Abstract
Summary: There are several experimental contexts in which it is important to identify DNA integration sites, such as insertional mutagenesis screens, gene and enhancer trap applications, and gene therapy. We previously developed an assay to identify millions of integrations in multiplexed barcoded samples at base-pair resolution. The sheer amount of data produced by this approach makes the mapping of individual sites non-trivial without bioinformatics support. This article presents the Genomic Integration Site Tracker (GeIST), a command-line pipeline designed to map the integration sites produced by this assay and identify the samples from which they came. GeIST version 2.1.0, a more adaptable version of our original pipeline, can identify integrations of murine leukemia virus, adeno-associated virus, Tol2 transposons or Ac/Ds transposons, and can be adapted for other inserted elements. It has been tested on experimental data for each of these delivery vectors and fine-tuned to account for sequencing and cloning artifacts.
Availability and implementation: GeIST uses a combination of Bash shell scripting and Perl. GeIST is available at http://research.nhgri.nih.gov/software/GeIST/.
Contact: burgess@mail.nih.gov
1 Introduction
The technique of identifying the sites at which foreign DNA integrates into a genome has many uses in genomic research, with the approach for identifying these sites taking numerous forms. Insertional mutagenesis can be used as a powerful forward genetic screen (Bard-Chapeau et al., 2014; Golling et al., 2002; Kleckner et al., 1977; Varshney et al., 2013). Gene therapy vectors can be tested to establish the expected level of insertional mutagenic activity (Li et al., 2011). Gene and enhancer traps are important tools for tagging and studying genomic elements (Clark et al., 2011; O’Kane and Gehring, 1987).
In each case, it is important to have a means of identifying the integration sites, preferably at as high a resolution as possible. For small-scale studies, this may be accomplished through inverse PCR, or through linker-mediated PCR (LM-PCR), subcloning, and Sanger sequencing (Devon et al., 1995; Ochman et al., 1988; Wu et al., 2003). At a larger scale, a high-throughput approach is preferable. We developed a method capable of identifying millions of integrations from a single run of high-throughput LM-PCR sequencing (Varshney et al., 2013). To maximize efficiency, the method uses 6-bp DNA barcodes that can be used to multiplex samples or discriminate between independent integration events (LaFave et al., 2014).
The large volume of data produced by this approach means that an unassisted interpretation of the sequencing output is non-trivial. Therefore, we have developed the Genomic Integration Site Tracker (GeIST) to address this. GeIST accepts a BAM or FASTQ file of paired-end LM-PCR sequences and a file indicating the association between samples and barcodes. The software returns a BAM file of the sequences at each integration junction, a Browser Extensible Data (BED) file for easy visualization of the integration patterns, and a summary of how often each barcode was detected.
2 Methods
2.1 Features
We previously reported the process of our LM-PCR analysis pipeline used to identify integrations of murine leukemia virus (MLV) (LaFave et al., 2014) and adeno-associated virus (AAV) (Chandler et al., 2015; Walia et al., 2014), providing software that was capable of re-running those specific experiments. We have since expanded the functionality of the GeIST workflow, most notably by allowing the user to choose between four types of integrated elements: MLV, AAV, Tol2 transposon vectors and Ac/Ds vectors. GeIST 2.1.0 also streamlines the addition of new elements; a user with modest programming skills could use GeIST to recover essentially any integrating DNA element. The mapping of the four aforementioned elements has been tested with experimental data run on an Illumina MiSeq sequencer. This experimental validation step has allowed us to fine-tune the workflow to account for idiosyncrasies that arise in real experiments, such as misleading base quality scores and cloning artifacts. GeIST version 2.1.0 explicitly indicates the group and individual sample of each integration in the output, as defined by the barcode file created by the user. This modification is well-suited to applying the assay to many samples or to strains composed of multiple individuals, but is flexible enough to work with a single sample. Users can assign barcodes into groups, which affects whether integrations at the same genomic position, but with different barcodes, are considered to be independent. This allows the user to distinguish between integrations that co-localize due to multiple independent events versus those due to shared ancestry.
2.2 Workflow
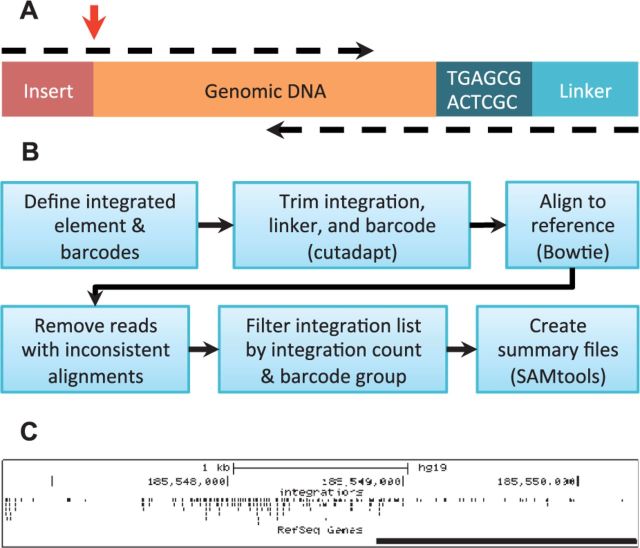
GeIST is designed to run in a Unix environment and requires cutadapt, Bowtie and SAMtools (Langmead et al., 2009; Li et al., 2009; Martin 2011); if the input is in the BAM format, BamTools is also required (Barnett et al., 2011). It is designed to process paired-end reads of LM-PCR amplicons (Fig. 1A). GeIST first identifies the fragments that contain the sequence of the integrated element at the start of a read, and the linker and barcode at the start of the other (Fig. 1B). Cutadapt is used to trim non-genomic sequences and low-quality bases, and a Perl script is used to track each fragment’s barcode. If the LM-PCR primer was upstream of the 3′ end of the integrated element, the cutadapt step also removes spurious amplifications. Trimmed sequences are aligned to a user-supplied genome index via Bowtie.
Fig. 1.
(A) The structure of the LM-PCR amplicons produced by the barcoding assay. Paired-end reads are represented as dashed arrows. GeIST is designed to identify the junction of the insert and the genomic DNA, indicated by the red arrow. (B) The GeIST workflow. (C) Example output of GeIST, in which the BED file is visualized on the UCSC genome browser (http://genome.ucsc.edu). This example shows the MLV integrations detected in human HepG2 cells in the span chr4:185,546,724–185,550,338
Two alignments are performed: the first uses reads that span the insert-genome junction (Fig. 1A, upper dashed arrow), and the second uses the reads paired with those from the first alignment (lower dashed arrow). The second set of reads is aligned without trimming the low-quality bases. Our rationale for this approach comes from our use of experimental data in designing GeIST. We observed that bases called as low quality were often still accurate, in that they overlapped and were consistent with the high-quality paired read. The reads containing the insert-genome junction are therefore trimmed for quality, but the pairs of these reads are not.
Reads that are not properly paired are removed. The remaining fragments are filtered to remove false positives: integration sites within 5 bp of and on the same strand as an integration site with a higher integration count are discarded, provided both sites share the same barcode group. GeIST produces a BAM file of the reads that span the insert-genome junction, while the position and barcode data is used to derive the other output files. In our experience, the BED file is invaluable for visualizing integration patterns with tools such as the UCSC browser (Kent et al., 2002; Fig. 1C).
GeIST contains additional steps to accommodate the specifics of certain integrated elements. For AAV data, GeIST makes use of an extra Perl script and an adjusted trimming strategy to account for the variable region of the inverted terminal repeat used for amplification (Linden et al., 1996). Similarly, an extra call to cutadapt is used to filter out MLV reads that arise from internal amplification of the provirus.
3 Conclusion
GeIST is an important and simple-to-use complement to our high-throughput barcoded sequencing and mapping strategy. It bridges the gap between raw sequence data and identifying genomic DNA integration sites in multiple samples or parsing independent events in a single sample. GeIST is also suited to identify multiple types of integrations. In principle, it can be further modified to identify any type of element that can be amplified via LM-PCR.
Conflict of Interest: none declared.
References
- Bard-Chapeau E.A., et al. (2014) Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nat. Genetics, 46, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett D.W., et al. (2011) BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics, 27, 1691–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler R.J., et al. (2015) Vector design influences hepatic genotoxicity after adeno-associated virus gene therapy. J. Clin. Invest., 125, 870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K.J., et al. (2011) In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat. Methods, 8, 506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devon R.S., et al. (1995) Splinkerettes–improved vectorettes for greater efficiency in PCR walking. Nucleic Acids Res., 23, 1644–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golling G., et al. (2002) Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 31, 135–140. [DOI] [PubMed] [Google Scholar]
- Kent W.J., et al. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., et al. (1977) Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J. Mol. Biol., 116, 125–159. [DOI] [PubMed] [Google Scholar]
- LaFave M.C., et al. (2014) MLV integration site selection is driven by strong enhancers and active promoters. Nucleic Acids Res., 42, 4257–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., et al. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., et al. (2011) Assessing the potential for AAV vector genotoxicity in a murine model. Blood, 117, 3311–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden R.M., et al. (1996) Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA, 93, 11288–11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J., 17, 10–12. [Google Scholar]
- O’Kane C.J., Gehring W.J. (1987) Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl. Acad. Sci. USA, 84, 9123–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., et al. (1988) Genetic applications of an inverse polymerase chain reaction. Genetics, 120, 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney G.K., et al. (2013) A large-scale zebrafish gene knockout resource for the genome-wide study of gene function. Genome Res., 23, 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia J.S., et al. (2014) Long-term correction of sandhoff disease following intravenous delivery of rAAV9 to mouse neonates. Mol. Ther., 23, 414–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., et al. (2003) Transcription start regions in the human genome are favored targets for MLV integration. Science, 300, 1749–1751. [DOI] [PubMed] [Google Scholar]