Abstract
Currently in the field of vascular biology, the role of epigenetics in endothelial cell biology and vascular disease has attracted more in-depth study. Using both in vitro and in vivo models of blood flow, investigators have recently begun to reveal the underlying epigenetic regulation of endothelial gene expression. Recently, our group, along with two other independent groups, have demonstrated that blood flow controls endothelial gene expression by DNA methyltransferases (DNMT1 and 3A). Disturbed flow (d-flow), characterized by low and oscillating shear stress (OS), is pro-atherogenic and induces expression of DNMT1 both in vivo and in vitro. D-flow regulates genome-wide DNA methylation patterns in a DNMT-dependent manner. The DNMT inhibitor 5-Aza-2’deoxycytidine (5Aza) or DNMT1 siRNA reduces OS-induced endothelial inflammation. Moreover, 5Aza inhibits the development of atherosclerosis in ApoE-/- mice. Through a systems biological analysis of genome-wide DNA methylation patterns and gene expression data, we found 11 mechanosensitive genes which were suppressed by d-flow in vivo, experienced hypermethylation in their promoter region in response to d-flow, and were rescued by 5Aza treatment. Interestingly, among these mechanosensitive genes, the two transcription factors HoxA5 and Klf3 contain cAMP-response-elements (CRE), which may indicate that methylation of CRE sites could serve as a mechanosensitive master switch in gene expression. These findings provide new insight into the mechanism by which flow controls epigenetic DNA methylation patterns, which in turn alters endothelial gene expression, regulates vascular biology, and induces atherosclerosis. These novel findings have broad implications for understanding the biochemical mechanisms of atherogenesis and provide a basis for identifying potential therapeutic targets for atherosclerosis.
Keywords: Epigenetic DNA methylation, DNMT, gene expression, flow, shear stress, endothelial function, atherosclerosis
1. Blood Flow Regulates Endothelial Cell Biology and Atherosclerosis
Atherosclerosis, an inflammatory disease of the arterial wall, is the major cause of heart attack, stroke, and peripheral arterial disease. Atherosclerosis typically occurs in curves or branches in the vasculature due to the effect of disturbed blood flow (d-flow) on endothelial gene expression, which naturally arises as a consequence of the geometry and is known to induce endothelial cell (EC) dysfunction.
Blood flow generates shear stress on vascular endothelial cells. Unidirectional, laminar shear stress (LS, or s-flow) is crucial for normal vascular function, whereas d-flow, characterized as low and oscillatory shear stress (OS), causes vascular dysfunction and disease (1-5). When compared to s-flow, ECs have drastically altered gene expression patterns both in vivo and in vitro (6-11). Atherosclerosis preferentially develops in areas of d-flow, where the dysfunctional EC phenotype initiates and perpetuates plaque accumulation (12). Epigenetics control aberrant gene expression in many diseases including cancer and cardiovascular disease, but the mechanism of flow-induced epigenetic gene regulation via DNA methylation has not been well studied until very recently.
2. Mechanosensitive Endothelial Gene Expression
High throughput, genome-wide gene expression studies have shown that shear stress regulates endothelial gene expression in vitro and in vivo (2, 4, 13-17). These studies have identified numerous shear-responsive regulatory pathways as well as novel mechanosensitive genes and functional gene clusters (18, 19).
Shear stress is able to be translated from the cell surface (luminal, junctional, and basal) through a variety of mechanosensors, including ion channels, nicotinamide adenine dinucleotide phosphate (NADPH) oxidase and xanthine oxidase (XO), receptor tyrosine kinases, G proteins, cell/cell and cell/matrix (integrins) adhesion complexes, the glycocalyx, caveolae, and cytoskeletal elements (20). Once the shear stress stimulus is applied to the cell, various intracellular pathways are triggered in a process known as mechanotransduction. Interestingly, many of these pathways converge on common signaling pathways, such as the mitogen-activated protein kinase (MAPKs) pathway and the phosphatidylinositol-3-OH kinase PI3K /Akt pathway (21). The MAPK pathway in particular can be activated through integrins, NADPH oxidase, or ion channels, among others. Integrins activated by mechanical stimuli phosphorylate and activate a complex of kinases (FAK, c-Src, Shc, paxillin, and p130CAS), adaptor proteins (Grb2, Crk), and guanine nucleotide exchange factors (Sos, C3G), which ultimately lead to the activation of Ras. When Ras becomes activated, this leads to the activation of MAPKs. The extracellular signal-regulated kinases (ERK1/2), members of the MAP kinase family, then activate transcription factors (c-myc, c-jun, c-fos, p62TCF) and/or eNOS (22). Furthermore, ion channels activate the MAPK pathway through protein kinase C (PKC) (22) whereas NADPH oxidase activates the MAPK pathway through ROS (23). An additional example of such is the activation of platelet endothelial cell adhesion molecule-1 (PECAM-1). PECAM-1 directly transmits mechanical force at the junctional surface and VEGFR2 (in complex with PECAM-1 and VE-cadherin) activates PI3K. After PI3K is activated, Akt is also activated, and this leads to cytoskeletal arrangement so that ECs exposed to anti-atherogenic LS align in the direction of flow and ECs exposed to pro-atherogenic OS have activation of Rac1, which in turn leads to increased ROS and NF-κB becomes activated. Thus, in OS, these same mechanosensors lead to the expression of pro-atherosclerotic genes such as ICAM1 by activating NF-κB. (24, 25). Furthermore, the PI3K/Akt pathway can converge with the same integrins that the MAPKs interact with and can lead to activation of eNOS (26).
These shear-responsive pathways often activate the MAPK and PI3K/Akt pathways differentially in response to LS vs. OS (13), Gimbrone 2000). In laminar shear, atheroprotective genes such as eNOS (11), the Kruppel-like factor family (Klf2 and Klf4) (27), Nrf2 (28), and superoxide dismutases (Mn-SOD, EC-SOD) (11), become upregulated. eNOS becomes phosphorylated and activated by Akt via a PI3K-dependent pathway (29) and leads to an anti-atherogenic phenotype in ECs by producing NO, which prevents EC expression of MCP-1 and plays a role in maintaining vessel tone (30). Klf2 also plays a role as a potent anti-atherogenic protein. It has been reported to prevent thrombin-mediated induction of multiple cytokines (such as MCP-1), as well as inhibiting inflammation and apoptosis, maintaining cell shape, and maintaining vessel tone (31). Klf2 becomes upregulated by LS through myocyte enhancer factor 2 (MEF2) transcription, which in turn is upregulated either directly through HDAC4/5 inhibition (32) or indirectly through LS effect on the MAPK pathway (namely MEK5 and ERK5) (33). Finally, Nrf2, another anti-atherogenic transcription factor, becomes upregulated in LS through the negative regulation of Keap1. Nrf2 exerts its anti-atherogenic effects by upregulating cytoprotective genes that contain antioxidant response elements (AREs) (28). As opposed to LS, in oscillatory shear stress, ECs express pro-inflammatory cytokines such as MCP-1 (34) and inflammatory cell adhesion molecules such as VCAM1 and ICAM1 (15). Furthermore, ECs express other pro-inflammatory proteins such as NADPH oxidase (28)(35), which mediates its effects through ROS, and bone morphogenetic protein (BMP4) (36). MCP-1 contains a phorbol ester (TPA)-responsive element (TRE) in its promoter region, which was also found to be shear-sensitive. Shyy et al discovered that MAPKs can control AP-1 mediated transcriptional control of this element (37). Similarly, VCAM1 and ICAM1 also contain these shear-responsive elements in their promoters(15). Finally, NADPH oxidase is in part regulated through the MAPK pathway as well via c-Jun NH2 kinase (JNK) (28)(35). Ultimately, these findings have demonstrated that laminar, unidirectional s-flow or LS upregulates “atheroprotective” genes and downregulates “pro-atherogenic” genes while disturbed, reversing, or stagnant d-flow or OS results in the opposite phenomenon of enhancing pro-atherogenic genes and suppressing atheroprotective genes.
2.3. Overview of Epigenetic Modifications
The latest advance in the vascular mechanobiology field has been the discovery of key regulators that control a large network of genes. This discovery has made a large impact in the field due to the fact that these key regulators may be therapeutic targets for the treatment and prevention of atherosclerosis. Therefore, key regulators of global transcription and translation, such as direct DNA structural modifiers, microRNAs, and transcription factors have been gaining importance. Direct DNA structural modifications such as DNA methylation, histone modifications, and chromatin remodeling complexes alter the structure of genomic DNA in order to control accessibility of DNA sequences (38), whereas transcription factors are recruited to specific regions of a gene promoter to form a transcriptional complex that assists RNA polymerase to bind and transcribe the gene to messenger RNA (mRNA). Unlike the aforementioned regulators, microRNAs (miRNAs) control genetic regulation at the RNA level. Through complementary binding of the seed region, a 7 or 8 nucleotide region near the 5’ region of the mature miRNA, to the 3’UTR of the mRNA, miRNAs can either block mRNA translation or cause mRNA degradation. However, due to the small region of complementarity, a single miRNA can regulate hundreds of mRNAs, and thus genes (39). Finally, once mRNA has been successfully translated into protein, post-translational modifications control their activity and stability.
3. Shear-responsive epigenetics
3.1. Epigenetics
Epigenetics is defined as the modification of genetic information in a sequence-independent manner, which is achieved through alteration of the DNA structure. Genomic DNA in an open, relaxed conformation is known as euchromatin and is associated with acetylated histones and unmethylated DNA, whereas condensed genomic DNA is defined as heterochromatin and is associated with methylated histones (trimethylated-histone 3 lysine 9 (H3K9) and trimethylated-histone 3 lysine 27 (H3K27)) and methylated DNA (40). Euchromatic DNA is usually transcriptionally active, whereas heterochromatin generally contains repeat elements and transcriptionally silent genes. Heterochromatin compaction predominantly occurs during development and differentiation (41-45).
3.2. Histone modifications and chromatin remodeling
Although many investigators have confirmed that blood flow regulates histone modifications and chromatin remodeling, it is still controversial as to whether these events are pro-atherogenic or anti-atherogenic. Early reports of shear-responsive epigenetic modifications described novel pathways by which LS mediates chromatin remodeling in cultured ECs by inducing histone H3/H4 acetylation and H3 phosphorylation (46, 47). LS activates genes with intrinsic histone acetyltransferase (HAT) activities, such as ribosomal S6 kinase-2 (RSK-2), mitogen- and stress-activated kinase-1 (MSK-1), and cAMP-responsive element–binding protein (CREB)/CREB-binding protein (CBP) complexes, and also deactivates histone deacetylases (HDACs). HDACs are categorized into class I (HDAC 1/2/3) and class II (HDAC5/7). LS blocks class II HDAC-mediated inhibition of myocyte enhancer factor-2 (MEF2), thus enabling MEF2-mediated KLF2 and eNOS expression (32). In contrast, OS induces both class I and II HDAC overexpression and accumulation in the nucleus via the phosphatidylinositol 3-kinase (PI3K)/Protein Kinase B (PKB/Akt) signaling pathway, thus blocking the anti-inflammatory transcription factors NF-E2-related factor 2 (Nrf2) and MEF2. It was later found that class I HDAC inhibition by siRNA or the class I-specific HDAC inhibitor valproic acid prevented OS-induced endothelial proliferation both in vivo and in vitro (48). Taken together, these studies suggest that acetylation of specific histones may be athero-protective.
Several groups have demonstrated that the HDACs play a functional role in EC inflammation and atherosclerosis, but the role of HDAC3 is unclear, as both pro-and anti-atherogenic effects have been reported. Inoe et. al first described that inhibition of HDAC3, which is highly expressed in endothelial cells, suppresses endothelial inflammation by decreasing TNFα-induced VCAM1 expression and monocyte adhesion (43). It was later shown that HDAC3 inhibition with trichostatin A (TSA) can prevent both TNFα and LPS-induced tissue factor expression and promoter activity in endothelial cells through reduced activity of the NF-kappaB-related “TF-kappaB” promoter (49, 50). HDAC3 also represses eNOS expression through deacetylation of the eNOS promoter (51).
Zampetaki et al. directly demonstrated that HDAC3 plays a role in endothelial cell survival and atherosclerosis development (46). ApoE-/- mice showed increased expression of HDAC3 in regions of d-flow and that HDAC3 protein was upregulated in endothelial cells exposed to d-flow in vitro. They showed that HDAC3 is phosphorylated by d-flow and has a strong interaction with Akt, causing Akt phosphorylation. However, knockdown of HDAC3 led to a marked increase in apoptosis in endothelial cells. Furthermore, in aortic isografts of mice treated with lentiviral shHDAC3, robust atherosclerotic lesions were observed. Thus, moderate levels of HDAC3 appears to promote endothelial survival and integrity, but its overexpression in d-flow regions may be pro-atherogenic(52). Fitting with this idea, the complete ablation of HDAC activity by TSA treatment exacerbated vascular inflammation and atherosclerosis in low-density lipoprotein (LDL) receptor-deficient (LDLR-/-) mice fed an atherogenic diet (53).
Furthermore, HDAC7 was shown to suppress endothelial proliferation by preventing the nuclear translocation of β-catenin by direct binding and downregulation of TCF-1/Id2 and cyclin D1. (54).
As opposed to deacetylation, histone acetylation has been reported to modulate oxLDL-induced cytokine release by ECs. OxLDL increases H3/4 acetylation and H3 phosphorylation on a global scale and specifically in the promoter region of oxLDL-related interleukin 8 (IL8) and monocyte-chemoattractant protein-1 (MCP-1) in cultured human ECs (HUVECs). Additionally, HDAC inhibition increases endothelial expression and secretion of these cytokines (55). Also, HDAC2 expression is reduced in human coronary atherosclerotic lesions (55). Taken together, these reports clearly indicate that HDACs are regulated by flow, but its pathophysiological roles may vary depending on specific HDACs and cell types involved.
4. Endothelial-specific gene expression mediated by epigenetics
Early epigenetic studies in the endothelium were mainly focused on studying the status of single genes and using this status to define markers of cellular differentiation towards the endothelial lineage. Understanding the function and regulation of genes that have EC-specific expression is of particular importance because the loss of constitutively active EC genes is frequently associated with endothelial dysfunction.
Histone acetylation is important for both LS-induced and endothelial-specific expression of eNOS. An HDAC inhibitor-induced increase in eNOS expression was linked to decreased DNA methylation of the eNOS promoter, and TSA/5Aza combination treatment was found to synergistically increase eNOS in non-ECs (56). This demonstrates the complementary interactions of histone modifications and DNA methylation. It was later discovered that 5Aza treatment alone could induce eNOS expression in non-ECs, and further that the eNOS promoter is heavily methylated in non-ECs but demethylated in ECs. Methylation of the eNOS promoter blocks binding of the transcription factors Sp1, Sp3, and Ets1, and thus eNOS transcription (57).
In addition to eNOS, Roundabout 4 (Robo4) is an endothelial-specific gene whose expression is controlled by promoter DNA methylation in non-endothelial cells, which prevents binding of essential transcription factor SP1. Both the methylation and gene expression status of Robo4 in non-endothelial cells can be reversed by treatment with 5Aza. Interestingly, while Robo4 is suppressed by LS and eNOS by OS, neither gene has shear-responsive promoter DNA methylation changes (58-61). The lineage-specific epigenetic changes of both eNOS and Robo4 have been carefully dissected to show that Robo4-specific methylation patterns are established during the transition from embryonic stem cells to Flk1+ mesodermal cells, while eNOS DNA methylation and concomitant histone modifications appear in early proangiogenic cells (58, 62).
Studies of epigenetically-modulated endothelial-specific gene expression were expanded to a repertoire of endothelial-specific genes, such as PECAM1 (CD31), von Willebrand factor (vWF), VE-cadherin, and intercellular adhesion molecule-2 (ICAM2) which were found to have endothelial-specific promoter demethylation patterns. As with the other genes, 5Aza induced their expression in non-endothelial cells and TSA had a synergistic effect (63). These studies, taken together, demonstrate that DNA methylation is a key mechanism for endothelial-specific gene expression.
5. DNA methylation is a novel epigenetic mechanism that regulates endothelial cell responses to shear stress
Recently, several groups independently converged on the seminal finding that DNA methyltransferases (DNMTs) are shear responsive proteins that regulate flow-mediated endothelial gene expression programs (61),(64), (65).
5.1. Kruppel like factor 4 (Klf4) suppression by disturbed flow in ECs is mediated by promoter DNA hypermethylation in a DNMT3a-dependent manner
Klf4 is a key mediator of endothelial function and has been well documented to maintain an anti-inflammatory, quiescent endothelial state in unidirectional flow conditions (66-69). Jiang et al. discovered that DNA methylation is a novel, flow-mediated mechanism of endothelial Klf4 transcriptional regulation both in vitro and in vivo (61). In this study, both DNMT3A expression and DNMT3A binding to the Klf4 promoter were found to increase due to d-flow. This led to DNA hypermethylation and decreased MEF2 binding. MEF2 is a key transcription factor that controls Klf4 upregulation in response to resveratrol in ECs (70). DNMT inhibition by 5Aza or RG108 rescued Klf4 expression and reversed the d-flow-induced suppression of the downstream targets eNOS and thrombomodulin. Additionally, the d-flow-induced overexpression of the proinflammatory monocyte chemoattractant protein-1 (MCP-1) was blunted. Thus, the methylation status of key genes such as Klf4 and eNOS can be a crucial indicator of endothelial phenotype.
5.2. D-flow upregulates DNMT1 expression in ECs, which alters DNA methylation patterns and endothelial gene expression.
Genome-wide studies of DNA methylation and gene expression, recently reported by our lab and others, provided a much broader view of flow-mediated DNA methylation changes and the relation of methylation status to atherosclerosis development in vivo. Our DNA methylome and transcriptome studies, using reduced representation bisulfite sequencing (RRBS) and microarray, respectively, revealed that d-flow regulates global DNA methylation patterns in a DNMT-dependent manner. Using a murine model of d-flow, which involves partial ligation of the left carotid artery (LCA), while keeping the contralateral right carotid artery (RCA) untouched as an internal control, we found a dramatic increase in DNMT1 expression in the LCA. Further, DNMT1 expression was induced by OS in cultured endothelial cells, and inhibition of DNMT with either 5Aza or DNMT1 siRNA markedly reduced OS-induced endothelial inflammation. Moreover, 5Aza treatment reduced lesion formation in both acute and chronic mouse models of atherosclerosis. Genome-wide RRBS and microarray studies from animals treated with 5Aza demonstrated that DNMT1 inhibition in d-flow regions reverts DNA methylation and gene expression back to the healthy s-flow condition (64).
These DNA methylome and transcriptome results initially were used to identify mechanosensitive genes that were hypermethylated in the promoter regions and were also downregulated by d-flow. These in silico analyses led us to discover 11 genes with the most dramatic d-flow-induced, 5Aza-reversible promoter hypermethylation and gene suppression. They were HoxA5, Tmem184b, Adamtsl5, Klf3, Cmklr1, Pkp4, Acvrl1, Dok4, Spry2, and Zfp46. Interestingly, 5 of those 11 genes (HoxA5, Klf3, Cmklr1, Acvrl1, and Spry2) contain CRE in their promoters. The pathophysiological relevance of those genes, especially the CRE-containing genes, is currently unknown and is under investigation. Of the mechanosensitive CRE containing genes, HoxA5 and Klf3 encode transcription factors and thus the methylation status of these loci could serve as a mechanosensitive master switch in gene expression (64).
Further systems biological analysis revealed that CRE methylation is regulated genome-wide in a mechanosensitive manner. CREs located specifically in gene promoters on the genome-scale are hypermethylated by d-flow in a 5Aza-preventable manner, suggesting a potential mechanism by which d-flow regulates gene expression by genome-wide CRE methylation. These CRE-containing mechanosensitive genes are the target of future studies (64).
More recently, Zhou et al. also reported that d-flow causes DNMT1 overexpression (65). Comparing OS to pulsatile, unidirectional LS in HUVECs, they found that OS increases DNMT1 mRNA and protein expression, DNMT1 nuclear translocation, and 5-methylcytosine (5mC) content. 5Aza treatment inhibited the OS-induced DNMT1 expression and prevented increases in 5mC. Using a rat partial carotid ligation model, they demonstrated that d-flow also induced DNMT1 protein expression and increased 5mC content in vivo.
These studies of shear-responsive DNA methylation regulators, global DNA methylation responses, and the functional importance of site-specific DNA methylation changes caused by d-flow demonstrated, for the first time, the key importance of DNA methylation in controlling global gene expression in endothelial dysfunction and atherosclerosis in d-flow regions.
6. Hox are novel shear-sensitive endothelial gene family regulated by promoter DNA methylation
Hox genes are homeobox transcription factors whose homeodomains recognize and bind to specific DNA sequences, enabling the coordinate regulation of sets of genes. Hox genes exist in four separate clusters on distinct chromosomes (HoxA, HoxB, HoxC and HoxD) and often have complementary functionality. Hox genes and their associated microRNAs are highly conserved developmental master regulators with tight tissue-specific, spatiotemporal control. These genes are known to be dysregulated in several cancers and are often controlled by DNA methylation (71-76).
Specific members of the Hox family have been implicated in vascular remodeling, angiogenesis, and disease by orchestrating changes in matrix degradation, integrins, and components of the extracellular matrix (77). HoxD3 and HoxB3 are pro-invasive, angiogenic genes that upregulate β3 and α5 integrins and Efna1 in endothelial cells, respectively (78-81). HoxA3 induces endothelial migration by upregulating metalloproteinase-14 (MMP14) and plasminogen activator urokinase receptor (uPAR) (82). Conversely, HoxD10 and HoxA5 have the opposite effect of suppressing endothelial migration and angiogenesis, and stabilizing adherens junctions by upregulating TIMP1, downregulating uPAR and MMP14, and by upregulating TSP2 and downregulating VEGFR2, Efna1, Hif1α and COX-2, respectively (83, 84).
HoxA5 also upregulates the tumor suppressor p53 and Akt1 by downregulation of PTEN (85). Suppression of HoxA5 has been shown to attenuate hemangioma growth (86). HoxA5 has far-reaching effects on gene expression, causing ~300 genes to become upregulated upon its induction in breast cancer cell lines (87). HoxA5 protein transduction domain overexpression prevents inflammation as shown by inhibition of TNFα-inducible monocyte binding to HUVECs (88, 89). Consistent with this finding, HoxA5 knockdown induced endothelial inflammation in LS-exposed cells (64).
The Hox families exhibit a high level of self-interaction, forming chromatin conformations known as topological domains (90). This global method of establishing a higher order genomic structure, that is evident here specifically within the Hox domains, suggests a novel mechanism by which shear regulates chromosomal conformation, thereby modulating DNA domain interactions by epigenetic mechanisms.
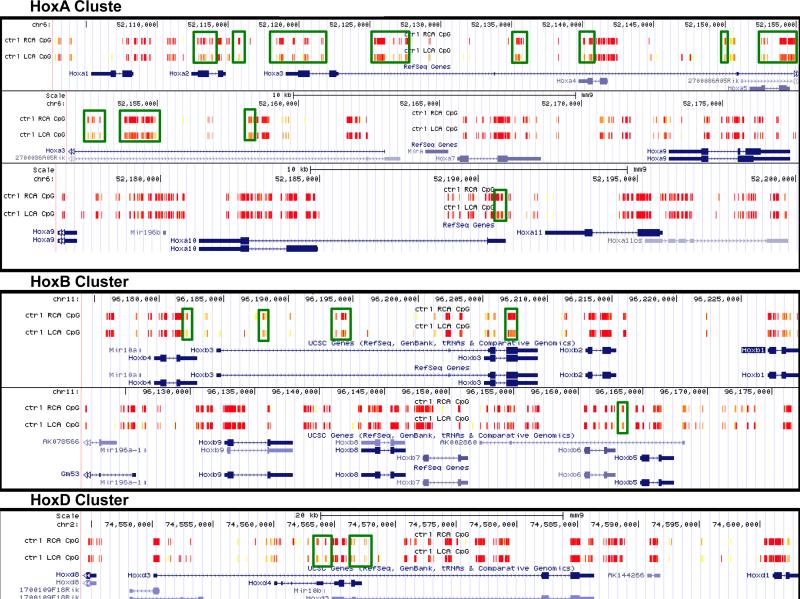
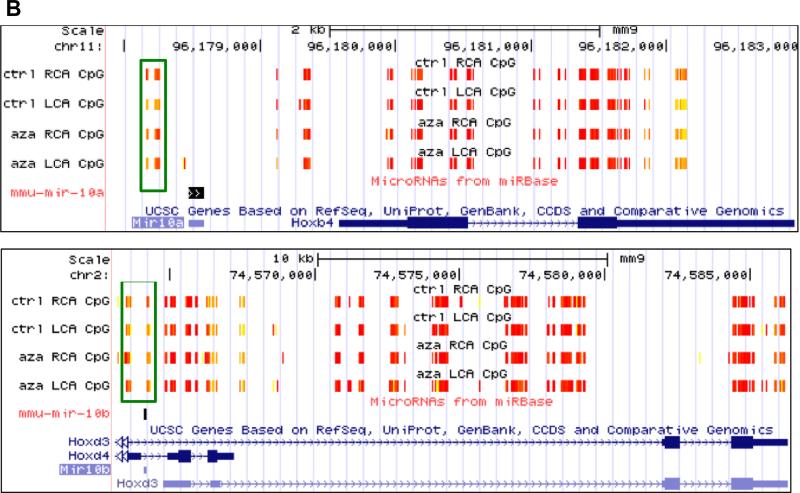
Some of the most prominent changes in DNA methylation in our RRBS dataset occur in the Hox gene family. Overall, there was very high coverage of CG sites localized to the Hox family gene clusters by our RRBS assay, and many of these sites exhibit strong changes in methylation in the d-flow LCA as compared to the s-flow RCA (indicated in green boxes in Figure 1). These methylation changes occur mainly at key functional regions, including promoters where methylation is associated with gene suppression, intron/exon boundaries where methylation may be associated with alternative transcript expression (Figure 1A). Many of these changes are also 5Aza-reversible (64).
Figure 1. Flow regulates DNA methylation of Hox family.
HoxA, HoxB, and HoxD gene family DNA methylation changes in endothelial cells due to disturbed flow are seen by reduced representation bisulfite sequencing (RRBS) in the UCSC Genome Browser from our mouse partial carotid ligation model (A). RRBS data in the UCSC Genome Browser showing 5Aza-reversible, d-flow-induced Hox promoter hypermethylation regions. Differentially methylated regions by shear stress are boxed in green (B). High methylation is denoted in yellow and low methylation is denoted in red.
Although the different Hox family subsets are located across different chromosomes, their methylation pattern is similarly regulated (Figure 1A). In mouse, the HoxA, B, and D families are located on chromosomes 6, 11, and 2, respectively. While the HoxA cluster shows the most flow-sensitive differential methylation, HoxB and HoxD clusters also show some mechanosensitive methylation patterns.
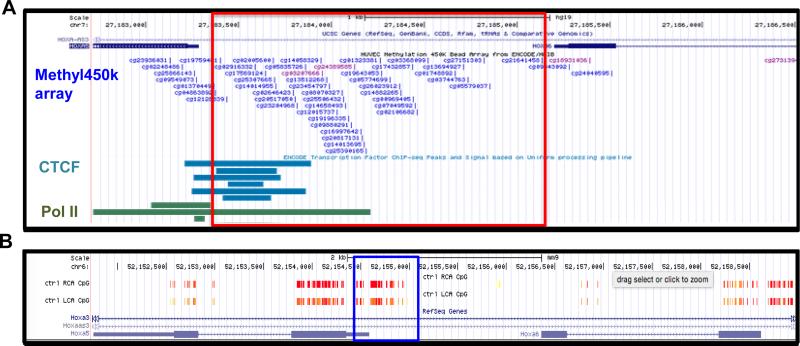
The functional importance of the HoxA5 promoter region in transcriptional regulation is demonstrated in Figure 2A and 2B. Although mouse endothelial Encyclopedia of DNA Elements (ENCODE) data is not yet available, the Hox clusters genomic sequences are highly conserved between mouse and human. Therefore we used ENCODE data obtained in human endothelial cells (HUVEC) to determine locations of transcription factor/complex binding to DNA in the homologous regions from mouse that we found to be differentially methylated by flow (Figure 2A). This analysis showed enriched Pol2 and CTCF binding at the CpG-dense HoxA5 promoter in HUVECs, suggesting its active transcriptional status. The HoxA locus is known to contain an insulator characterized by several CTCF binding sites that separates the co-regulation of expression of HoxA1-7 and HoxA9-13 (91). CTCF is a DNA insulator binding protein that marks the transition from tightly packed heterochromatin to relaxed euchromatin. Pol2 contains the active subunit for synthesizing RNA from the DNA template, and in combination with other RNA polymerase subunits, forms the DNA groove binding domain.
Figure 2. Flow- and differentiation-dependent methylation changes in the HoxA5 promoter.
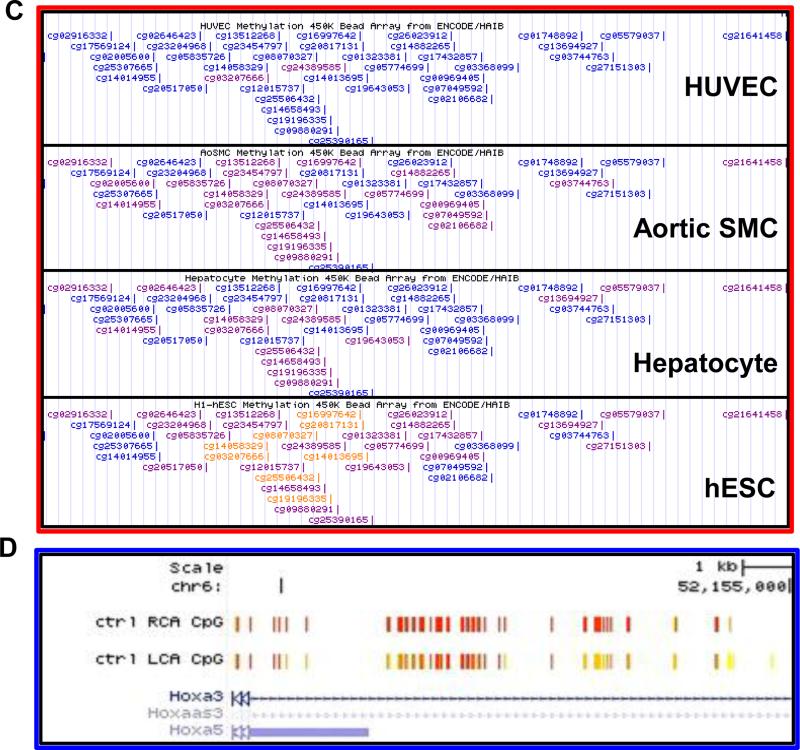
UCSC Genome Browser view of the RRBS data showing flow-dependent methylation changes in the HoxA5 promoter (A). The ENCODE consortium ChIP-Seq and Methyl450k array datasets for HUVEC show that the transcription factor CTCF and RNA Polymerase II are enriched at the HoxA5 promoter where there is a high density of CpG sites (B). Zoomed-in view of the red boxed area from panel A (C). UCSC Genome Browser screenshot of the ENCODE Methyl450k array datasets shows a comparison of the HoxA5 promoter methylation profile across cell types from the least differentiated (human embryonic stem cells) to the most endothelial-like (HUVECs). Highly methylated regions in orange, partially methylated regions in purple, and unmethylated regions in blue. (D).
Previously, Shirodkar et al. showed that the endothelial specific genes eNOS and VE-cadherin are heavily methylated in their promoter region in non-differentiated cells such as human embryonic stem cells, but as they become differentiated towards the endothelial cells, they become demethylated and genes highly expressed (59). Interestingly, we find that in the same region that has mechanosensitive DNA methylation in the HoxA5 promoter, the same phenomenon occurs (Figure 2C and D).
Comparison of the HoxA5 promoter methylation profile across cell types from the least differentiated (human embryonic stem cells) to endothelial cells (HUVECs) with hepatocytes and aortic smooth muscle cells as intermediate types using ENCODE datasets, shows that the HoxA5 promoter is heavily methylated in non-differentiated cells and becomes demethylated as cells differentiate down the endothelial lineage (Figure 2A) (63, 92). While it is known that Hox genes' DNA methylation status is a useful marker to distinguish between cell types for differentiation, we demonstrate that this is indeed the case in the transition of cell type-specific differentiation (Figure 2A).
Our RRBS data indicates the key importance of genome-wide CREB binding sites (CRE) as a potential global regulator of gene expression. We expect that the HoxA5 promoter CRE methylation status regulates CREB binding and downstream expression of HoxA5, but unfortunately neither CREB nor its binding partners p300/CBP were covered by the ENCODE consortium endothelial Chromatin-IP datasets.
Interestingly, both HoxA5 and Klf3 are suppressed in acute myeloid leukemia (AML), which is treated by the DNMT inhibitor 5Aza. While HoxA5 is known to be hypermethylated in AML, it has not yet been shown whether 5Aza directly reverses the methylation of this gene (93, 94). Of additional interest, AML is also treated with alltrans retinoic acid, a drug known to influence DNA methylation of a small number of specific genes. Furthermore, this drug was shown to rescue Klf3 expression by an unknown mechanism (95, 96).
7. Perspectives
Emerging findings reveal that epigenetic mechanisms, DNA methylation, HDAC-mediated histone modifications, and miRNAs, play a critical role in regulating the expression of genes involved in vascular biology and disease. While DNA methylation, especially 5mC is the most studied covalent DNA modification, other modifications exist. These include 5-hydroxymethylcytosine (5hC), 5-formylcytosine (5fC) and 5-carboxycytosine (5cC). The corollary finding that ten-eleven-translocation (TET) enzymes convert 5mC into 5hmC also introduces the important concept of site-specific DNA demethylation, which could play an important role in both regulating DNA methylation patterns as well as in establishing aberrant demethylation in disease (97).
Thus far, DNA methylation in atherosclerosis has been mainly studied in the context of CpG methylation found in the promoter region of genes, especially in dense CpG-rich regions known as CpG islands. However, DNA methylation in other locales, including coding or noncoding gene regulatory regions, is known to play a role in controlling gene expression, splice variant expression, and in chromatin conformation and genome stabilization (98-100). It would be interesting to examine whether there is d-flow-induced differential endothelial DNA methylation in locations including enhancers, promoters, intron/exon boundaries, transposable elements, 3’UTRs, transcription factor binding sites, and across the genome either sporadically or in the context of specific sequence motifs.
Finally,it was recently shown that the anti-atherosclerotic effect of DNMT inhibition by 5Aza also has a mechanistic basis in the immune milieu (101). Because atherosclerosis is a systemic disease caused by multi-cell type dysfunctions, including immune cells, smooth muscle cells, and fibroblasts, a systems wide approach will be necessary to understand the role of global methylation changes and crosstalk between cell types via circulating miRNAs in modulating disease development in order to fully realize the therapeutic potential of epigenetic interventions.
Drawing upon epigenetics-based cancer diagnostics currently in use, it is foreseeable that epigenetic targets such as DNMTs, specific gene promoter DNA methylation, histone modifications, and miRNAs could serve as biomarkers for cardiovascular disease diagnosis as well as targets for therapeutic intervention.
ACKNOWLEDGMENTS
This work was supported by funding from National Institutes of Health grants HL119798, HL113451, HL095070 and HL124879 to HJ. HJ is John and Jan Portman Professor. JD is a National Science Foundation pre-doctoral fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Akimoto S, Mitsumata M, Sasaguri T, Yoshida Y. Laminar shear stress inhibits vascular endothelial cell proliferation by inducing cyclin-dependent kinase inhibitor p21(Sdi1/Cip1/Waf1). Circ Res. 2000;86:185–190. doi: 10.1161/01.res.86.2.185. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Cardena G, Comander JI, Blackman BR, Anderson KR, Gimbrone MA. Mechanosensitive endothelial gene expression profiles: scripts for the role of hemodynamics in atherogenesis? Ann N Y Acad Sci. 2001;947:1–6. [PubMed] [Google Scholar]
- 3.Chatzizisis YS, Coskun AU, Jonas M, Edelman ER, Feldman CL, Stone PH. Role of endothelial shear stress in the natural history of coronary atherosclerosis and vascular remodeling: molecular, cellular, and vascular behavior. J Am Coll Cardiol. 2007;49:2379–2393. doi: 10.1016/j.jacc.2007.02.059. [DOI] [PubMed] [Google Scholar]
- 4.Chien S, Shyy JY. Effects of hemodynamic forces on gene expression and signal transduction in endothelial cells. Biol Bull. 1998;194:390–391. doi: 10.2307/1543122. discussion 392-393. [DOI] [PubMed] [Google Scholar]
- 5.He XH, Wu GF, Zhang Y, Chen XL, Zhang ZS, Zhan CY, Liu J, He JG, Xiong Y, Fang DQ, et al. [Effect of chronic enhanced external counterpulastion on gene expression profiles of arterial endothelial cells of pigs fed with high-cholesterol diet]. Nan Fang Yi Ke Da Xue Xue Bao. 2008;28:1195–1197. [PubMed] [Google Scholar]
- 6.White SJ, Hayes EM, Lehoux S, Jeremy JY, Horrevoets AJ, Newby AC. Characterization of the differential response of endothelial cells exposed to normal and elevated laminar shear stress. Journal of cellular physiology. 2011;226:2841–2848. doi: 10.1002/jcp.22629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gimbrone MA, Jr., Topper JN, Nagel T, Anderson KR, Garcia-Cardena G. Endothelial dysfunction, hemodynamic forces, and atherogenesis. Ann N Y Acad Sci. 2000;902:230–239. doi: 10.1111/j.1749-6632.2000.tb06318.x. discussion 239-240. [DOI] [PubMed] [Google Scholar]
- 8.Chappell DC, Varner SE, Nerem RM, Medford RM, Alexander RW. Oscillatory shear stress stimulates adhesion molecule expression in cultured human endothelium. Circ Res. 1998;82:532–539. doi: 10.1161/01.res.82.5.532. [DOI] [PubMed] [Google Scholar]
- 9.Dhawan SS, Avati Nanjundappa RP, Branch JR, Taylor WR, Quyyumi AA, Jo H, McDaniel MC, Suo J, Giddens D, Samady H. Shear stress and plaque development. Expert Rev Cardiovasc Ther. 2010;8:545–556. doi: 10.1586/erc.10.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abumiya T, Sasaguri T, Taba Y, Miwa Y, Miyagi M. Shear stress induces expression of vascular endothelial growth factor receptor Flk-1/KDR through the CT-rich Sp1 binding site. Arterioscler Thromb Vasc Biol. 2002;22:907–913. doi: 10.1161/01.atv.0000018300.43492.83. [DOI] [PubMed] [Google Scholar]
- 11.Topper JN, Cai J, Falb D, Gimbrone MA., Jr. Identification of vascular endothelial genes differentially responsive to fluid mechanical stimuli: cyclooxygenase-2, manganese superoxide dismutase, and endothelial cell nitric oxide synthase are selectively up-regulated by steady laminar shear stress. Proc Natl Acad Sci U S A. 1996;93:10417–10422. doi: 10.1073/pnas.93.19.10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Cardena G, Comander J, Anderson KR, Blackman BR, Gimbrone MA., Jr. Biomechanical activation of vascular endothelium as a determinant of its functional phenotype. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4478–4485. doi: 10.1073/pnas.071052598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chien S. Mechanotransduction and endothelial cell homeostasis: the wisdom of the cell. Am J Physiol Heart Circ Physiol. 2007;292:H1209–1224. doi: 10.1152/ajpheart.01047.2006. [DOI] [PubMed] [Google Scholar]
- 14.Davies PF. Flow-mediated endothelial mechanotransduction. Physiol Rev. 1995;75:519–560. doi: 10.1152/physrev.1995.75.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagel T, Resnick N, Atkinson WJ, Dewey CF, Jr., Gimbrone MA., Jr. Shear stress selectively upregulates intercellular adhesion molecule-1 expression in cultured human vascular endothelial cells. J Clin Invest. 1994;94:885–891. doi: 10.1172/JCI117410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skogsberg J, Lundstrom J, Kovacs A, Nilsson R, Noori P, Maleki S, Kohler M, Hamsten A, Tegner J, Bjorkegren J. Transcriptional profiling uncovers a network of cholesterol-responsive atherosclerosis target genes. PLoS Genet. 2008;4:e1000036. doi: 10.1371/journal.pgen.1000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen BP, Li YS, Zhao Y, Chen KD, Li S, Lao J, Yuan S, Shyy JY, Chien S. DNA microarray analysis of gene expression in endothelial cells in response to 24-h shear stress. Physiol Genomics. 2001;7:55–63. doi: 10.1152/physiolgenomics.2001.7.1.55. [DOI] [PubMed] [Google Scholar]
- 18.Tarbell JM, Shi ZD, Dunn J, Jo H. Fluid Mechanics, Arterial Disease, and Gene Expression. Annu Rev Fluid Mech. 2014;46:591–614. doi: 10.1146/annurev-fluid-010313-141309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwak BR, Back M, Bochaton-Piallat ML, Caligiuri G, Daemen MJ, Davies PF, Hoefer IE, Holvoet P, Jo H, Krams R, et al. Biomechanical factors in atherosclerosis: mechanisms and clinical implications. Eur Heart J. 2014;35:3013–3020. 3020a–3020d. doi: 10.1093/eurheartj/ehu353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatzizisis YS, Coskun AU, Jonas M, Edelman ER, Feldman CL, Stone PH. Role of endothelial shear stress in the natural history of coronary atherosclerosis and vascular remodeling: molecular, cellular, and vascular behavior. Journal of the American College of Cardiology. 2007;49:2379–2393. doi: 10.1016/j.jacc.2007.02.059. [DOI] [PubMed] [Google Scholar]
- 21.Li YS, Haga JH, Chien S. Molecular basis of the effects of shear stress on vascular endothelial cells. J Biomech. 2005;38:1949–1971. doi: 10.1016/j.jbiomech.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 22.Traub O, Berk BC. Laminar shear stress: mechanisms by which endothelial cells transduce an atheroprotective force. Arterioscler Thromb Vasc Biol. 1998;18:677–685. doi: 10.1161/01.atv.18.5.677. [DOI] [PubMed] [Google Scholar]
- 23.Hwang J, Saha A, Boo YC, Sorescu GP, McNally JS, Holland SM, Dikalov S, Giddens DP, Griendling KK, Harrison DG, et al. Oscillatory shear stress stimulates endothelial production of O2- from p47phox-dependent NAD(P)H oxidases, leading to monocyte adhesion. J Biol Chem. 2003;278:47291–47298. doi: 10.1074/jbc.M305150200. [DOI] [PubMed] [Google Scholar]
- 24.Tzima E, Del Pozo MA, Kiosses WB, Mohamed SA, Li S, Chien S, Schwartz MA. Activation of Rac1 by shear stress in endothelial cells mediates both cytoskeletal reorganization and effects on gene expression. EMBO J. 2002;21:6791–6800. doi: 10.1093/emboj/cdf688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tzima E, Irani-Tehrani M, Kiosses WB, Dejana E, Schultz DA, Engelhardt B, Cao G, DeLisser H, Schwartz MA. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- 26.Shyy JY, Chien S. Role of integrins in endothelial mechanosensing of shear stress. Circ Res. 2002;91:769–775. doi: 10.1161/01.res.0000038487.19924.18. [DOI] [PubMed] [Google Scholar]
- 27.Wang N, Miao H, Li YS, Zhang P, Haga JH, Hu Y, Young A, Yuan S, Nguyen P, Wu CC, et al. Shear stress regulation of Kruppel-like factor 2 expression is flow pattern-specific. Biochem Biophys Res Commun. 2006;341:1244–1251. doi: 10.1016/j.bbrc.2006.01.089. [DOI] [PubMed] [Google Scholar]
- 28.Takabe W, Jen N, Ai L, Hamilton R, Wang S, Holmes K, Dharbandi F, Khalsa B, Bressler S, Barr ML, et al. Oscillatory shear stress induces mitochondrial superoxide production: implication of NADPH oxidase and c-Jun NH2-terminal kinase signaling. Antioxid Redox Signal. 2011;15:1379–1388. doi: 10.1089/ars.2010.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimmeler S, Fleming I, Fisslthaler B, Hermann C, Busse R, Zeiher AM. Activation of nitric oxide synthase in endothelial cells by Akt-dependent phosphorylation. Nature. 1999;399:601–605. doi: 10.1038/21224. [DOI] [PubMed] [Google Scholar]
- 30.Zeiher AM, Fisslthaler B, Schray-Utz B, Busse R. Nitric oxide modulates the expression of monocyte chemoattractant protein 1 in cultured human endothelial cells. Circ Res. 1995;76:980–986. doi: 10.1161/01.res.76.6.980. [DOI] [PubMed] [Google Scholar]
- 31.Nayak L, Lin Z, Jain MK. “Go with the flow”: how Kruppel-like factor 2 regulates the vasoprotective effects of shear stress. Antioxid Redox Signal. 2011;15:1449–1461. doi: 10.1089/ars.2010.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W, Ha CH, Jhun BS, Wong C, Jain MK, Jin ZG. Fluid shear stress stimulates phosphorylation-dependent nuclear export of HDAC5 and mediates expression of KLF2 and eNOS. Blood. 2010;115:2971–2979. doi: 10.1182/blood-2009-05-224824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parmar KM, Larman HB, Dai G, Zhang Y, Wang ET, Moorthy SN, Kratz JR, Lin Z, Jain MK, Gimbrone MA, Jr., et al. Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2. J Clin Invest. 2006;116:49–58. doi: 10.1172/JCI24787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shyy YJ, Hsieh HJ, Usami S, Chien S. Fluid shear stress induces a biphasic response of human monocyte chemotactic protein 1 gene expression in vascular endothelium. Proc Natl Acad Sci U S A. 1994;91:4678–4682. doi: 10.1073/pnas.91.11.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takabe W, Warabi E, Noguchi N. Anti-atherogenic effect of laminar shear stress via Nrf2 activation. Antioxid Redox Signal. 2011;15:1415–1426. doi: 10.1089/ars.2010.3433. [DOI] [PubMed] [Google Scholar]
- 36.Rhee WJ, Ni CW, Zheng Z, Chang K, Jo H, Bao G. HuR regulates the expression of stress-sensitive genes and mediates inflammatory response in human umbilical vein endothelial cells. Proc Natl Acad Sci U S A. 2010;107:6858–6863. doi: 10.1073/pnas.1000444107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shyy JY, Lin MC, Han J, Lu Y, Petrime M, Chien S. The cis-acting phorbol ester “12-O-tetradecanoylphorbol 13-acetate”-responsive element is involved in shear stress-induced monocyte chemotactic protein 1 gene expression. Proc Natl Acad Sci U S A. 1995;92:8069–8073. doi: 10.1073/pnas.92.17.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson KD. DNA methylation and chromatin - unraveling the tangled web. Oncogene. 2002;21:5361–5379. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- 39.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mason K, Liu Z, Aguirre-Lavin T, Beaujean N. Chromatin and epigenetic modifications during early mammalian development. Anim Reprod Sci. 2012;134:45–55. doi: 10.1016/j.anireprosci.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 41.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 42.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 43.de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 44.Fahrner JA, Baylin SB. Heterochromatin: stable and unstable invasions at home and abroad. Genes Dev. 2003;17:1805–1812. doi: 10.1101/gad.1123303. [DOI] [PubMed] [Google Scholar]
- 45.Ragoczy T, Telling A, Sawado T, Groudine M, Kosak ST. A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res. 2003;11:513–525. doi: 10.1023/a:1024939130361. [DOI] [PubMed] [Google Scholar]
- 46.Chen W, Bacanamwo M, Harrison DG. Activation of p300 histone acetyltransferase activity is an early endothelial response to laminar shear stress and is essential for stimulation of endothelial nitric-oxide synthase mRNA transcription. J Biol Chem. 2008;283:16293–16298. doi: 10.1074/jbc.M801803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Illi B, Nanni S, Scopece A, Farsetti A, Biglioli P, Capogrossi MC, Gaetano C. Shear stress-mediated chromatin remodeling provides molecular basis for flow-dependent regulation of gene expression. Circ Res. 2003;93:155–161. doi: 10.1161/01.RES.0000080933.82105.29. [DOI] [PubMed] [Google Scholar]
- 48.Lee DY, Lee CI, Lin TE, Lim SH, Zhou J, Tseng YC, Chien S, Chiu JJ. Role of histone deacetylases in transcription factor regulation and cell cycle modulation in endothelial cells in response to disturbed flow. Proc Natl Acad Sci U S A. 2012;109:1967–1972. doi: 10.1073/pnas.1121214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inoue K, Kobayashi M, Yano K, Miura M, Izumi A, Mataki C, Doi T, Hamakubo T, Reid PC, Hume DA, et al. Histone deacetylase inhibitor reduces monocyte adhesion to endothelium through the suppression of vascular cell adhesion molecule-1 expression. Arterioscler Thromb Vasc Biol. 2006;26:2652–2659. doi: 10.1161/01.ATV.0000247247.89787.e7. [DOI] [PubMed] [Google Scholar]
- 50.Wang J, Mahmud SA, Bitterman PB, Huo Y, Slungaard A. Histone deacetylase inhibitors suppress TF-kappaB-dependent agonist-driven tissue factor expression in endothelial cells and monocytes. J Biol Chem. 2007;282:28408–28418. doi: 10.1074/jbc.M703586200. [DOI] [PubMed] [Google Scholar]
- 51.Zhang MX, Zhang C, Shen YH, Wang J, Li XN, Chen L, Zhang Y, Coselli JS, Wang XL. Effect of 27nt small RNA on endothelial nitric-oxide synthase expression. Mol Biol Cell. 2008;19:3997–4005. doi: 10.1091/mbc.E07-11-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zampetaki A, Zeng L, Margariti A, Xiao Q, Li H, Zhang Z, Pepe AE, Wang G, Habi O, deFalco E, et al. Histone deacetylase 3 is critical in endothelial survival and atherosclerosis development in response to disturbed flow. Circulation. 2010;121:132–142. doi: 10.1161/CIRCULATIONAHA.109.890491. [DOI] [PubMed] [Google Scholar]
- 53.Choi JH, Nam KH, Kim J, Baek MW, Park JE, Park HY, Kwon HJ, Kwon OS, Kim DY, Oh GT. Trichostatin A exacerbates atherosclerosis in low density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2005;25:2404–2409. doi: 10.1161/01.ATV.0000184758.07257.88. [DOI] [PubMed] [Google Scholar]
- 54.Margariti A, Zampetaki A, Xiao Q, Zhou B, Karamariti E, Martin D, Yin X, Mayr M, Li H, Zhang Z, et al. Histone deacetylase 7 controls endothelial cell growth through modulation of beta-catenin. Circ Res. 2010;106:1202–1211. doi: 10.1161/CIRCRESAHA.109.213165. [DOI] [PubMed] [Google Scholar]
- 55.Dje N'Guessan P, Riediger F, Vardarova K, Scharf S, Eitel J, Opitz B, Slevogt H, Weichert W, Hocke AC, Schmeck B, et al. Statins control oxidized LDL-mediated histone modifications and gene expression in cultured human endothelial cells. Arterioscler Thromb Vasc Biol. 2009;29:380–386. doi: 10.1161/ATVBAHA.108.178319. [DOI] [PubMed] [Google Scholar]
- 56.Gan Y, Shen YH, Wang J, Wang X, Utama B, Wang J, Wang XL. Role of histone deacetylation in cell-specific expression of endothelial nitric-oxide synthase. J Biol Chem. 2005;280:16467–16475. doi: 10.1074/jbc.M412960200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan Y, Fish JE, D'Abreo C, Lin S, Robb GB, Teichert AM, Karantzoulis-Fegaras F, Keightley A, Steer BM, Marsden PA. The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J Biol Chem. 2004;279:35087–35100. doi: 10.1074/jbc.M405063200. [DOI] [PubMed] [Google Scholar]
- 58.Okada Y, Funahashi N, Tanaka T, Nishiyama Y, Yuan L, Shirakura K, Turjman AS, Kano Y, Naruse H, Suzuki A, et al. Endothelial cell-specific expression of roundabout 4 is regulated by differential DNA methylation of the proximal promoter. Arterioscler Thromb Vasc Biol. 2014;34:1531–1538. doi: 10.1161/ATVBAHA.114.303818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park KW, Morrison CM, Sorensen LK, Jones CA, Rao Y, Chien CB, Wu JY, Urness LD, Li DY. Robo4 is a vascular-specific receptor that inhibits endothelial migration. Dev Biol. 2003;261:251–267. doi: 10.1016/s0012-1606(03)00258-6. [DOI] [PubMed] [Google Scholar]
- 60.Suchting S, Heal P, Tahtis K, Stewart LM, Bicknell R. Soluble Robo4 receptor inhibits in vivo angiogenesis and endothelial cell migration. FASEB J. 2005;19:121–123. doi: 10.1096/fj.04-1991fje. [DOI] [PubMed] [Google Scholar]
- 61.Jiang YZ, Jimenez JM, Ou K, McCormick ME, Zhang LD, Davies PF. Hemodynamic Disturbed Flow Induces Differential DNA Methylation of Endothelial Kruppel-Like Factor 4 (KLF4) Promoter In Vitro and In Vivo. Circ Res. 2014 doi: 10.1161/CIRCRESAHA.115.303883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohtani K, Vlachojannis GJ, Koyanagi M, Boeckel JN, Urbich C, Farcas R, Bonig H, Marquez VE, Zeiher AM, Dimmeler S. Epigenetic regulation of endothelial lineage committed genes in pro-angiogenic hematopoietic and endothelial progenitor cells. Circ Res. 2011;109:1219–1229. doi: 10.1161/CIRCRESAHA.111.247304. [DOI] [PubMed] [Google Scholar]
- 63.Shirodkar AV, St Bernard R, Gavryushova A, Kop A, Knight BJ, Yan MS, Man HS, Sud M, Hebbel RP, Oettgen P, et al. A mechanistic role for DNA methylation in endothelial cell (EC)-enriched gene expression: relationship with DNA replication timing. Blood. 2013;121:3531–3540. doi: 10.1182/blood-2013-01-479170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dunn J, Qiu H, Kim S, Jjingo D, Hoffman R, Kim CW, Jang I, Son DJ, Kim D, Pan C, et al. Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis. J Clin Invest. 2014;124:3187–3199. doi: 10.1172/JCI74792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou J, Li YS, Wang KC, Chien S. Epigenetic Mechanism in Regulation of Endothelial Function by Disturbed Flow: Induction of DNA Hypermethylation by DNMT1. Cell Mol Bioeng. 2014;7:218–224. doi: 10.1007/s12195-014-0325-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerzsten RE, Edelman ER, Jain MK. Kruppel-like factor 4 regulates endothelial inflammation. The Journal of biological chemistry. 2007;282:13769–13779. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- 67.Wu W, Xiao H, Laguna-Fernandez A, Villarreal G, Jr., Wang KC, Geary GG, Zhang Y, Wang WC, Huang HD, Zhou J, et al. Flow-Dependent Regulation of Kruppel-Like Factor 2 Is Mediated by MicroRNA-92a. Circulation. 2011;124:633–641. doi: 10.1161/CIRCULATIONAHA.110.005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamik A, Jain MK. MiRrored regulation of KLF2 and KLF4. Arteriosclerosis, thrombosis, and vascular biology. 2012;32:839–840. doi: 10.1161/ATVBAHA.112.245563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huddleson JP, Ahmad N, Srinivasan S, Lingrel JB. Induction of KLF2 by fluid shear stress requires a novel promoter element activated by a phosphatidylinositol 3-kinase-dependent chromatin-remodeling pathway. J Biol Chem. 2005;280:23371–23379. doi: 10.1074/jbc.M413839200. [DOI] [PubMed] [Google Scholar]
- 70.Villarreal G, Jr., Zhang Y, Larman HB, Gracia-Sancho J, Koo A, Garcia-Cardena G. Defining the regulation of KLF4 expression and its downstream transcriptional targets in vascular endothelial cells. Biochem Biophys Res Commun. 2010;391:984–989. doi: 10.1016/j.bbrc.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanzer A, Amemiya CT, Kim CB, Stadler PF. Evolution of microRNAs located within Hox gene clusters. J Exp Zool B Mol Dev Evol. 2005;304:75–85. doi: 10.1002/jez.b.21021. [DOI] [PubMed] [Google Scholar]
- 72.Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rossig L, Urbich C, Bruhl T, Dernbach E, Heeschen C, Chavakis E, Sasaki K, Aicher D, Diehl F, Seeger F, et al. Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. J Exp Med. 2005;201:1825–1835. doi: 10.1084/jem.20042097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han L, Witmer PD, Casey E, Valle D, Sukumar S. DNA methylation regulates MicroRNA expression. Cancer Biol Ther. 2007;6:1284–1288. doi: 10.4161/cbt.6.8.4486. [DOI] [PubMed] [Google Scholar]
- 75.Pei L, Choi JH, Liu J, Lee EJ, McCarthy B, Wilson JM, Speir E, Awan F, Tae H, Arthur G, et al. Genome-wide DNA methylation analysis reveals novel epigenetic changes in chronic lymphocytic leukemia. Epigenetics : official journal of the DNA Methylation Society. 2012;7:567–578. doi: 10.4161/epi.20237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci U S A. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gorski DH, Walsh K. The role of homeobox genes in vascular remodeling and angiogenesis. Circ Res. 2000;87:865–872. doi: 10.1161/01.res.87.10.865. [DOI] [PubMed] [Google Scholar]
- 78.Boudreau N, Andrews C, Srebrow A, Ravanpay A, Cheresh DA. Induction of the angiogenic phenotype by Hox D3. J Cell Biol. 1997;139:257–264. doi: 10.1083/jcb.139.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boudreau NJ, Varner JA. The homeobox transcription factor Hox D3 promotes integrin alpha5beta1 expression and function during angiogenesis. J Biol Chem. 2004;279:4862–4868. doi: 10.1074/jbc.M305190200. [DOI] [PubMed] [Google Scholar]
- 80.Myers C, Charboneau A, Boudreau N. Homeobox B3 promotes capillary morphogenesis and angiogenesis. J Cell Biol. 2000;148:343–351. doi: 10.1083/jcb.148.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen Y, Xu B, Arderiu G, Hashimoto T, Young WL, Boudreau N, Yang GY. Retroviral delivery of homeobox D3 gene induces cerebral angiogenesis in mice. J Cereb Blood Flow Metab. 2004;24:1280–1287. doi: 10.1097/01.WCB.0000141770.09022.AB. [DOI] [PubMed] [Google Scholar]
- 82.Mace KA, Hansen SL, Myers C, Young DM, Boudreau N. HOXA3 induces cell migration in endothelial and epithelial cells promoting angiogenesis and wound repair. J Cell Sci. 2005;118:2567–2577. doi: 10.1242/jcs.02399. [DOI] [PubMed] [Google Scholar]
- 83.Myers C, Charboneau A, Cheung I, Hanks D, Boudreau N. Sustained expression of homeobox D10 inhibits angiogenesis. Am J Pathol. 2002;161:2099–2109. doi: 10.1016/S0002-9440(10)64488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rhoads K, Arderiu G, Charboneau A, Hansen SL, Hoffman W, Boudreau N. A role for Hox A5 in regulating angiogenesis and vascular patterning. Lymphat Res Biol. 2005;3:240–252. doi: 10.1089/lrb.2005.3.240. [DOI] [PubMed] [Google Scholar]
- 85.Arderiu G, Cuevas I, Chen A, Carrio M, East L, Boudreau NJ. HoxA5 stabilizes adherens junctions via increased Akt1. Cell Adh Migr. 2007;1:185–195. doi: 10.4161/cam.1.4.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu Y, Cuevas IC, Gabriel RA, Su H, Nishimura S, Gao P, Fields A, Hao Q, Young WL, Yang GY, et al. Restoring transcription factor HoxA5 expression inhibits the growth of experimental hemangiomas in the brain. J Neuropathol Exp Neurol. 2009;68:626–632. doi: 10.1097/NEN.0b013e3181a491ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen H, Rubin E, Zhang H, Chung S, Jie CC, Garrett E, Biswal S, Sukumar S. Identification of transcriptional targets of HOXA5. J Biol Chem. 2005;280:19373–19380. doi: 10.1074/jbc.M413528200. [DOI] [PubMed] [Google Scholar]
- 88.Lee JY, Park KS, Cho EJ, Joo HK, Lee SK, Lee SD, Park JB, Chang SJ, Jeon BH. Human HOXA5 homeodomain enhances protein transduction and its application to vascular inflammation. Biochem Biophys Res Commun. 2011;410:312–316. doi: 10.1016/j.bbrc.2011.05.139. [DOI] [PubMed] [Google Scholar]
- 89.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim YJ, Cecchini KR, Kim TH. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc Natl Acad Sci U S A. 2011;108:7391–7396. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Raney BJ, Cline MS, Rosenbloom KR, Dreszer TR, Learned K, Barber GP, Meyer LR, Sloan CA, Malladi VS, Roskin KM, et al. ENCODE whole-genome data in the UCSC genome browser (2011 update). Nucleic Acids Res. 2011;39:D871–875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Strathdee G, Sim A, Soutar R, Holyoake TL, Brown R. HOXA5 is targeted by cell-type-specific CpG island methylation in normal cells and during the development of acute myeloid leukaemia. Carcinogenesis. 2007;28:299–309. doi: 10.1093/carcin/bgl133. [DOI] [PubMed] [Google Scholar]
- 94.Kim SY, Hwang SH, Song EJ, Shin HJ, Jung JS, Lee EY. Level of HOXA5 hypermethylation in acute myeloid leukemia is associated with short-term outcome. Korean J Lab Med. 2010;30:469–473. doi: 10.3343/kjlm.2010.30.5.469. [DOI] [PubMed] [Google Scholar]
- 95.Miftakhova R, Sandberg T, Hedblom A, Nevzorova T, Persson JL, Bredberg A. DNA methylation in ATRA-treated leukemia cell lines lacking a PML-RAR chromosome translocation. Anticancer Res. 2012;32:4715–4722. [PubMed] [Google Scholar]
- 96.Humbert M, Halter V, Shan D, Laedrach J, Leibundgut EO, Baerlocher GM, Tobler A, Fey MF, Tschan MP. Deregulated expression of Kruppel-like factors in acute myeloid leukemia. Leuk Res. 2011;35:909–913. doi: 10.1016/j.leukres.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 97.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 100.Jjingo D, Conley AB, Yi SV, Lunyak VV, Jordan IK. On the presence and role of human gene-body DNA methylation. Oncotarget. 2012;3:462–474. doi: 10.18632/oncotarget.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cao Q, Wang X, Jia L, Mondal AK, Diallo A, Hawkins GA, Das SK, Parks JS, Yu L, Shi H, et al. Inhibiting DNA Methylation by 5-aza-2'-deoxycytidine Ameliorates Atherosclerosis Through Suppressing Macrophage Inflammation. Endocrinology. 2014 doi: 10.1210/en.2014-1595. en20141595. [DOI] [PMC free article] [PubMed] [Google Scholar]