Abstract
To search for reliable biomarkers and drug targets for management of hepatocellular carcinoma (HCC), we performed a global proteomic analysis of a pair of HCC cell lines with distinct differentiation statuses using 2-DE coupled with MALDI-TOF MS. In total, 106 and 55 proteins were successfully identified from the total cell lysate and the cytosolic, nuclear and membrane fractions in well-differentiated (HepG2) and poorly differentiated (SK-Hep–1) HCC clonal variants, respectively. Among these proteins, nine spots corresponding to proteins differentially expressed between HCC cell types were selected and confirmed by immunofluorescence staining and western blotting. Notably, Annexin 1 (ANX1), ANX–2, vimentin and stress-associated proteins, such as GRP78, HSP75, HSC–70, protein disulfide isomerase (PDI), and heat shock protein–27 (HSP27), were exclusively up-regulated in SK-Hep–1 cells. Elevated levels of ANX–4 and antioxidant/metabolic enzymes, such as MnSOD, peroxiredoxin, NADP-dependent isocitrate dehydrogenase, α-enolase and UDP-glucose dehydrogenase, were observed in HepG2 cells. We functionally demonstrated that ANX1 and HSP27 were abundantly overexpressed only in highly invasive types of HCC cells, such as Mahlavu and SK-Hep–1. Knockdown of ANX1 or HSP27 in HCC cells resulted in a severe reduction in cell migration. The in-vitro observations of ANX1 and HSP27 expressions in HCC sample was demonstrated by immunohistochemical stains performed on HCC tissue microarrays. Poorly differentiated HCC tended to have stronger ANX1 and HSP27 expressions than well-differentiated or moderately differentiated HCC. Collectively, our findings suggest that ANX1 and HSP27 are two novel biomarkers for predicting invasive HCC phenotypes and could serve as potential treatment targets.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies in the world, with a mortality rate of approximately one million each year [1, 2]. The prognosis of HCC remains poor even with a combination of chemotherapies and radiation therapies because of intrinsic and/or acquired treatment resistance and a high rate of metastasis [3, 4]. Thus, a better understanding of the biochemical and molecular properties of HCC may lead to the development of biomarkers and therapeutic strategies. Differentiation is an important cellular process that regulates the clonal increase of the cell population, and the differentiation status of a cancer cell is known to play a pivotal role in the extent of carcinogenesis and its metastatic propensity [5]. Thus, the identification of molecules that determine the differentiation status (i.e., mesenchymal or epithelial) of HCC may provide important clues for drug development.
The differentiation of human hepatocytes is particularly interesting because varieties of plasma protein markers have been well characterized [6–8]. Because HCC is a hepatocyte malignancy, Chang et al. previously proposed that the expression patterns of plasma proteins and/or plasma membrane protein markers could be used as an approach for studying human HCC differentiation status [9]. However, this technique, although specific, is laborious and time-consuming because of the necessity of analyzing at least 15 different plasma proteins secreted in the culture medium. Subsequently, some independent differentiation-associated biomarkers have been discovered [10–13], but their clinical significance has not been verified thus far.
The current interest in proteomics has arisen in part because of the prospect that a proteomic approach to disease investigation may overcome some of the limitations encountered by other methodologies [14, 15]. With this premise in mind, we aimed to identify protein biomarkers in different components of HCC cells with distinct disparities in differentiation status. The rationale for this approach is that protein expression during cell differentiation may vary among different compartments (cytosol, nucleus and membrane fractions) of HCC cells. Some of these proteins may play pivotal roles in controlling the proliferative capability and metastatic behaviors. Furthermore, the translocation of proteins to the nucleus may also be crucial in initiating various biological events. In this study, we examined the protein expression in different cellular compartments and identified candidate proteins that were overexpressed or down-regulated in two HCC cell lines with distinct differentiation states. The identified proteins and their proposed functions may provide important information for therapeutic designs and may serve as potential biomarkers for predicting disease progression or treatment responses.
Materials and Methods
Origin and characteristics of HCC cells used in this study
A panel of five HCC subline variants was selected for this study, and their differentiation statuses were established based on their morphological characteristics, secreted plasma protein profiles, pattern of lactate dehydrogenase (LD) isoenzyme expression[11], pattern of thyroid hormone β1 nuclear receptor (h-TRβ1) expression and pattern of hepatocyte-derived growth factor (HDGF) expression [16]. The HepG2 subline, a well-differentiated HCC variant, and the SK-Hep–1 subline, a poorly-differentiated HCC variant, were selected for proteomic analysis.
Cell culture
HepG2, Hep3B, and SK-Hep–1 cells were purchased from ATCC. HepJ5 and Mahlavu cell were gifted from Dr. C.S Yang, National Taiwan University and Dr. C. P. Hu, Veterans General Hospital, Taiwan [17, 18]. Those cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (Sigma Chemical Co., St. Louis. MO, USA) supplemented with 10% fetal bovine serum (FBS) (Hyclone Laboratories, Logan, UT, USA) and incubated at 37°C in a humidified atmosphere with 5% CO2.
Two-dimensional electrophoresis
Cultured cells were solubilized in lysis buffer containing 7 M urea, 2 M thiourea, 4% CHAPS, 1% IPG buffer, pH3-10 (GE Healthcare Life Sciences, Piscataway, NJ, USA), 65 mM DTT, and 10 nM PMSF. The protein concentration was measured using the Bradford assay (Bio-Rad, Hercules, CA, USA). Proteins were applied onto isofocusing gels (13-cm IPG linear strip, GE Healthcare Life Sciences, Piscataway, NJ, USA). The running conditions for the IEF were as follows: 30 V for 12 h, 100 V for 0.5 h, 250 V for 0.5 h, 500 V for 0.5 h, 1000 V for 0.5 h, 2000 V for 0.5 h, 4000 V for 0.5 h, and 8000 V for 0.5 h, up to 70 kV-h. IPG strips were equilibrated for 10 min in a solution containing 50 mM Tris-HCl (pH8.8), 6 M urea, 20% SDS, 30% glycerol, 2% DTT, and a trace of bromophenol blue, followed by 10 min in the same solution except that DTT was replaced with 2.5% iodoacetamide. The IPG gel strips were embedded in gels containing 0.5% agarose. 2-D SDS-PAGE was performed on 12% acrylamide gels (GE Healthcare, USA) at 6 mA/gel until the bromophenol blue dye front reached the bottom of the gel. After approximately 16–18 h, all gels were visualized using a mass-compatible silver stain and scanned using an image scanner (GE Healthcare, USA). Protein spots were quantified using Nonlinear Progenesis software (technical support provided by J&H Technology, Taipei, Taiwan). All experiments were repeated at least three times.
Tryptic in-gel digestion of proteins and MALDI-TOF MS
Selected protein targets of approximately 1 mm in diameter on the 2-D gels were manually excised. Spots excised from the stained gels were processed according to the standard MS sample preparation protocol [19, 20]. In-gel digestion of proteins was performed using MS-grade Trypsin Gold (Promega, Madison, WI) overnight at 37°C. Tryptic digests were extracted using 10 μL Milli-Q water initially, followed by two extractions with a total of 20 μL of 50% acetonitrile / 0.1% trifluoroacetic acid. The combined extracts were dried in a vacuum concentrator at room temperature and dissolved in 1 μL of 5% acetonitrile with 0.5% trifluoroacetic acid. The ESI-MS/MS mass spectrometer utilized for protein analysis was a Thermo LTQ-Orbitrap (Thermo Scientific, UK). The MS/MS signal was analyzed using the MASCOT search engine (www.matrixscience.com). The search parameters were defined as follows: database, NCBInr 20120129; taxonomy, viridiplantae (Green Plants); enzyme, trypsin; fixed modification, carbamidomethylation; variable modifications, oxidation; peptide MS tolerance, ± 0.5 Da; and fragment MS tolerance, ± 0.5 Da and allowance of one missed cleavage.
Separation of proteins from HCC compartments and western blotting
Subcellular fractions were prepared using the Pierce Cytoplasmic Nuclear extraction kit (Thermo, USA). In brief, harvested cells were resuspended with CERI and CERII and centrifuged at 16000x g for 5 min, and the cytoplasmic fraction (supernatant) was separated from the nuclei (pellet). Subsequently, the pellet was resuspended with NER. After centrifugation at 16000x g for 10 min, the supernatant was collected as the nuclear fraction. 2-DE lysis buffer with 8 M urea and 4% CHAPS was added to the remaining pellet to isolate the membrane fraction. The protein samples were separated using SDS-PAGE and analyzed via immunoblotting. The primary antibodies used targeted vimentin, Annexin1 (ANX1), ANX4, and heat shock protein–27 (HSP27) (1:1000; Santa Cruz Biotech., Santa Cruz, CA, USA). Enhanced chemiluminescence (Immobilon Western Chemiluminescent AP substrate or ECL reagent, Millipore, Billerica, MA, USA) was used for detection. Expression of β-actin was used to control for equal gel loading.
Immunofluorescence staining
Approximately 1×105 cells were seeded in 6-well culture plates. When the cells were 50–60% confluent, they were fixed in an iced acetone and methanol 1:1 (v/v) mixed solution for 15 min. The cells were permeabilized with PBS containing 1% (v/v) Triton X–100 and incubated with primary antibodies. To detect primary antibody binding sites, the plates were washed and stained with FITC-conjugated secondary antibodies. After washing in PBS, propidium iodide (PI) was added as a nuclear counterstain. The cells were visualized using a fluorescence microscope (Nikon).
Gene silencing
Expression of ANX1 and HSP27 in HCC cells was ablated using Mission shRNA clones from Sigma Chemical Co. (St. Louis, MO). Mission shRNA clones are sequence-verified shRNA lentiviral plasmids purchased from the National RNAi Core Facility (Taiwan, ROC) for gene silencing in mammalian cells. The parental vector (pLKO.1<-puro) enables transient transfection or stable selection through puromycin resistance. The target sequence for the human ANXA1 mRNA (NM_000700.1) gene was 5’-CATAAGGCCATAATGGTTAAA–3’. The target sequence for the human HSP27 mRNA (NM_001540.3) gene was 5’-CCGATGAGACTGCCGCCAAGT–3’. The non-target shRNA control vector (SHC002) was purchased from Sigma Chemical Co. (St. Louis, MO), and the sequence of scrambled shRNA was 5’-CAACAAGATGAAGAGCACCAA–3’. Briefly, 1.5×105 cells were washed twice with PBS and mixed with 0.5 μg of the plasmid. One pulse was applied for 20 ms under a fixed voltage of 1.4 kV on a pipette-type Neon microporator (Life Technologies). Stably transfected cells were selected by puromycin for 2 weeks. The expression level of ANX1 and HSP27 was determined by real-time PCR and western blotting.
Transwell migration assay
In vitro cell migration studies were performed using a BD Falcon cell culture insert (BD Biosciences, Durham, NC) as previously described [21]. Briefly, we suspended 1×105 cells in 500 μL of serum-free DMEM, and the cells were seeded into the upper part of each chamber. The lower compartment of each chamber was filled with 1 mL of DMEM that contained a 10% FCS serum gradient. After incubation for 24 hours at 37°C in 5% CO2, the non-migrating cells were removed from the upper surface of the membrane by scraping. Cells on the reverse side of the membrane were stained with 0.1% crystal violet. The migrating cells were counted under a microscope at 100-fold magnification.
Real-time quantitative reverse transcription-PCR (qRT-PCR) analysis
Total RNA was isolated from colon cancer cells using Trizol reagent according to the manufacturer’s instructions (Invitrogen Life Technologies). The cDNA was amplified from 2 μg of total RNA in a final volume of 20 μL using Moloney murine leukemia virus (M-MLV) reverse transcriptase at 37°C for 90 min. The sequences of the qPCR primers were 5’-CCCCCATGCCCAAGCTA -3’ (forward) and 5’-TCGAAGGTGACTGGGATGGT -3’ (reverse) for HSP27, 5’-AAAGGTGGTCCCGGATCAG -3’ (forward) and 5’-CATCCACACCTTTAACCATTATgg -3’ (reverse) for ANX1, 5'-AGCGCGGCTACAGCT–3'(forward) and 5’-GGCCATCTCTTGCTCGAAGT–3’ (reverse) for β-actin. The quantitative RT-PCR reaction was performed using ABI SYBR Green Master Mix using in an ABI StepOne system (Applied Biosystems, Grand Island, NY). Thermal cycling was performed in the ABI StepOne system. The quantitative PCR conditions were 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The cycle parameters were 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, 60°C for 1 minute, and a final extension at 72°C for 10 minutes.
Tissue microarray staining by HSP27 and ANX1
Tissue microarray sets totally including 80 cases of primary HCC (catalog No. CSA4 and CS4) were purchased from SuperBioChips Laboratories. (Seoul, South Korea). The pathologic diagnosis and tumor grading of these cases were microscopically reconfirmed by a pathologist. The grading system of World Health Organization was used. HCC was divided into well-differentiated (WD), moderately differentiated (MD) and poorly differentiated (PD). An immunohistochemical stain with HSP27 (catalog No.: ab2790. 1:750; Abcamplc, Cambridge, United Kingdom) was performed. Expression levels of HSP27 were scored semiquantitatively as weakly positive (1+), moderately positive (2+) and strongly positive (3+). To understand the correlation between pathologic characteristics and expression levels of ANX1, two tissue microarray sets totally including 88 cases of HCC (catalog No. CSA4 and CS5) were purchased from SuperBioChips Laboratories. An immunohistochemical stain with ANX1 (clone: MRG3. 1:150; Cell Marque Corp., Rocklin, California, USA) was performed. Expression levels of ANX1 were scored semiquantitatively as negative, weakly positive and strongly positive. The association between pathologic characteristics and ANX1 expression was analyzed.
Statistical analysis
All experiments were repeated a minimum of three times. All data collected from real-time RT-PCR analysis, MTT assays and migration assays are expressed as the mean ± SD. The data presented in some figures are from a single experiment that was quantitatively similar to the replicate experiments. Statistical significance was determined using Student’s t test (two-tailed) or chi-square test to compare two groups of data sets.
Results
Identification of abundantly expressed proteins in HCC cells
Approximately 450 μg of the total protein obtained from total cell lysates of either HepG2 or SK-Hep–1 cells was focused by IEF on pH 3–10 nonlinear IPG strips before being separated on a 12% polyacrylamide gel (PAGE). The spots were visualized by Coomassie blue G250 staining. Among the 60 spots that were identified, 46 were associated with HepG2 cells, and 14 spots belonged to SK-Hep–1 cells. Protein spots of interest were analyzed using MALDI-TOF and the MALDI spectra followed by identification using the MASCOT search engine. The MALDI-TOF identification of the protein spots from the lysates of HCC sublines is presented in Fig 1A and Table 1. We also isolated proteins from various cellular compartments of HepG2 and SK-Hep–1 cells. Cytosolic (500 μg), nuclear (250 μg) and membrane (250 μg) proteins were then subjected to IEF followed by PAGE. Protein spots were identified by MALDI-TOF, as indicated in Fig 1B, 1C and 1D and Tables 2, 3 and 4. Among the abundantly expressed protein spots on the 2-DE map (161 spots), a total of 106 spots (64%) belonged to the well-differentiated HepG2 cells, with the following order of distribution in each cellular compartment: total cell lysate (46; 46%)>cytosol (35; 33%)>membrane (13; 12%)>nucleus (12; 11%) (Fig 2 and Tables 2, 3 and 4). Conversely, a total of 55 spots (36%) belonged to the poorly differentiated SK-Hep–1 cells, with the following order of abundance in each cellular compartment: nucleus (18; 33%)>cytosol (16; 29%)>total cell lysate (14; 25%)>membrane (7; 13%) (Fig 2 and Tables 2, 3 and 4). Notably, the well-differentiated HCC cells overexpressed antioxidant/metabolic enzymes, such as MnSOD, peroxiredoxin (Prdx), NADP-dependent isocitrate dehydrogenase (ICDH), α-enolase and UDP-glucose dehydrogenase. In contrast, poorly differentiated HCC cells exhibited high levels of stress-associated proteins, such as Grp78, HSP75, HSC–70, protein disulfide isomerase (PDI), and HSP27 (Tables 5 and 6).
Fig 1. Representative 2D gel images of various cellular compartments depicting identified protein spots that were differentially expressed between HepG2 and SK-Hep–1 clonal variants.
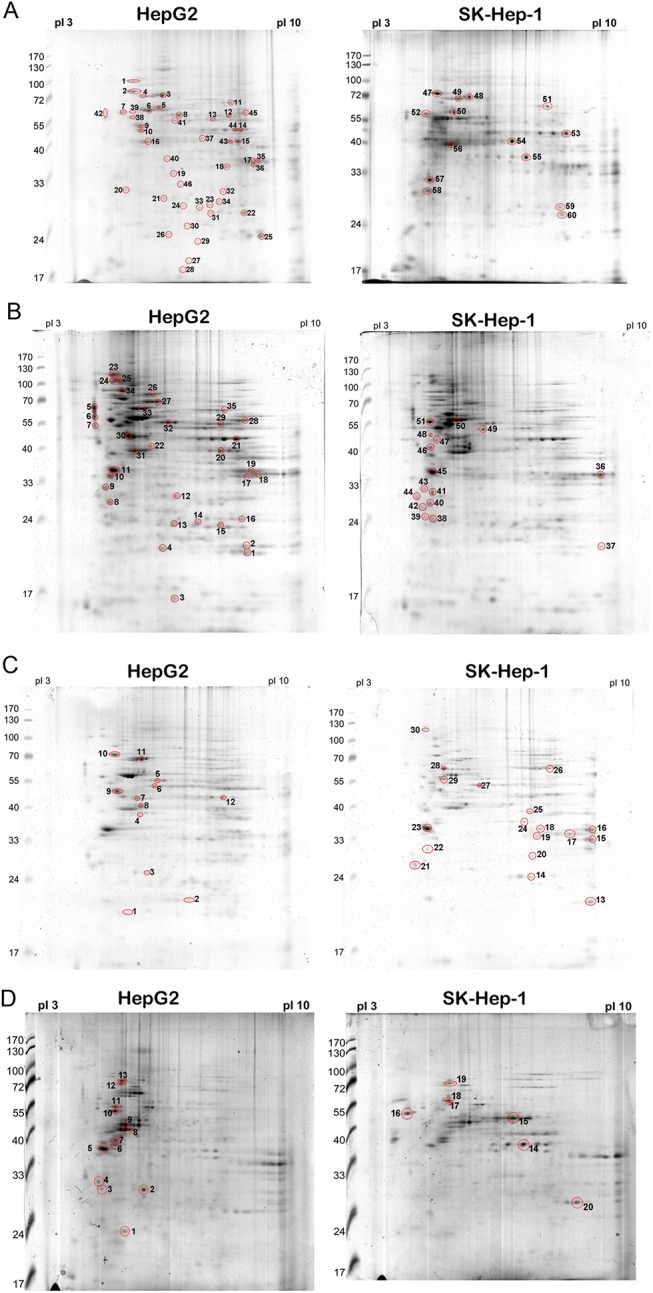
(A) Total cell lysate, (B) cytosol, (C) nucleus, and (D) membrane. Differentially expressed proteins are numbered and boxed. Reference proteins are indicated by arrowheads.
Table 1. Total proteins of HepG2 and SK-Hep–1 cells were identified by MALDI-TOF.
| Cells | Spot NO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| HepG2 | ||||||||
| 1 | Dnak-type molecular chaperone HSPA5 | 72.185/5.03 | 188(61%) | Chaperone | b | |||
| 2 | Dnak-type molecular chaperone HSPA5 | 72.185/5.03 | 256(62%) | Chaperone | b | |||
| 3 | Dnak-type chaperone precursor | 73.92/5.87 | 200(61%) | Chaperone | b | |||
| 4 | Dnak-type chaperone precursor | 71.082/5.37 | 141(56%) | Chaperone | b | |||
| 5 | P61978 | Heterogeneous nuclear ribonucleoprotein K | 47.756/5.46 | 102(46%) | Nucleus/ cytoplasm | Regulation of transcription | d | |
| 6 | P10809 | HSP 60A | 61.157/5.70 | 188(71%) | Mitochondrial matrix | Transport the mitochondrial proteins | b | |
| 7 | NP 00684 | Protein disulfide-isomerase (ER60) | 57.48/4.76 | 179(64%) | ER lumen | Folding of proteins containing disulfide bonds in ER | h, i | |
| 8 | NP 00684 | Protein disulfide-isomerase (ER60) | 57.146/5.98 | 189(60%) | ER lumen | Folding of proteins containing disulfide bonds in ER | i | |
| 9 | NP 057078 | H+-transporting two-sector ATPase | 56.525/5.26 | 145(77%) | Mitochondrion | ATP synthesis coupled proton transport | c, l | |
| 10 | P17980 | Proteasome subunit p50 | 49.458/5.13 | 88(60%) | Nucleus/ cytoplasm | Ubiquitin-dependent Protein catabolism | i | |
| Cells | Spot NO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| HepG2 | 11 | P31948 | Stress induced phosphoprotein1 (STI 1) | 65.089/6.27 | 139(66%) | Nucleus/ cytoplasm | Modulation the molecular chaperones HSC 70 and HSP 90 | b |
| 12 | NP 000680 | Aldehyde dehydrogenase 1A1 | 55.454/6.30 | 106(67%) | Cytoplasm | Alcohol metabolic enzyme | g | |
| 13 | P78371 | T complex protein 1 subunit beta | 57.794/6.01 | 185(79%) | Cytoplasm | Chaperone | b | |
| 14 | P06733 | α-enolase | 47.35/6.99 | 230(76%) | Cytoplasm | Metabolic enzyme (Glycolysis) | g | |
| 15 | NP 005887 | NADP-dependent isocitrate dehydrogenase | 46.944/6.34 | 149(75%) | Mitochondrial matrix | Metabolic enzyme (TCA cycle) | g | |
| 16 | NP 001606 | Actin-gamma | 42.108/5.31 | 124(82%) | Cytoplasm | Cytoskeleton, cell motility | c, l | |
| 17 | P42330 | Aldo-keto reductase | 37.23/8.06 | 81(61%) | Cytoplasm | Aldehyde metabolism | g | |
| 18 | P42330 | Aldo-keto reductase family 1 | 36.20/6.76 | 62(61%) | Cytoplasm | Aldehyde metabolism | g | |
| 19 | P50225 | Aryl sulfotransferase | 34.288/5.68 | 127(84%) | Cytoplasm | Amine biosynthesis | g | |
| 20 | P07951 | Tropomyosin | 28.89/4.76 | 52(20%) | Cytoplasm | Cytoskeleton, regulation of muscle contraction | g | |
| 21 | P35232 | Prohibitin | 29.589/5.57 | 214(83%) | Mitochondria inner membrane | Gene regulation and modulate the cell proliferation | d | |
| 22 | P60174 | Triosephosphate isomerase | 26.807/6.51 | 127(84%) | Metabolic enzyme (Glycolysis) | g | ||
| 23 | ER lumenal 28 | 29.032/6.77 | 80(61%) | m | ||||
| 24 | ER lumenal 28 | 29.032/6.77 | 75(57%) | m | ||||
| 25 | P30086 | PEBP | 21.027/7.42 | 81(81%) | Cytoplasm | Regulation of central nervous system | k | |
| Cells | Spot NO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| HepG2 | 26 | P32119 | Peroxiredoxin II | 21.92/5.67 | 74(49%) | Cytoplasm | Antioxidant enzyme | h |
| 27 | NP 000260 | NM23-H1 | 19.869/5.42 | 113(60%) | Nucleus/ cytoplasm | Modulate the cell proliferation and nucleotide metabolism | c, d | |
| 28 | ND | |||||||
| 29 | P52945 | Insulin activator factor | 83.906/5.6 | 69(20%) | Nucleus | Activate insulin | g | |
| 30 | ND | |||||||
| 31 | P60174 | Triosephosphate isomerase (TIM) | 26.807/6.51 | 150(76%) | Cytoplasm | Metabolic enzyme (Glycolysis) | g | |
| 32 | P25786 | Proteasome (macropain), α-type 1 | 29.864/6.15 | 86(40%) | Nucleus/ cytoplasm | Ubiquitin-dependent protein catabolism | i | |
| 33 | Enoyl-CoA hydratase | 31.807/8.34 | 167(73%) | m | ||||
| 34 | NP 002119 | High-mobility group box 1 | 18.470/9.72 | 72(44%) | Nucleus | Gene regulation | d, h | |
| 35 | P52895 | 3-α-hydroxysteroid/ dihydrodiol dehydrogenase | 37.221/8.02 | 239(85%) | Cytoplasm | Steroid metabolism | g | |
| 36 | P42330 | Aldo-keto reductase family 1 | 36.226/7.12 | 155(70%) | Cytoplasm | Aldehyde metabolism | g | |
| 37 | ND | |||||||
| 38 | P10809 | GroEL (HSP 60) | 61.187/5.70 | 139(54%) | Mitochondrial matrix | Transport the mitochondrial proteins | b | |
| 39 | P10809 | GroEL (HSP 60) | 61.187/5.70 | 139(54%) | Mitochondrial matrix | Transport the mitochondrial proteins | b | |
| 40 | NP 777480 | TGF-β receptor interacting protein 1 | 36.878/5.38 | 103(57%) | Nucleus | Gene regulation | d | |
| Cells | Spot NO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| HepG2 | 41 | NP 002264 | Keratin 8 | 41.083/4.94 | 159(72%) | Intermediate filament | Cytoskeleton organization and biogenesis | c |
| 42 | P27797 | Calreticulin variant | 47.061/4.30 | 120(38%) | ER | Protein folding | i | |
| 43 | NP 005887 | NADP-dependent isocitrate dehydrogenase | 46.931/6.53 | 144(66%) | Mitochondrial matrix | Metabolic enzyme (TCA cycle) | g | |
| 44 | P06733 | α-enolase | 47.35/6.99 | 288(73%) | Cytoplasm | Metabolic enzyme (Glycolysis) | g | |
| 45 | NP 003350 | UDP-glucose dehydrogenase | 55.674/6.73 | 225(77%) | Cytoplasm | Metablolic enzyme (Glycosaminoglycan biosynthesis) | g | |
| 46 | P09525 | Annexin A4 | 33.759/5.64 | 60(32%) | Mitochondrion | Calcium binding protein | e, f, l | |
| SK-Hep–1 | ||||||||
| 47 | P11021 | Grp 78 | 72.402/5.07 | 250(57%) | ER lumen | Protein folding in ER | i | |
| 48 | Q12931 | HSP 75 | 74.02/5.97 | 274(64%) | Mitochondrion | Expression an ATPase activity | b | |
| 49 | P11142 | Heat-shock cognate protein 70 | 71.082/5.37 | 119(53%) | Nucleus/ cytoplasm | Chaperone | b | |
| 50 | P10809 | Chaperonin | 61.187/5.70 | 212(77%) | Mitochondrial matrix | Transport the mitochondrial proteins | b | |
| 51 | P31948 | Stress induced phosphoprotein1 (STI 1) | 63.227/6.40 | 164(56%) | Nucleus/ cytoplasm | Modulation the molecular chaperones HSC 70 and HSP 90 | b | |
| 52 | NP 00684 | Protein disulfide isomerase (PDI) | 57.48/4.76 | 208(65%) | ER lumen | Folding of proteins containing disulfide bonds in ER | h, i | |
| Cells | Spot NO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| SK-Hep–1 | 53 | P06733 | α-enolase | 47.35/6.99 | 219(72%) | Cytoplasm | Metabolic enzyme (Glycolysis) | g |
| 54 | AAH01751 | Chromosome 20 open reading frame 4 | 43.843/5.66 | 59(31%) | Cytoplasm | cell growth | d | |
| 55 | P33992 | Replication licensing factor, MCM5 | 82.990/8.56 | 83(33%) | Nucleus | Initiation of DNA replication | d | |
| 56 | NP 001606 | Actin-gamma | 42.108/5.31 | 118(72%) | Cytoplasm | Cytoskeleton, cell motility | c, l | |
| 57 | P06753 | Tropomyosin3 | 32.856/4.72 | 87(58%) | Cytoplasm | Cytoskeleton, regulation of muscle contraction | c, l | |
| 58 | P06753 | Tropomyosin3 | 27.386/4.77 | 100(51%) | Cytoplasm | Cytoskeleton, regulation of muscle contraction | c, l | |
| 59 | P18669 | Phosphoglycerate mutase B | 28.90/6.67 | 141(81%) | Cytoplasm | Metabolic enzyme (Glycolysis) | g | |
| 60 | P60174 | Triosephosphate isomerase (TIM) | 26.807/6.51 | 112(82%) | Cytoplasm | Metabolic enzyme (Glycolysis) | g |
ND: None detected.
The functional classification (Class) of identified proteins is shown a ~ m. a: cell cycle; b: chaperone/ stress response; c: cytoskeleton/ cell mobility; d: DNA replication/ gene regulation/ cell proliferation; e: ion channels; f: membrane proteins; g: metabolic enzyme; h: protection and detoxification; i: protein synthesis and degradation; j: signal transduction; k: transport/ binding proteins; l: intermediate filaments; m: unannotated/ function inferred.
Table 2. Cytosolic fraction proteins of Hep G2 and SK-Hep–1 cells were identified by MALDI-TOF.
| Cells | Spot NO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| HepG2 | ||||||||
| 1 | NP 00067 | Mn SOD | 22.304/6.86 | 105(64%) | Mitochondrion | Antioxidant activity | h | |
| 2 | NP 859048 | Peroxiredoxin I | 14.054/6.25 | 85(73%) | Mitochondrion | Antioxidant activity | h | |
| 3 | ND | |||||||
| 4 | P32119 | Peroxiredoxin II | 21.918/5.67 | 106(62%) | Cytoplasm | Antioxidant activity | h | |
| 5 | P27797 | Calreticulin variant | 47.061/4.30 | 181(60%) | ER | Protein folding | i | |
| 6 | P27797 | Calreticulin variant | 47.061/4.30 | 248(67%) | ER | Protein folding | i | |
| 7 | P27797 | Calreticulin variant | 47.061/4.30 | 182(54%) | ER | Protein folding | i | |
| 8 | P06753 | Tropomyosin | 27.387/4.71 | 156(52%) | Cytoplasm | Cytoskeleton, regulation of muscle contraction | c, l | |
| 9 | P12004 | PCNA | 29.092/4.57 | 91(63%) | Nucleus | Cell cycle regulation | a, d | |
| 10 | P06748 | Nucleophosmin, B23 | 31.090/4.71 | 68(40%) | Nucleus | Regulate cell proliferation | a | |
| 11 | P52895 | 3-α-hydroxysteroid/ dihydrodiol dehydrogenase | 37.221/8.02 | 114(66%) | Cytoplasm | Steroid metabolism | g | |
| 12 | P09525 | Annexin A4 | 35.957/5.85 | 151(67%) | Mitochondrion | Calcium binding protein | e, f, l | |
| 13 | P30041 | Peroxiredoxin VI | 25.002/6.02 | 92(61%) | Cytoplasm | Antioxidant activity | h | |
| 14 | P04792 | HSP 27 | 22.427/7.83 | 99(63%) | Nucleus/ Cytoplasm | Protein folding | b | |
| 15 | P60174 | Triosephosphate isomerase (TIM) | 26.807/6.51 | 169(79%) | Cytoplasm | Metabolic enzyme (Glycolysis) | g | |
| Cells | Spot NO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| HepG2 | 16 | P18669 | Phosphoglycerate mutase 1 | 28.769/6.75 | 153(79%) | Cytoplasm | Metabolic enzyme (Glycolysis) | g |
| 17 | P42330 | Aldo keto reductase | 36.226/7.12 | 115(67%) | Cytoplasm | Aldehyde metabolism | g | |
| 18 | NP 002037 | Glucose 3 phosphate dehydrogenase | 36.202/8.26 | 112(64%) | Cytoplasm | Metabolic enzyme (Glycolysis) | g | |
| 19 | P52895 | 3-α-hydroxysteroid/ dihydrodiol dehydrogenase | 37.221/8.02 | 114(66%) | Cytoplasm | Steroid metabolism | g | |
| 20 | NP 005887 | Isocitrate dehydrogenase | 46.944/6.34 | 147(72%) | Mitochondrial matrix | Metabolic enzyme (TCA cycle) | g | |
| 21 | P06733 | α-enolase | 47.35/6.99 | 288(73%) | Cytoplasm | Metabolic enzyme (Glycolysis) | g | |
| 22 | P05783 | Cytokeratin 18 | 47.305/5.27 | 268(69%) | Cytoplasm | Cytoskeleton | c | |
| 23 | NP 003290 | Tumor rejection antigen, gp96 | 92.567/4.77 | 244(55%) | Cytoplasm | Protein folding and transport | h | |
| 24 | P19338 | Nucleolin | 58.576/4.57 | 96(32%) | Nucleus | DNA/ RNA binding | d | |
| 25 | NP 003290 | Tumor rejection antigen, gp96 | 92.567/4.77 | 244(55%) | Cytoplasm | Protein folding and transport | h | |
| Cells | Spot NO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| HepG2 | 26 | P38646 | Grp 75 | 74.019/5.97 | 271(58%) | Mitochondrion | Cell proliferation and Cellular aging | d |
| 27 | NP 004125 | HSP 70k 9B (mortalin–2) | 74.093/6.04 | 268(58%) | Cytoplasm | Anti-apoptosis, protein folding | b | |
| 28 | NP 003350 | UDP-glucose dehydrogenase | 55.674/6.73 | 298(75%) | Cytoplasm | Metablolic enzyme (Glycosaminoglycan biosynthesis) | g | |
| 29 | P00352 | Retinal dehydrogenase (AL1A1) | 55.323/6.29 | 112(53%) | Cytoplasm | Metabolic enzyme (retinoic acid biosynthesis) | g | |
| 30 | P60709 | β-actin | 42.08/5.37 | 87(66%) | Cytoskeleton | Cytoskeleton | c, l | |
| 31 | NP 001606 | gamma-actin | 42.108/5.31 | 165(77%) | Cytoplasm | Cytoskeleton, cell motility | c, l | |
| 32 | NP 00684 | Protein disulfide-isomerase (ER 60) | 57.146/5.98 | 301(67%) | ER lumen | Folding of proteins containing disulfide bonds in ER | h, i | |
| 33 | P10809 | Chaperonin GroEL procursor | 61.187/5.70 | 162(57%) | Mitochondrial matrix | Transport the mitochondrial proteins | b, k | |
| 34 | P11021 | Grp 78 | 72.407/5.07 | 188(55%) | ER lumen | Protein folding in ER | k | |
| 35 | P31939 | Bifunctional purine biosynthesis protein, PURH | 64.938/6.39 | 268(74%) | Nucleus/ cytoplasm | Nucleic acid metabolism | g | |
| Cells | Spot NO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| SK-Hep–1 | ||||||||
| 36 | P04406 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | 36.202/8.26 | 112(64%) | Cytoplasm | Metabolic enzyme (Glycolysis) | g | |
| 37 | P56876 | Probable thioredoxin peroxidase (PAGA) | 22.324/8.27 | 165(71%) | Nucleus/ cytoplasm | Antioxidant enzyme | h | |
| 38 | P63104 | 14.3.3 protein zeta/delta | 30.10/4.72 | 149(58%) | Cytoplasm | Signal transduction | j | |
| 39 | P31947 | 14.3.3 protein sigma (Stratifin) | 27.871/4.68 | 96(44%) | Cytoplasm | Cell proliferation, signal transduction | a, d, j | |
| 40 | P06753 | Tropomyosin | 27.387/4.71 | 120(50%) | Cytoplasm | Cytoskeleton, regulation of muscle contraction | c, l | |
| 41 | P07951 | Tropomyosin | 32.856/4.72 | 98(52%) | Cytoplasm | Cytoskeleton, regulation of muscle contraction | c, l | |
| 42 | P62258 | 14.3.3 protein epsilon | 29.326/4.63 | 137(66%) | Cytoplasm | Signal transduction | j | |
| 43 | P12004 | PCNA | 29.092/4.57 | 91(63%) | Nucleus | Cell cycle regulation | a, d | |
| 44 | BAA82513 | Pre-mRNA splicing factor SP2p32 | 31.287/4.74 | 73(61%) | Cytoplasm | mRNA processing | d | |
| 45 | P06748 | B23 | 31.090/4.71 | 82(43%) | Nucleus | Regulate cell proliferation | a | |
| 46 | P08670 | Vimentin | 53.545/5.06 | 283(72%) | Cytoskeleton | Cytoskeleton, cell motility | c, f, l | |
| 47 | P08670 | Vimentin | 53.545/5.06 | 306(71%) | Cytoskeleton | Cytoskeleton, cell motility | c, f, l | |
| 48 | P08670 | Vimentin | 53.464/4.99 | 281(80%) | Cytoskeleton | Cytoskeleton, cell motility | c, f, l | |
| 49 | NP 002264 | Keratin 8 | 30.802/5.02 | 177(83%) | Intermediate filament | Cytoskeleton organization and biogenesis | c | |
| Cells | Spot NO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| SK-Hep–1 | 50 | P10809 | Chaperonin GroEL procursor | 61.187/5.70 | 162(57%) | Mitochondrial matrix | Transport the mitochondrial proteins | b, k |
| 51 | NP 00684 | Protein disulfide-isomerase | 57.480/4.76 | 267(69%) | ER lumen | Folding of proteins containing disulfide bonds in ER | h, i |
ND: None detected.
The functional classification (Class) of identified proteins is shown a ~ m. a: cell cycle; b: chaperone/ stress response; c: cytoskeleton/ cell mobility; d: DNA replication/ gene regulation/ cell proliferation; e: ion channels; f: membrane proteins; g: metabolic enzyme; h: protection and detoxification; i: protein synthesis and degradation; j: signal transduction; k: transport/ binding proteins; l: intermediate filaments; m: unannotated/ function inferred.
Table 3. Nuclear fraction proteins of Hep G2 and SK-Hep–1 cells were identified by MALDI-TOF.
| Cells | Spot NO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| HepG2 | ||||||||
| 1 | P56876 | Probable thioredoxin peroxidase (PAGA) | 22.324/8.27 | 165(71%) | Nucleus/ cytoplasm | Antioxidant enzyme | h | |
| 2 | NP 002119 | High-mobility group box 1 | 18.470/9.72 | 48(52%) | Nucleus | Cell proliferation, anti-apoptosis | d, h | |
| 3 | NP 112533 | Heterogeneous nuclear ribonucleoprotein B1 | 37.464/8.97 | 182(61%) | Nucleus | Regulation of transcription | d | |
| 4 | P04406 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | 36.07/8.58 | 98(58%) | Cytoplasm | Metabolic enzyme (Glycolysis) | g | |
| 5 | P07355 | Annexin A2 (lipocortin II) | 38.677/7.56 | 216(67%) | Plasma membrane | Calcium binding protein | e, f, l | |
| 6 | P04083 | Annexin A1 (lipocortin1) | 38.787/6.64 | 202(64%) | Plasma membrane | Calcium binding protein | e, f, l | |
| 7 | P07355 | Annexin A2 (lipocortin II) | 38.677/7.56 | 216(67%) | Plasma membrane | Calcium binding protein | e, f, l | |
| 8 | NP 003657 | Basic leucine zipper nuclear factor 1 (JEM–1) | 13.750/9.36 | 60(65%) | Nucleus/ cytoplasm | Cell proliferation and gene regulation | d | |
| 9 | P 24534 | Elongation factor 1-β | 24.788/4.50 | 56(41%) | Cytoplasm | Regulate translation | d | |
| 10 | ND | |||||||
| Cells | Spot NO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| HepG2 | 11 | P06748 | Nucleophosmin, B23 | 29.617/4.47 | 69(38%) | Nucleus | Regulate cell proliferation | a |
| 12 | P33991 | DNA replication licensing factor, MCM4 | 97.068/6.28 | 43(13%) | Nucleus | Initiation of DNA replication | d | |
| SK-Hep–1 | ||||||||
| 13 | NP 006146 | Septin 2 (NEDD5) | 36.824/6.85 | 74(33%) | Cytoplasm | GTP binding | a, d | |
| 14 | P02545 | Lamin C | 65.153/6.40 | 254(67%) | Nucleus | Protein binding | l | |
| 15 | NP 002264 | Keratin 8 | 30.802/5.02 | 203(81%) | Intermediate filament | Cytoskeleton organization and biogenesis | c | |
| 16 | NP 001034267 | ATP sythase D chain, mitochondria | 18.405/5.22 | 62(53%) | Mitochondrion | ATP synthesis | g | |
| 17 | NP 031478 | Peroxiredoxin III | 28.047/7.11 | 63(52%) | Mitochondrion | Antioxidant activity | h | |
| 18 | P35232 | Prohibitin | 29.859/5.57 | 184(79%) | Mitochondrial inner membrane | Gene regulation and modulate the cell proliferation | d | |
| 19 | NP 038470 | Somatin-like protein 2 (SLP–2) | 38.642/6.88 | 132(69%) | Cytoskeleton | Receptor binding | c, f | |
| 20 | NP 00684 | Protein disulfide-isomerase(ER 60) | 57.146/5.98 | 301(67%) | ER lumen | Folding of proteins containing disulfide bonds in ER | h, i | |
| Cells | Spot NO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| SK-Hep–1 | 21 | NP 002264 | Keratin 8 (fragment) | 41.083/4.94 | 127(51%) | Intermediate filament | Cytoskeleton organization and biogenesis | c |
| 22 | NP 002264 | Keratin 8 (fragment) | 41.083/4.94 | 127(51%) | Intermediate filament | Cytoskeleton organization and biogenesis | c | |
| 23 | P05783 | Cytokeratin 18 | 47.305/5.27 | 268(69%) | Cytoplasm | Cytoskeleton | c | |
| 24 | NP 001677 | H+-transporting two-sector ATPase, β chain | 56.525/5.26 | 250(69%) | Mitochondrion | ATP synthesis coupled proton transport | k | |
| 25 | Dnak-type molecular chaperone precursor HSPA5 precursor | 72.185/5.03 | 288(50%) | Chaperone | b | |||
| 26 | P38646 | Grp 75 | 74.019/5.97 | 355(63%) | Mitochondrion | Cell proliferation and Cellular aging | d | |
| 27 | P08670 | Vimentin | 53.545/5.06 | 306(71%) | Cytoskeleton | Cytoskeleton, cell motility | c, f, l | |
| 28 | P08670 | Vimentin | 53.545/5.06 | 306(71%) | Cytoskeleton | Cytoskeleton, cell motility | c, f, l | |
| 29 | P06733 | α-enolase | 47.35/6.99 | 128(58%) | Cytoplasm | Metabolic enzyme (Glycolysis) | g | |
| 30 | P11021 | Grp 78 | 72.402/5.07 | 223(53%) | ER lumen | Protein folding in ER | i |
ND: None detected.
The functional classification (Class) of identified proteins is shown a ~ m. a: cell cycle; b: chaperone/ stress response; c: cytoskeleton/ cell mobility; d: DNA replication/ gene regulation/ cell proliferation; e: ion channels; f: membrane proteins; g: metabolic enzyme; h: protection and detoxification; i: protein synthesis and degradation; j: signal transduction; k: transport/ binding proteins; l: intermediate filaments; m: unannotated/ function inferred.
Table 4. Membrane fraction proteins of Hep G2 and SK-Hep–1 cells were identified by MALDI-TOF.
| Cells | Spot NO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| HepG2 | ||||||||
| 1 | NP 001034267 | ATP synthase D chain | 18.405/5.22 | 80(68%) | Mitochondrion | ATP synthesis | g | |
| 2 | P35232 | Prohibitin | 29.859/5.57 | 135(73%) | Mitochondrial inner membrane | Gene regulation and modulate the cell proliferation | c, l | |
| 3 | P06576 | ATP synthase β-subunit | 34.026/4.90 | 61(50%) | Mitochondrion | ATP synthesis | g | |
| 4 | P06753 | Tropomyosin3 | 27.386/4.77 | 86(42%) | Cytoplasm | Cytoskeleton, regulation of muscle contraction | c, l | |
| 5 | P06748 | Nucleophosmin, B23 | 33.026/4.6 | 54(34%) | Nucleus | Regulate cell proliferation | a | |
| 6 | NP 004491 | Heterogeneous nuclear ribonucleoprotein C | 27.861/4.55 | 117(59%) | Nucleus | mRNA processing | d, h | |
| 7 | P60709 | Actin β subunit | 40.536/5.55 | 63(40%) | Cytoskeleton | Cytoskeleton | c, l | |
| 8 | P60709 | Actin β chain | 41.321/5.56 | 114(58%) | Cytoskeleton | Cytoskeleton | c, l | |
| 9 | NP 001606 | Actin-gamma | 42.108/5.31 | 57(39%) | Cytoplasm | Cytoskeleton, cell motility | c, l | |
| 10 | NP 057078 | H+-transporting two-sector ATPase | 56.525/5.26 | 132(53%) | Mitochondrion | ATP synthesis coupled proton transport | k | |
| 11 | NP 001677 | H+-transporting two-sector ATPase β chain | 56.525/5.26 | 170(65%) | Mitochondrion | ATP synthesis coupled proton transport | k | |
| Cells | SpotNO. | Accession NO. | Identification | Theoretical M.W./ PI | Score (Seq Cov) | Cellular localization | Molecular function | Class. |
| 12 | P20100 | Lamin B1 | 66.522/5.11 | 273(53%) | Nucleus | Structure protein | l | |
| 13 | P20100 | Lamin B1 | 66.522/5.11 | 256(58%) | Nucleus | Structure protein | l | |
| SK-Hep–1 | ||||||||
| 14 | AAK01919 | mAb 3H11 antigen | 69.954/5.19 | 66(24%) | Nucleus/ cytoplasm | Protein binding | k | |
| 15 | ND | |||||||
| 16 | ND | |||||||
| 17 | P08670 | Vimentin | 49.680/5.19 | 211(62%) | Cytoskeleton | Cytoskeleton, cell motility | c, f, l | |
| 18 | P08670 | Vimentin | 49.680/5.19 | 204(66%) | Cytoskeleton | Cytoskeleton, cell motility | c, f, l | |
| 19 | P20100 | Lamin B1 | 66.522/5.11 | 186(54%) | Nucleus | Structure protein | l | |
| 20 | NP 112533 | Heterogeneous nucleus ribonucleoprotein B1 | 37.464/8.97 | Nucleus | mRNA processing | d, h |
ND: None detected.
The functional classification (Class) of identified proteins is shown a ~ m. a: cell cycle; b: chaperone/ stress response; c: cytoskeleton/ cell mobility; d: DNA replication/ gene regulation/ cell proliferation; e: ion channels; f: membrane proteins; g: metabolic enzyme; h: protection and detoxification; i: protein synthesis and degradation; j: signal transduction; k: transport/ binding proteins; l: intermediate filaments; m: unannotated/ function inferred.
Fig 2. Protein classification.
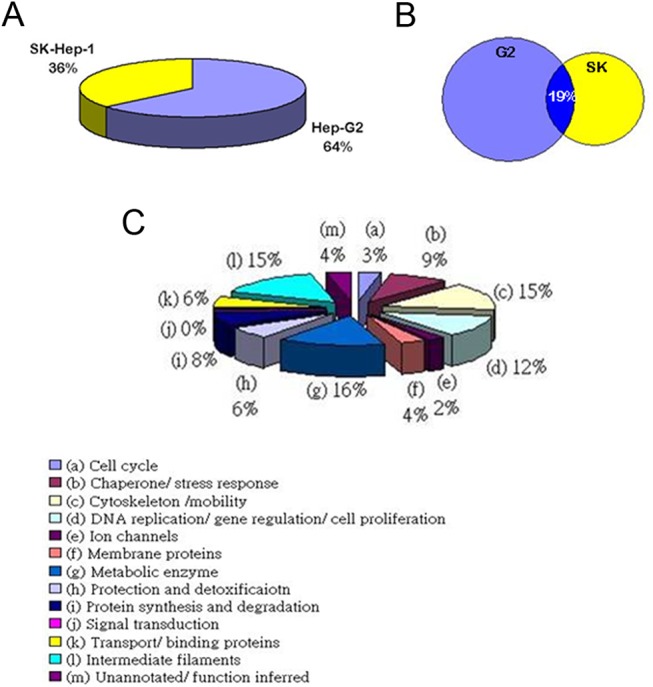
The functional classifications of the identified proteins.a: Cell cycle: 3%; b: chaperone/stress response: 9%; c: cytoskeleton/ cell mobility: 15%; d: DNA replication/gene regulation/cell proliferation: 12%; e: ion channels: 2%; f: membrane proteins: 4%; g: metabolic enzyme: 16%; h: protection and detoxification: 6%; i: protein synthesis and degradation: 8%; j: signal transduction: 8%; k: transport/binding proteins: 6%; l: intermediate filaments: 15%; and m: unannotated/ function inferred: 4%.
Table 5. List of unregulated proteins in the 2DE map of either the well-differentiated HepG2 cells or the poorly differentiated SK-Hep–1 cells identified by MALDI-TOF MS.
| Cellular compartemts | Spot No | Association code | Name | Theoretical (kDa)/pI | HepG2 | SK-Hep–1 | Fold | Cellular localization | Function |
|---|---|---|---|---|---|---|---|---|---|
| Total Cell lysate | 1 | NP005887 | NADP+-dependent isocitrate dehydrogenase | 46.944/6.34 | ↑ | ↓ | 0.01 | Mitochondria | Metabolic enzyme in TCA cycle |
| 2 | P32119 | Peroxiredoxin II | 21.918/5.67 | ↑ | ↓ | 0.27 | Cytoplasm | Antioxidant enzyme | |
| 3 | NP003350 | UDP-glucose dehydrogenase | 55.674/6.73 | ↑ | ↓ | 0.19 | Cytoplasm | Metabolic enzyme in Glycosaminoglycan biosynthesis | |
| 4 | P09525 | Annexin 4 | 33.757/5.64 | ↑ | ↓ | 0.34 | Mitochondria | Calcium binding protein | |
| 5 | P33992 | Replication licensing factor MCM5 | 82.990/8.56 | ↓ | ↑ | 4.09 | Nucleus | Initiation of DNA replication | |
| 6 | P06753 | Tropomyosin3 | 27.386/4.77 | ↓ | ↑ | 4.1 | Cytoplasm | Cytoskeleton | |
| Cytosol | 1 | P09525 | Annexin 4 | 35.957/5.85 | ↑ | ↓ | 0.45 | Mitochondria | Calcium binding protein |
| 2 | NP005887 | NADP+-dependent isocitrate dehydrogenase | 46.944/6.34 | ↑ | ↓ | 0.01 | Mitochondria | Metabolic enzyme in TAC cycle | |
| 3 | NP00067 | MnSOD | 22.304/6.86 | ↑ | ↓ | 0.2 | Mitochondria | Antioxidant enzyme | |
| 4 | NP859048 | Peroxiredoxin I | 14.054/6.25 | ↑ | ↓ | 0.22 | Mitochondria | Antioxidant enzyme | |
| 5 | P32119 | Peroxiredoxin II | 21.918/5.67 | ↓ | ↑ | 0.6 | Cytoplasm | Antioxidant enzyme | |
| 6 | NP003350 | UDP-glucose dehydrogenase | 55.674/6.73 | ↑ | ↓ | 0.34 | Cytoplasm | Glycosaminoglycan | |
| 7 | P04792 | Heat Shock Protein 27 | 22.427/7.83 | ↓ | ↑ | 1.03 | Nucleus/ Cytoplasm | Protein folding | |
| 8 | P08670 | Vimentin | 53.545/5.06 | ↓ | ↑ | 1.23 | Cytoplasm | Cell motility | |
| Nucleus | 1 | P04083 | Annexin 1 | 38.787/6.64 | ↓ | ↑ | 4.53 | Plasm membrane | Calcium binding protein |
| 2 | P07355 | Annexin 2 | 38.677/7.56 | ↓ | ↑ | 3.27 | Plasm membrane | Calcium binding protein | |
| 3 | P33991 | Replication Licensing factor,MCM4 | 97.068/6.28 | ↓ | ↑ | 1.17 | Nucleus | Initiation of DNA replication biosynthesis | |
| 4 | NP031478 | Peroxiredoxin III | 28.047/6.73 | ↑ | ↓ | 0.08 | Mitochondria | Antioxidant enzyme | |
| Membrane | 1 | P08670 | Vimentin | 49.680/5.19 | ↓ | ↑ | 3.57 | Cytoplasm | Cell motility |
| 2 | P35232 | Prohibitin | 29.859/5.57 | ↑ | ↓ | 0.14 | Mitochondria | Cell proliferation |
Table 6. Summary of the differentially expressed proteins in HepG2 and SK-Hep–1 cells using MALDI-TOF MS and categorized according to their functional roles.
| Classification | Identified protein | Hep-G2 | SK-Hep–1 |
|---|---|---|---|
| Group Ⅰ Intermediate filament | Annexin–1 | + | +++++ In nucleus |
| Annexin–2 | - | ++++ In nucleus | |
| Annexin–4 | ++++ In cytoplasm | + | |
| Group Ⅱ Protection and detoxification | PeroxiredoxinⅠ | ++++ In cytoplasm | + |
| Peroxiredoxin Ⅱ | ++++ In cytoplasm | - | |
| Peroxiredoxin Ⅲ | +++ In nucleus | - | |
| Group Ⅲ Cytoskeleton protein | Vimentin | - | ++++ In membrane |
| Group Ⅳ Metabolic enzyme | NADP-dependent Isocitrate dehydrogenase | ++++ In cytoplasm | - |
Note: “+” sign denotes positive expression; “-” denotes negative expression
Differential protein spot analysis
Comparative proteome analysis of various cellular compartments between HepG2 and SK-Hep–1 cells was performed, and 8 protein spots were successfully identified in both cell types. These proteins were classified according to their functional attributes (Tables 5 and 6). ANX1 and ANX2 were differentially overexpressed in the nucleus of SK-Hep–1 cells. Conversely, ANX4 was only differentially expressed in the well-differentiated HepG2 cells. Prdx I, II, and III and ICDH were preferentially expressed in the well-differentiated HepG2 cells. Vimentin, an EMT marker, was detected more prominently in SK-Hep–1 cells.
Confirmation of differential expression by immunofluorescence staining and western blotting
Using actin, PCNA and Na-K+ ATPase as internal controls for the cytosolic, nuclear and membrane fractions, respectively, we confirmed the differential expression of the proteins in various cellular compartments by western blotting. As shown in Fig 3, ANX4 was highly expressed in the cytosolic and nuclear fractions of the well-differentiated HepG2 cells. In contrast, ANX1 and vimentin were overexpressed in the poorly differentiated SK-Hep–1 cells (Fig 3). Immunofluorescence analysis was used to confirm the results of the western blots (Figs 4 and 5).
Fig 3. Validation of the differential protein expression between HepG2 and SK-Hep–1 cells by western blot.
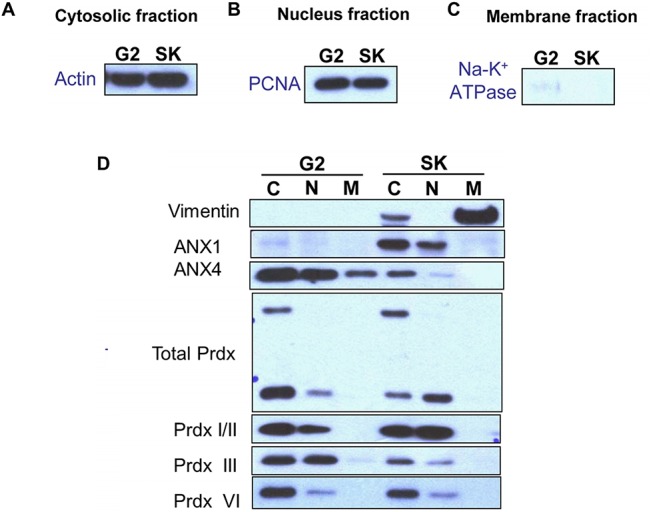
Actin was used as the internal control in the cytosolic fraction (A), PCNA served as the internal control in the nuclear fraction (B), and Na-K+ ATPase was used as the internal control in the membrane fractions (C). (D) C: cytosolic fraction protein, N: nuclear fraction protein, M: membrane fraction protein.
Fig 4. Validation of the differential protein expression between HepG2 and SK-Hep–1 cells by immunofluorescence staining.
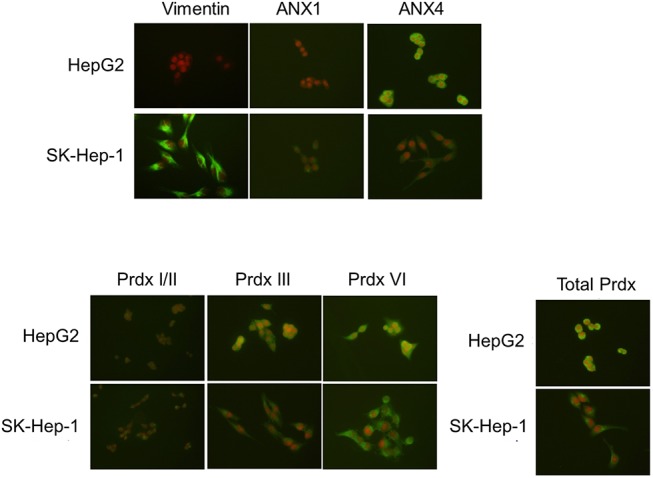
The expression patterns of ANX1, ANX4 and Prdx were detected by immunofluorescence staining as described in the Methods and Materials.
Fig 5. Expression patterns of differentially expressed proteins identified by comparative proteomic analysis in a panel of five HCC sublines with established differentiation/de-differentiation status.
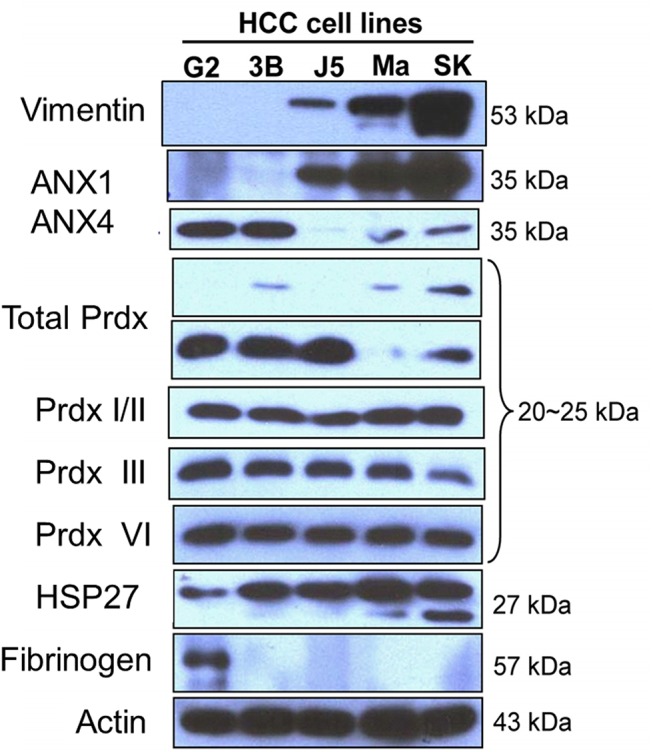
The differentiation status (well-differentiated to poorly differentiated) of these five HCC cell lines was on the order of HepG2, Hep3B, HepJ5, Mahlavu, and SK-Hep–1. The expression patterns of ANX1, ANX4, HSP27, Prdx isoforms, vimentin, and fibrinogen were detected by western blotting.
ANX1 is a biomarker for metastatic potential in HCC cell lines
The differentially expressed protein candidates identified in HepG2 and SK-Hep–1 cells via our proteomic approach are molecular biomarkers of differentiation status. We validated the expression profiles of these proteins using a panel of HCC cells in a descending order of differentiation based on our previous studies[11, 12]. First, ANX1 expression levels progressively increased as HCC cells became less differentiated (Fig 6). The expression profile of vimentin, an EMT marker, was similar to that of ANX1 (Fig 6). In contrast, ANX4 was only overexpressed in the well-differentiated HCC sublines, HepG2 and Hep3B. Because vimentin overexpression is generally recognized as a metastatic phenotype indicator in cancer cells, we hypothesized that ANX1 overexpression may serve a similar functional role in promoting metastasis in HCC cells. To test our hypothesis, we silenced ANX1 expression in HepJ5 cells and demonstrated that this manipulation severely impeded the migration of these cells (Fig 6). This finding strongly suggests that ANX1 and vimentin are involved in the regulation of the metastatic potential of poorly differentiated HCC cells.
Fig 6. Knockdown of AXN1 expression levels suppressed cell migration.
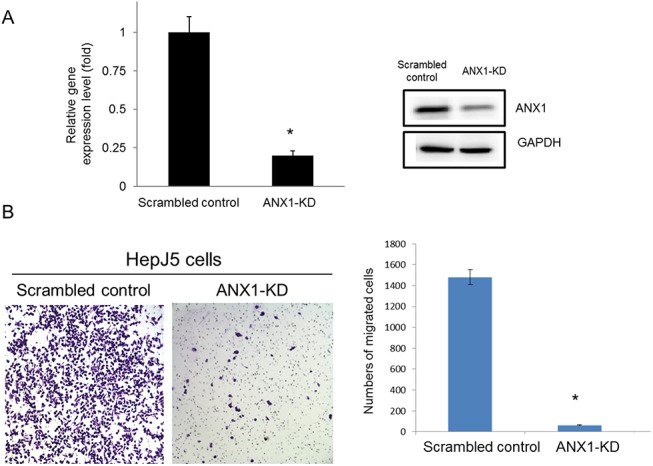
AXN1 expression levels were knocked down by transfection of AXN1-specific shRNA into HepJ5 cells as described in the Methods and Materials. (A) The expression levels of AXN1 in control and AXN1-KD cells were determined by real-time PCR and western blotting. (B) The migration of scrambled control and AXN1-KD cells was determined by the transwell migration assay as described in the Methods and Materials.
Higher HSP27 expression is correlated with a highly metastatic potential in HCC cells
Consistent with these observations, we also discovered that high expression of HSP27 was correlated with the less differentiated cell types. To confirm the role of HSP27 in HCC cells, we further silenced HSP27 expression by shRNA in SK-Hep–1 cells and performed migration assays. As shown in Fig 7, silencing HSP27 dramatically reduced the migration of SK-Hep–1 cells, indicating that HSP27 expression is critical in modulating the differentiation status of HCC cells.
Fig 7. Knockdown of HSP27 expression levels suppressed cell migration.
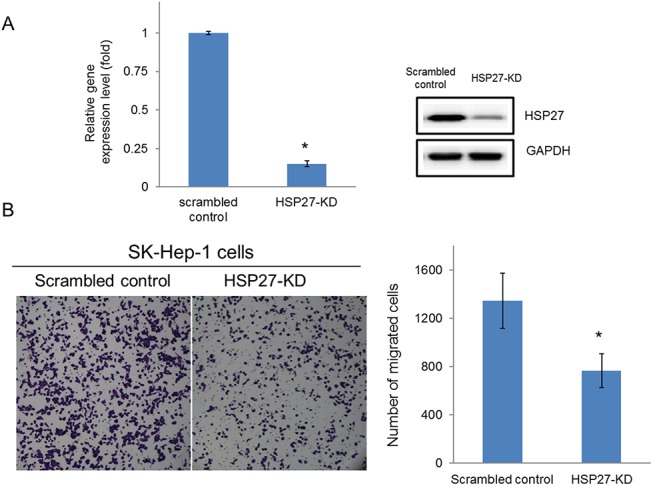
HSP27 expression levels were knocked down by transfection with HSP27-specific shRNA into SK-Hep–1 cells as described in the Methods and Materials. (A) The expression levels of HSP27 in control and HSP27-KD cells were determined by real-time PCR and western blotting. (B) The migration of scrambled control and HSP27-KD cells was determined by transwell migration assayas described in the Methods and Materials.
The pathologic characteristics and expression levels of HSP27 and ANX1
To understand the correlation between pathologic characteristics and expression levels of HSP 27 and ANX1, the immunohistochemical stains with HSP 27 and ANX1 in tissue microarray sets were performed. As shown in Fig 8, HCC was divided into well-differentiated (WD), moderately differentiated (MD) and poorly differentiated (PD). HSP27 expression levels were scored semiquantitatively as weakly positive (1+), moderately positive (2+) and strongly positive (3+). PD HCC tended to express stronger HSP27 than WD HCC (p<0.001) (Fig 8A). MD HCC also revealed stronger HSP27 expression than WD HCC (p<0.05). HSP27 expression was not statistically different between PD HCC and MD HCC. In addition, strong ANX1 expression was more commonly found in PD group compared with WD and MD group (p<0.05 and p<0.01, respectively, Fig 8B).
Fig 8. The pathologic characteristics and expression levels of HSP 27 and ANX1.
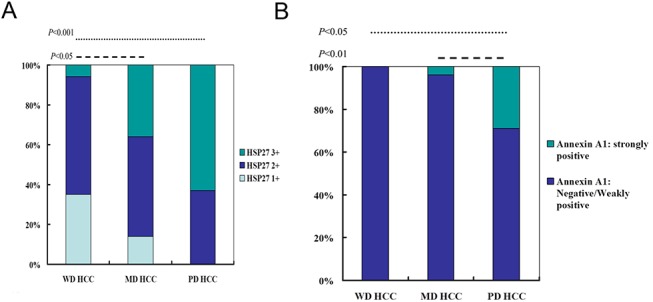
To understand the correlation between pathologic characteristics and expression levels of HSP 27 and ANX1, the immunohistochemical stains with HSP 27 and ANX1 in HCC tissue microarray were performed. HCC was divided into well-differentiated (WD), moderately differentiated (MD) and poorly differentiated (PD). (A) HSP27 expression levels were scored semiquantitatively as weakly positive (1+), moderately positive (2+) and strongly positive (3+). (B) Expression levels of ANX1 were scored semiquantitatively as negative, weakly positive and strongly positive. The association between pathologic characteristics and ANX1 expression was analyzed.
Discussion
Previous studies have indicated that the levels of reactive oxygen species (ROS) are significantly higher in many cancer cells than in normal cells. Consequently, cancer cells exhibit higher intrinsic oxidative stress [13, 22–25]. The increased ROS stress in cancer cells is associated with de-differentiation and the acquisition of an invasive phenotype [26, 27]. In our study, well-differentiated HCC cells overexpressed antioxidant/metabolic enzymes, such as MnSOD, Prdx, ICDH, α-enolase and UDP-glucose dehydrogenase (Tables 5 and 6). In contrast, poorly differentiated HCC cells exhibited high levels of PDI. Therefore, the intrinsic oxidative stress of HCC cells was modulated through the regulation of antioxidant enzymes and was differentiation status-dependent.
Characteristic changes during EMT include the down-regulation of epithelial markers (e.g., E-cadherin) and the up-regulation of mesenchymal markers, such as vimentin and N-cadherin [28–30]. In this study, we initially established that membranous vimentin is abundantly expressed in poorly differentiated SK-Hep–1 cells. In addition, using a panel of five HCC sublines, we demonstrated that vimentin was abundantly expressed in poorly differentiated HCC sublines, including Mahlavu, HepJ5 and SK-Hep–1 cells (Fig 5). These data suggest that the de-differentiation of HCC cells is associated with oxidative stress, which may be the driving force and/or protective mechanism for EMT induction and invasive potential. This notion is supported by the up-regulation in many different cancer cell types of NF-κB, which is responsive to stimuli generated during high intrinsic oxidative stress conditions, such as H2O2 and GSSG [30, 31]. De-differentiated HCC cells, such as the Mahlavu and SK-Hep–1 sublines, exhibited substantial up-regulation of NFκB (data not shown).
The elevated level of ANX1 detected in poorly differentiated HCC cells, such as Mahlavu and SK-Hep–1 cells, attracted our attention. ANX1 belongs to a family of calcium/phospholipid-binding and actin regulatory proteins. Graauw et al.[32]reported that ANX1 and its family member ANX2 are candidate regulators of the oncogene-induced cell morphology switch. During such a switch, tumor cells change from an epithelial to a more migratory, mesenchymal-like phenotype, thus leading to metastasis and the progression of cancer. Our data are consistent with these findings, as the expression pattern of ANX1 was similar to that of vimentin in HCC cells. Increased expression of ANX1 was only observed in poorly differentiated sublines, such as Mahlavu and SK-Hep–1 cells, suggesting that ANX1 is associated with the invasive phenotype (Fig 5). Consistently, down-regulation of ANX1 significantly reduced the migration of invasive HepJ5 cells (Fig 6) and strong ANX1 expression was more commonly found in PD group compared with WD and MD group (Fig 8B). Those results demonstrates that ANX1 may play a pivotal pole in increasing the metastatic potential of cancer cells and that ANX1 may serve as a biomarker for the aggressive phenotype of HCC cells in conjunction with vimentin.
In contrast to ANX1, ANX4 was uniquely overexpressed in only well-differentiated HCC cells, including the HepG2 and Hep3B sublines. Although the exact role of ANX4 in HCC carcinogenesis remains unclear, Han et al. has suggested that overexpression of ANX4 is associated with paclitaxel resistance in cancer cells [33]. Whether ANX4 can also contribute to the acquisition of chemoresistance in HCC cells warrants further investigation.
Finally, among the differentially expressed proteins, the elevated expression of HSP27 was associated with the less differentiated cellular phenotype (Fig 5). Previous studies have indicated that phosphorylation plays a key role in the regulation of HSP27 function and may contribute to the survival of cells during oxidative stress and apoptosis [34, 35]. In addition, HSP27 participates in maintaining GSH in its reduced form during oxidative stress [36]. Another study demonstrated that elevated expression of HSP27 is correlated with the enhanced migration of endothelial cells [37]. Silencing of HSP27 significantly reduced the migration of invasive SK-Hep–1 cells (Fig 7). Besides, we also demonstrated that PD HCC tended to expression strong HSP27 than WD HCC (p<0.001) (Fig 8A). MD HCC also revealed stronger HSP27 expression than WD HCC (p<0.05). Collectively, these observations suggest that overexpression of HSP27 in de-differentiated HCC cells may be a molecular indicator for HCC metastatic potential.
In conclusion, we have unveiled a group of biomarkers that are relevant to the metastatic status of HCC cells using a proteomic approach and subsequent validation in a panel of five HCC sublines with varying degrees of differentiation. Collectively, a panel of biomarkers, including vimentin, ANX1 and HSP27, were identified as metastatic indicators for HCC cells in our study and may be considered potential therapeutic targets.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors received no specific funding for this work.
References
- 1. Bosch FX, Ribes J, Cleries R, Diaz M. Epidemiology of hepatocellular carcinoma. Clinics in liver disease. 2005;9(2):191–211, v. 10.1016/j.cld.2004.12.009 . [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61(2):69–90. 10.3322/caac.20107 . [DOI] [PubMed] [Google Scholar]
- 3. Thomas MB, Zhu AX. Hepatocellular carcinoma: the need for progress. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23(13):2892–9. 10.1200/JCO.2005.03.196 . [DOI] [PubMed] [Google Scholar]
- 4. Zhu AX. Systemic therapy of advanced hepatocellular carcinoma: how hopeful should we be? The oncologist. 2006;11(7):790–800. 10.1634/theoncologist.11-7-790 . [DOI] [PubMed] [Google Scholar]
- 5. King KL, Li AF, Chau GY, Chi CW, Wu CW, Huang CL, et al. Prognostic significance of heat shock protein–27 expression in hepatocellular carcinoma and its relation to histologic grading and survival. Cancer. 2000;88(11):2464–70. . [DOI] [PubMed] [Google Scholar]
- 6. Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209(4455):497–9. . [DOI] [PubMed] [Google Scholar]
- 7. Kaighn ME, Prince AM. Production of albumin and other serum proteins by clonal cultures of normal human liver. Proceedings of the National Academy of Sciences of the United States of America. 1971;68(10):2396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corlu A, Kneip B, Lhadi C, Leray G, Glaise D, Baffet G, et al. A plasma membrane protein is involved in cell contact-mediated regulation of tissue-specific genes in adult hepatocytes. The Journal of cell biology. 1991;115(2):505–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang C, Lin Y, TW OL, Chou CK, Lee TS, Liu TJ, et al. Induction of plasma protein secretion in a newly established human hepatoma cell line. Molecular and cellular biology. 1983;3(6):1133–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xanthopoulos KG, Mirkovitch J. Gene regulation in rodent hepatocytes during development, differentiation and disease. European journal of biochemistry / FEBS. 1993;216(2):353–60. . [DOI] [PubMed] [Google Scholar]
- 11. Liu TZ, Chen PY, Chiu DT, Wei JS, Chang KS, Lin KH. Detection of a novel lactate dehydrogenase isozyme and an apparent differentiation-associated shift in isozyme profile in hepatoma cell lines. Cancer letters. 1994;87(2):193–8. . [DOI] [PubMed] [Google Scholar]
- 12. Lin KH, Lin YW, Lee HF, Liu WL, Chen ST, Chang KS, et al. Increased invasive activity of human hepatocellular carcinoma cells is associated with an overexpression of thyroid hormone beta 1 nuclear receptor and low expression of the anti-metastatic nm23 gene. Cancer letters. 1995;98(1):89–95. . [PubMed] [Google Scholar]
- 13. Liu TZ, Hu CC, Chen YH, Stern A, Cheng JT. Differentiation status modulates transcription factor NF-kappaB activity in unstimulated human hepatocellular carcinoma cell lines. Cancer letters. 2000;151(1):49–56. Epub 2000/04/15. . [DOI] [PubMed] [Google Scholar]
- 14. Phizicky E, Bastiaens PI, Zhu H, Snyder M, Fields S. Protein analysis on a proteomic scale. Nature. 2003;422(6928):208–15. 10.1038/nature01512 . [DOI] [PubMed] [Google Scholar]
- 15. Petricoin EF, Zoon KC, Kohn EC, Barrett JC, Liotta LA. Clinical proteomics: translating benchside promise into bedside reality. Nature reviews Drug discovery. 2002;1(9):683–95. 10.1038/nrd891 . [DOI] [PubMed] [Google Scholar]
- 16. Hu TH, Huang CC, Liu LF, Lin PR, Liu SY, Chang HW, et al. Expression of hepatoma-derived growth factor in hepatocellular carcinoma. Cancer. 2003;98(7):1444–56. 10.1002/cncr.11653 . [DOI] [PubMed] [Google Scholar]
- 17. Huang YH, Lin KH, Chen HC, Chang ML, Hsu CW, Lai MW, et al. Identification of postoperative prognostic microRNA predictors in hepatocellular carcinoma. PloS one. 2012;7(5):e37188 Epub 2012/05/26. 10.1371/journal.pone.0037188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hsu ML, Chen SW, Lin KH, Liao SK, Chang KS. Cytokine regulation of HIV–1 LTR transactivation in human hepatocellular carcinoma cell lines. Cancer letters. 1995;94(1):41–8. Epub 1995/07/20. . [DOI] [PubMed] [Google Scholar]
- 19. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical chemistry. 1996;68(5):850–8. Epub 1996/03/01. . [DOI] [PubMed] [Google Scholar]
- 20. Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20(3):601–5. Epub 1999/04/27. . [DOI] [PubMed] [Google Scholar]
- 21. Huang MT, Wei PL, Liu JJ, Liu DZ, Huey-Chun H, An J, et al. Knockdown of thrombomodulin enhances HCC cell migration through increase of ZEB1 and decrease of E-cadherin gene expression. Annals of surgical oncology. 2010;17(12):3379–85. Epub 2010/07/14. 10.1245/s10434-010-1163-4 . [DOI] [PubMed] [Google Scholar]
- 22. Toyokuni S, Okamoto K, Yodoi J, Hiai H. Persistent oxidative stress in cancer. FEBS letters. 1995;358(1):1–3. Epub 1995/01/16. . [DOI] [PubMed] [Google Scholar]
- 23. Kumar B, Koul S, Khandrika L, Meacham RB, Koul HK. Oxidative stress is inherent in prostate cancer cells and is required for aggressive phenotype. Cancer research. 2008;68(6):1777–85. Epub 2008/03/15. 10.1158/0008-5472.CAN-07-5259 . [DOI] [PubMed] [Google Scholar]
- 24. Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer research. 1991;51(3):794–8. Epub 1991/02/01. . [PubMed] [Google Scholar]
- 25. Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nature reviews Drug discovery. 2009;8(7):579–91. Epub 2009/05/30. 10.1038/nrd2803 . [DOI] [PubMed] [Google Scholar]
- 26. Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436(7047):123–7. Epub 2005/07/08. 10.1038/nature03688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nishikawa M. Reactive oxygen species in tumor metastasis. Cancer letters. 2008;266(1):53–9. Epub 2008/03/26. 10.1016/j.canlet.2008.02.031 . [DOI] [PubMed] [Google Scholar]
- 28. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nature reviews Cancer. 2002;2(6):442–54. 10.1038/nrc822 . [DOI] [PubMed] [Google Scholar]
- 29. Solanas G, Porta-de-la-Riva M, Agusti C, Casagolda D, Sanchez-Aguilera F, Larriba MJ, et al. E-cadherin controls beta-catenin and NF-kappaB transcriptional activity in mesenchymal gene expression. Journal of cell science. 2008;121(Pt 13):2224–34. 10.1242/jcs.021667 . [DOI] [PubMed] [Google Scholar]
- 30. Yang MH, Chen CL, Chau GY, Chiou SH, Su CW, Chou TY, et al. Comprehensive analysis of the independent effect of twist and snail in promoting metastasis of hepatocellular carcinoma. Hepatology. 2009;50(5):1464–74. 10.1002/hep.23221 . [DOI] [PubMed] [Google Scholar]
- 31. Min C, Eddy SF, Sherr DH, Sonenshein GE. NF-kappaB and epithelial to mesenchymal transition of cancer. Journal of cellular biochemistry. 2008;104(3):733–44. 10.1002/jcb.21695 . [DOI] [PubMed] [Google Scholar]
- 32. Lim LH, Pervaiz S. Annexin 1: the new face of an old molecule. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2007;21(4):968–75. 10.1096/fj.06-7464rev . [DOI] [PubMed] [Google Scholar]
- 33. Han EK, Tahir SK, Cherian SP, Collins N, Ng SC. Modulation of paclitaxel resistance by annexin IV in human cancer cell lines. British journal of cancer. 2000;83(1):83–8. 10.1054/bjoc.2000.1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Benndorf R, Hayess K, Ryazantsev S, Wieske M, Behlke J, Lutsch G. Phosphorylation and supramolecular organization of murine small heat shock protein HSP25 abolish its actin polymerization-inhibiting activity. The Journal of biological chemistry. 1994;269(32):20780–4. . [PubMed] [Google Scholar]
- 35. Lavoie JN, Lambert H, Hickey E, Weber LA, Landry J. Modulation of cellular thermoresistance and actin filament stability accompanies phosphorylation-induced changes in the oligomeric structure of heat shock protein 27. Molecular and cellular biology. 1995;15(1):505–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Preville X, Salvemini F, Giraud S, Chaufour S, Paul C, Stepien G, et al. Mammalian small stress proteins protect against oxidative stress through their ability to increase glucose-6-phosphate dehydrogenase activity and by maintaining optimal cellular detoxifying machinery. Experimental cell research. 1999;247(1):61–78. 10.1006/excr.1998.4347 . [DOI] [PubMed] [Google Scholar]
- 37. Piotrowicz RS, Hickey E, Levin EG. Heat shock protein 27 kDa expression and phosphorylation regulates endothelial cell migration. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 1998;12(14):1481–90. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.


