Abstract
Background
Although the complete genome sequences of a large number of organisms have been determined, the exact proteomes need to be characterized. More specifically, the extent to which post-translational processes such as proteolysis affect the synthesized proteins has remained unappreciated. We examined this issue in selected protein phosphatases of the protease-rich malaria parasite, Plasmodium falciparum.
Results
P. falciparum encodes a number of Ser/Thr protein phosphatases (PP) whose catalytic subunits are composed of a catalytic core and accessory domains essential for regulation of the catalytic activity. Two examples of such regulatory domains are found in the Ca+2-regulated phosphatases, PP7 and PP2B (calcineurin). The EF-hand domains of PP7 and the calmodulin-binding domain of PP2B are essential for stimulation of the phosphatase activity by Ca+2. We present biochemical evidence that P. falciparum generates these full-length phosphatases as well as their catalytic cores, most likely as intermediates of a proteolytic degradation pathway. While the full-length phosphatases are activated by Ca+2, the processed cores are constitutively active and either less responsive or unresponsive to Ca+2. The processing is extremely rapid, specific, and occurs in vivo.
Conclusions
Post-translational cleavage efficiently degrades complex full-length phosphatases in P. falciparum. In the course of such degradation, enzymatically active catalytic cores are produced as relatively stable intermediates. The universality of such proteolysis in other phosphatases or other multi-domain proteins and its potential impact on the overall proteome of a cell merits further investigation.
Background
The cellular proteome, the end product in the flow of genetic information in biology [1], is generally well predicted from the genome sequence [2]. The initially translated polypeptides, however, may undergo a variety of post-translational modifications that must be determined experimentally [3,4]. A major post-translation event in the life of a protein is proteolytic processing [5]. Although proteolysis is undoubtedly responsible for the general degradation and turn over of all proteins, it often constitutes a physiologically important mechanism of biological regulation for specific proteins and pathways. Among the best studied examples are: the production of enzymes from zymogens (such as trypsin from trypsinogen) [6], generation of active caspases from procaspases [7], and cleavage of viral polyproteins to generate functional proteins [8]. Clearly, in such cases, the processed products have a half-life long enough to be physiologically useful. The existence of multiple proteases in all living cells that differ in specificity, kinetics and regulation [5] further suggests that their roles may extend beyond being purely degradative. In spite of its potential importance, however, the impact of proteolytic processing on the global proteome has remained under-investigated.
P. falciparum is a protozoan parasite of the Apicomplexan family and causative agent of deadly malaria [9]. Based on its genome sequence, P. falciparum is predicted to contain only about 5,500 proteins, as compared to about 40,000 of its human victim [10]. The parasite goes through a complex series of life stages spanning asexual and sexual stages in the human and mosquito hosts, respectively, and marked by stage-specific up- and down-regulation of many genes. Recent DNA microarray studies have revealed an intricate continuum of transcriptional regulation of the vast majority of parasitic genes over the 48-hour developmental cycle in human erythrocytes [11,12]. Because of the high activity of proteases in the parasite, we reasoned that proteolytic processing may represent a major post-translational event in this organism.
In this communication, we document post-translational processing for two Ca+2-activated serine/threonine (Ser/Thr) protein phosphatases of P. falciparum, namely PP2B/calcineurin and PP7/RdgC. These two enzymes possess common as well as contrasting features. Protein phosphatase 2B, also known as calcineurin (and called protein phosphatase 3 in the new nomenclature), is a Ca+2-stimulated Ser/Thr phosphatase that plays important roles in various biochemical pathways such as T-cell signalling [13,14]. The holoenzyme is a heterodimer composed of the catalytic subunit A (calcineurin A or CnA) and the regulatory subunit B (CnB). In addition to the catalytic domain, CnA contains a CaM (calmodulin)-binding domain, a CnB-binding domain and an autoinhibitory domain [15-21]. The CnB subunit itself is a CaM-like Ca+2-binding protein that contains four high-affinity Ca+2-binding EF hands [15,16]. Binding of Ca+2 to CnB stimulates phosphatase activity, although to a lesser extent than when CaM is also present [15,16,19,22]. Finally, CnB is also required for the inhibition of calcineurin by cyclophilin-cyclosporin A (Cyp-CsA) complex [23]. Crystal structures of calcineurin and its complex with CyP-CsA have been solved [15,16,24,25] and thus, the mutual interacting domains and amino acid residues are well-defined. PP7 is a Ca+2-activated EF-hand phosphatase, orthologs of which have been characterized in Drosophila and human [26-28].
Our results presented here suggest that the catalytic subunits of both phosphatases are produced as active full-length polypeptides but are subsequently trimmed to shorter active proteins that are also enzymatically active but have lost important regulatory domains.
Results
Characterization and recombinant expression of PfPP2B (calcineurin)
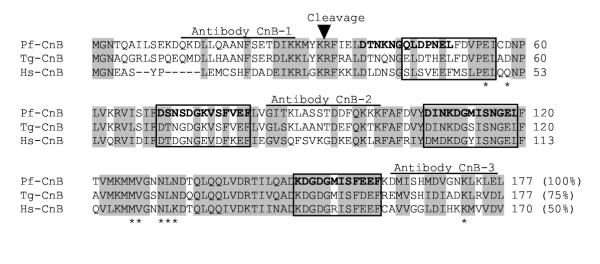
The P. falciparum sequence database (PlasmoDB) shows that the putative homologs of both subunits of calcineurin exist in the parasite, although the predicted CnA sequence has undergone revisions over the last few years. We will refer to the P. falciparum subunits with a Pf prefix, viz., PfCnA and PfCnB. Using specific oligonucleotide primers both cDNAs were amplified by RT-PCR, cloned and sequenced; the accession numbers for PfCnA and PfCnB were PF08_0129 (PlasmoDB) and AAO33818 (GenBank) respectively. Multiple alignments of selected CnA and CnB homologs are presented in Figs. 1 and 2, respectively, which show the high homology of the P. falciparum sequences with the others including those of Toxoplasma gondii, especially in the functionally conserved domains. The Apicomplexan enzymes are homologous to the α and γ isoforms of CnA rather than β, because they lack the N-terminal polyproline motif that is a conserved feature of the β isoform. The minimal catalytic core of Ser/Thr phosphatases of the PPP family (i.e., essentially all Ser/Thr phosphatases except PP2C) is about 230 amino acid long and is equivalent to the bacteriophage lambda phosphatase [29,30]. In PfCnA this region roughly corresponds to residue 43–285 and contains all the PPP signature motifs and conserved residues known to be important in catalysis (Fig. 1). Based on homology to the human CnA, the putative CnB-binding and CaM-binding regions of PfCnA span residues 328–390 and 390–414 respectively (Fig. 1). Interestingly, the PfCnA sequence has an approximately 130-residue insert between these two regions. The only other well-characterized Apicomplexan CnA, that of Toxoplasma gondii, also contains an insert in the same position that is about 38-residue long (Fig. 1). Obviously, regardless of their role, these peptide insertions do not disrupt the functions of the CnB- and CaM-binding domains.
Figure 1.
Sequence similarity and cleavage sites of the plasmodial calcineurin A (CnA) subunit. The P. falciparum (Pf) CnA sequence has been described in the Results. The accession numbers of other CnA sequences are: T. gondii (Tg) AAM97278, Paramecium tetraurelia (Pa) AF014922, H. sapiens (Hs) NP_000935. The sequences were aligned using the ClustalW program at the European Bioinformatics Institute (EMBL) server, and later refined by visual inspection. The numbers of amino acid residues are shown on the right. Residues that are identical or conservatively replaced (ST, AGILMV, DE, HKR, NQ, FWY) in all four sequences are highlighted in gray. The catalytic core, the CnB-binding, CaM-binding and the autoinhibitory domains are so marked based on studies of human CnA [15, 16, 19, 21, 25]. The catalytic domain is boxed. Residues that interact with the CsA-CyP complex are asterisked [24, 25]. The cleavage site of the plasmodial enzyme is marked by the closed triangle (between E527 and R528). Antibodies were made against synthetic peptides of the sequence TEEPPDYKALRDHLKKEGR, NCSMNDTDEGINDIV and RTLRKENELIVQLKGCSPG as shown, and used in immunoblot analysis in Fig. 3. The insertions in the Plasmodium and Toxoplasma sequences have been deleted for the sake of multiple alignment and space, but are shown on the bottom; the open triangle marks their location. The % numbers at the end represent amino acid identities of the Plasmodium sequence with the others.
Figure 2.
Sequence similarity and cleavage sites of the plasmodial calcineurin B (CnB) subunit. The accession number of the CnB subunits are: P. falciparum (Pf) AAO33818, T. gondii (Tg) AAM97279, H. sapiens (Hs) NP_000936. The cleavage site is indicated by the closed triangle (between K32 and R33). The four EF-hand motifs are boxed. Antibodies were made against synthetic peptides of the sequence QKDLLQAANFSETDIK, GITKLASSTDDFQKKK and DMISHMDVGNKLKLE as shown, and used in immunoblot analysis in Fig. 4. Residues that contact CyP-CsA are asterisked [24, 25]. The % numbers at the end represent amino acid identities of the Plasmodium sequence with the others.
To characterize the biochemistry of the P. falciparum calcineurin, both subunits were expressed in the E. coli-pET system and purified by affinity chromatography as described in Methods. The relative mobility (Mr) of PfCnA and PfCnB in SDS-PAGE were 69 k and 21 k, respectively, which roughly matched their predicted sizes of 70 kDa and 20 kDa plus about 2 kDa added for the His tags at the N- and C-terminus, respectively (Figs. 3,4; lane 3).
Figure 3.
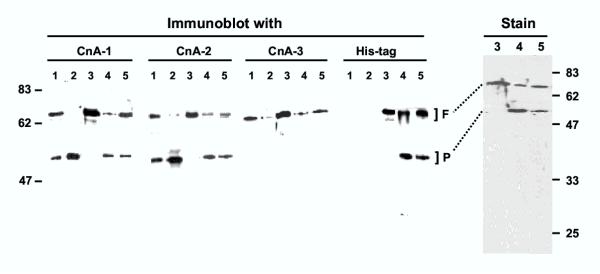
Processing of PfCnA in vivo and in vitro. Immunoblot analysis was performed with peptide antibodies CnA-1, -2 and -3 (described in Fig. 1) and with the anti-His tag antibody. The lanes contained the following samples: 1 = 50 μg total protein from freshly isolated parasite; 2 = same as 1 except that the parasite was incubated at room temperature for 5 min; 3 = 120 ng of purified His-tagged (recombinant) PfCnA; 4 = as in lane 3 but following incubation with parasite extract; 5 = as in lane 4 except that the incubation mixture also contained protease inhibitor cocktail. In the right panel (Stain) higher quantities (5 μg) of the same recombinant protein samples of the corresponding lane numbers were analyzed and the gel stained with Coomassie Blue R250. The full-length and processed bands are indicated as F and P, respectively. Size markers are indicated on the two sides. Details are given in Methods.
Figure 4.
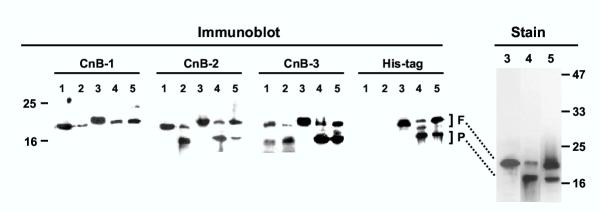
Processing of PfCnB in vivo and in vitro. Immunoblot analysis was performed with the antibodies CnB-1, -2 and -3 (described in Fig. 2) and the anti-His tag antibody on the following samples. 1 = total protein (60 μg) from freshly isolated parasite; 2 = same as lane 1 except that the parasite was incubated at room temperature for 5 min; 3 = purified recombinant His-tagged PfCnB (120 ng); 4 = as in lane 3 but after incubation with parasite extract; 5 = as in lane 4 except that the incubation mixture also contained protease inhibitor cocktail. In the right panel (Stain) higher quantities (5 μg) of the same recombinant protein samples of the corresponding lane numbers were analyzed and the gel stained with Coomassie Blue R250. The full-length and processed bands are indicated as F and P, respectively. Size markers are indicated on the two sides. Details are given in Methods.
Proteolytic processing of P. falciparum calcineurin in vivo and in vitro
We previously reported a protein phosphatase activity purified from erythrocyte-grown P. falciparum that exhibited calcineurin-like properties [31]. The activity was stimulated by Ca+2 and calmodulin, and was inhibited by the cyclophilin-CsA (cyclosporin A) complex, which are hallmarks of calcineurin. Strangely, however, the polypeptide pattern of the parasitic preparation, as reported before [29], does not match the sizes of the full-length recombinant PfCnA and PfCnB described here. Whereas the recombinant PfCnA and PfCnB are 70 and 20 kDa, respectively (Figs. 1,2,3,4), the previously published calcineurin preparation was composed of 55 kDa and 16 kDa polypeptides on the basis of mobility in SDS-PAGE [31]. To determine whether we were dealing with multiple species of calcineurin in P. falciparum, we subjected each of these proteins to peptide mass spectroscopic analysis followed by trypsinization. The 55 kDa band and the recombinant PfCnA indeed showed common peptides, as did the 16 kDa band and recombinant PfCnB, suggesting that these proteins had common sequences. The simplest possibility at this point was that the 55 kDa and the 16 kDa bands were fragments of full-length PfCnA and PfCnB, respectively, perhaps generated by proteolytic processing. In what follows, we provide further evidence to confirm this.
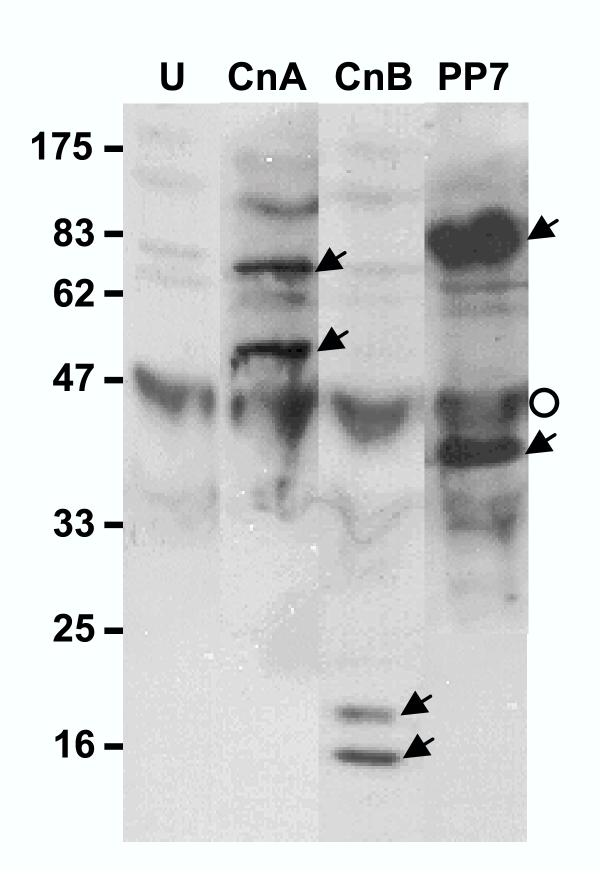
We first developed three peptide antibodies each for PfCnA and PfCnB proteins. The peptides corresponded to N-terminal, internal and C-terminal sequences – and were designated 1, 2 and 3, respectively. For example, in PfCnA, they were named CnA-1, CnA-2 and CnA-3, etc. These peptides are marked in the PfCnA and PfCnB sequences in Fig. 1 and Fig. 2, respectively. We reasoned that if proteolysis was the underlying mechanism, then it had to be catalyzed by parasitic proteases, since the recombinants expressed in E. coli were full-length. To investigate if the conversion was time-dependent, the parasitic pellet obtained after saponification of the infected RBC was held at room temperature for increasing lengths of time, and then boiled in SDS sample buffer [32], followed by immunoblot analysis using the three antibodies. These results are shown in the immunoblot panels of Figs. 3 and 4, for PfCnA and PfCnB, respectively. It is clear that for both proteins, all three antibodies were able to detect the full-length PfCnA and PfCnB (70 and 20 kDa, respectively) (in lanes numbered 1), demonstrating the activity of the antibodies in immunoblot. Even the freshest parasites, however, additionally contained significant amounts of the processed proteins. The processed 55 kDa band of PfCnA could be detected with N-terminal CnA-1 and internal CnA-2 antibodies (Fig. 3) but not with the C-terminal CnA-3 antibody. Reciprocally, the processed 16 kDa fragment of PfCnB could be detected with the internal CnB-2 and the C-terminal CnB-3 antibodies but not with the N-terminal CnB-1 antibody (Fig. 4). The ratio of intensities of the full-length and the processed bands from multiple experiments revealed that 40–50% of CnA and 20–35% of CnB in the freshly isolated parasite could be found processed. These results suggested that most of the processing had already occurred in vivo. Essentially all the full-length protein disappeared by 5 min with concomitant increase of the processed bands, attesting to the high efficiency of processing. As shown in the immunoblot of Fig. 5 (lanes CnA and CnB), the processed fragments could be detected even when the infected erythrocytes were directly boiled in SDS sample buffer and analyzed in SDS-PAGE without wasting any time for the isolation of the parasite. We infer that the processing of PfCnA and PfCnB, regardless of its mechanism, occurs in vivo.
Figure 5.
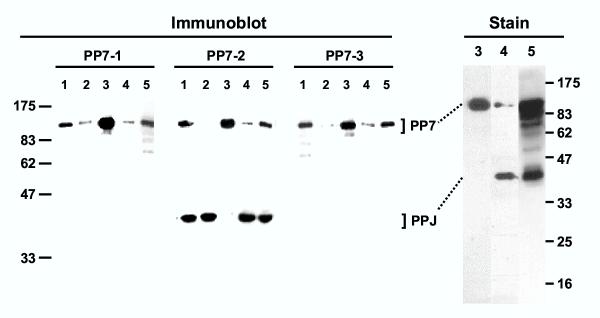
Processed phosphatases in vivo. P. falciparum-infected erythrocytes (RBC) at 15% parasitemia were centrifuged at 12,000 × g in cold for 1 min and the pellet resuspended in 50 μl SDS sample buffer [32], followed by boiling (5 min). The undissolved material (mostly RBC skeleton and parasitic hemozoin) was removed by centrifugation and the supernatant (250 μg protein per lane) analyzed by immunoblot using internal peptide antibodies against the indicated proteins (CnA-2, CnB-2 and PP7-2, respectively; compare with the bands in Fig. 3, 4, and 7, respectively). The two bands (full-length and processed) in each lane are indicated by arrowheads. Lane U represents a similarly processed sample from uninfected RBC, probed with a 1:1:1 mixture of all three antibodies. The background bands are due to nonspecific binding of the IgG's to various RBC components, the darkest of which is marked by the open circle. The size markers are shown on the left.
To pave the way to the mechanism of the processing, we attempted to establish an in vitro processing reaction. We reasoned that if the processing is a post-translational event, it may be possible to reproduce it in vitro using the bacterial recombinant proteins and catalytic amounts of P. falciparum extract as the potential source of the processing activity. This was indeed found to be true. As shown in the immunoblots in Figs. 3 and 4 (lanes numbered 4), substantial portions of the recombinant PfCnA and PfCnB were converted to the shorter fragments whose size and antibody reactivity matched to those of the corresponding in vivo processed bands (compare lane 1 or 2 with lane 4 in a given antibody panel). The His-tags added ~2 kDa to the molecular weight of the recombinants; hence their slower mobility as observed. The presence of the tags in the processed recombinant bands was confirmed by their reactivity to the respective anti-tag antibodies (panels labeled "His-tag" in Fig. 3 and 4), which further proved that the N-terminus of PfCnA and C-terminus of PfCnB were intact in the processed fragments. In summary, the in vitro cleavage appeared to mimic in vivo processing with respect to Mr in SDS-PAGE and antibody cross-reactivity.
To gain a preliminary knowledge of the nature of the putative protease, we attempted to inhibit the in vitro processing by a cocktail of protease inhibitors described under Methods. This caused a retardation of the processing, although a complete inhibition could not be achieved (Figs. 3 and 4, all lanes numbered 5; compare with unprocessed bands in lanes numbered 4 in the same panel). As antibodies may vary in efficiency and may not provide a good quantitation of the antigens, the cleavage product of the recombinant proteins were also analyzed by direct staining. Results in Fig. 3 and 4 (Panels labeled "Stain") showed that the stained pattern strongly matched the immunoblot of the corresponding samples, thus validating the conclusions. It is also to be noted that for both PfCnA and PfCnB, the other half of the protein could not be detected, suggesting that it is rapidly proteolyzed into smaller fragments. Together, these results strongly suggest that PfCnA and PfCnB are processed by a highly efficient parasitic protease or proteases in vivo.
The reactivity of the processed bands to the peptide antibodies provided an approximate location of the processing sites (Fig. 1 and 2). The availability of relatively large quantities of the bacterial recombinant proteins prompted us to determine the processing site more accurately by either mass spectrometric analyses or N-terminal sequence analyses of the C-terminal fragments. For PfCnA, where the 55 kDa processed product corresponded to the N-terminus (Fig. 1), the best choice was to subject it to an accurate mass determination. This led to the identification of the major cleavage site to be between Glu527 and Arg528 (Fig. 1). For PfCnB, the processed product was the C-terminal fragment (Fig. 2), which was subjected to N-terminal sequence analysis. This led to the placement of the major cleavage site between Lys32 and Arg33 (Fig. 2), which was also confirmed by the determination of mass.
Thus, processing of PfCnA occurred between the putative CnB-binding and CaM-binding domains (Fig. 1). In other words, the processing led to the loss of the putative CaM-binding domain, while the rest of the protein remained intact.
Altered regulation of parasitic calcineurin as a result of processing
Once we roughly mapped the processing sites, we set out to determine the potential effects of processing on enzymatic activity. The phosphatase activities of the full-length and cleaved recombinants were determined under various combinations of CaM and Ca+2, and presented in Table 1. The properties of the purified parasitic enzyme (processed) were described previously [31]; they were further confirmed here (data not shown) and found to be essentially identical to those of the processed recombinant enzyme. The following important points emerge from these results. First, the full-length holoenzyme had a slightly lower basal activity than the processed one (1.2 U vs. 2.6 U). We believe that this is because PfCnA has an autoinhibitory (AI) domain at the C-terminus (Fig. 1). Although the exact length of the AI domain in PfCnA needs to be determined, deletion of the corresponding region from human CnA led to increased activity [19,21]. We then tested the effect of proteolysis on the responsiveness of the recombinant enzyme to Ca+2 and CaM, and in general, the holoenzyme was found to be more responsive than the processed one. Thus, while the processed enzyme was unresponsive to Ca+2, the full-length holoenzyme was stimulated about 7-fold (Table 1). CaM stimulated the full-length enzyme nearly 24-fold, while stimulating the processed enzyme by about 5-fold. Although the putative CaM-binding domain of PfCnA was lost upon processing (Fig. 1), the processed holoenzyme obviously retained some ability to interact with CaM. The residual interaction may be mediated by another uncharacterized domain of PfCnA or by PfCnB. A combination of Ca+2 and CaM led to a 9-fold stimulation of the processed enzyme but an 80-fold stimulation of the full-length enzyme. Thus, the full-length enzyme showed a much greater activation by Ca+2 and CaM. Although the regulation of calcineurin by Ca+2 and CaM is a complex process [19], the substantially lower response of the processed enzyme is most easily explained by the loss of one Ca+2-binding EF hand in the processed PfCnB subunit and by the loss of the CaM-binding domain of the PfCnA subunit. Essentially similar relative activities before and after processing were obtained when the calcineurin-specific peptide substrate RII was used, demonstrating that the substrate-specificity of the enzyme was left intact through processing (data not shown). In summary, the proteolytic loss of selected regulatory domains of the P. falciparum calcineurin led to altered regulation while keeping the basic enzymatic function intact.
Table 1.
Altered regulation of P. falciparum phosphatases upon processing
| Additionsa | Full-length enzymeb | Processed enzymec |
| Calcineurin | ||
| None | 1.2 ± 0.5 U | 2.6 ± 0.6 U |
| Ca+2 | 8.2 ± 1.0 U | 2.8 ± 0.8 U |
| CaM | 25.4 ± 4.8 U | 12.5 ± 2.0 U |
| Ca+2, CaM | 84.6 ± 8.0 U | 21.5 ± 2.8 U |
| Ca+2, CaM, Mg+2 | 88.0 ± 9.5 U | 23.6 ± 2.9 U |
| Ca+2, CaM, OA | 82.0 ± 9.6 U | 20.0 ± 3.0 U |
| Ca+2, CaM, CyP, CsA | 4.5 ± 0.8 U | 6.1 ± 0.7 U |
| PP7 | ||
| None | 5.2 ± 0.9 U | 4.8 ± 0.6 U |
| Mg+2 | 8.5 ± 1.1 U | 5.1 ± 0.7 U |
| Mn+2 | 14.6 ± 1.8 U | 68.5 ± 8.5 U |
| Ca+2 | 6.4 ± 0.8 U | 4.8 ± 0.8 U |
| Mn+2, Ca+2 | 60.0 ± 8.0 U | 70.8 ± 9.0 U |
| Mn+2, Ca+2, CaM | 60.8 ± 7.5 U | 69.2 ± 8.2 U |
| Mn+2, Ca+2, OA | 68.8 ± 8.1 U | 70.6 ± 9.5 U |
aThe phosphatase assays using recombinant enzymes and 32P-labeled phosphohistone substrate were carried out as described under Methods. The numbers represent nanomoles of 32Pi liberated per minute per mg enzyme (protein). For calcineurin, essentially similar relative response was observed using the RII peptide as substrate (data not shown). Where indicated, the following additions were made at the indicated concentrations: Ca+2, Mg+2, Mn+2 (2 mM each), calmodulin (CaM, 40 units per ml), CyP (recombinant PfCyP19, 1 μM), cyclosporin A (CsA, 1 μM), OA (0.2 μM) [30]. bThe full-length enzymes were purified recombinant proteins; for calcineurin, a 1:1 molar mixture of recombinant tagged PfCnA and PfCnB was used. cThe processed calcineurin was a 1:1 molar mixture of the 55 kDa and 16 kDa fragments of PfCnA and PfCnB, respectively, obtained after proteolytic processing (Fig. 3, 4). Similarly, the processed PfPP7 was the ~38 kDa product obtained by proteolysis of recombinant PfPP7 (Fig. 7). The activity values of the native (processed) enzymes, purified from the parasite [31, 33], were very similar to the processed recombinant enzymes and, therefore, were not shown to conserve space. Each value is a mean from three experiments with the standard error as shown.
Finally, the full-length recombinant P. falciparum holoenzyme was found to be sensitive to cyclophilin-CsA using recombinant Plasmodium cyclophilin 19 (Cyp19) in assays described previously [31] and its Ki for Cyp19 was measured to be 360 nM, which was comparable to that of the native processed enzyme (410 nM) [31]. PfCnB was absolutely needed for this sensitivity as recombinant PfCnA alone was resistant, consistent with similar results with mammalian calcineurin [23]. Thus, processing had little effect on the CsA-sensitivity of the plasmodial enzyme.
P. falciparum protein phosphatase PP7 and its processed form, PPJ
In this section we report the biochemical characterization and processing of a second Ca+2-stimulated phosphatase in P. falciparum. Sequence homology (not shown) placed it in the Drosophila RdgC/ human PP7 phosphatase family and therefore, we named it PfPP7 [7,8,33]. Using RT-PCR with nested primers, we confirmed that the cDNA sequence predicted in PlasmoDB was correct (Accession number PF14_0224), and thus, we only show the conceptually translated protein sequence and its relevant domains (Fig. 6). As shown, the cDNA is made up of 16 exons whereby the largest exon encodes most of the catalytic core. We cloned the full-length cDNA in the pET-15b vector and expressed the 959 amino acid long protein in E. coli essentially as described earlier for PfCnA. As detailed later, the 112 kDa recombinant PfPP7 protein exhibited phosphatase activity in vitro against a variety of substrates, including 32P-phosphohistone and pNPP, and required Mn+2 and Ca+2 for optimal activity, supporting its PP7 identity.
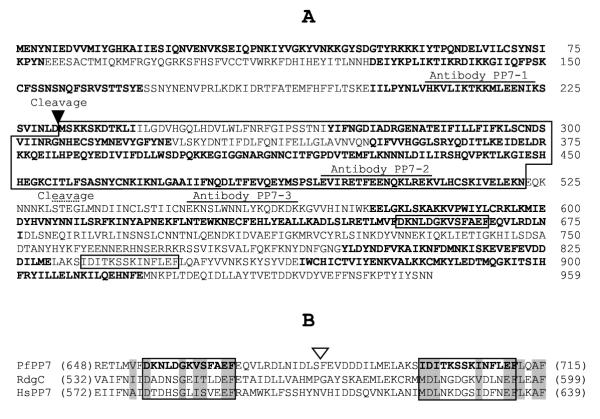
Figure 6.
The sequence and cleavage sites of PfPP7. Panel A: The complete sequence of P. falciparum PP7 is shown. Peptides coded for by alternate exons are in bold. The PfPPJ sequence is boxed. Antibodies PP7-1, -2 and -3 were made against synthetic peptides of the sequence HKVLIKTKKMLEENIK, EVIRETFEENQKLREK, EKNSLWNNLYKQDKDK, respectively (as shown) and used in immunoblot analysis in Fig. 7. The two proteolytic cleavage sites are indicated; one between D231 and M232 (closed triangle), and the other and less precise site within the STEG sequence (broken line). Note that the cleavage essentially produces PPJ from PP7. The two EF-hand motifs are boxed in both panels A and B. Panel B: Sequence homology between the three PP7 homologs of P. falciparum, Drosophila (RdgC) and human origin. The location of a 141-residue insert (not shown) in the Plasmodium sequence is indicated by the open triangle; it is absent in the other homologs.
In previous work [33], we had cloned a portion of this cDNA and showed that the corresponding protein, expressed in E. coli, was an active protein Ser/Thr phosphatase, which we named PfPPJ (Accession No. AAF19176). The PfPPJ sequence is demarcated within PfPP7 in Fig. 6, and the biochemical properties of the two are compared in Table 1. The starting Met of the PfPPJ ORF was in fact the internal in-frame Met232 of PfPP7. Because the draft Plasmodium genome sequence available at the time was missing a stop codon for the PfPP7 open reading frame, one had to be added into the PCR primer to generate the recombinant PfPPJ clone. While PfPP7 contains 959 amino acid residues, PfPPJ had only 294 residues; thus, recombinant PfPPJ in effect represented the catalytic core of PfPP7, and lacked the regulatory EF hands in the C-terminal half of PfPP7 (Fig. 6).
Interestingly, a parasitic protein band resembling the recombinant PfPPJ was previously detected in P. falciparum lysates [33], which cross-reacted with an antibody against the peptide EVIRETFEENQKLREK (PP7-2 in Fig. 6) near the C-terminus of PfPPJ, corresponding to residues 494–509 of PfPP7. To explain this, we hypothesized that PfPPJ in the parasite might have been produced by a post-translational processing of PfPP7, perhaps by a proteolytic mechanism similar to the one seen for calcineurin. To test this, we performed an essentially similar series of experiments. First, the PP7-2 antibody [33] was reused because the peptide is common to both PfPP7 and PfPPJ sequences. Additional antibodies (PP7-1 and PP7-2) were made against peptides that were in the PfPP7 sequences just outside of PfPPJ; these are all marked in Fig. 6. Freshly harvested parasitic cell pellet was then probed with these antibodies in immunoblot. Results (Fig. 7) show that PfPP7 could indeed be detected at the earliest time point, although PfPPJ was much more abundant. The ratio of the PP7 and PPJ bands in lane 1 of PP7-2 panel was 28:72, indicating that only about a third of the total PP7 remained unprocessed. Upon further incubation of the parasite pellet, PfPP7 completely disappeared and produced PfPPJ that could be detected by the internal peptide antibody (PP7-2). Essentially similar processing could be reproduced by incubating recombinant PfPP7 with small amounts of parasitic extracts as a potential source of proteases (Fig. 7, lanes 3–5). Use of protease inhibitor cocktail inhibited the processing to some extent (ranging from 30–50%) but could not completely prevent it (Fig. 7, lanes numbered 5). The stained gel pattern (Fig. 7, panel "Stain") was very similar to the immunoblot with PP7-2, thus validating each other. The cleavage was observed even when the infected erythrocytes were quickly boiled in SDS buffer and analyzed by immunoblot (Fig. 5), confirming that the processing is physiological and occurs in vivo.
Figure 7.
Processing of PfPP7 to PfPPJ in vivo and in vitro. Immunoblot analysis was performed with the antibodies PP7-1, -2 and -3 (Fig. 6) and anti-His tag antibody on the following samples: 1 = total protein (60 μg) from freshly isolated parasite; 2 = same as lane 1 except that the parasite was incubated at room temperature for 5 min; 3 = purified His-tagged recombinant PfPP7 (120 ng); 4 = as in lane 3 but after incubation with parasite extract; 5 = as in lane 4 except that the incubation mixture also contained protease inhibitor cocktail. The "Stain" panel analyzed reactions of the same lane numbers as above with higher amounts of protein (5 μg in lanes 3 and 4; 10 μg in lane 5); the gel was stained with Coomassie Blue. As indicated, the 112 kDa band is PfPP7 and the 38 kDa band approximates PfPPJ. The size markers are presented on the left and right. Details are given in Methods.
As with PfCnA and PfCnB, the approximate location of the PfPP7 cleavage sites could be gleaned from the reactivity pattern of the processed fragments with the peptide antibodies and the His-tag antibody (Fig. 6 and 7). The N-terminal cleavage site was then accurately determined by N-terminal sequencing of the PfPPJ polypeptide obtained by in vitro cleavage of the recombinant PfPP7 (Fig. 6; "Stain" panel). This sequence matched that of native PfPPJ determined previously [33], and thus, the cleavage site is between Asp231 and Met232 (Fig. 6). We then attempted to determine the C-terminal cleavage site by mass spectra of the PfPPJ produced by in vitro processing. This produced a heterogeneous data (not shown), indicating that the cleavage occurred at multiple peptide bonds between Ser531 and Gly534. This is marked in Fig. 6, showing that the processing resulted in the loss of the EF-hand motifs that are considered important for Ca+2 binding.
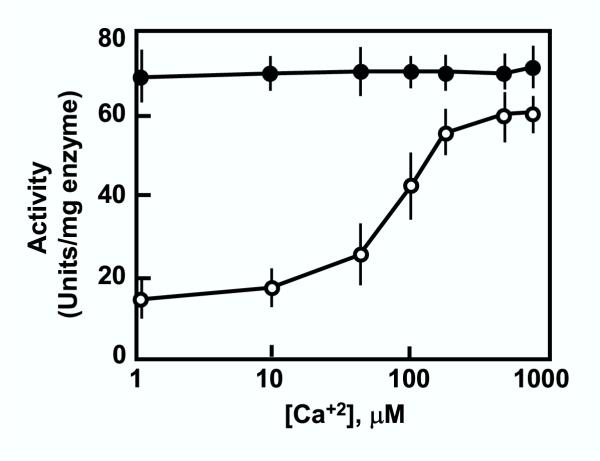
When the catalytic properties of PfPP7 and PfPPJ were compared important similarities and differences emerged. While PfPPJ was able to dephosphorylate phosphohistone as well as phosphorylase a [33], PfPP7 preferred phosphohistone. In the presence of Mn+2 and Ca+2 (1 mM each), the specific activities of PfPPJ and PfPP7 were respectively 205 and 18 units per mg enzyme with phosphorylase a as substrate, whereas the corresponding numbers were 72 and 60 with phosphohistone as substrate. For this reason, we used phosphohistone as the substrate in the rest of the paper, such as in Table 1 and Fig. 8. With both PfPP7 and PfPPJ, very little activity was seen in the absence of a divalent cation. With PfPPJ, Mn+2 alone was sufficient to activate the enzyme to the fullest extent (Table 1); Ca+2 did not cause any further activation. In sharp contrast, PfPP7 had a lower basal activity that was dependent on Mn+2 but was stimulated nearly 4-fold by Ca+2 (Fig. 7); in other words, PfPP7 was a Mn+2-dependent Ca+2-stimulated phosphatase. Mg+2 could not substitute for Mn+2. While confirming the previous results of PfPPJ [33], these results showed that unlike PfPPJ, the full-length PfPP7 is indeed a Ca+2-stimulated enzyme, true to this family of phosphatases [26]. It thus appears that Mn+2 is essential to impart proper folding to both enzymes for their basal activity and that in PP7 further stimulation of the Mn+2-containing enzyme is achieved by Ca+2 through an allosteric mechanism. The lack of Ca+2-response in PfPPJ is most easily explained by the loss of the proposed EF hand sequences upon proteolytic cleavage (Fig. 6). We do not know why the constitutively high Ca+2-independent activity of PfPPJ practically equals the Ca+2-stimulated activity of PfPP7 (Table 1). However, it is possible that as in the case of CnA, the C-terminus of PfPP7 has an autoinhibitory effect on the phosphatase activity, and that the binding of Ca+2 serves to change the conformation of this domain such that the autoinhibitory property is lost. Clearly, it will be interesting to determine whether PP7 has such an autoinhibitory domain.
Figure 8.
Stimulation of the phosphatase activity of PfPP7, but not PfPPJ, by Ca+2. The histone dephosphorylase activities of recombinant PfPP7 (open circle) and PfPPJ (closed circle) were measured as described in Methods. Each data point is a mean of three measurements with the error bar as shown.
Discussion
In this paper, we focus on two Ca+2-activated phosphatases of P. falciparum, namely calcineurin/PP2B and PP7. Biochemical studies of recombinant enzymes showed that both were highly Ca+2 dependent. The identities of the enzymes were established by sequence homology as well as general family-specific biochemical traits. For calcineurin, the diagnostic properties were: the ability to dephosphorylated the RII peptide, Ca+2-dependence, sensitivity to the CyP-CsA complex, and resistance to OA. For PP7, the most relevant properties were: stimulation by Ca+2 and resistance to OA. PP7 also needed additional divalent cation for basal activity: while the human PP7 was shown to require Mg+2, the recombinant PfPP7 required Mn+2 instead. Interestingly, the catalytic subunits of both the plasmodial enzymes contained large insertions outside the catalytic cores and between two regulatory modules. In PfCnA, the 131-residue insertion was located roughly between the CnB-binding and the CaM-binding domains (Fig. 1); and in PP7, the 141-residue insertion was located between two EF hand motifs (Fig. 6). Although the three-dimensional structure of PP7 has not been solved, the crystal structure of human calcineurin suggests that there is enough space to accommodate the insertion. In PfCnA, the cleavage occurs within the insertion, suggesting that this stretch of peptide may have a disordered solvent-exposed structure, rendering it particularly susceptible to proteolytic attack. As discussed below, a significant finding of our study is the generation of shorter, constitutively active and specific fragments of these enzymes in vivo.
Most enzymes, including protein phosphatases, contain defined catalytic and regulatory domains that are modular in nature. Protein Ser/Thr phosphatases, in particular, contain a highly conserved central catalytic core that is necessary and sufficient for phosphatase activity [30,34,35]. Most of the eukaryotic phosphatases contain additional N-terminal or C-terminal extensions that serve in regulatory roles [30,34-37]. Previously, we used recombinant DNA technology to define the minimum catalytic core of protein Ser/Thr phosphatases and showed that it is roughly equivalent to the bacteriophage lambda phosphatase [30]. Recombinantly expressed catalytic cores of PP1 and PP5 indeed exhibited unregulated enzymatic activities resembling those of the lambda phosphatase, i.e., they retained phosphatase activity but lost interaction with effectors and inhibitors [30]. For example, while PP1 is OA-sensitive, a truncated PP1 containing exclusively the lambda-homology region, is resistant [30]. The PP5 phosphatases contain a TPR domain near the N-terminus that is autoinhibitory; however, the enzyme could be activated by the interaction of unsaturated fatty acids with the TPR domain, which is a distinguishing characteristic of PP5 [38,39]. P. falciparum also has a PP5 ortholog with N-terminal TPR domains, deletion of which indeed resulted in elevated phosphatase activity accompanied by loss of activation by fatty acid [40,41].
Similar conclusions were reached by proteolytic analysis of phosphatases. The lambda phosphatase, representing the catalytic core with a compact structure [29], was highly resistant to trypsin in vitro [30]. In PP5, removal of the TPR domain by protease treatment resulted in increased activity [40]. A large body of literature also exists on the proteolysis of calcineurin, arriving at a similar conclusion. Tryptic digestion of human calcineurin in vitro led to the loss of the 17 kDa C-terminus of the CnA subunit including the CaM-binding and autoinhibitory domains [14,18,42]. The resultant fragment was essentially equivalent to the catalytic core and was highly active; however, it was no longer stimulated by Ca+2 and CaM. The tryptic cleavage in human CnA occurred after residue 392 [42], which is almost exactly the same region where PfCnA is also cleaved (Fig. 1). As shown in Fig. 1, the CnB-binding domain is contained in the processed PfCnA, which explains why the processed calcineurin purified from the parasite retained its heterodimeric nature [31]. Finally, as mentioned before, crystal structure of the trypsin-resistant human CnA-CnB in complex with cyclophilin-cyclosporin (CyP-CsA) was determined, which led to the identification of amino acid residues of CnA and CnB that play an important role in CyP-CsA interaction [24]. These residues are marked with asterisk in Fig. 1 and 2, respectively, and it is clear that the processed P. falciparum subunits retain all such residues. This explains why the processed Pf CnA-CnB heterodimer dimer retained the CyP-CsA sensitivity of the full-length enzyme (Table 1). Enzymatic assays showed that the analogous trypsin-resistant core of the human CnA-CnB also retained CyP-CsA sensitivity [43]. Clearly, constitutively active, unregulated catalytic cores could be produced from full-length enzymes.
In this paper, we provide evidence that this indeed happens in vivo. Nearly two decades ago, a preliminary report suggested processing of CnA in T cells by a Ca+2-dependent protease [44]; however, the generality or the exact mechanism of such processing was not investigated further. While this paper was in preparation, evidence was presented that during excitotoxicity in human neuronal cells, calcineurin A (CnA) is directly cleaved by the Ca+2-dependent cysteine protease, calpain. The in vivo cleavage could be inhibited by calpain inhibitors, calpastatin and ALLM (N-acetyl-Leu-Leu-methioninal) [45]. Although multiple products were produced by cleavage at C-terminal sites that were close to each other, the smallest fragment was a result of cleavage between the two basic residues, Arg392 and Lys393 (Fig. 1). As mentioned, processing in this region produces a Ca+2-independent phosphatase with higher basal activity. Recombinant overexpression of this processed fragment or constitutively active CnA in the neuronal cells led to apoptosis, characteristic of excitotoxic neurodegeneration [45]. The proteolytic activation of a Ca+2-activated phosphatase by a Ca+2-dependent protease was proposed as an intriguing regulatory pathway, responding to certain pathological states.
Thus, the plasmodial CnA is processed in a region where proteolytic processing has been known to occur in other CnA homologs. We have tentatively assumed that the processing observed for plasmodial CnA, CnB and PP7 are proteolytic phenomena based on the facts that the processing increases with time and can be partially inhibited by traditional protease inhibitors. As revealed in these immunoblots, significant processing occurred even in freshly isolated parasites and in parasites that are still inside the host RBC.
Unraveling of the exact mechanism of the processing event(s) will clearly require more extensive studies. Autoproteolysis or self-cleavage is ruled out because the purified recombinant proteins did not undergo self-processing in vitro (lane 3 in Fig. 3,4,7). A role of the Plasmodium proteasome [46] seems unlikely because neither MG132 nor lactacystin inhibited the processing in vivo or in vitro (data not shown). The particular protease inhibitor cocktail used here (see Methods) is claimed to inhibit serine, cysteine, aspartic and metalloproteases as well as calpain, the protease responsible for the processing of human CnA mentioned above. However, we have not been able to completely stop the processing of the plasmodial enzymes with this cocktail in vitro as well as in vivo (Figs. 3,4,7). At this time, we tend to believe that the processing is catalyzed by plasmodial protease(s) that may be at least partially resistant to the traditional protease inhibitors. A PlasmaDB query using the term "protease" reveals nearly two dozen proteases; however, a recent study has suggested the existence of as many as 92 protease genes in P. falciparum by sequence homology, at least 88 of which have been validated at the transcriptional level [47]. One of these is a potentially novel calpain-like protease that lacks a Ca+2-binding EF hand and therefore, appears to be Ca+2-indepenedent [47]. It is also to be noted that multiple proteases often act in concert to extensively cleave a substrate protein. For example, cleavage by acidic cysteine, aspartic, and metalloproteases followed by a neutral aminopeptidase results in the complete hydrolysis of hemoglobin into single amino acids [48]. Proteolytic cleavage of cytoskeletal proteins by cysteine and aspartic proteases is also important in erythrocytic rupture and invasion [49]. Based on these findings, we propose that an atypical calpain and other unique plasmodial proteases may be responsible for the processing of parasitic CnA, CnB and PP7 phosphatases, perhaps through a combination of endo- and exo-proteolytic reactions. A number of cysteine proteases of P. falciparum, including a calpain-like protease, are expressed mainly in the schizont stage. As schizont is a stage of high metabolic activity and a prelude to gametogenesis, it raises the intriguing possibility that the processing of calcineurin and PP7 occurs mainly at this stage. In any case, it is unlikely that this processing simply reflected a universal protein degradation as it happened in specific regions of the proteins and was not seen in many other plasmodial proteins that we and others have studied over the years. PfPP1 [50] was retested as one such negative control; self-incubation of the parasite or incubation of the recombinant PP1 with parasitic extract did not reveal any degradation of PP1 (data not shown).
In summary, we have shown that specific post-translational cleavage of plasmodial protein phosphatases in vivo can generate kinetically stable products that are more active and less regulated than their unprocessed precursors. Whether the processing itself is regulated during the erythrocytic development cycle of the parasite would be an interesting query to answer.
Conclusions
Protease treatment of phosphatases in vitro is known to result in the loss of regulatory domains and production of protease-resistant catalytic cores. We provide biochemical evidence of such post-translational processing in the protease-rich parasite, P. falciparum, whereby smaller phosphatases were rapidly generated from larger phosphatase, most likely through proteolytic cleavage. While the larger enzymes had regulatory domains that allowed their activation by Ca+2, the smaller counterparts were devoid of such domains and hence, were constitutively active. The processed enzymes approximated the protease-resistant catalytic cores of the phosphatases generated in vitro. The rapidity of the processing strongly suggested that this occurred in the parasite cells in vivo and was, therefore, physiologically relevant. It will be interesting to investigate the generality of such post-translational processing and its effect on the cellular proteome.
Methods
Recombinant proteins, peptide antibodies and immunoblot
The open reading frames (cDNA) of the proteins were amplified by RT-PCR using the primers shown in Table 2. The N-terminally His-tagged P. falciparum CnA and PP7 proteins were expressed using the pET15b vector, and the C-terminally His-tagged CnB was expressed from the pET23a vector (Novagen). All recombinants were expressed in E. coli BL21(DE3) containing pMICO by IPTG induction as described [51] and the soluble fraction of the bacterial extract was used as the source of the recombinant proteins. All three recombinants were purified by Ni+2-agarose chromatography as described in the manufacturer's protocol (Novagen); the His-tag antibody was also purchased from the same manufacturer.
Table 2.
Primers used for amplification and cloning of the phosphatase cDNAs
| cDNA cloned | Primer pair |
| PfCnA | N6CTCGAGGAACCACTGCCTGATCCGAAG |
| N6GGATCCTTATTCGTTGGATGGCCTCTTTTCGT | |
| PfCnB | N6GCTAGCATGGGAAACACACAAGCGATATTATC |
| N6CTCGAGTAATTCTAGCTTCAATTTATTTCCAAC | |
| PfPP7 | N6CTCGAGGAAAATTACAACATTGAAGATGTTG |
| N6GGATCCTTAATTATTTGAATATATATATGTAGGC |
In each pair, the first primer is the 5' sense primer, and second one is the 3' antisense primer. All primers contained a random hexanucleotide sequence (N6) for efficient restriction of the PCR product, followed by restriction sites (underlined): XhoI (CTCGAG), BamHI (GGATCC), NheI (GCTAGC). All sequences are written 5' → 3'. Note that the natural start codons (ATG) of both PfCnA and PfPP7 were removed because translation would originate from the upstream ATG of the pET-15b vector (in order to add the N-terminal His-tag). In PfCnB, the ATG was left intact but the stop codon was removed so that translation could continue into the pET-23a vector to add the C-terminal His-tag.
Peptide sequences marked in the respective Figures were synthesized and antibodies were raised in rabbits using standard procedures as described [31]. SDS-PAGE and immunoblot (Western) using chemiluminescence-based detection were performed essentially as described [40]. Mass spectrometry (MALDI-TOF) was carried out at the Proteomics Core facility and N-terminal microsequencing were performed as described [31,33]. Where mentioned, immunoblot bands were digitally scanned in a Fujifilm LAS-1000 Intelligent Dark Box II Image Reader (Fujifilm, Edison, NJ), and the intensities integrated and added using the Image Gauge v4.0 software.
Growth, lysis and analysis of P. falciparum
P. falciparum (3D7) was grown in human A-positive erythrocytes and infected cells were harvested at about 15% parasitemia and lysed by the saponin method essentially as described [51]. The processed PfPP2B (calcineurin) and PPJ were purified from the parasites using procedures established previously [31,33]. In brief, the parasites were either processed immediately or held at room temperature for various times, and unless otherwise mentioned, all subsequent fractionation procedures were performed in cold or with ice-cold buffers. The parasite cell pellet was resuspended in buffer A (50 mM Tris-Cl, pH 7.5, 10% glycerol, 1 mM DTT, 20 mM NaCl) plus 1% Triton X-100 and the "complete protein inhibitor cocktail" (Roche); one mini tablet was dissolved per 10 ml of buffer A as prescribed by the manufacturer. The suspension was sonicated and the lysate clarified by a brief centrifugation at 15,000 × g for 5 min in a refrigerated centrifuge. For SDS-PAGE and immunoblot, about 50 μg of this lysate was analyzed in each lane.
To test for parasitic proteolytic activity in vitro, a similar extract was made except that the protease inhibitor was omitted. About 100 ng of the appropriate recombinant protein was incubated with 1 μl of 1:10 dilution of the uninhibited extract in buffer A for 10 min at room temperature followed by SDS-PAGE and immunoblot analysis.
Phosphatase assay
32P-labeled phosphohistone was made by phosphorylation of histone with the catalytic subunit of protein kinase A and was used as substrate for phosphatase assay essentially as described previously [40]. Standard assays were performed in buffer A, and additional activators or inhibitors were added only where mentioned. Reactions were incubated at 37°C. Reactions were carried out in 80 μl followed by quantification of the liberated 32Pi by phosphomolybdate extraction assay. Reactions were followed with time to ensure linearity, and results were corrected by subtraction of the corresponding values from an enzyme-free reaction. Phosphatase reactions were routinely initiated with the addition of substrate. Where indicated, the following additions were made at the indicated concentrations 10 minutes prior to initiating the reaction with the addition of substrate: Ca+2, Mg+2, Mn+2 (2 mM each), calmodulin (CaM, 1 μM,), CyP (recombinant PfCyP19, 1 μM), cyclosporin A (CsA, 1 μM), OA (0.2 μM) [31,33]. Use of cyclosporin A (CsA) and recombinant P. falciparum cyclophilin 19 (CyP19) for calcineurin inhibition assay has been described before [31]. One unit of phosphatase activity was defined as nanomoles of phosphate liberated per min at 37°C. The processed recombinants were obtained by complete proteolytic digestion as in Fig. 3,4,6, except that incubation was carried out for 20 min to ensure maximum proteolysis; the preparations were essentially free of full-length proteins as monitored by SDS-PAGE (data not shown). The endogenous (processed) calcineurin was purified from the parasite essentially as described [31] and was at least 80% pure with no single contaminant exceeding 10% as judged by SDS-PAGE and staining.
Authors' contributions
RK was involved in parasite growth and provided all the parasitic materials; AM did the cloning work; AO carried out the immunoblots; BA contributed to the phosphatase assays; SB conceived, designed and coordinated the study, contributed to the phosphatase assays, and wrote most of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This research was supported in part by NIH grant AI045803 from the National Institute of Allergy and Infectious Diseases (to S. B.).
Contributor Information
Rajinder Kumar, Email: rkumar@usamail.usouthal.edu.
Alla Musiyenko, Email: musiyenkoalla@hotmail.com.
Anja Oldenburg, Email: anja.oldenburg@gmx.net.
Brian Adams, Email: badams@bbl.usouthal.edu.
Sailen Barik, Email: sbarik@jaguar1.usouthal.edu.
References
- Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- The International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Whelan SA, Hart GW. Proteomic approaches to analyze the dynamic relationships between nucleocytoplasmic protein glycosylation and phosphorylation. Circ Res. 2003;93:1047–1058. doi: 10.1161/01.RES.0000103190.20260.37. [DOI] [PubMed] [Google Scholar]
- Schweppe RE, Haydon CE, Lewis TS, Resing KA, Ahn NG. The characterization of protein post-translational modifications by mass spectrometry. Acc Chem Res. 2003;36:453–461. doi: 10.1021/ar020143l. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Kassell B, Kay J. Zymogens of proteolytic enzymes. Science. 1973;180:1022–1027. doi: 10.1126/science.180.4090.1022. [DOI] [PubMed] [Google Scholar]
- Donepudi M, Grutter MG. Structure and zymogen activation of caspases. Biophys Chem. 2002;101–102:145–153. doi: 10.1016/S0301-4622(02)00151-5. [DOI] [PubMed] [Google Scholar]
- Khan AR, Khazanovich-Bernstein N, Bergmann EM, James MN. Structural aspects of activation pathways of aspartic protease zymogens and viral 3C protease precursors. Proc Natl Acad Sci U S A. 1999;96:10968–10975. doi: 10.1073/pnas.96.20.10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouniotis DS, Proudfoot O, Minigo G, Hanley JL, Plebanski M. Malaria parasite interactions with the human host. J Postgrad Med. 2004;50:30–34. [PubMed] [Google Scholar]
- Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu Y, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, Winzeler EA. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay J, Lee J, Lee J, Lee JI, Yu JR, Jee C, Cho JH, Jung S, Lee MH, Zannoni S, Singson A, Kim do H, Koo HS, Ahnn J. Calcineurin, a calcium/calmodulin-dependent protein phosphatase, is involved in movement, fertility, egg laying, and growth in Caenorhabditis elegans. Mol Biol Cell. 2002;13:3281–3293. doi: 10.1091/mbc.E02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankunas K, Graef IA, Neilson JR, Park SH, Crabtree GR. Signaling through calcium, calcineurin, and NF-AT in lymphocyte activation and development. Cold Spring Harb Symp Quant Biol. 1999;64:505–516. doi: 10.1101/sqb.1999.64.505. [DOI] [PubMed] [Google Scholar]
- Kissinger CR, Parge HE, Knighton DR, Lewis CT, Pelletier LA, Tempczyk A, Kalish VJ, Tucker KD, Showalter RE, Moomaw EW, et al. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature. 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- Griffith JP, Kim JL, Kim EE, Sintchak MD, Thomson JA, Fitzgibbon MJ, Fleming MA, Caron PR, Hsiao K, Navia MA. X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex. Cell. 1995;82:507–522. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- Yang SA, Klee C. Study of calcineurin structure by limited proteolysis. Methods Mol Biol. 2002;172:317–334. doi: 10.1385/1-59259-183-3:317. [DOI] [PubMed] [Google Scholar]
- Milan D, Griffith J, Su M, Price ER, McKeon F. The latch region of calcineurin B is involved in both immunosuppressant-immunophilin complex docking and phosphatase activation. Cell. 1994;79:437–447. doi: 10.1016/0092-8674(94)90253-4. [DOI] [PubMed] [Google Scholar]
- Klee CB, Ren H, Wang X. Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J Biol Chem. 1998;273:13367–13370. doi: 10.1074/jbc.273.22.13367. [DOI] [PubMed] [Google Scholar]
- Yang SA, Klee CB. Low affinity Ca2+-binding sites of calcineurin B mediate conformational changes in calcineurin A. Biochemistry. 2000;39:16147–16154. doi: 10.1021/bi001321q. [DOI] [PubMed] [Google Scholar]
- Perrino BA. Regulation of calcineurin phosphatase activity by its autoinhibitory domain. Arch Biochem Biophys. 1999;372:159–165. doi: 10.1006/abbi.1999.1485. [DOI] [PubMed] [Google Scholar]
- Feng B, Stemmer PM. Ca2+ binding site 2 in calcineurin-B modulates calmodulin- dependent calcineurin phosphatase activity. Biochemistry. 2001;40:8808–8814. doi: 10.1021/bi0025161. [DOI] [PubMed] [Google Scholar]
- Haddy A, Swanson SKH, Born TL, Rusnak F. Inhibition of calcineurin by cyclosporin A-cyclophilin requires calcineurin B. FEBS Lett. 1992;314:37–40. doi: 10.1016/0014-5793(92)81456-V. [DOI] [PubMed] [Google Scholar]
- Huai Q, Kim HY, Liu Y, Zhao Y, Mondragon A, Liu JO, Ke H. Crystal structure of calcineurin-cyclophilin-cyclosporin shows common but distinct recognition of immunophilin-drug complexes. Proc Natl Acad Sci U S A. 2002;99:12037–12042. doi: 10.1073/pnas.192206699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Harrison SC. Crystal structure of human calcineurin complexed with cyclosporin A and human cyclophilin. Proc Natl Acad Sci U S A. 2002;99:13522–13526. doi: 10.1073/pnas.212504399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Honkanen RE. Molecular cloning, expression, and characterization of a novel human serine/threonine protein phosphatase, PP7, that is homologous to Drosophila retinal degeneration C gene product (rdgC) J Biol Chem. 1998;273:1462–1468. doi: 10.1074/jbc.273.3.1462. [DOI] [PubMed] [Google Scholar]
- Sherman PM, Sun H, Macke JP, Williams J, Smallwood PM, Nathans J. Identification and characterization of a conserved family of protein serine/threonine phosphatases homologous to Drosophila retinal degeneration C. Proc Natl Acad Sci U S A. 1997;94:11639–11644. doi: 10.1073/pnas.94.21.11639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva AV, Kutuzov MA. RdgC/PP5-related phosphatases: novel components in signal transduction. Cell Signal. 1999;11:555–562. doi: 10.1016/S0898-6568(99)00032-7. [DOI] [PubMed] [Google Scholar]
- Barik S. Expression and biochemical properties of a protein serine/threonine phosphatase encoded by bacteriophage lambda. Proc Natl Acad Sci USA. 1993;90:10633–10637. doi: 10.1073/pnas.90.22.10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansai T, Dupuy LC, Barik S. Interactions between a minimal protein serine/threonine phosphatase and its phosphopeptide substrate sequence. J Biol Chem. 1996;271:24401–24407. doi: 10.1074/jbc.271.40.24401. [DOI] [PubMed] [Google Scholar]
- Dobson S, May T, Berriman M, Del Vecchio C, Fairlamb AH, Chakrabarti D, Barik S. Characterization of protein Ser/Thr phosphatases of the malaria parasite, Plasmodium falciparum: inhibition of the parasitic calcineurin by cyclophilin-cyclosporin complex. Mol Biochem Parasitol. 1999;99:167–181. doi: 10.1016/S0166-6851(99)00010-9. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Dobson S, Bracchi V, Chakrabarti D, Barik S. Characterization of a novel serine/threonine protein phosphatase (PfPPJ) from the malaria parasite, Plasmodium falciparum. Mol Biochem Parasitol. 2001;115:29–39. doi: 10.1016/S0166-6851(01)00260-2. [DOI] [PubMed] [Google Scholar]
- Wera S, Hemmings BA. Serine/threonine protein phosphatases. Biochem J. 1995;311:17–29. doi: 10.1042/bj3110017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen PT. Novel protein serine/threonine phosphatases: variety is the spice of life. Trends Biochem Sci. 1997;22:245–251. doi: 10.1016/S0968-0004(97)01060-8. [DOI] [PubMed] [Google Scholar]
- Connor JH, Quan HQ, Ramaswamy NT, Zhang L, Barik S, Zheng J, Cannon JP, Lee EYC, Shenolikar S. Inhibitor-1 interaction domain that mediates the inhibition of protein phosphatase-1. J Biol Chem. 1998;273:27716–27724. doi: 10.1074/jbc.273.42.27716. [DOI] [PubMed] [Google Scholar]
- Connor JH, Kleeman T, Barik S, Honkanen RE, Shenolikar S. Importance of the β 12-β 13 loop in protein phosphatase-1 catalytic subunit for inhibition by toxins and mammalian protein inhibitors. J Biol Chem. 1999;274:22366–22372. doi: 10.1074/jbc.274.32.22366. [DOI] [PubMed] [Google Scholar]
- Sinclair C, Borchers C, Parker C, Tomer K, Charbonneau H, Rossie S. The tetratricopeptide repeat domain and a C-terminal region control the activity of Ser/Thr protein phosphatase 5. J Biol Chem. 1999;274:23666–23672. doi: 10.1074/jbc.274.33.23666. [DOI] [PubMed] [Google Scholar]
- Kang H, Sayner SL, Gross KL, Russell LC, Chinkers M. Identification of amino acids in the tetratricopeptide repeat and c-terminal domains of protein phosphatase 5 involved in autoinhibition and lipid activation. Biochemistry. 2001;40:10485–10490. doi: 10.1021/bi010999i. [DOI] [PubMed] [Google Scholar]
- Dobson S, Kar B, Kumar R, Adams B, Barik S. A novel tetratricopeptide repeat (TPR) containing PP5 serine/threonine protein phosphatase in the malaria parasite, Plasmodium falciparum. BMC Microbiol. 2001;1:31. doi: 10.1186/1471-2180-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenthal C, Klinkert MQ. Identification and biochemical characterisation of a protein phosphatase 5 homologue from Plasmodium falciparum. Mol Biochem Parasitol. 2002;120:257–268. doi: 10.1016/S0166-6851(02)00007-5. [DOI] [PubMed] [Google Scholar]
- Manalan AS, Klee CB. Activation of calcineurin by limited proteolysis. Proc Natl Acad Sci U S A. 1983;80:4291–4295. doi: 10.1073/pnas.80.14.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson SK, Born T, Zydowsky LD, Cho H, Chang HY, Walsh CT, Rusnak F. Cyclosporin-mediated inhibition of bovine calcineurin by cyclophilins A and B. Proc Natl Acad Sci U S A. 1992;89:3741–3745. doi: 10.1073/pnas.89.9.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallant EA, Brumley LM, Wallace RW. Activation of a calmodulin-dependent phosphatase by a Ca2+-dependent protease. Biochemistry. 1988;27:2205–2211. doi: 10.1021/bi00406a059. [DOI] [PubMed] [Google Scholar]
- Wu HY, Tomizawa K, Oda Y, Wei FY, Lu YF, Matsushita M, Li ST, Moriwaki A, Matsui H. Critical role of calpain-mediated cleavage of calcineurin in excitotoxic neurodegeneration. J Biol Chem. 2004;279:4929–4940. doi: 10.1074/jbc.M309767200. [DOI] [PubMed] [Google Scholar]
- Paugam A, Bulteau AL, Dupouy-Camet J, Creuzet C, Friguet B. Characterization and role of protozoan parasite proteasomes. Trends Parasitol. 2003;19:55–59. doi: 10.1016/S1471-4922(02)00064-8. [DOI] [PubMed] [Google Scholar]
- Wu Y, Wang X, Liu X, Wang Y. Data-mining approaches reveal hidden families of proteases in the genome of malaria parasite. Genome Res. 2003;13:601–616. doi: 10.1101/gr.913403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PJ. Hydrolysis of erythrocyte proteins by proteases of malaria parasites. Curr Opin Hematol. 2002;9:140–145. doi: 10.1097/00062752-200203000-00010. [DOI] [PubMed] [Google Scholar]
- Rosenthal PJ, Sijwali PS, Singh A, Shenai BR. Cysteine proteases of malaria parasites: targets for chemotherapy. Curr Pharm Des. 2002;8:1659–1672. doi: 10.2174/1381612023394197. [DOI] [PubMed] [Google Scholar]
- Kumar R, Adams B, Oldenburg A, Musiyenko A, Barik S. Characterisation and expression of a PP1 serine/threonine protein phosphatase (PfPP1) from the malaria parasite, Plasmodium falciparum : demonstration of its essential role using RNA interference. Malar J. 2002;1:5. doi: 10.1186/1475-2875-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Musiyenko A, Barik S. The heat shock protein 90 of Plasmodium falciparum and antimalarial activity of its inhibitor, geldanamycin. Malar J. 2003;2:30. doi: 10.1186/1475-2875-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]