Abstract
The mosquito, Culex pipiens pallens (L.), is an important vector of encephalitis and filariasis in northern China. The control of these mosquitoes occurs primarily via the use of pyrethroid insecticides, such as deltamethrin. The widespread and improper application of pyrethroid has resulted in the evolution of pyrethroid resistance amongst many mosquito populations, including Cx. pipiens pallens. Previous studies using high-throughput transcriptome sequencing have identified that the venom allergen 5 gene is differentially expressed between deltamethrin-susceptible and deltamethrin-resistant Cx. pipiens pallens. In this study, quantitative real-time polymerase chain reaction analyses revealed that venom allergen 5 was significantly overexpressed in adult females of both deltamethrin-resistant laboratory populations and two field populations. The transcriptional level of venom allergen 5 in the laboratory populations was elevated as the levels of deltamethrin resistance increased. Full-length cDNAs of the venom allergen 5 gene were cloned from Cx. pipiens pallens, and contained an open reading frame of 765 bp, encoding a protein with 254 amino acids. The deduced amino acid sequence shared 100% identity with the ortholog in Culex quinquefasciatus Say. The overexpression of venom allergen 5 decreased the susceptibility of mosquito cells to deltamethrin, while knockdown of this gene by RNAi increased the susceptibility of mosquitoes to deltamethrin. This study provides the first evidence of the association between the venom allergen 5 gene and deltamethrin resistance in mosquitoes.
Keywords: Culex pipiens pallens, pyrethroid, quantitative real-time PCR, venom allergen 5, RNAi
Mosquitoes are important vectors of numerous infectious diseases of humans, including malaria (Mitri and Vernick 2012), dengue fever (Jeffery et al. 2009), yellow fever (Barrett and Higgs 2007), West Nile fever (Styer et al. 2011), encephalitis, and filariasis (Gambhir and Michael 2008, Smith et al. 2008). The chemical control of mosquitoes, via the use of insecticides, is the primary means of managing the spread of these diseases, as it is simple, rapid, and economical (Hemingway et al. 2006, Mahande et al. 2012). Since the insecticide, dichlorodiphenyltrichloroethane (DDT), was introduced in 1939, chemical insecticides have undergone four generations of development: DDT and other organochlorines, organophosphorus, carbamates, and pyrethroids. At present, pyrethroids are the most widely used of these (van den Berg et al. 2012); however, their widespread and improper use has resulted in the evolution of pyrethroid resistance (Casimiro et al. 2006, World Health Organization [WHO] 2012), which has become a major obstacle for mosquito-borne diseases management (Jinfu 1999, Hemingway et al. 2002).
To date, three main types of pyrethroid-resistant mechanisms have been recognized. Target resistance occurs via mutations in the voltage-gated sodium channel target sites of the insect nervous system (Dong et al. 2014). Metabolic resistance occurs via increases in either protein levels or the activity of detoxification enzymes that resist insecticides. For example, esterases (Wu et al. 2011), glutathione S-transferases (Lumjuan et al. 2011), and cytochrome P450 (CYP or P450; Edi et al. 2014) have all been demonstrated to be associated with metabolic resistance. In addition, insecticides induce physiological changes in mosquitoes such as a thickening of the epidermis, which subsequently lowers their permeability to insecticides (Wood et al. 2010). Studies investigating the mechanisms of pyrethroid resistance have identified other resistance-related genes, such as UDP-glycosyltransferases (Vontas et al. 2005, Bozzolan et al. 2014) and serine proteases (Reid et al. 2012, Strachecka et al. 2013).
Pyrethroid resistance in mosquitoes is common and broadly distributed in China (Wang et al. 2012). Many pyrethroid resistance-related genes have been found in Cx. pipiens pallens, the dominant species of mosquito in northern China, such as CYP6F1 (Gong et al. 2005), opsin (Hu et al. 2007), arrestin (Sun et al. 2012), and PSMB6 (Sun et al. 2013). However, pyrethroid resistance is actually a complex phenotype of polygenic inheritance (Ffrench-Constant et al. 2004). Indeed, none of the currently known genes can entirely explain the molecular basis for pyrethroid resistance in Cx. pipiens pallens. Hence, identifying novel genes associated with pyrethroid resistance and elucidating their regulatory mechanisms is critical for the effective control of Cx. pipiens pallens.
The known insect venom allergens are proteins of 10–50 kDa containing 100–400 amino acid residues, many of which have been expressed in bacteria, insect, or yeast cells (King and Spangfort 2000). Some of the venom allergen-like proteins in Schistosoma mansoni Sambon are suggested to be immune modulators and vaccine candidates (Chalmers et al. 2008, Farias et al. 2012). The venom allergen 5 gene is one of the major allergens identified in many insect venoms (King and Spangfort 2000) and is often associated with allergic responses in humans (Muller et al. 2009). Patients show varying degrees of cross-reactivity to the related venom allergen 5 genes of species such as yellow jackets, hornets, and paper wasps (Henriksen et al. 2001). Within blood-feeding ticks (Mans et al. 2008), flies (Charlab et al. 1999), and mosquitoes (Calvo et al. 2007), the venom allergen 5 proteins are part of a cocktail of salivary proteins believed to function either in the suppression of the host immune system or in the prevention of clotting to prolong feeding (Ribeiro and Francischetti 2003, Dos Santos-Pinto et al. 2014).
The venom allergen 5 protein contains a sperm-coating protein (SCP)-like extracellular protein domain, and belongs to the SCP superfamily. This family includes plant pathogenesis-related protein 1 (Kitajima and Sato 1999), mammalian cysteine-rich secretory proteins (Da Ros et al. 2007), and allergen 5 from insect venom. One member of this superfamily, Tex31, from the venom duct of Conus textile L., has been shown to possess proteolytic activity sensitive to serine protease inhibitors (Milne et al. 2003). Currently, little is known about the biological functions of the SCP superfamily in mosquitoes.
Previously, we undertook a large-scale transcription profiling study using high-throughput transcriptome sequencing in deltamethrin-susceptible (DS) and deltamethrin-resistant (DR) strains of Cx. pipiens pallens, from which we identified many differentially expressed genes, including venom allergen 5 (Y. L. et al., unpublished data). In this study, we first use quantitative real-time polymerase chain reaction (qRT-PCR) to quantify the expression levels of several genes that are differentially expressed between susceptible and resistant mosquitos. We quantified the relative expression levels of these genes in adult females of DS and DR strains from both a laboratory population and two field populations. The laboratory population included mosquitoes at different developmental stages and different levels of deltamethrin resistance. We then cloned the full-length cDNAs of venom allergen 5 in Culex pipiens pallens (L.). Subsequently, to further investigate the role of venom allergen 5 in deltamethrin resistance, we verified its functionality both in vitro and in vivo. We provide the first evidence for the association of venom allergen 5 with deltamethrin resistance in mosquitoes.
Materials and Methods
Mosquito Strains
The DS strain of Cx. pipiens pallens was collected from Tangkou town of Shandong Province (35.12 N; 116.50 E) in 2009 and reared in our laboratory without exposure to any insecticide. The DR strains (DR1, DR2, and DR3) were selected from early fourth-instar larvae of the DS strain exposed to deltamethrin for >30 generations. Before selection, the 50% larval lethal concentration (LC50) to deltamethrin was determined by larval bioassay (Chen et al. 2010) and used as the screening concentration. All laboratory populations were maintained at 28°C, 70–80% humidity, and a photoperiod of 16:8 (L:D) h. The two field populations of Cx. pipiens pallens were collected from Shanghe (37.31 N; 117.16 E) and Gudao (37.85 N; 118.81 E) towns of Shandong Province in 2011. To distinguish the susceptible and the resistant strains, nonblood-fed adult females 2–3 d postemergence were exposed to 0.05% deltamethrin-impregnated drug membranes by the WHO susceptibility tube bioassay (WHO 1998, 2013). The mosquitoes that survived the 24-h recovery period were classified as deltamethrin resistant, while those knocked down early during the bioassay were classified as deltamethrin susceptible. The knocked-down mosquitoes were immediately preserved in Eppendorf tubes without allowing a 24-h recovery period to prevent postmortem RNA degradation (Bonizzoni et al. 2012, Zhu et al. 2014).
RNA Extraction and cDNA Synthesis
Total RNA was extracted from all mosquitoes by RNAiso plus (Takara, Tokyo, Japan). DNase I (Takara, Tokyo, Japan) was used to remove potential contaminant genomic DNA. Total RNA integrity was assessed by denaturing agarose gel electrophoresis and purity and concentration were authenticated using a NanoDrop spectrophotometer (NanoDrop, Wilmington, DE). The first-strand cDNA was synthesized from total RNA with the PrimeScript RT Master Mix (Takara, Tokyo, Japan).
Quantitative RT-PCR Analyses
The qRT-PCR reactions were performed with a 7300 FAST Real-Time PCR System (Applied Biosystems, Foster City, CA) using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Gene-specific qRT-PCR primers were designed by Primer Premier 3.0 (Premier Biosoft International). The PCR conditions were as follows: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, 60°C for 1 min. The PCR products were used for melting curve and gel electrophoresis analysis to confirm their amplification specificity. The data were analyzed with 7300 System SDS Software v1.2.1 (Applied Biosystems, Foster City, CA). We chose β-actin (Canales et al. 2009) and RsP7 (Leal et al. 2013) as the internal normalization. The relative expression levels of each gene were calculated using the 2 -(ΔΔCt) method (Livak and Schmittgen 2001). We used qRT-PCR to detect the expression levels of the venom allergen 5 gene in adult females of both the DS and DR strains of the laboratory and field populations. The laboratory populations included mosquitoes with different levels of deltamethrin resistance (DS, DR1, DR2, and DR3 strains) and at different development stages (eggs, first- to fourth-instar larvae, pupae, female adults, and male adults). Three technical and three biological replicates were performed for qRT-PCR analyses. All primer sequences are presented in Table 1.
Table 1.
List of the primers for qRT-PCR
| Gene | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | Length |
|---|---|---|---|
| β-actin | AGCGTGAACTGACGGCTCTG | ACTCGTCGTACTCCTGCTTGG | 153 bp |
| RsP7 | CCTGGAGCTGGAGATGAACT | ACGATGGCCTTCTTGTTGTT | 99 bp |
| CPIJ800157 | CTACGACATTCCCAAGGATACAA | CATCATTTCGCCCATACAGC | 178 bp |
| CPIJ000835 | TTAACGAAAACTACGTGCTGACTG | GGGCTGACTCCACCCTGATA | 172 bp |
| CPIJ013082 | TGACCTGGGCTGTCGATATG | GCCGTGGTCGTTGGTTTT | 235 bp |
| CPIJ013725 | GACAGGAGGAGATGGAACAGGAT | TCGGGCCAAATCATCGTAGT | 125 bp |
| CPIJ002786 | AGCTGGATGGTGCTCTGGAA | AGCCGTAGTTGCCGCAGATA | 120 bp |
| CPIJ015028 | GGACTACTGCAACCCGGACTT | TTTTCGCATCCACGCAATC | 95 bp |
| CPIJ009877 | CGGTTCTGTAATCGTTTGGG | CGTATCGTTGTCCGTCCTGTT | 165 bp |
| CPIJ007024 | GACCTTAATCCGAACTTTGCC | GCCTCCCTAGCCAACTGACC | 175 bp |
| CPIJ802088 | TTCCAGAAGCGTAACATCATCA | TCTTGCGTTGGTTCAGCCT | 184 bp |
| CPIJ009326 | CCCAAGGTCGCTTCTCGTA | GGCTGGAATCCGTTCTCGT | 82 bp |
| CPIJ800292 | GAACAACGAATCCATAGCGG | GGTGCATTGCGAGCATGAG | 104 bp |
Full-length Cloning and Sequencing
The full-length cDNA of venom allergen 5 in Cx. pipiens pallens was amplified in three sections: the open reading frame (ORF) and the 5′ and 3′-cDNA ends (5′ and 3′-RACE). The ORF was amplified using 2× Taq Plus Master Mix (Vazyme, Nanjing, China). Templates of 5′-RACE and 3′-RACE were prepared with a SMART RACE cDNA Amplification Kit (Clontech, Mountain View, CA). The PCR reactions were carried out using Advantage 2 Polymerase Mix (Clontech, Mountain View, CA) according to the manufacturer’s protocol. PCR products were separated on a 1% agarose gel and then purified with a QIA quick Gel Extraction Kit (QIAGEN, GmbH, Germany). The purified products were then inserted into the pMD-19 T simple vector (Takara, Tokyo, Japan) to be sequenced at the Beijing Genomics Institute. The three sections were assembled to generate the full-length cDNA. All primer sequences for the 5′ and 3′ RACE are presented in Table 2.
Table 2.
List of the primers for RACE
| Fragment | Sense primer (5′ to 3′) | Antisense primer (5′ to 3′) |
|---|---|---|
| ORF | ATGTCGATCAACCGTAAAGTTC | TCAAGCTCCCACATCCACCG |
| 5′-RACE | Universal Primer A Mixa | CGCATCCACGCAATCCGACGACAGC |
| 3′-RACE | CACTTTGCCACGCCGAAGCTGATGA | Universal Primer A Mixa |
aThe Universal Primer A Mix (UPM) was a mixture of 5′-CTAATACGACTCACTATAGGGC AAGCAGTGGTATCAACGCAGAGT-3′ and 5′-CTAATACGACTCACTATAGGGC-3′.
Sequence Alignment and Phylogenetic Tree
The standard protein–protein BLAST sequence comparison programs (http://beta.uniprot.org/?tab0blast) were used to search for sequences in the SWISSPROT databases with similarities to the translated sequences of venom allergen 5. Similar amino acid sequences were aligned using the ClustalW2 computer program (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The phylogenetic tree was constructed using the neighbor-joining method of the MEGA5.1 program (Tamura et al. 2011). Structure analysis (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used to search for conserved domains within the protein.
Construction of the Eukaryotic Expression Plasmid
The ORF of venom allergen 5 was inserted into the eukaryotic expression vector pIB/V5-His (Invitrogen, Carlsbad, CA) and amplified with specific primers: forward primer 5′-GGACTAGTGAGATGGAA ATGTCGATCAACCGTAAAGTTCAAG-3′ (containing the SpeI recognition site, ACTAGT); reverse primer 5′-CCGCTCGAGCGAGCTCCCACATCCACCG-3′ (containing the XhoI recognition site, CTCGAG). A Kozak sequence (GAGATGG) was added before the start codon ATG, and two additional AA were added after it to avoid a frameshift mutation (Kozak 1986). To ensure the fusion expression of the His tag protein, the stop codon TGA was removed and two additional CG were added to avoid a frameshift mutation. The PCR-amplified product and the pIB/V5-His expression vector were both digested by SpeI and XhoI. The two expected bands were then purified using the QIA quick Gel Extraction Kit (QIAGEN, GmbH, Germany) and ligated with T4 DNA Ligase. The ligated product was transformed into TOP10 competent cells (Tiangen, Beijing, China). After overnight culture, positive clones were identified by PCR and sequenced by Beijing Genomics Institute.
Plasmid Transfection and Cytotoxicity Assay
An Aedes albopictus (Skuse) C6/36 cell line was obtained from the Chinese Center for Type Culture Collection (CCTCC, Wuhan, China). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and with antibiotics (called DMEM-complete), and were maintained in a 5% CO2 humidified incubator at 28°C.
One aliquot of C6/36 cells was preincubated in a six-well plate (4 × 105 cells per well) overnight until the cell confluency achieved 80%. Then, 2 μg pIB/V5-His-venom allergen 5 plasmid DNA and 5 μl FuGENE HD transfection reagent (Promega, Madison, WI) were added to 100 μl DMEM-complete, mixed gently, and incubated at room temperature for 30 min. The plasmid DNA of the pIB/V5-His empty vector was the negative control (NC). The mixture was then added to the six-well plate for transient transfection experiments. At 48 h posttransfection, we collected the cells and carried out western blot analysis to confirm successful transient transfection and protein expression. Protein was extracted from C6/36, C6/36-NC, and C6/36-venom allergen 5 cells using RIPA lysis buffer (Beyotime, Shanghai, China). We determined protein concentrations using the Enhanced BCA Protein Assay Kit (Beyotime, Shanghai, China). Up to 40 μg proteins were analyzed by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel for 30 min at 80 V and 120 min at 100 V. Then, we transferred the protein to a NC membrane for 50 min at 300 mA using the Trans-Blot SD Cell and Systems (Bio-Rad, Richmond, CA). The His-tag fusion protein was detected using an anti-His monoclonal antibody (1:500; ABGENT, San Diego, CA) and a peroxidase-conjugated goat anti-mouse secondary antibody (1:2000; Bioworld, Louis Park, MN). Finally, the bound antibodies were recognized with a SuperSignal West Pico Chemiluminescent Substrate Kit (Thermo, Rockford, IL), and the signals were detected using a Bio-Rad ChemiDoc XRS scanner and the Quantity One software (Bio-Rad, Richmond, CA). β-actin was used as the internal control.
The other aliquot of C6/36 cells was preincubated in a 96-well plate (8,000 cells per well). After transfection with pIB/V5-His-venom allergen 5 or pIB/V5-His empty vector (NC) plasmid DNA for 48 h, the C6/36 cells were treated with a series of concentrations of deltamethrin (0, 100.5, 101.0, 101.5, 102.0, and 102.5 μg/ml). Before the experiment, 0.1 g of deltamethrin was completely dissolved in 1.0 ml of dimethyl sulfoxide (DMSO; Sigma, St. Louis, MO). Then, we diluted the different concentrations of deltamethrin with DMEM-complete. The wells of different concentrations of deltamethrin had the same final concentration of DMSO (0.5%, v/v). After 43 h, we added 10 μl of CCK-8 solution (Dojindo, Kumamoto, Japan) to each well. Five hours later, we measured the absorbance at 450 and 630 nm using a microplate reader (Biotek, Winooski, VT). Then, the absorbance at 630 nm was subtracted from the absorbance of the same well measured at 450 nm to eliminate any background signal. The percentage of cell viability at each concentration was calculated relative to the cell viability at 0 μg/ml concentration.
RNAi and WHO Susceptibility Tube Bioassay
According to the full-length sequence of venom allergen 5, we designed and synthesized the siRNA40 (GenePharma, Shanghai, China). Nonblood-fed adult female mosquitoes from the DR1 strain that had emerged 1 d before were used for the microinjection experiments. According to standard methodology (Blandin et al. 2002), we microinjected 0.07 μl of NC RNA or siRNA40 (5 μg/μl) into the thorax of mosquitoes. The sequences of NC and siRNA40 are provided in Table 3. The wild-type (WT) mosquitoes that were not microinjected were used as the noninjected control. Three days later, we verified the RNAi efficiency using qRT-PCR, and assessed the viability of adult female mosquitoes by a WHO susceptibility tube bioassay with 0.05% deltamethrin-impregnated drug membranes. Three days after microinjection, 20–25 female mosquitoes were transferred into each tube and exposed to the 0.05% deltamethrin-impregnated drug membranes for 1 h and were then transferred into recovery tubes. The number of alive and dead mosquitoes in each tube after a 24-h recovery period was recorded to determine their viability (the ratio of the number alive after the 24-h recovery period to the total number of mosquitoes before the 1-h exposure period in each tube; WHO 2013). The nonpyrethroid control membrane (PY control) was simultaneously used to test the mosquitoes with siRNA40 microinjection.
Table 3.
List of the siRNA sequences for RNAi
| Name | Sense (5′ to 3′) | Antisense (5′ to 3′) |
|---|---|---|
| siRNA-40 | GAUCAACCGUAAAGUUCAATT | UUGAACUUUACGGUUGAUCTT |
| NC | GCGACGAUCUGCCUAAGAUdTdT | AUCUUAGGCAGAUCGUCGCdTdT |
Statistics
All data are presented as mean ± SD of three independent experiments. Mosquito viability was analyzed using the chi-square test. The Student’s t-test was used to analyze the data of qRT-PCR and cell viability. The statistical significance level was set at P < 0.05.
Results
Screening Candidate Genes
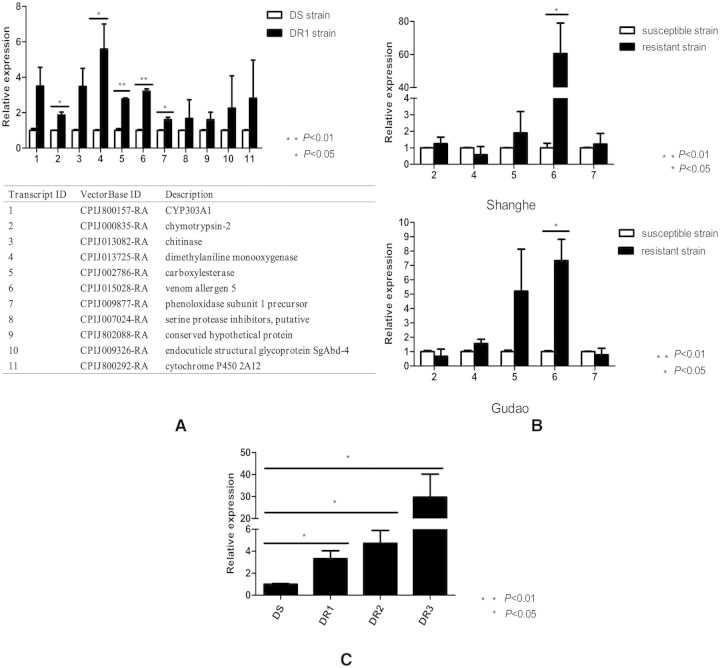
Referring to the comparative transcriptome sequencing results, we selected 11 genes to test their relative expression levels at the adult female stage of the DS and the DR1 strains. As shown in Fig. 1A, 5 of the 11 genes (chymotrypsin-2, dimethylaniline monooxygenase, carboxylesterase, venom allergen 5, and phenoloxidase subunit 1 precursor) were significantly overexpressed in the DR1 strain compared with the DS strain (P < 0.05). Then, we verified the expression levels of these five genes in the susceptible and resistant strains of the two field populations (Shanghe and Gudao). The results indicated that the gene expression levels of venom allergen 5 were 60.58- and 7.34-fold higher in the resistant strains than the susceptible strains (P < 0.05; Fig. 1B). Next, we quantified the gene expression level of venom allergen 5 in four strains (DS, DR1, DR2, and DR3) with different levels of deltamethrin resistance. The LC50 of the DS, DR1, DR2, and DR3 strains were 0.03, 0.85, 3.7, and 7.0 mg/liter, respectively. As shown in Fig. 1C, the venom allergen 5 expression levels were 3.3-, 4.7-, and 29.7-fold higher in the DR1, DR2, and DR3 strains, respectively, than the DS strain. Thus, we selected venom allergen 5 as our candidate gene for subsequent verification of whether it was associated with deltamethrin resistance or not.
Fig. 1.
Screening candidate genes via quantitative real-time PCR. (A) The relative expression levels of 11 genes in adult female mosquitoes of the DS and the DR1 strains. Five of the 11 genes (chymotrypsin-2, dimethylaniline monooxygenase, carboxylesterase, venom allergen 5, and phenoloxidase subunit 1 precursor) were significantly overexpressed in the DR1 strain compared with the DS strain. (B) The relative expression levels of five genes in adult female mosquitoes of the susceptible and resistant strains of two field populations (Shanghe and Gudao). The relative expression levels of the venom allergen 5 were 60.58- and 7.34-fold higher in the resistant strains than the susceptible strains. (C) The relative expression levels of venom allergen 5 in adult female mosquitoes with different levels of deltamethrin resistance (DS, DR1, DR2, and DR3). The LC50 of the DS, DR1, DR2, and DR3 strains were 0.03, 0.85, 3.7, and 7.0 mg/liter, respectively. The venom allergen 5 expression levels were 3.3-, 4.7-, and 29.7-fold higher in the DR1, DR2, and DR3 strains, respectively, than the DS strain. Figures show the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 compared with the DS strain.
Cloning the Full-length cDNA of Venom allergen 5 in Cx. pipiens pallens
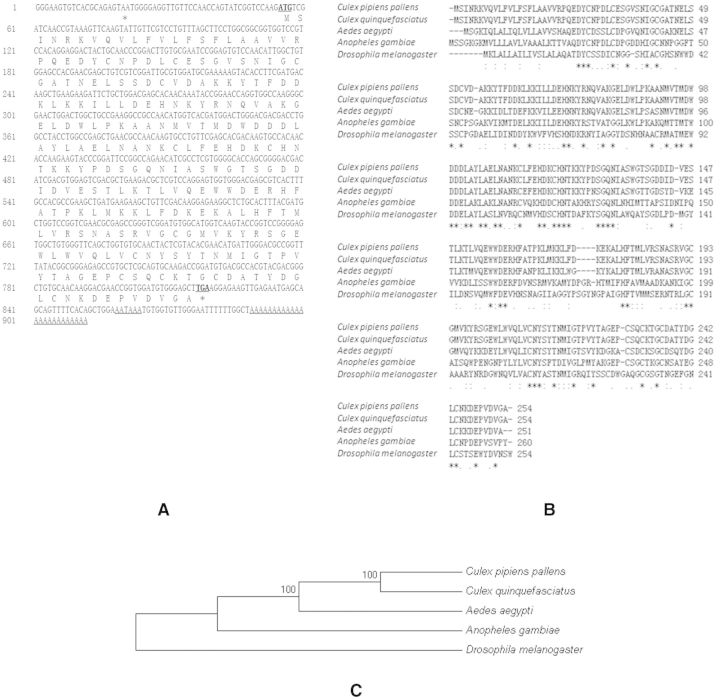
The full-length cDNA of venom allergen 5 contained 912 bp. The ORF region had 765 bp and encoded a protein with 254 amino acids. The start codon ATG was found at positions 55–57, while the same frame stop codon TAA was at positions 817–819 with a polyadenylation signal sequence “AATAAA” and a poly (A) tail present at the 3′-untranslated region (Fig. 2A). The full-length cDNA sequence of venom allergen 5 in Cx. pipiens pallens has been entered into GenBank (accession number: KF723295.1). The deduced amino acid of venom allergen 5 in Cx. pipiens pallens shared 100% identity with the ortholog in Culex quinquefasciatus Say, 70.8% identity with Aedes aegypti (L.), 43.85% identity with Anopheles gambiae Giles, and 32.95% identity with Drosophila melanogaster Meigen, according to the ClustalW2 software (Fig. 2B). Phylogenetic relationships showed that venom allergen 5 in Cx. pipiens pallens has the highest homology with Cx. quinquefasciatus (Fig. 2C). Structure analysis showed that the protein contains a SCP-like extracellular protein domain, as found primarily in eukaryotes (http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=240180).
Fig. 2.
Full-length cDNA of venom allergen 5 in Cx. pipiens pallens. (A) The nucleotide and deduced amino acid sequences of venom allergen 5 in Cx. pipiens pallens. The initial codon “ATG” and the termination codon “TGA” are labeled in bold letters and underlined. The asterisk indicates the stop codon. The polyadenylation signal sequence “AATAAA” and a poly (A) in the 3′-untranslated region are underlined. (B) Amino acid alignment of the venom allergen 5 gene in Cx. pipiens pallens and four other species. Asterisks indicate identical amino acids and dots indicate similar amino acids. (C) Phylogenetic relationships of the venom allergen 5 in Cx. pipiens pallens and four other species. Species name and GenBank Accession No.: Cx. pipiens pallens, KF723295.1; Cx. quinquefasciatus, XP_001865176.1; Ae. aegypti, XP_001661917.1; An. gambiae XP_314254.5; D. melanogaster, AAD03844.1.
In Vitro Validation by Plasmid Transfection and Cyotoxicity Assay
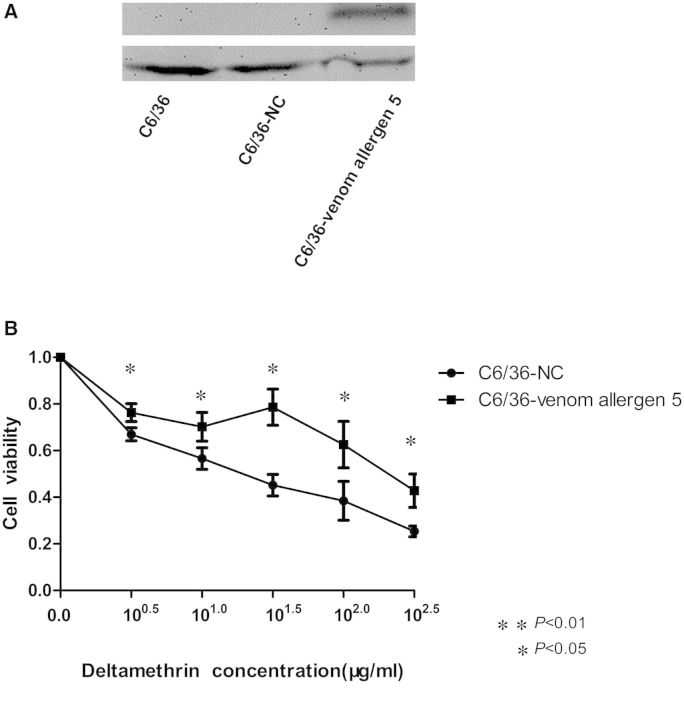
To further validate the functionality of venom allergen 5 in pyrethroid resistance in vitro, we transiently transfected the pIB/V5-His-venom allergen 5 and pIB/V5-His (NC) plasmid DNA into the C6/36 cells. As shown in Fig. 3A, western blot analysis revealed that C6/36 cells with pIB/V5-His-venom allergen 5 transfection expressed the corresponding protein of ∼32 kDa, which confirmed the successful expression of the exogenous venom allergen 5 in C6/36 cells. We measured cell viability over a wide range of concentrations (0 to 102.5 μg/ml) of deltamethrin using the CCK-8 kit as a cyotoxicity assay (Fig. 3B). The cell viability of transiently transfected cells decreased as the concentration of deltamethrin increased. The significantly increased viability of C6/36-venom allergen 5 compared with the C6/36-NC cells with each of the five deltamethrin concentrations (P < 0.05) suggested that the overexpression of venom allergen 5 might decrease deltamethrin cyotoxicity in vitro.
Fig. 3.
In vitro validation by plasmid transfection and cytotoxicity assay. (A) Results of western blot analysis of the protein expression of venom allergen 5 in the C6/36, C6/36-NC, and C6/36-venom allergen 5 cells. β-actin was used as the internal control. The C6/36-venom allergen 5 cells expressed the corresponding protein of ∼32 kDa. The experiment was repeated three times. (B) Cytotoxicity assay using a CCK-8 kit. The x-axis shows the five concentrations of deltamethrin. The y-axis gives cell viability (as a proportion of the total cells relative to a 0 μg/ml concentration). The viability of the C6/36-venom allergen 5 cells was significantly higher than the C6/36-NC cells with each of the five deltamethrin concentrations. Figures show the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 compared with the C6/36-NC cells.
In Vivo Validation by RNAi and WHO Susceptibility Tube Bioassay
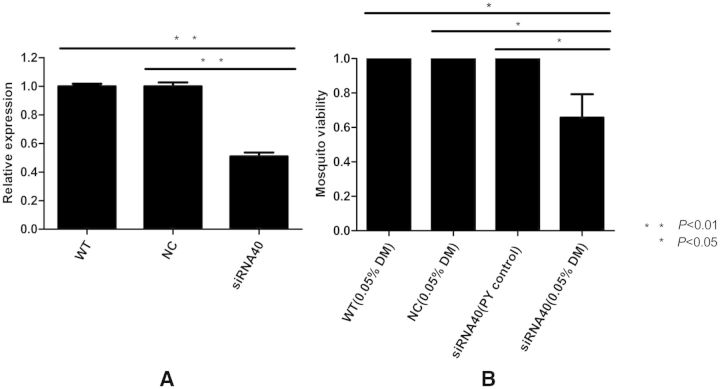
To validate the relationship of venom allergen 5 to pyrethroid resistance in vivo, we microinjected designed siRNA40 into the thorax of female adult mosquitoes from the DR1 strain to knock down the expression of venom allergen 5. As shown in Fig. 4A, siRNA40 could reduce the gene expression of venom allergen 5 by ∼50%, compared with the WT and NC groups (P < 0.01). The results of a WHO susceptibility tube bioassay with 0.05% deltamethrin-impregnated drug membranes suggested that the knockdown expression of venom allergen 5 decreased the viability of female adult mosquitoes by 30–40%, compared with the control groups (P < 0.05; Fig. 4B). The knockdown of venom allergen 5 by RNAi increased deltamethrin toxicity in vivo.
Fig. 4.
In vivo validation by RNAi and WHO susceptibility tube bioassay. (A) RNAi efficiency verification of venom allergen 5 by qRT-PCR. The NC RNA and siRNA40 were microinjected into the thorax of adult female mosquitoes. The WT mosquitoes that were not microinjected were used as a noninjected control. The siRNA40 reduced the expression of venom allergen 5 by ∼50% compared with the WT and the NC groups. (B) The viability of microinjected mosquitoes following a WHO susceptibility tube bioassay with 0.05% deltamethrin-impregnated drug membranes. The knockdown expression of venom allergen 5 by siRNA40 decreased the viability of adult female mosquitoes by 30–40% compared with the WT and the NC groups. All mosquitoes in the PY control (nonpyrethroid control membrane) tubes were alive. Figures show the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01.
Expression Level of the Venom Allergen 5 Gene at Different Developmental Stages
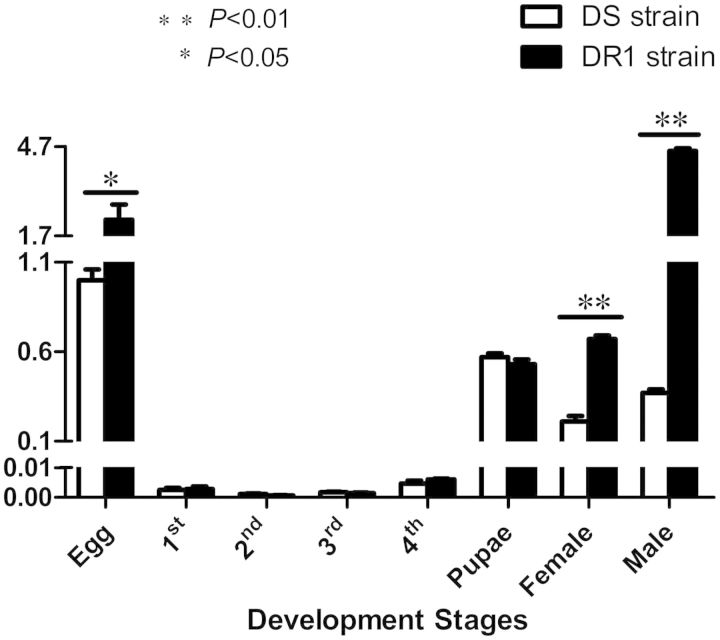
We used qRT-PCR to test the expression levels of venom allergen 5 at different developmental stages in the DS and the DR1 strains of Cx. pipiens pallens. As shown in Fig. 5, venom allergen 5 was transcribed at all developmental stages. The transcriptional levels of venom allergen 5 were relatively higher in egg, pupae, and adults than in fist- to fourth-instar larvae. The transcriptional levels of this gene were 2.24-, 3.3-, and 12.3-fold higher in the DR1 strain than in the DS strain at the egg, female adult, and male adult stages (P < 0.05).
Fig. 5.
Quantitative real-time PCR analysis of venom allergen 5 at different developmental stages of Cx. pipiens pallens. The venom allergen 5 gene was transcribed at all developmental stages. The transcriptional levels of venom allergen 5 were 2.2-, 3.3-, and 12.3-fold higher in the DR1 strain than in the DS strain at the egg, female, and male adult stages. The expression level of mosquitoes at the egg stage of the DS strain was considered to be 1. The figures show the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 compared with the DS strain.
Discussion
It is widely accepted that the development of pyrethroid resistance in Culex mosquitoes occurs via multiple mechanisms and interacting genes (Liu et al. 2011). Hence, identifying novel resistance genes and elucidating their mechanisms are long-term focal issues in pyrethroid resistance research. In this study, we selected 11 candidate genes for qRT-PCR analysis based on results of comparative transcriptome sequencing results. We found that while transcript 1 (CYP303A1) and transcript 11 (cytochrome P450 2A12) were 3.5- and 2.28-fold higher in the DR1 strain than in the DS strain, the difference was not statistically significant (P > 0.05). These two transcripts belong to the CYP2 clan of the cytochrome P450 superfamily that contains many putative genes thought to be involved in pyrethroid resistance (Yang and Liu 2011). However, by far, the majority of pyrethroid resistance-associated genes belong to the CYP6 family of the CYP3 clan (Edi et al. 2014), and the CYP4 family of the CYP4 clan (Reid et al. 2014). In contrast, little is currently known about the relationship between the CYP2 clan and insecticide resistance. We found no significant difference in CYP303A1 and cytochrome P450 2A12 between the DS strain and the DR1 strain. Similarly, the majority of the P450 genes were not associated with pyrethroid resistance in Cx. quinquefasciatus (Yang and Liu 2011). Thus, these two P450 genes were not chosen for further examination in the two field populations.
However, venom allergen 5 was significantly overexpressed in the adult females of DR laboratory populations and the two field populations. Moreover, we found that the transcriptional level of venom allergen 5 in laboratory populations was elevated as the levels of deltamethrin resistance increased. To further investigate the role of venom allergen 5 in deltamethrin resistance, we verified its functionality both in vitro and in vivo. We report that the overexpression of venom allergen 5 decreased the susceptibility of mosquito cells to deltamethrin treatment in vitro, mainly presented as decreased deltamethrin cytotoxicity. On the other hand, the knockdown of venom allergen 5 by RNAi increased the susceptibility of mosquitoes to deltamethrin exposure in vivo, mainly presented as increased deltamethrin toxicity. These two verifications of the functionality of this candidate gene strongly suggest that venom allergen 5 is associated with deltamethrin resistance in mosquitoes. Hence, this study further supports the idea that pyrethroid resistance is a complicated genetic phenomenon and is involved in a multimechanism or interaction of several genes (David et al. 2005, Liu et al. 2007).
The venom allergen 5 gene was expressed at all developmental stages of Cx. pipiens pallens and its expression levels were significantly higher in the eggs, females, and males of the DR1 strain than in those stages of the DS strain. These results suggest that venom allergen 5 might play a role in deltamethrin resistance at multiple developmental stages. Interestingly, we also found that the expression levels of this gene were extremely low in the first- to fourth-instar larval stages. We speculate that venom allergen 5 is important for cell growth and development. This gene may be a multifunctional protein, performing different functions at different stages, as do the insect UDP-glycosyltransferases that play important roles in detoxication of xenobiotics, cuticle formation, pigmentation, and olfaction (Huang et al. 2008, Bozzolan et al. 2014). The specific biological function of venom allergen 5 in Cx. pipiens pallens warrants further study.
The putative protein of venom allergen 5 contains an SCP-like extracellular protein domain. It has been proposed that the SCP domain may function as an endopeptidase. Endopeptidases are capable of hydrolyzing peptide bonds in the intermediate portion of proteins and play important roles in many biological processes. The differential transcription of genes coding for peptidase activity has been reported in insecticide-resistant Drosophila strains (Pedra et al. 2004). Indeed, peptidases are thought to drive insecticide resistance via their roles in the induction of protein biosynthesis, the modification of enzyme conformation (Ahmed et al. 1998), or in meeting energy demands during stress in protein degradation (Pedra et al. 2004). In addition, the SCP domain has also been proposed to be a Ca2+ chelating serine protease. Serine proteases, such as trypsin and chymotrypsin, have been recognized as pyrethroid metabolism-associated genes (Yang et al. 2008, Xiong et al. 2014). Different trypsin-like serine proteinases may favor energy accumulation and amino acid provision, possibly mitigating the fitness costs associated with insecticide resistance in some strains of maize weevil (Silva et al. 2010).
Venom allergen 5 is a SCP superfamily member. Helothermine, another SCP superfamily member found in the venom of the Mexican beaded lizard (Heloderma horridum horridum), blocks both voltage-gated Ca2+ and K+ channels (Nobile et al. 1994, 1996). It is known that pyrethroids cause paralysis in insects by preventing the closure of the voltage-gated sodium channels (Hemingway et al. 2004, Dong et al. 2014). However, the role of venom allergen 5 in modulating voltage-gated sodium channels remains unknown, and a subject worthy of further study.
The results of this study provide the first evidence indicating that the venom allergen 5 gene with a SCP domain is associated with pyrethroid resistance in mosquitoes. This gene is expected to become a target gene in field detection of resistance.
Acknowledgments
This work was supported by the National Institutes of Health of the United States (NIH) (grant 2R01AI075746), the National Natural Science Foundation of China (grants 81171900, 81101279, and 81301458), the National S & T Major Program (grants 2012ZX10004-219 and 2012ZX10004-220), the Specialized Research Fund for the Doctoral Program of Higher Education of China (grant 20113234120007), and the Natural Science Foundation of Jiangsu Province (grant 81101279).
References Cited
- Ahmed S., Wilkins R. M., Mantle D. 1998. Comparison of proteolytic enzyme activities in adults of insecticide resistant and susceptible strains of the housefly M. domestica L. Insect. Biochem. Mol. Biol. 28: 629–639. [DOI] [PubMed] [Google Scholar]
- Barrett A. D., Higgs S. 2007. Yellow fever: a disease that has yet to be conquered. Annu. Rev. Entomol. 52: 209–229. [DOI] [PubMed] [Google Scholar]
- Blandin S., Moita L. F., Kocher T., Wilm M., Kafatos F. C., Levashina E. A. 2002. Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Reports 3: 852–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonizzoni M., Afrane Y., Dunn W. A., Atieli F. K., Zhou G., Zhong D., Li J., Githeko A., Yan G. 2012. Comparative transcriptome analyses of deltamethrin-resistant and -susceptible Anopheles gambiae mosquitoes from Kenya by RNA-Seq. PLoS ONE 7: e44607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzolan F., Siaussat D., Maria A., Durand N., Pottier M. A., Chertemps T., Maibeche-Coisne M. 2014. Antennal uridine diphosphate (UDP)-glycosyltransferases in a pest insect: Diversity and putative function in odorant and xenobiotics clearance. Insect. Mol. Biol. 23: 539–549. [DOI] [PubMed] [Google Scholar]
- Calvo E., Dao A., Pham V. M., Ribeiro J. M. 2007. An insight into the sialome of Anopheles funestus reveals an emerging pattern in anopheline salivary protein families. Insect. Biochem. Mol. Biol. 37: 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales M., Naranjo V., Almazan C., Molina R., Tsuruta S. A., Szabo M. P., Manzano-Roman R., Perez de la Lastra J. M., Kocan K. M., et al. 2009. Conservation and immunogenicity of the mosquito ortholog of the tick-protective antigen, subolesin. Parasitol. Res. 105: 97–111. [DOI] [PubMed] [Google Scholar]
- Casimiro S., Coleman M., Hemingway J., Sharp B. 2006. Insecticide resistance in Anopheles arabiensis and Anopheles gambiae from Mozambique. J. Med. Entomol. 43: 276–282. [DOI] [PubMed] [Google Scholar]
- Chalmers I. W., McArdle A. J., Coulson R. M., Wagner M. A., Schmid R., Hirai H., Hoffmann K. F. 2008. Developmentally regulated expression, alternative splicing and distinct sub-groupings in members of the Schistosoma mansoni venom allergen-like (SmVAL) gene family. BMC Genomics 9: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlab R., Valenzuela J. G., Rowton E. D., Ribeiro J. M. 1999. Toward an understanding of the biochemical and pharmacological complexity of the saliva of a hematophagous sand fly Lutzomyia longipalpis. Proc. Natl. Acad. Sci. USA. 96: 15155–15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhong D., Zhang D., Shi L., Zhou G., Gong M., Zhou H., Sun Y., Ma L., He J., et al. 2010. Molecular ecology of pyrethroid knockdown resistance in Culex pipiens pallens mosquitoes. PLoS ONE 5: e11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Ros V., Busso D., Cohen D. J., Maldera J., Goldweic N., Cuasnicu P. S. 2007. Molecular mechanisms involved in gamete interaction: Evidence for the participation of cysteine-rich secretory proteins (CRISP) in sperm-egg fusion. Soc. Reprod. Fertil 65: 353–356. [PubMed] [Google Scholar]
- David J. P., Strode C., Vontas J., Nikou D., Vaughan A., Pignatelli P. M., Louis C., Hemingway J., Ranson H. 2005. The Anopheles gambiae detoxification chip: a highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proc. Natl. Acad. Sci. USA. 102: 4080–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong K., Du Y., Rinkevich F., Nomura Y., Xu P., Wang L., Silver K., Zhorov B. S. 2014. Molecular biology of insect sodium channels and pyrethroid resistance. Insect. Biochem. Mol. Biol. 50C: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos-Pinto J. R., Dos Santos L. D., Andrade Arcuri H., Castro F. M., Kalil J. E., Palma M. S. 2014. Using proteomic strategies for sequencing and post-translational modifications assignment of antigen-5, a major allergen from the venom of the social wasp Polybia paulista. J. Proteome. Res. 13: 855–865. [DOI] [PubMed] [Google Scholar]
- Edi C. V., Djogbenou L., Jenkins A. M., Regna K., Muskavitch M. A., Poupardin R., Jones C. M., Essandoh J., Ketoh G. K., Paine M. J., et al. 2014. CYP6 P450 enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet. 10: e1004236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias L. P., Rodrigues D., Cunna V., Rofatto H. K., Faquim-Mauro E. L., Leite L. C. 2012. Schistosoma mansoni venom allergen like proteins present differential allergic responses in a murine model of airway inflammation. PLoS Negl. Trop. Dis. 6: e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ffrench-Constant R. H., Daborn P. J., Le Goff G. 2004. The genetics and genomics of insecticide resistance. Trends Genet 20: 163–170. [DOI] [PubMed] [Google Scholar]
- Gambhir M., Michael E. 2008. Complex ecological dynamics and eradicability of the vector borne macroparasitic disease, lymphatic filariasis. PLoS ONE 3: e2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong M. Q., Gu Y., Hu X. B., Sun Y., Ma L., Li X. L., Sun L. X., Sun J., Qian J., Zhu C. L. 2005. Cloning and overexpression of CYP6F1, a cytochrome P450 gene, from deltamethrin-resistant Culex pipiens pallens. Acta Biochim. Biophys. Sin. (Shanghai) 37: 317–326. [DOI] [PubMed] [Google Scholar]
- Hemingway J., Field L., Vontas J. 2002. An overview of insecticide resistance. Science 298: 96–97. [DOI] [PubMed] [Google Scholar]
- Hemingway J., Hawkes N. J., McCarroll L., Ranson H. 2004. The molecular basis of insecticide resistance in mosquitoes. Insect. Biochem. Mol. Biol. 34: 653–665. [DOI] [PubMed] [Google Scholar]
- Hemingway J., Beaty B. J., Rowland M., Scott T. W., Sharp B. L. 2006. The innovative vector control consortium: improved control of mosquito-borne diseases. Trends Parasitol. 22: 308–312. [DOI] [PubMed] [Google Scholar]
- Henriksen A., King T. P., Mirza O., Monsalve R. I., Meno K., Ipsen H., Larsen J. N., Gajhede M., Spangfort M. D. 2001. Major venom allergen of yellow jackets, Ves v 5: structural characterization of a pathogenesis-related protein superfamily. Proteins 45: 438–448. [DOI] [PubMed] [Google Scholar]
- Hu X., Sun Y., Wang W., Yang M., Sun L., Tan W., Sun J., Qian J., Ma L., Zhang D. 2007. Cloning and characterization of NYD-OP7, a novel deltamethrin resistance associated gene from< i> Culex pipiens pallens. Pestic. Biochem. Physiol. 88: 82–91. [Google Scholar]
- Huang F. F., Chai C. L., Zhang Z., Liu Z. H., Dai F. Y., Lu C., Xiang Z. H. 2008. The UDP-glucosyltransferase multigene family in Bombyx mori. BMC Genomics 9: 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery J. A., Thi Yen N., Nam V. S., Nghia le T., Hoffmann A. A., Kay B. H., Ryan P. A. 2009. Characterizing the Aedes aegypti population in a Vietnamese village in preparation for a Wolbachia-based mosquito control strategy to eliminate dengue. PLoS Negl. Trop. Dis. 3: e552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinfu W. 1999. Resistance to deltamethrin in Culex pipiens pallens (Diptera: Culicidae) from Zhejiang, China. J. Med. Entomol. 36: 389–393. [DOI] [PubMed] [Google Scholar]
- King T. P., Spangfort M. D. 2000. Structure and biology of stinging insect venom allergens. Int. Arch. Allergy Immunol. 123: 99–106. [DOI] [PubMed] [Google Scholar]
- Kitajima S., Sato F. 1999. Plant pathogenesis-related proteins: molecular mechanisms of gene expression and protein function. J. Biochem. 125: 1–8. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292. [DOI] [PubMed] [Google Scholar]
- Leal W. S., Choo Y. M., Xu P., da Silva C. S., Ueira-Vieira C. 2013. Differential expression of olfactory genes in the southern house mosquito and insights into unique odorant receptor gene isoforms. Proc. Natl. Acad. Sci. USA. 110: 18704–18709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Liu H., Zhu F., Zhang L. 2007. Differential expression of genes in pyrethroid resistant and susceptible mosquitoes, Culex quinquefasciatus (S.). Gene 394: 61–68. [DOI] [PubMed] [Google Scholar]
- Liu N., Li T., Reid W. R., Yang T., Zhang L. 2011. Multiple Cytochrome P450 genes: Their constitutive overexpression and permethrin induction in insecticide resistant mosquitoes, Culex quinquefasciatus. PLoS ONE 6: e23403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lumjuan N., Rajatileka S., Changsom D., Wicheer J., Leelapat P., Prapanthadara L. A., Somboon P., Lycett G., Ranson H. 2011. The role of the Aedes aegypti Epsilon glutathione transferases in conferring resistance to DDT and pyrethroid insecticides. Insect. Biochem. Mol. Biol. 41: 203–209. [DOI] [PubMed] [Google Scholar]
- Mahande A. M., Dusfour I., Matias J. R., Kweka E. J. 2012. Knockdown resistance, rdl alleles, and the annual entomological inoculation rate of wild mosquito populations from lower Moshi, Northern Tanzania. J. Glob. Infect. Dis. 4: 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans B. J., Andersen J. F., Francischetti I. M., Valenzuela J. G., Schwan T. G., Pham V. M., Garfield M. K., Hammer C. H., Ribeiro J. M. 2008. Comparative sialomics between hard and soft ticks: Implications for the evolution of blood-feeding behavior. Insect. Biochem. Mol. Biol. 38: 42–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne T. J., Abbenante G., Tyndall J. D., Halliday J., Lewis R. J. 2003. Isolation and characterization of a cone snail protease with homology to CRISP proteins of the pathogenesis-related protein superfamily. J. Biol. Chem. 278: 31105–31110. [DOI] [PubMed] [Google Scholar]
- Mitri C., Vernick K. D. 2012. Anopheles gambiae pathogen susceptibility: The intersection of genetics, immunity and ecology. Curr. Opin. Microbiol. 15: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U. R., Johansen N., Petersen A. B., Fromberg-Nielsen J., Haeberli G. 2009. Hymenoptera venom allergy: Analysis of double positivity to honey bee and Vespula venom by estimation of IgE antibodies to species-specific major allergens Api m1 and Ves v5. Allergy 64: 543–548. [DOI] [PubMed] [Google Scholar]
- Nobile M., Noceti F., Prestipino G., Possani L. D. 1996. Helothermine, a lizard venom toxin, inhibits calcium current in cerebellar granules. Exp. Brain Res. 110: 15–20. [DOI] [PubMed] [Google Scholar]
- Nobile M., Magnelli V., Lagostena L., Mochca-Morales J., Possani L. D., Prestipino G. 1994. The toxin helothermine affects potassium currents in newborn rat cerebellar granule cells. J. Membr. Biol. 139: 49–55. [DOI] [PubMed] [Google Scholar]
- Pedra J. H., McIntyre L. M., Scharf M. E., Pittendrigh B. R. 2004. Genome-wide transcription profile of field- and laboratory-selected dichlorodiphenyltrichloroethane (DDT)-resistant Drosophila. Proc. Natl. Acad. Sci. USA. 101: 7034–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid W. R., Zhang L., Liu F., Liu N. 2012. The transcriptome profile of the mosquito Culex quinquefasciatus following permethrin selection. PLoS ONE 7: e47163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid W. R., Thornton A., Pridgeon J. W., Becnel J. J., Tang F., Estep A., Clark G. G., Allan S., Liu N. 2014. Transcriptional analysis of four family 4 P450s in a Puerto Rico strain of Aedes aegypti (Diptera: Culicidae) compared with an Orlando strain and their possible functional roles in permethrin resistance. J. Med. Entomol. 51: 605–615. [DOI] [PubMed] [Google Scholar]
- Ribeiro J. M., Francischetti I. M. 2003. Role of arthropod saliva in blood feeding: Sialome and post-sialome perspectives. Annu. Rev. Entomol. 48: 73–88. [DOI] [PubMed] [Google Scholar]
- Silva L. B., Reis A. P., Pereira E. J., Oliveira M. G., Guedes R. N. 2010. Partial purification and characterization of trypsin-like proteinases from insecticide-resistant and -susceptible strains of the maize weevil, Sitophilus zeamais. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 155: 12–19. [DOI] [PubMed] [Google Scholar]
- Smith D. R., Adams A. P., Kenney J. L., Wang E., Weaver S. C. 2008. Venezuelan equine encephalitis virus in the mosquito vector Aedes taeniorhynchus: Infection initiated by a small number of susceptible epithelial cells and a population bottleneck. Virology 372: 176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachecka A., Borsuk G., Olszewski K., Paleolog J., Lipinski Z. 2013. Proteolysis on the body surface of pyrethroid-sensitive and resistant Varroa destructor. Acta Parasitol. 58: 64–69. [DOI] [PubMed] [Google Scholar]
- Styer L. M., Lim P. Y., Louie K. L., Albright R. G., Kramer L. D., Bernard K. A. 2011. Mosquito saliva causes enhancement of West Nile virus infection in mice. J. Virol. 85: 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zou P., Yu X. Y., Chen C., Yu J., Shi L. N., Hong S. C., Zhou D., Chang X. L., Wang W. J., et al. 2012. Functional characterization of an arrestin gene on insecticide resistance of Culex pipiens pallens. Parasit. Vectors 5: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Ye Y., Sun H., Yu J., Zhang L., Sun Y., Zhang D., Ma L., Shen B., Zhu C. 2013. Identification of proteasome subunit beta type 6 (PSMB6) associated with deltamethrin resistance in mosquitoes by proteomic and bioassay analyses. PLoS ONE 8: e65859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg H., Zaim M., Yadav R. S., Soares A., Ameneshewa B., Mnzava A., Hii J., Dash A. P., Ejov M. 2012. Global trends in the use of insecticides to control vector-borne diseases. Environ. Health Perspect. 120: 577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vontas J., Blass C., Koutsos A. C., David J. P., Kafatos F. C., Louis C., Hemingway J., Christophides G. K., Ranson H. 2005. Gene expression in insecticide resistant and susceptible Anopheles gambiae strains constitutively or after insecticide exposure. Insect Mol. Biol. 14: 509–521. [DOI] [PubMed] [Google Scholar]
- Wang Z. M., Li C. X., Xing D., Yu Y. H., Liu N., Xue R. D., Dong Y. D., Zhao T. Y. 2012. Detection and widespread distribution of sodium channel alleles characteristic of insecticide resistance in Culex pipiens complex mosquitoes in China. Med. Vet. Entomol. 26: 228–232. [DOI] [PubMed] [Google Scholar]
- (WHO) World Health Organization. 1998. Test procedures for insecticide resistance monitoring in malaria vectors, bio-efficacy and persistence of insecticides on treated surfaces. WHO, Geneva, Switzerland. [Google Scholar]
- (WHO) World Health Organization. 2012. Global plan for insecticide resistance management in malaria vectors. World Health Organization, Geneva, Switzerland. [Google Scholar]
- (WHO) World Health Organization. 2013. Test procedures for insecticide resistance monitoring in malaria vector mosquitoes. World Health Organization, Geneva, Switzerland. [Google Scholar]
- Wood O., Hanrahan S., Coetzee M., Koekemoer L., Brooke B. 2010. Cuticle thickening associated with pyrethroid resistance in the major malaria vector Anopheles funestus. Parasit. Vectors 3: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Yang Y., Yuan G., Campbell P. M., Teese M. G., Russell R. J., Oakeshott J. G., Wu Y. 2011. Overexpressed esterases in a fenvalerate resistant strain of the cotton bollworm, Helicoverpa armigera. Insect. Biochem. Mol. Biol. 41: 14–21. [DOI] [PubMed] [Google Scholar]
- Xiong C., Fang F., Chen L., Yang Q., He J., Zhou D., Shen B., Ma L., Sun Y., Zhang D., et al. 2014. Trypsin-catalyzed deltamethrin degradation. PLoS ONE 9: e89517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Liu N. 2011. Genome analysis of cytochrome P450s and their expression profiles in insecticide resistant mosquitoes, Culex quinquefasciatus. PLoS ONE 6: e29418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Zhou D., Sun L., Zhang D., Qian J., Xiong C., Sun Y., Ma L., Zhu C. 2008. Expression and characterization of two pesticide resistance-associated serine protease genes (NYD-tr and NYD-ch) from Culex pipiens pallens for metabolism of deltamethrin. Parasitol. Res. 103: 507–516. [DOI] [PubMed] [Google Scholar]
- Zhu G., Zhong D., Cao J., Zhou H., Li J., Liu Y., Bai L., Xu S., Wang M. H., Zhou G., et al. 2014. Transcriptome profiling of pyrethroid resistant and susceptible mosquitoes in the malaria vector, Anopheles sinensis. BMC Genomics 15: 448. [DOI] [PMC free article] [PubMed] [Google Scholar]