Abstract
Homoplasmy, the occurrence of a single mitochondrial DNA haplotype within an individual, has been the accepted condition across most organisms in the animal kingdom. In recent years, a number of exceptions to this rule have been reported, largely due to the ease with which single nucleotide polymorphisms can be detected. Evidence of heteroplasmy—two or more mitochondrial variants within a single individual—has now been documented in a number of invertebrates; however, when present, heteroplasmy usually occurs at low frequencies both within individuals and within populations. The implications of heteroplasmy may be far reaching, both to the individual in relation to its health and fitness, and when considering the evolutionary dynamics of populations. We present novel evidence for frequent mtDNA heteroplasmy in the bed bug, Cimex lectularius L. (Hemiptera: Cimicidae). Our findings show that heteroplasmy is common, with 5 of 29 (17%) populations screened exhibiting two mitochondrial variants in a ∼1:2 ratio within each individual. We hypothesize that the mechanism underlying heteroplasmy in bed bugs is paternal leakage because some haplotypes were shared among unrelated populations and no evidence for nuclear mitochondrial DNA sequences was detected.
Keywords: cytochrome oxidase I, paternal leakage, Cimex lectularius, Hemiptera, Cimicidae
Mitochondrial heteroplasmy is the joint occurrence of two or more mtDNA haplotypes within a single individual (Avise 2000). Classical theory asserts that an organism’s mitochondrial genome is inherited clonally from its mother, with instances of heteroplasmy efficiently removed through vegetative segregation (Birky 1978). As a result, heteroplasmy is expected to be uncommon and likely short-lived (Gyllensten et al. 1991), but see Solignac et al. (1984). In recent years, however, the ease with which nucleotide polymorphisms are detected within the genome has facilitated a growing body of evidence suggesting that these laws governing mtDNA inheritance are in fact greatly over-simplified (White et al. 2008). As such, heteroplasmy has been documented across a variety of invertebrate organisms (Solignac et al. 1983, Paduan and Ribolla 2008, Magnacca and Brown 2009).
Heteroplasmy has been linked to a number of potential routes, which may relate to the inherently high mutation rate of mtDNA, the error-prone nature of DNA polymerase, and the lack of DNA repair mechanisms within mitochondria (Avise 2000, Chinnery et al. 2000). Therefore, mutations may arise during gametogenesis and embryonic development, generating unique haplotypes. If generated within a female’s germ-line cells, these novel mitochondrial haplotypes may be passed on to the offspring, assuming they persist through the germ-line bottleneck associated with oocyte formation and escape selection against deleterious mutations (Chinnery et al. 2000, White et al. 2008). Alternatively, mitochondrial heteroplasmy may arise through the failure of the machinery, which prevents the paternal mitochondria housed within the male gamete from entering the oocyte cytoplasm (Sutovsky et al. 2000). In contrast to mutation, paternal mtDNA leakage (Lansman et al. 1983) may result in the co-existence of unique maternally and paternally derived mtDNA lineages within the zygote (White et al. 2008). Paternal mtDNA leakage is considered extremely rare because several stochastic and molecular mechanisms efficiently exclude paternal mtDNA. These mechanisms include its dilution by female mtDNA in the zygote (Wolff and Gemmell 2008), selective tagging of paternal mtDNA and its destruction upon fertilization, and a second dilution through the bottleneck that occurs in the early stages of embryogenesis. Alternatively, instances of pseudo-heteroplasmy may result from the translocation of mtDNA sequences into the nuclear genome, resulting in nonfunctional nuclear copies of mitochondrial genes, termed “NUMTs;” identical priming sites may amplify both (Smith et al. 1992). As nuclear inserts should evolve independently of the mitochondrial genes, and are subject to different selection forces and recombination, mutations may be predicted to occur with equal likelihood across all codon positions under this pseudo-heteroplasmy scenario. In contrast, mutations within the mitochondrial copies of the same gene sequences are expected to be more common in the third codon position, and thus largely synonymous.
Mitochondrial heteroplasmy is typically infrequent even within organisms in which it has been reported, and heteroplasmy is rarely documented in natural populations, with most examples coming from experimental hybrids and natural hybrid zones (Kvist et al. 2003, Fontaine et al. 2007); but see (Paduan and Ribolla 2008, Fonseca et al. 2009). Heteroplasmy often poses considerable problems to the health of the organism (Russell and Turnbull 2014) because the introgression of paternal haplotypes may disrupt nuclear–mitochondrial interactions and introduce male-specific deleterious mutations. Moreover, heteroplasmy can introduce significant ambiguities to the interpretation of phylogenetic, phylogeographic, and population genetic data (White et al. 2008, Magnacca and Brown 2009).
It has been suggested that heteroplasmy may in fact be more common than previously recognized (White et al. 2008), yet few studies have characterized its occurrence and frequency at the intra-specific level. Our observations of recurring ambiguous mitochondrial sequences in the bed bug, Cimex lectularius L. (Hemiptera: Cimicidae) (Booth et al. 2015), led us to undertake a preliminary investigation of mtDNA heteroplasmy and investigate its frequency among samples collected in two south-central U.S. states, Oklahoma and Missouri. The bed bug, an obligate ectoparasite of humans and a species of significant human health concern, exhibits a cosmopolitan distribution following a dramatic global resurgence over the past two decades. Mitochondrial and nuclear DNA studies have revealed discordant patterns of infestation, with the former suggesting that infestations are commonly formed from multiple unrelated individuals (Szalanski et al. 2008), and the latter the opposite (Booth et al. 2012, Saenz et al. 2012, Fountain et al. 2014). After polymerase chain reaction (PCR) amplification and cloning of a 508-bp fragment of the cytochrome oxidase I (COI) subunit, we present several instances of high frequency heteroplasmy within multiple populations over a broad geographic range.
Materials and Methods
Bed bugs were collected live from 29 geographically distinct infested structures prior to treatment by pest management services in Oklahoma and Missouri. Specimens were immediately preserved in 95% ethanol. Genomic DNA was extracted using GeneJET Genomic DNA Purification Kit (Thermo Scientific Inc., Hundson, NH). Individual insects were screened for potential heteroplasmy in a 508-bp fragment of the COI subunit amplified using newly designed primers BB-COI-F (5′-AACTTAGACAAACCTGGCTCA-3′) and BB-COI-R2 (5′-GTGTTGGTAAAGTACAGGATCTY-3′). PCR was performed in 18-µl volumes containing: 1× PCR Buffer, 2 mM MgCl2, 100 mM dNTPs, 2.5 µM of each primer, 0.5 U Taq DNA polymerase (Apex, Genesee Scientific, San Diego), ∼50 ng of DNA template, and ddH2O to make 18 µl. PCR cycling conditions were composed of an initial denaturation stage of 5 min at 95°C, followed by 35 cycles each consisting 1 min at 95°C, 1 min at 58°C, and 1 min at 72°C. This was followed by a final extension stage of 72°C for 5 min.
Five samples where chromatograms revealed clear secondary overlapping peaks (see Fig. 1, for example) at isolated positions, indicative of heteroplasmy, were selected for cloning (Table 1). These samples were re-amplified to confirm the presence of double peaks and thus eliminate the likelihood that replication error during PCR producing these results. Additionally, during COI primer development, these five samples were among samples selected to screen three newly designed primer sets (additional primer information available upon request from W. Booth). All primer pairs consistently amplified the double peaks in the expected positions. PCR amplicons were ligated into One Shot TOP10 chemically competent E. coli vectors and cloned using the TOPO-TA cloning kit (Life Technologies Corporation, Carlsbad, CA). Cells were plated on LB agar containing 100 mg/µl ampicillin (Gentrox, Worcester, MA). Colony PCR was performed directly on unique clones using standard M13 primers. In total, 63 unique colony PCR products were purified using ExoSAP-IT (Affymetrix Inc., Santa Clara, CA) and bi-directionally sequenced on an ABI PRISM 3100xl Genetic Analyzer (Applied Biosystems, Foster City, CA) using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA; see Table 1). Sequences were visualized and edited in CLC Main Workbench (CLC bio, Aarhus, Denmark).
Fig. 1.
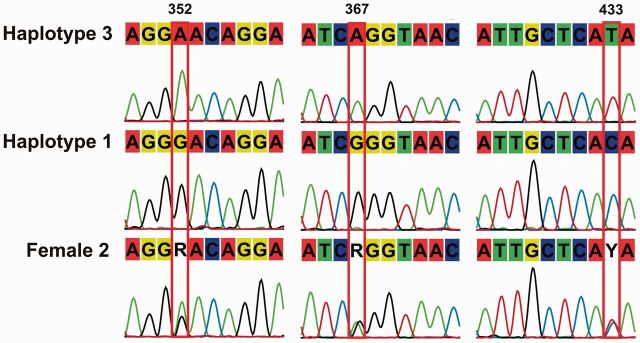
Representative COI chromatograms of a heteroplasmic female and her mtDNA variants as determined through cloning.
Table 1.
Sequence ID, sample locations, GenBank accessions numbers, polymorphism position, number cloned, and proportion of each haplotype in C. lectularius
| Sequence ID | Sample collection | COI accession no. | 244 | 322 | 352 | 367 | 433 | 547 | 595 | No. of clones | Proportion of clones |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AR5-1 | Tulsa, OK | KR002575 | Y | G | G | G | C | A | T | ||
| Haplotype 1 | KR002573 | C | – | – | – | – | – | – | 11 | 0.69 | |
| Haplotype 2 | KR002574 | T | – | – | – | – | – | – | 5 | 0.31 | |
| AR7-1 | Tulsa, OK | KR002580 | C | G | R | R | Y | A | T | ||
| Haplotype 1 | KR002578 | – | – | G | G | C | – | – | 2 | 0.29 | |
| Haplotype 3 | KR002579 | – | – | A | A | T | – | – | 5 | 0.71 | |
| AR19-1 | Muskogee, OK | KR002584 | C | R | G | G | C | R | Y | ||
| Haplotype 1 | KR002571 | – | G | – | – | – | A | T | 9 | 0.76 | |
| Haplotype 4 | KR002585 | – | A | – | – | – | G | C | 3 | 0.24 | |
| RC2-1 | St Louis, MO | KR002576 | Y | G | G | G | C | A | T | ||
| Haplotype 1 | KR002572 | C | – | – | – | – | – | – | 4 | 0.29 | |
| Haplotype 2 | KR002583 | T | – | – | – | – | – | – | 10 | 0.71 | |
| RC3-1 | St Louis, MO | KR002577 | Y | G | G | G | C | A | T | ||
| Haplotype 1 | KR002581 | C | – | – | – | – | – | – | 5 | 0.36 | |
| Haplotype 2 | KR002582 | T | – | – | – | – | – | – | 9 | 0.64 |
Nucleotide ambiguity codes based on IUPAC designations. Y represents cytosine (C) and thymine (T); R represents adenine (A) and guanine (G).
Results and Discussion
Each of the five specimens selected for cloning exhibited prominent secondary peaks. These secondary peaks were consistently re-amplified using multiple primer combinations and sample repetition (Fig. 1). These secondary peaks were not considered to result from NUMTs given that no significant sequence homology was found following a BLAST search of the primers or the amplified products against the bed bug genome (NCBI BioProject PRJNA167477) (performed 23 September 2014). Following cloning, each specimen was found to possess two distinct mitochondrial haplotypes with a total of four unique haplotypes recovered overall (Fig. 1; Table 1). A common haplotype previously reported in European populations found infesting both humans and bats, here denoted as haplotype 1 (Balvin et al. 2012, Booth et al. 2015), was shared by all. In all instances, mtDNA haplotypes appeared in a ratio of 1:2, but haplotype 1 was not always the most abundant (Table 1). The second haplotype in each specimen differed from haplotype 1 by either one or three nucleotide substitutions. Haplotype 2, which differs from haplotype 1 at a single base, has previously been reported in C. lectularius (Balvin et al. 2012) and was found in three of the five specimens analyzed; the remaining two haplotypes had not been reported in C. lectularius. Each distinct haplotype resulted from synonymous third position codon mutations. We did not determine the sensitivity of the PCR for rare haplotypes. However, because mtDNA haplotypes did not occur at unusually low frequencies (generally 1:2), we likely captured all the heteroplasmic variation within individual bed bugs. While instances of single nucleotide polymorphisms were found across a number of clones, these were not found to generate secondary peaks in the original specimen sequence, nor were they observed more than once in the specific individuals’ clones. The frequency of detection was consistent with Taq copy errors (Keohavong and Thilly 1989).
With the unambiguous detection of heteroplasmy in C. lectularius, the question turns to its origin. It is highly unlikely that heteroplasmy results either from mutations early in the insects’ development, or from more recent germ line mutations drifting to high copy numbers. Rather, our results suggest that paternal mtDNA leakage is likely responsible for heteroplasmy because in three populations (AR5-1, RC2-1, and RC3-1), identical heteroplasmic mtDNA haplotypes are present. Furthermore, these haplotypes have been recorded previously in samples sequenced within Europe (Balvin et al. 2012, Booth et al. 2015). Additionally, some haplotypes in this study differed by three mutations at the region of COI sequenced, each time with the mutation present in the third codon position. The likelihood that the same haplotypes would arise independently in multiple populations on multiple continents is negligible.
Where it appears, heteroplasmy generally occurs at very low frequencies both intra-individual and intra-population (Paduan and Ribolla 2008). In contrast, within C. lectularius, it appears much more frequently, with the original five specimens selected from only 29 bed bugs sequenced (i.e., 17%). We may, therefore, speculate that this high frequency of heteroplasmy in C. lectularius may also result from a relaxed mitochondrial bottleneck during oogenesis, allowing heteroplasmy to persist through a large number of generations following paternal leakage (Lutz et al. 2000). The high prevalence of mitochondrial heteroplasmy and the high frequency of the alternate haplotypes in individuals within bed bug populations appear uncommon in other species, and may represent a unique opportunity for the study of heteroplasmy.
The confirmation of heteroplasmy has broad implications to the biology of C. lectularius and inferences about its phylogeny and demographics. It questions the validity of the application of mtDNA to address the infestation dynamics of this species. Szalanski et al. (2008) reported multiple instances of more than one mtDNA haplotype within single infestations, inferred as resulting from introductions from multiple source populations. In contrast, three studies using nuclear microsatellite data revealed genetically depauperate infestations with four or fewer alleles per infestation, suggesting that infestations are likely founded by a single gravid female or a small group of highly related individuals (Booth et al. 2012, Saenz et al. 2012, Fountain et al. 2014). mtDNA sequencing of one such infestation revealed individuals both heteroplasmic and homoplasmic (see Fig. 2). It is clear that caution must be exercised when using only mtDNA to infer infestation dynamics and population-level relationships.
Fig. 2.
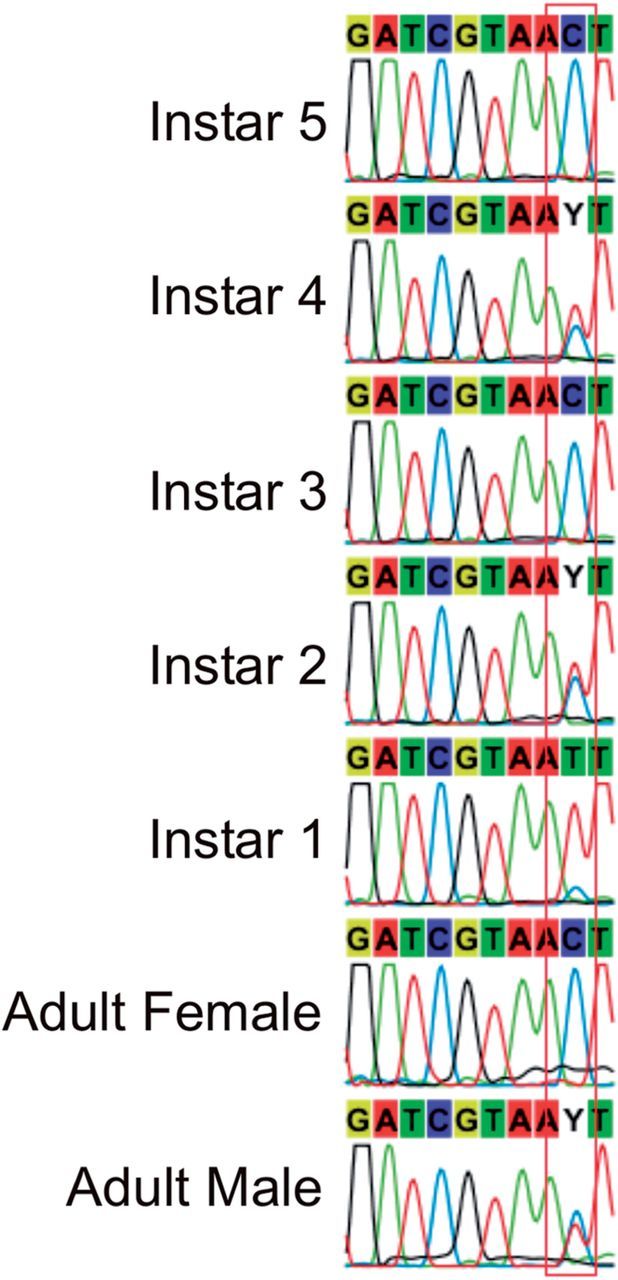
Representative COI chromatograms of two adult and five instar bed bugs revealing both heteroplasmic and homoplasmic individuals within a single infestation. The parentage of the instars presented is unknown and cannot be assumed to be the adults shown.
While more work is necessary to tease apart the mechanisms contributing to the high frequency of mitochondrial heteroplasmy in bed bugs, based on our findings, we hypothesize that paternal leakage may be common and its effects long lasting. We further suggest that mitochondrial heteroplasmy may be a relatively recent event in C. lectularius. This species all but disappeared from temperate homes following extensive insecticide-based interventions in the 1940s to the 1960s. An ongoing global resurgence of bed bugs might be re-uniting historically allopatric lineages, resulting in “hybridization-like” events. We suggest that in allopatry, molecular mechanisms that prevent transmission of paternal mtDNA to the oocyte may have been relaxed, leading to extensive paternal mtDNA leakage in inter-population “hybrids.” The ease of maintaining bed bugs in laboratory cultures, and the availability of isolated populations and museum specimens makes C. lectularius an excellent model organism for the study of the origins and maintenance of mitochondrial heteroplasmy. C. lectularius may also contribute to a better understanding of the roles of environmental factors (e.g., temperature, starvation, Wolbachia infection; Nikoh et al. 2014] in facilitating, maintaining, and purging paternal mtDNA.
Acknowledgments
We thank Arrow Exterminators (Oklahoma) and Rottler Pest Control (Missouri) for their continued support in providing field-collected bed bug samples. We thank two anonymous reviewers for their valuable feedback. We thank the i5k Initiative at Baylor College of Medicine (supported by the Human Genome Sequencing Center Grant ID 2U54HG003273-09, NIH/NHGRI) for access to the bed bug genome sequence. This research was supported by grants from the Oklahoma Center for the Advancement of Science and Technology (OCAST) (HR13-211) and The University of Tulsa Summer faculty development program to WB.
References Cited
- Avise J. C. 2000. Phylogeography: The history and formation of species. Harvard University Press, Cambridge, MA. [Google Scholar]
- Balvin O., Munclinger P., Kratochvil L., Vilimova J. 2012. Mitochondrial DNA and morphology show independent evolutionary histories of bedbug Cimex lectularius (Heteropera: Cimicidae) on bats and humans. Parasitol. Res. 111: 457–469. [DOI] [PubMed] [Google Scholar]
- Birky C. W. 1978. Transmission genetics of mitochondria and chloroplast. Annu. Rev. Genet. 12: 471–512. [DOI] [PubMed] [Google Scholar]
- Booth W., Balvin O., Vargo E. L., Vilímova J., Schal C. 2015. Host association drives genetic divergence in the bed bug, Cimex lectularius. Mol. Ecol. 24: 980–992. [DOI] [PubMed] [Google Scholar]
- Booth W., Saenz V. L., Santangelo R. G., Wang C., Schal C., Vargo E. L. 2012. Molecular markers reveal infestation dynamics of the bed bug (Hemiptera: Cimicidae) within apartment buildings. J. Med. Entomol. 49: 535–546. [DOI] [PubMed] [Google Scholar]
- Chinnery P. F., Thorburn D. R., Samuels D. C., White S. L., Dahl H. H., Turnbull D. M., Lightowlers R. N., Howell N. 2000. The inheritance of mitochondrial DNA heteroplasmy: Random drift, selection, or both? Trends. Genet. 16: 500–505. [DOI] [PubMed] [Google Scholar]
- Fonseca M. M., Brito J. C., Paulo O. S., Carretero M. A., Harris D. J. 2009. Systematic and phylogeographic assessment of the Acanthodactilus erythrurus group (Reptilia: Lacertidae) based on phylogenetic analyses of mitochondrial and nuclear DNA. Mol. Phylogenet. Evol. 51: 131–142. [DOI] [PubMed] [Google Scholar]
- Fontaine K. M., Cooley J. R., Simon C. 2007. Evidence for paternal leakage in hybrid periodical cicadas (Hemitpera: Magicicada spp.). PLoS ONE 2: e892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain T., Davaux L., Horsburgh G., Reinhardt K., Butlin R. K. 2014. Human-facilitated metapopulation dynamics in an emerging pest species, Cimex lectularius. Mol. Ecol. 23: 1071–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllensten U., Wharton D., Josefsson A., Wilson A. C. 1991. Paternal inheritance of mitochondrial DNA in mice. Nature 352: 255–257. [DOI] [PubMed] [Google Scholar]
- Keohavong P., Thilly W. G. 1989. Fidelity of DNA polymerase in DNA amplification. Proc. Natl. Acad. Sci. USA. 86: 9253–9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist L., Martens J., Nazarenko A. A., Orel M. 2003. Paternal leakage of mitochondrial DNA in the Great tit (Parsus major). Mol. Biol. Evol. 20: 243–247. [DOI] [PubMed] [Google Scholar]
- Lansman R. A., Avise J. C., Huettel M. D. 1983. Critical experimental test of the possibility of “paternal leakage” of mitochondrial DNA. P. Nat. Acad. Sci. USA. 80: 1969–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz S., Weisser H. J., Heizmann J., Pollak S. 2000. Mitochondrial heteroplasmy among maternally related individuals. Int. J. Legal Med. 113: 155–161. [DOI] [PubMed] [Google Scholar]
- Magnacca K. N., Brown M.J.F. 2009. Tissue segregation of mitochondrial haplotypes in heteroplasmic Hawaiian bees: Implications for DNA barcoding. Mol. Ecol. Resour. 10: 60–68. [DOI] [PubMed] [Google Scholar]
- Nikoh N., Hosokawa T., Moriyama M., Oshima K., Hattori M., Fukatsu T. 2014. Evolutionary – origin of insect-Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. USA 111: 10257–10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paduan K.D.S., Ribolla P.E.M. 2008. Mitochondrial DNA polymorphism and heteroplasmy in populations of Aedes aegypti in Brazil. J. Med. Entomol. 45: 59–67. [DOI] [PubMed] [Google Scholar]
- Russell O., Turnbull D. 2014. Mitochondrial DNA disease—molecular insights and potential routes to a cure. Exp.Cell Res. 325: 38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz V. L., Booth W., Schal C., Vargo E. L. 2012. Genetic analysis of bed bug populations reveals small propagule size within individual infestations but high genetic diversity across infestations from the eastern United States. J. Med. Entomol. 49: 865–875. [DOI] [PubMed] [Google Scholar]
- Smith M., Thomas W., Patton J. 1992. Mitochondrial DNA-like sequence in the nuclear genome of an akodontine rodent. Mol. Biol. Evol. 9: 204–215. [DOI] [PubMed] [Google Scholar]
- Solignac M., Monnerot M., Mounolou J. C. 1983. Mitochondrial DNA heteroplasmy in Drosophila mauritiana. Proc. Natl. Acad. Sci. USA 80: 6942–6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solignac M., Génermont J., Monnerot M., Mounolou J. C. 1984. Genetics of mitochondria in Drosophila: mtDNA inheritance in heteroplasmic strains of D. mauritiana . Mol. Genet. Genomics 197: 183–188. [Google Scholar]
- Sutovsky P., Moreno R. D., Ramalho-Santos J., Dominko T., Simerly C., Schatten G. 2000. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol. Reprod. 63: 582–590. [DOI] [PubMed] [Google Scholar]
- Szalanski A. L., Austin J. W., McKern J. A., Steelman C. D., Gold R. E. 2008. Mitochondrial and ribosomal internal transcribed spacer 1 diversity of Cimex lectularius (Hemiptera: Cimicidae). J. Med. Entomol. 45: 229–236. [DOI] [PubMed] [Google Scholar]
- White J. D., Wolff J. N., Pierson M., Gemmell N. J. 2008. Revealing the hidden complexities of mtDNA inheritance. Mol. Ecol. 17: 4925–4942. [DOI] [PubMed] [Google Scholar]
- Wolff J. N., Gemmell N. J. 2008. Lost in the zygote: The dilution effect of paternal mtDNA upon fertilization. Heredity 101: 429–434. [DOI] [PubMed] [Google Scholar]


