening method and identified a PP2C-PYL interaction by using this PP2C to screen a yeast cDNA library.
PeHAB1 gained its name because of its high sequence similarity to AtHAB1. BLAST research analysis based on the poplar database revealed more than 76 PP2Cs in poplar. Thus, we preferred the overexpression method instead of constructing knock-down or knock-out lines to avoid the defect of mutation analysis that may be caused by functional redundancy. In Arabidopsis, the phenotype of hab1-1 plants and HAB1-overexpressing plants are consistent, showing the function of HAB1 as a negative regulator of ABA signaling [26–28]. Here, ectopic overexpression of PeHAB1 in Arabidopsis showed an ABA-insensitive phenotype, including reduction of the negative effects of ABA on germination and root growth, as well as the reduction of the positive effects that ABA signaling normally has on mounting appropriate responses to drought/dehydration stress. All these results indicated a negative regulator PeHAB1 function in ABA signaling. Very recently, a predicted PP2CA ortholog named G059200 was cloned and functionally characterized in poplar [29]. The performance of G059200 transgenic lines exhibited reduced responses to exogenous ABA and reduced tolerance to salt stress, indicating that the ABA signal mediated by Clade A PP2Cs may be conserved in poplar. However, whether the pathway of PYL-PP2C interactions is conserved remain unknown. With the question of which PP2C member may be crucial for ABA response and whether the member could function through physical interaction with the PYL protein, we focused our study on a PP2C member that was found in a drought-treated RNA library. The expression of PeHAB1 is induced by drought and ABA but not by high salt. This expression pattern is similar to BolABI1 [30]. We compared the expression pattern of PeHAB1 with several Clade A PP2Cs retrieved from P. trichocarpa, and found its accumulation pattern is similar to that of Potri.018G060300. Sequence alignment revealed that PeHAB1 and Potri.018G060300 exhibited 87.07% identity at the nucleic acid level and 92.16% identity at the amino acid level. Thus, they were supposed to be orthologs.
PP2C may interact with specific PYL members with or without ABA supplementation. For example, AtHAB1 interacted with AtPYL5 without the added ABA, and AtHAI3 exhibited strong interaction with AtPYL7 and weak interaction with AtHAI1 [31]. Here, we proved that PeHAB1 could interact with PePYL4 in the absence of ABA. However, PeHAB1 exhibited low-affinity with PePYL4 through the introduction of Trp420Ala mutation, indicating that the tryptophan residue located in the flap region of PeHAB1 is important for PYL4 interaction. Moreover, P124S mutation in the gate region of PePYL4 will destroy the affinity with PeHAB1 to a considerable extent, and this result is similar to that of AaPYL9 [25]. These results provided evidence that PeHAB1 may achieve its function of negative regulation in ABA signaling by interacting specifically with PYL(s) through gate-latch-lock mechanism.
ABA-agonists have the possibility to improve plant yield or any other properties under advance abiotic stress. Therefore, identification of ABA-agonists is important as a fundamental research [24]. We have previously identified several abiotic stress regulators in P. euphratica, certain of which are involved in ABA signaling (data not shown). However, core ABA signaling components have not been identified in P. euphratica. Characterization of PP2C genes may explore the ABA signaling study in this high abiotic stress tolerant tree.
Materials and Methods
Ethics approval
All P. euphratica seeds were collected in a public area where a natural P. euphratica population distributed along the Tarim River (40°51′N, 80°10′ E; Altitude, 972 m, 100–1500 m away from the river). P. euphratica is a protected species, but no permission for seed collection is required in China. The experiments for seed germination and seedling growth were conducted in the National Engineering Laboratory for Tree Breeding Lab (40°0′9′′ N, 80°20′15′′ E). This laboratory is owned and managed by Beijing Forestry University.
Plant materials and treatments
Two-year-old P. euphratica seedlings were planted in individual pots (15 L) containing loam soil and placed in a greenhouse at Beijing Forestry University. Arabidopsis (Columbia ecotype) used for transformation were grown in 9-cm pots filled with customized soil mixture under cool-white light conditions (16 h light/8 h dark, 18,500 lux, 22°C, 70% relative humidity) until the flowers bloomed. For ABA and NaCl treatment, plants of P. euphratica were sprayed with 200 μM ABA solution or 300 μM NaCl. For drought treatment, P. euphratica were transferred from well-saturated soil to filter paper. Cold treatment was conducted by transferring young plants to a growth chamber set at 4°C under a 16 light/8 dark cycle. For all treatments, leaves were collected at the points of 0, 1, 3, 6, and 12 h and immediately frozen in liquid nitrogen. One-year-old seedlings of P. tricorcarpa were used to detect the expression levels of group A PP2C poplar candidates and the treatments were performed as described above.
cDNA library construction
SMART cDNA library kit (Clontech, CA) was employed to construct a cDNA library for P. euphratica. Total RNA was extracted from drought-treated leaves collected at 3, 6, 9 h from two-year-old P. euphratica by using CTAB method [32]. After removing the contaminating genomic DNA with turbo DNA-free kit (Ambion), Oligo dT cellulose chromatography was used to enrich mRNA. After purification, cDNA was normalized and digested with SfiI, ligated into the SfiI predigested DNR-LIB vector, and transformed into Escherichia coli DH10B.The cDNA library was plated on LB plates with X-gal, isopropyl-D-thiogalactopyranoside and ampicillin. Approximately 1500 white colonies were randomly picked and sequenced.
PP2C gene searching and PeHAB1 isolation
Nucleotide homology searching was conducted by using BLAST program on Phytozome network. We define a match if a putative PP2C hit with an E-value ≤ 1e-10. Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html) combined with Popgenie databases (http://popgenie.org/) were also used to search for PP2C genes in poplar. The full-length cDNA was isolated by polymerase chain reaction (PCR) with the primers 5’-ATGGAGGAGATGTATCCGG-3’ and 5’-TCATGTTTTGGTTTTGAACTTCC-3’. The phylogenetic tree was constructed by neighbor-joining bootstrap method (bootstrap analysis with 1,000 replicates). We used MEGA 6.0 software [33] based on multiple alignments of the amino acid sequences of 27 representative AtPP2Cs retrieved from the TAIR database (https://www.arabidopsis.org/) and all annotated poplar PP2Cs retrieved from Phytozome and Popgenie databases.
Real-time quantitative reverse transcription PCR analysis
Quantitative PCR (qPCR) was performed to determine the gene expression level of ABA-related group A PP2C candidates from P. trichocarpa and PeHAB1 isolated from the drought-treated cDNA library of P. euphratica. qPCR was conducted using a power SYBR Green PCR Kit (Applied Biosystems) with a StepOnePlus™ real-time PCR system [34]. The relative quantification value was calculated by the 2−ΔΔCt method using respective PtEF1a and PeActin as internal control [35,36]. A multiple abiotic stress inducible gene PeDREB2 was employed as experimental control in PeHAB1 expression pattern analysis. All primers used in qPCR analysis are given in S1 Table. Each PCR assay was conducted for three biological replicates, and for each replicate, three technological replicates were repeated.
Identification of PeHAB1-interacting proteins in poplar by yeast two-hybrid screening
The MoBiTec Grow'n'Glow two-hybrid system (MoBiTec, Goettingen, Germany) based on the Brent LexA Interaction Trap method was applied using the PeHAB1 as a bait [37]. Full-length PeHAB1 were cloned into the pEG202 vector to fuse to the LexA DNA-binding domain. To moderate the inherent transactivation potential of full-length PeHAB1, we utilized the low sensitivity Saccharomyces cerevisiae strain EGY188, which contains two copies of the LexA operator upstream of the LEU2 reporter integrated in the yeast genome. pGNG1 vector was used as the reporter plasmid with a URA3 selectable marker and the GFP reporter gene, allowing for simple screening by UV transillumination. A pJG4-5 plasmid containing TRP1 selectable marker and SV40 unclear localization sequence inserted with P. euphratica cDNAs in the EcoRI and XhoI sites was employed in the screening. Plasmids were sequentially transformed into EGY188 by lithium acetate method. Colonies grown in DOBA-HIS-LEU-URA-TRP plate and showing green fluorescence when exposed to standard UV-light in darkroom were proposed to be putative colonies.
Interaction confirmation
Potential positive transformants were confirmed by retransforming the yeast reporter strain with the cDNA of each potential candidate, streaking onto new DOBA-HIS-LEU-URA-TRP plates, and incubation for 24–72 h at 30°C. Colonies colonies were re-incubated in liquid medium and examined by a confocal microscope (Leica TCS SP5 II). The fusion of LexA and mouse p53 (AA 72–390) protein was served as positive bait control with the prey control vector pJG4-5-LTA which carries the gene for LTA, a protein known to interact with p53. GAL4-based two-hybrid assay was also used to verify interaction using pGBKT7-PeHAB1 as bait and pGADT7-PePYL4 as prey according to the Clontech’s protocol. For colonies containing different bait and prey plasmids, β-galactosidase activity was analyzed by colony-fit filter assay.
Vector construction and plant transformation
Full-length PeHAB1 cDNA was inserted into the pBI121 vector downstream of the 35S promoter of Cauliflower mosaic virus (CaMV) to construct the plasmids for transformation. The plasmid was introduced into Agrobacterium tumefaciens strain GV3101. Plants were then transformed by using floral-dip method [38]. The pBI121 vector was also transformed as a control. The presence of T-DNA in transgenic plants was confirmed by kanamycin selection as well as PCR detection of the NPTII gene and HAB1 gene, as in a previous report [34].
Germination assay
To determine the sensitivity of seedling inhibition established by ABA, a germination assay was performed. Approximately 100 seeds from each genotype were sterilized and sown on MS medium in triplicate. The medium contained 3% sucrose and 0.8% agar at pH 5.8 and was supplemented with different concentrations of ABA (0, 0.5, 1, and 3 μM ABA). Seeds were incubated at 4°C for 3 days before being placed at 22°C. The percentage of seeds that had germinated and developed fully green expanded cotyledons was determined, and germination was scored at the indicated time points. The germination and development for seeds in the medium with 0.75 μM ABA concentration were also recorded.
Root growth assays
Root growth assay was also conducted to score ABA sensitivity. Sterilized seeds of T3 homozygous lines were first germinated and grown on MS agar medium for 5 days, and then transferred onto MS plates with or without 10 μM ABA. Plates were incubated in a vertical position in the growth chamber, and root length was investigated 5 days later.
Transpiration rate and stomatal aperture measurements
For transpiration rate measurement, leaves were detached from 3-week-old plants, weighed immediately on a piece of weighing paper, and then subjected to air-drying (the aerial relative humidity was 40%, and the temperature was between 22 and 23°C). Weights were measured for 10 plants per line at the designated time points of 20, 40, and 60 min. The percentage of initial fresh weight was calculated at the indicated periods of time. Stomatal aperture was measured as in a previous work [34].
Supporting Information
(A) cDNA and deduced amino acid Sequence of PePYL4. (B) Phylogenetic tree of PePYL4 and PYR/PYL/RCAR proteins from Arabidopsis. The tree was constructed by Neighbor–Joining method based on multiple alignments of the full-length amino acid sequences. The numbers next to each node give bootstrap values for 1000 replicates.
(TIF)
(XLSX)
Acknowledgments
This work was supported by National Natural Science Foundation of China (31370597 and 31100492) and Beijing Higher Education Young Elite Teacher Project (YETP0754).
Data Availability
All relevant data are within the paper or Supporting Information files.
Funding Statement
This work was supported by National Natural Science Foundation of China (31370597 and 31100492) and Beijing Higher Education Young Elite Teacher Project (YETP0754). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lee SC, Luan S (2012) ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ 35: 53–60. 10.1111/j.1365-3040.2011.02426.x [DOI] [PubMed] [Google Scholar]
- 2. Qin F, Shinozaki K, Yamaguchi-Shinozaki K (2011) Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol 52: 1569–1582. 10.1093/pcp/pcr106 [DOI] [PubMed] [Google Scholar]
- 3. Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, et al. (2002) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2: 282–291. [DOI] [PubMed] [Google Scholar]
- 4. Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, et al. (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis . Plant Cell Physiol 50: 2123–2132. 10.1093/pcp/pcp147 [DOI] [PubMed] [Google Scholar]
- 5. Fuchs S, Grill E, Meskiene I, Schweighofer A (2013) Type 2C protein phosphatases in plants. FEBS J 280: 681–693. 10.1111/j.1742-4658.2012.08670.x [DOI] [PubMed] [Google Scholar]
- 6. Tougane K, Komatsu K, Bhyan SB, Sakata Y, Ishizaki K, Yamato KT, et al. (2010) Evolutionarily conserved regulatory mechanisms of abscisic acid signaling in land plants: characterization of ABSCISIC ACID INSENSITIVE1-like type 2C protein phosphatase in the liverwort Marchantia polymorpha . Plant Physiol 152: 1529–1543. 10.1104/pp.110.153387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xue T, Wang D, Zhang S, Ehlting J, Ni F, Jakab S, et al. (2008) Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis . BMC Genomics 9: 550 10.1186/1471-2164-9-550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, et al. (2009) Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol 150: 1345–1355. 10.1104/pp.109.137174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reyes D, Rodriguez D, Gonzalez-Garcia MP, Lorenzo O, Nicolas G, García-Martínez JL et al. (2006) Overexpression of a protein phosphatase 2C from beech seeds in Arabidopsis shows phenotypes related to abscisic acid responses and gibberellin biosynthesis. Plant Physiol 141: 1414–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, et al. (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis . Plant J 61: 290–299. 10.1111/j.1365-313X.2009.04054.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, et al. (2009) Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol 16: 1230–1236. 10.1038/nsmb.1730 [DOI] [PubMed] [Google Scholar]
- 12. Miyakawa T, Fujita Y, Yamaguchi-Shinozaki K, Tanokura M (2013) Structure and function of abscisic acid receptors. Trends Plant Sci 18: 259–266. 10.1016/j.tplants.2012.11.002 [DOI] [PubMed] [Google Scholar]
- 13. Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soon FF, Ng LM, Zhou XE, West GM, Kovach A, Tan MH, et al. (2012) Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335: 85–88. 10.1126/science.1215106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, et al. (2009) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668. 10.1038/nature08591 [DOI] [PubMed] [Google Scholar]
- 16. Ma Y (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1266–1266. [DOI] [PubMed] [Google Scholar]
- 17. Ludwików A (2015) Targeting proteins for proteasomal degradation–a new function of Arabidopsis ABI1 protein phosphatase 2C. Front in Plant Sci 6: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma T, Wang J, Zhou G, Yue Z, Hu Q, Chen Y, et al. (2013) Genomic insights into salt adaptation in a desert poplar. Nat Commun 4:2797 10.1038/ncomms3797 [DOI] [PubMed] [Google Scholar]
- 19. Muhammad AJ, Mattsson (2014) A putative poplar PP2C-encoding gene negatively regulates drought and abscisic acid responses in transgenic Arabidopsis thaliana . TREES 28: 531–543. [Google Scholar]
- 20. Romero P, Lafuente MT, Rodrigo MJ (2012) The Citrus ABA signalosome: identification and transcriptional regulation during sweet orange fruit ripening and leaf dehydration. J Exp Bot 63: 4931–4945. 10.1093/jxb/ers168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen J, Xia X, Yin W (2009) Expression profiling and functional characterization of a DREB2-type gene from Populus euphratica . Biochem Biophys Res Commun 378: 483–487. 10.1016/j.bbrc.2008.11.071 [DOI] [PubMed] [Google Scholar]
- 22. Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, et al. (2009) Structural basis of abscisic acid signalling. Nature 462: 609–614. 10.1038/nature08583 [DOI] [PubMed] [Google Scholar]
- 23. Zhang F, Fu X, Lv Z, Shen Q, Yan T, Jiang W, et al. (2014) Type 2C Phosphatase 1 of Artemisia annua L. Is a Negative Regulator of ABA Signaling. Biomed Res Int 2014: 521794 10.1155/2014/521794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Santiago J, Dupeux F, Betz K, Antoni R, Gonzalez-Guzman M, Rodriguez L, et al. (2012) Structural insights into PYR/PYL/RCAR ABA receptors and PP2Cs. Plant Sci 182: 3–11. 10.1016/j.plantsci.2010.11.014 [DOI] [PubMed] [Google Scholar]
- 25. Zhang F, Lu X, Lv Z, Zhang L, Zhu M, Jiang W, et al. (2013) Overexpression of the Artemisia orthologue of ABA receptor, AaPYL9, enhances ABA sensitivity and improves artemisinin content in Artemisia annua L. PLoS One 8: e56697 10.1371/journal.pone.0056697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, et al. (2004) Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J 37: 354–369. [DOI] [PubMed] [Google Scholar]
- 27. Saez A, Robert N, Maktabi MH, Schroeder JI, Serrano R, Rodriguez PL (2006) Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol 141: 1389–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arshad M, Mattsson J (2014) A putative poplar PP2C-encoding gene negatively regulates drought and abscisic acid responses in transgenic Arabidopsis thaliana . TREES 28: 531–543. [Google Scholar]
- 30. Yuan FF, Wang MY, Hao HM, Zhang YF, Zhao HX, Xie CG (2013) Negative regulation of abscisic acid signaling by the Brassica oleracea ABI1 ortholog. Biochem Biophys Res Commun 442: 202–208. 10.1016/j.bbrc.2013.11.035 [DOI] [PubMed] [Google Scholar]
- 31. Bhaskara GB, Nguyen TT, Verslues PE (2012) Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol 160: 379–395. 10.1104/pp.112.202408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116. [Google Scholar]
- 33. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen J, Xue B, Xia X, Yin W (2013) A novel calcium-dependent protein kinase gene from Populus euphratica, confers both drought and cold stress tolerance. Biochem Biophys Res Commun 441: 630–636. 10.1016/j.bbrc.2013.10.103 [DOI] [PubMed] [Google Scholar]
- 35. Wang HL, Chen J, Tian Q, Wang S, Xia X, Yin W (2014) Identification and validation of reference genes for Populus euphratica gene expression analysis during abiotic stresses by quantitative real-time PCR. Physiol Plant 152: 529–545. 10.1111/ppl.12206 [DOI] [PubMed] [Google Scholar]
- 36. Xu M, Zhang B, Su XH, Zhang SG, Huang MR (2011) Reference gene selection for quantitative real-time polymerase chain reaction in Populus . Analy Biochem 408: 337–339. [DOI] [PubMed] [Google Scholar]
- 37. Lasi M, Pauly B, Schmidt N, Cikala M, Stiening B, Käsbauer T, et al. (2010) The molecular cell death machinery in the simple cnidarian Hydra includes an expanded caspase family and pro-and anti-apoptotic Bcl-2 proteins. Cell Res 20: 812–825. 10.1038/cr.2010.66 [DOI] [PubMed] [Google Scholar]
- 38. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) cDNA and deduced amino acid Sequence of PePYL4. (B) Phylogenetic tree of PePYL4 and PYR/PYL/RCAR proteins from Arabidopsis. The tree was constructed by Neighbor–Joining method based on multiple alignments of the full-length amino acid sequences. The numbers next to each node give bootstrap values for 1000 replicates.
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the paper or Supporting Information files.