Abstract
Recent years have witnessed a flurry of important technological and methodological developments in the discovery and analysis of copy number variations (CNVs), which are increasingly enabling the systematic evaluation of their impact on a broad range of phenotypes from molecular-level (intermediate) traits to higher-order clinical phenotypes. Like single nucleotide variants in the human genome, CNVs have been linked to complex traits in humans, including disease and drug response. These recent developments underscore the importance of incorporating complex forms of genetic variation into disease mapping studies and promise to transform our understanding of genome function and the genetic basis of disease. Here we review some of the findings that have emerged from transcriptome studies of CNVs facilitated by the rapid advances in -omics technologies and corresponding methodologies.
Keywords: copy number variation, transcriptome, gene expression, eQTLs, GWAS
Introduction
Although genome-wide association studies (GWAS) have been unsurpassed in identifying disease susceptibility and quantitative trait loci [1], these studies have primarily focused on single-nucleotide polymorphisms (SNPs). The SNP findings have been, on the whole, impressive, despite the fact that much more clearly remains to be done to identify additional sources of the missing heritability [2] and to assign a precise genetic variant and a causal mechanism to the growing number of discovered loci. Even with increased characterization of more complex forms of genetic variation [3], most prominently copy number variations (CNVs; usually defined as genomic segments of size ≥1 kb showing copy number variability among individuals with respect to a reference genome), it is clear that little is known about the overall contribution of structural variation to complex phenotypes. Certainly, CNVs are increasingly the focus of considerable research in medical genetics [4, 5], and their investigation has been particularly crucial in efforts to characterize the genetic underpinnings of psychiatric and neurodevelopmental phenotypes [6, 7]. Although generating reliable CNV data continue to be a primary challenge, it should be noted that every SNP-based GWAS conducted to date has concomitantly generated data that can enable detection of CNV [8, 9]. Furthermore, there have been key analytic advances [10] in genotype calling as well as validation of these more complex types of genetic variation, paving the way for a more systematic integration of CNVs into studies of genome function and disease mapping.
CNV and the transcriptome: disease susceptibility and genome function
Gene expression traits serve as a surrogate for the complexity and range of human phenotypic variation, and thus, a comprehensive survey of the impact of CNVs on variation in gene regulation can have profound consequences for our understanding of the genetic basis of complex traits. The connection between CNVs and disease susceptibility has of course long been the subject of active research, across a range of disease architecture from Mendelian disorders (such as Williams-Beuren syndrome [11], Potocki-Lupski syndrome [12, 13] or Charcot-Marie Tooth neuropathy Type 1A [14]) to common complex diseases such as cardiovascular disease and diabetes, whose genetic etiology is relatively less well-understood [15, 16]. Studies of neuropsychiatric phenotypes (e.g. schizophrenia [17, 18] and autism spectrum disorder [7, 19, 20]) have highlighted the important contribution of de novo CNV to disease pathogenesis. Nevertheless, a map of the genetic basis of gene expression variation in a comprehensive collection of tissues [21, 22] with a particular focus on CNVs can greatly expand our understanding of the context specificity of their effect on disease. Furthermore, as described in Maynard Olson’s ‘less-is-more’ hypothesis, deletions may be a driving force in genome evolution, with loss of gene function a common evolutionary response to a change in the environment and pattern of selective pressures [23, 24], thus coupling molecular evolution and function. More broadly, the enrichment of (human) genes, within CNVs, that impact inflammatory response, immunity, protein secretion, and olfaction may indeed indicate the adaptive benefit of gene dosage [25].
Comprehensive catalogs of CNVs [9, 26–28] among putatively phenotypically normal individuals have reinforced the finding of extensive genetic heterogeneity [29] and a highly dynamic structure in the genome. The International HapMap Consortium has facilitated large-scale CNV surveys of the human genome [27, 28] in world populations with ancestry from Europe, Asia and Africa, and these studies have shown that copy number variable regions cover substantially more nucleotide content than the more widely studied SNPs, highlighting the importance of incorporating CNVs into studies of human disease and genome function. The subsequent survey of genomic structural variants by the 1000 Genomes Project, which sought to discover and validate structural variants (of ≥50 bp in size), mapped approximately 15 000 structural variants at nucleotide resolution [30] using whole genome sequencing data. Nearly 20% of the genotyped deletions were not tagged by HapMap SNPs [30], suggesting the importance of directly interrogating some CNVs for use in association studies. The catalog of CNVs and genome-wide gene expression data in the HapMap populations has afforded opportunities for annotating the identified CNVs with information on expression quantitative trait loci (eQTLs) and for quantifying their contribution to gene expression variation relative to SNPs [31]. Stranger et al. [31] observed that CNVs (>100 kb in length) and SNPs captured approximately 18% and 84% respectively of the total variation in gene expression in lymphoblastoid cell lines (LCLs). Although this may underestimate the impact of CNVs on the transcriptome (because of the relatively greater completeness of SNP catalogs, the greater challenge of genotyping CNVs and the much larger number of CNVs of <100 kb in length that were excluded from the analysis), the study underscores the need to evaluate both types of variation to characterize the genetic basis of complex phenotypes. The authors also reported that of the >14 000 genes tested in LCLs derived from Utah residents with ancestry from northern and western Europe (CEU), Han Chinese in Beijing, China (CHB), Japanese in Tokyo, Japan (JPT), and Yoruba in Ibadan Nigeria (YRI), 85, 44, 58 and 96 genes, respectively, showed significant associations in expression with at least one of the nearly 25 000 autosomal comparative genomic hybridization (CGH) clones. Among these target genes, 12% replicated at the same significance level in at least one other population with 2% significant in all four populations. Of note, the CNV associations with gene expression reported in the study appeared to be more highly population-specific than the identified SNP associations, among which, for instance, a much larger proportion (8%) were significant in all populations. Early high-resolution population surveys of deletion sites indicated that such polymorphisms, consistent with similar findings on SNPs, show greater diversity among individuals of African descent, but also may be under more extreme selection than SNPs [3]. Certain CNVs show a highly unusual degree of population differentiation [32], and selection on a copy-number variable gene (such as diet-related selective pressures on the salivary amylase gene [AMY1] [33]) may offer insights into recent human evolutionary history. Additional in-depth studies on the contribution of CNVs to population differences in gene expression variation are therefore warranted.
Among the CNVs that show replicated associations with expression across all the HapMap populations is a deletion influencing UGT2B17—a gene involved in the metabolism of sex steroid hormones. The same deletion of the gene had been previously identified by an earlier study of common deletion polymorphisms in the human genome in a subset of the CEU [9] as one of the genes involved in olfaction, drug response, and steroid metabolism with coding exons that were found to be commonly deleted. Furthermore, the UGT2B17 CNV has been found to be associated with osteoporosis [34] and was also identified as a causal variant for graft-versus-host disease (GVHD) after hematopoietic stem cell transplantation [35], suggesting potential pleiotropy. The latter association with GVHD highlights the importance of assessing the effect of the deletion on the expression of the gene in multiple (GVHD-affected) tissues, including liver, intestine and skin. In a recent in-depth study of cis- and trans- acting factors influencing mRNA expression and catalytic activity of hepatic UDP-glucuronosyltransferases [36], the UGT2B17 CNV was found to significantly account for variability in UGT2B17 transcription and testosterone glucuronidation rate in human liver. Taken together, these results suggest that CNV may exert broad effects on complex traits and underscore the need for comprehensive assessment of the functionality of CNVs as regulatory variation (eQTLs) in primary tissues to elucidate disease mechanisms.
Analytic challenges in interpretation of CNV associations
The interpretation of CNV associations with phenotype is fraught with analytic challenges, not the least of which is that CNVs may be in linkage disequilibrium (LD), or share a common genealogical history, with SNPs [37]. The observation that common CNVs may be well-tagged by SNPs (r2 ≥ 0.80) as tCNVs [38] implies that their effect can be indirectly evaluated in SNP-based studies. However, although LD may enhance the effectiveness of GWAS, it also may severely limit their resolution, affecting the ability to fine-map causal loci. Indeed, in a large, direct genome-wide association study of eight common human diseases [39], most common CNVs were found to be tCNVs and have therefore been previously interrogated in SNP GWAS studies. Furthermore, the study identified artifacts that can generate false-positive associations in CNV studies, such as, for our purposes here, those of gene expression. In particular, the study found systematic CNV differences between cell lines and blood. The well-known example of a common deletion polymorphism 20 kb upstream of IRGM—a gene that has been shown to play a key role in autophagy and in the control of intracellular bacillary load [40]—that has been found to confer risk to Crohn’s disease [41] illustrates the difficulties in identifying the causal variant at a disease-associated locus. The CNV is in perfect LD (r2 = 1) with a variant (rs13361189) [42] in the region that was the most strongly associated SNP with Crohn’s disease; therefore, the causal association and any association induced by LD are difficult to distinguish statistically. Sequencing of the coding region of the gene demonstrated that the causal variants do not alter the amino acid sequence of the gene, hinting at a regulatory effect. The combination of Inflammatory Bowel Disease Genetics Consortium, HapMap and extended HapMap data did not allow resolution of the functional variant, as the SNP and the CNV were perfectly correlated in samples of various ancestries [41]. The deletion and protective haplotypes showed divergent expression patterns in a variety of heterozygous cell lines. HeLa cells, the hepatocellular carcinoma cell line SNU182 and LCLs showed higher expression levels of IRGM for the protective haplotype, whereas the colon carcinoma cell line HCT116 and primary smooth muscle cells from human bronchus showed higher expression from the deletion haplotype. Manipulation of IRGM expression in HeLa cells significantly regulated anti-bacterial autophagy, which suggests a link to the disease. Collectively, these results from McCarroll et al. [41] highlight the challenges in fine mapping causal variants in copy variable regions of the genome, but also the indispensability of transcriptome studies, in multiple tissues, for identifying the genetic mechanism(s) of disease.
CNVs as expression QTLs
The mapping of CNVs as eQTLs [38] can greatly facilitate the search for causal links between genetic variation and disease susceptibility. Gene expression is a key intermediate phenotype and thus genetic variants (e.g. SNPs or CNVs) associated with gene expression as eQTLs may underlie the genetic basis of higher-order traits such as disease risk. Although the Wellcome Trust Case Control Consortium (WTCCC) CNV study concluded that common CNVs appear unlikely to play a major role in the genetic basis of several complex diseases investigated [39] and that, furthermore, there is no enrichment of associations among CNVs involving exonic deletions, CNVs may still be causal for some of the observed SNP associations with complex disease. Indeed, as has been noted, tag SNPs for CNVs can facilitate SNP-level analyses and simulation studies (and also enable replication of CNV associations). Notably, tag SNPs (r2 ≥ 0.80) for tCNVs have been found to be significantly enriched for cis-eQTLs; furthermore, reproducible trait associations (in the National Human Genome Research Institute [NHGRI] catalog) show a significant overrepresentation for tCNVs [38]. It should be noted that the WTCCC CNV study has several important methodological shortcomings, which may apply more broadly to other CNV association studies. For example, a large proportion of the CNVs examined in the study could not be reliably genotyped. Moreover, although the MHC Haplotype Project [43] has identified numerous CNVs on human leukocyte antigen (HLA) haplotypes, LD in this complex region makes fine mapping of the causal variants for the known disease associations (with autoimmune disorders [39]) or the associations with gene expression extremely challenging. Furthermore, the WTCCC study focused primarily on common CNV, and the role of rare CNVs could not be systematically investigated. These limitations highlight the technical challenges in quantifying the contribution of CNVs to the genetic basis of common diseases as well as their regulatory impact on the transcriptome.
Identifying CNVs associated with gene expression is analogous to SNP-based eQTL mapping approaches, which seek to relate variation in gene expression levels to genotype. CNV genotype data are tested for association with gene expression traits, and, as in the case of SNPs, a multiple testing problem arises from the large number of resulting tests. There are, however, differences with approaches to identify SNP eQTLs. For example, multi-allelic CNVs [44] can have highly complex genotypes (with three or more segregating alleles). As the range of copy numbers increases, molecular discrimination of high copy numbers becomes more difficult [45], thereby reducing the accuracy and power of eQTL mapping. Because multi-allelic CNVs are the largest source of gene dosage variation in humans [44], this issue is particularly salient for mapping of CNV eQTLs. Furthermore, inferring accurate integer copy number states is analytically challenging; Sudmant et al. [46], using whole-genome sequence data to overcome the challenges of hybridization-based methods for copy number assay, therefore treated copy number genotype as a continuous variable. Indeed, mapping of CNV eQTLs may well assume a continuous genotype variable, in contrast to SNP eQTL mapping, which, even in the use of imputed dosages, assumes a very discrete genotype variable.
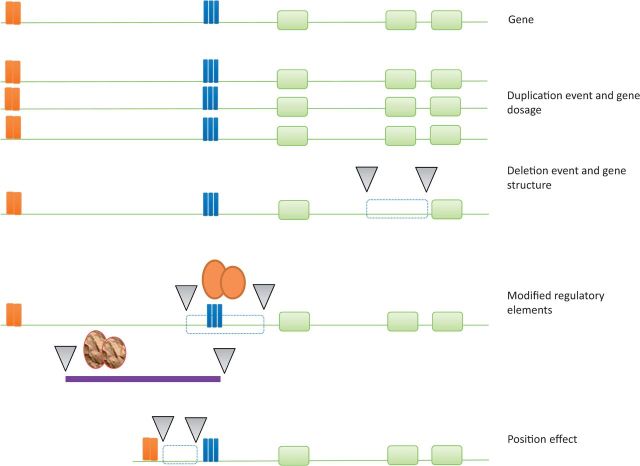
Structural variation can control phenotype, including gene expression, in several ways [47] (Figure 1). The expression of dosage-sensitive genes can be modified by a gene duplication or deletion event. CNVs that only partially overlap a dosage-sensitive gene can induce reduced expression, or by disrupting structure, lead to novel transcripts. CNVs can influence the transcriptome not merely by regulating the expression of strictly co-localizing genes, but through a more distal regulatory mechanism that can reach several hundred kilobases from the breakpoints. Indeed, CNVs have been found to regulate in cis normal-copy flanking genes (as in the case of a deletion that causes Williams-Beuren syndrome [48]), but CNVs can also mediate their phenotypic effect as trans regulators of gene expression [38]. Because deletion or duplication of regulatory SNPs can lead to altered transcription, the fact that a large proportion of CNVs harbor regulatory SNPs [38] implies that certain CNVs may regulate the expression of target genes at a distance [49] from the CNV through position effects (e.g. through the insertion or deletion of regulatory elements) [47, 50]. Indeed, Stranger et al. [31] reported that more than half of expression probes (in LCLs) that show association with a CGH clone do map outside the interval defined by the CNV, suggesting that CNVs may act as potentially distal regulators of gene expression by altering regulatory and other functional elements, as opposed to altering dosage, of the target gene. The presence, rather than the change in copy number, of the CNV interval can also have surprising effects on gene expression. Jacquemont et al. [51] investigated the phenotypic impact of the 16p11.2 CNV interval; notably, genes centromeric to the rearrangement interval displayed no significant difference between cases and controls, in marked contrast to genes telomeric to the interval, which showed significant variation. The latter genes were, however, similarly upregulated in both deletion and duplication carriers, suggesting the presence of the interval (rather than the change in copy number) caused the effects on transcript levels.
Figure 1.
CNVs may influence phenotype though several mechanisms. A gene duplication or deletion event can alter the expression of dosage-sensitive genes. CNVs that only partially overlap a dosage-sensitive gene can induce reduced expression, or by disrupting structure, lead to novel transcripts. CNVs can regulate normal flanking genes, often through a distal mechanism that can extend several megabases from the breakpoints, by modifying regulatory elements. CNVs can also regulate the expression of target genes through position effects (i.e. through the insertion or deletion of sequences leading to alterations in distance to regulatory elements). Green boxes represent exons. Blue boxes represent promoters while orange boxes represent distal enhancers. Triangles mark the breakpoints of CNVs. Circles represent transcription factors that may bind to regulatory elements. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
Mechanistically, structural variation can induce alterations in chromatin architecture that is consistent with long-range effects on global expression [52]. CNVs combined with epigenetic mechanisms can influence transcription beyond the effect obtained from the chromosomal gain or loss; for example, CNVs on imprinted loci may result in allele-specific differences in expression of target genes according to parental origin. This combination of copy number and additional epigenetic changes can then lead to downstream phenotypic outcomes (such as the observed clinical heterogeneity of 15q11-13 duplication syndromes [53]).
Furthermore, incorporating CNVs into disease association studies or eQTL mapping may unmask otherwise undetected SNP effects on phenotype. Besides regulating the transcription of dosage-sensitive genes, CNVs may influence the transcriptome via effects on dosage-insensitive genes by unmasking a functional SNP. Indeed, accounting for copy number status can be an approach to mapping variants associated with a variety of traits, most prominently regulatory variation [54]. In a recent study, annotation of SNPs located in CNV regions with information on chromatin state (e.g. enhancers, promoters, Polycomb-repressed regions, heterochromatic and repetitive regions), DNaseI hypersensitivity sites and transcription factor binding site (TFBS) regions in LCLs showed that the unmasked regulatory variants are highly enriched for enhancer elements (but not promoter elements) and accessible chromatin zones; furthermore, SNPs located in CNVs show significant differential allelic effect on TFBS [54]. These findings highlight the importance of an integrative approach to the functional analysis of CNVs and SNPs. Moreover, loss-of-function variants identified through the 1000 Genomes Project data [55] located in CNV and copy number stable regions appear to show differential effects on nonsense-mediated decay and on all known transcripts of a gene [54], suggesting potential rare-variant mechanisms through which CNVs may mediate global effects on the transcriptome.
Future directions
Transcriptome studies will continue to uncover the functional consequences of CNVs. To elucidate the molecular mechanisms underlying disease risk, it will be important to assess how CNVs influence gene expression in a comprehensive collection of tissues [22] such as now being facilitated by the GTEx Project [21]. Studies of epigenetic mechanisms such as encoded in the complex chromatin architecture and the three-dimensional nuclear organization, using multiple reference tissues, promise to yield important insights into the global effects of CNVs on regulatory function. The question of how CNVs modulate gene expression during development will require deep knowledge of tissue transcriptome dynamics and longitudinal analyses of their impact. Studies of potential interactions of CNVs with non-coding transcripts and pseudogenes [56] may help to dissect the multilayer regulatory circuitry of the human transcriptome. Furthermore, evaluation of the causal role of CNVs associated with gene expression and in strong LD with SNPs may be accelerated by the application of the CRISPR-Cas systems [57] for genome editing. Finally, more complete and accurate maps of structural variation in the genome will be needed to obtain accurate estimates of the relative (tissue-specific) contributions on gene expression of a widening spectrum of genetic variation.
Key points.
Genome-wide association studies have characterized the effect of common single-nucleotide polymorphisms on complex human traits, but have done a far less comprehensive interrogation of the effects of copy number variation.
Copy number polymorphisms have been associated in some cases with complex traits in humans. However, the mechanisms underlying those associations have not been fully elucidated; thus, a comprehensive interrogation of the effects on molecular phenotypes, including gene expression levels, is warranted.
Copy number polymorphisms can affect gene expression through complex mechanisms that extend beyond simple gene dosage effects, and include, for example, insertion and deletion of gene regulatory regions and alterations of physical proximity of genes and regulatory elements.
An understanding of the effects of copy number variation on gene expression, and how those effects relate to single-nucleotide polymorphism effects, is needed to understand the role of genetic variation in complex traits.
Funding
The authors would like to acknowledge support from the Genotype-Tissue Expression project (GTeX) grant (R01 MH090937).
Biographies
Eric R. Gamazon is a Bioinformatics Scientist in the Section of Genetic Medicine, Department of Medicine at the University of Chicago. His research focuses on the genetics of gene expression and complex traits.
Barbara E. Stranger is an Assistant Professor in the Section of Genetic Medicine, Department of Medicine and a Core Member of the Institute for Genomics and Systems Biology at the University of Chicago. She has a long-standing interest in population genetics and gene regulatory processes, and how these shape phenotypic variability.
References
- 1.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA 2009;106:9362–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature 2009;461:747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eichler EE. Widening the spectrum of human genetic variation. Nat Genet 2006;38:9–11. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 2005;307:1434–40. [DOI] [PubMed] [Google Scholar]
- 5.Aitman TJ, Dong R, Vyse TJ, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 2006;439:851–5. [DOI] [PubMed] [Google Scholar]
- 6. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 2008;455:237–41. [DOI] [PMC free article] [PubMed]
- 7.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 2008;82:477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad DF, Andrews TD, Carter NP, et al. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet 2006;38:75–81. [DOI] [PubMed] [Google Scholar]
- 9.McCarroll SA, Hadnott TN, Perry GH, et al. Common deletion polymorphisms in the human genome. Nat Genet 2006;38:86–92. [DOI] [PubMed] [Google Scholar]
- 10.McCarroll SA, Kuruvilla FG, Korn JM, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet 2008;40:1166–74. [DOI] [PubMed] [Google Scholar]
- 11.Bayes M, Magano LF, Rivera N, et al. Mutational mechanisms of Williams-Beuren syndrome deletions. Am J Hum Genet 2003;73:131–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potocki L, Chen KS, Park SS, et al. Molecular mechanism for duplication 17p11.2- the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet 2000;24:84–7. [DOI] [PubMed] [Google Scholar]
- 13.Ricard G, Molina J, Chrast J, et al. Phenotypic consequences of copy number variation: insights from Smith-Magenis and Potocki-Lupski syndrome mouse models. PLoS Biol 2010;8:e1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abe A, Nakamura K, Kato M, et al. Compound heterozygous PMP22 deletion mutations causing severe Charcot-Marie-Tooth disease type 1. J Hum Genet 2010;55:771–3. [DOI] [PubMed] [Google Scholar]
- 15.Blair DR, Lyttle CS, Mortensen JM, et al. A nondegenerate code of deleterious variants in Mendelian loci contributes to complex disease risk. Cell 2013;155:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell KJ. What is complex about complex disorders? Genome Biol 2012;13:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirov G, Pocklington AJ, Holmans P, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry 2012;17:142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy SE, Makarov V, Kirov G, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet 2009;41:1223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science 2007;316:445–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar RA, KaraMohamed S, Sudi J, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet 2008;17:628–38. [DOI] [PubMed] [Google Scholar]
- 21.The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henrichsen CN, Vinckenbosch N, Zollner S, et al. Segmental copy number variation shapes tissue transcriptomes. Nat Genet 2009;41:424–9. [DOI] [PubMed] [Google Scholar]
- 23.Olson MV. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet 1999;64:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko DC, Gamazon ER, Shukla KP, et al. Functional genetic screen of human diversity reveals that a methionine salvage enzyme regulates inflammatory cell death. Proc Natl Acad Sci USA 2012;109:E2343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen DQ, Webber C, Ponting CP. Bias of selection on human copy-number variants. PLoS Genet 2006;2:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science 2004;305:525–8. [DOI] [PubMed] [Google Scholar]
- 27.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature 2006;444:444–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conrad DF, Pinto D, Redon R, et al. Origins and functional impact of copy number variation in the human genome. Nature 2010;464:704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet 2004;36:949–51. [DOI] [PubMed] [Google Scholar]
- 30.Mills RE, Walter K, Stewart C, et al. Mapping copy number variation by population-scale genome sequencing. Nature 2011;470:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stranger BE, Forrest MS, Dunning M, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 2007;315:848–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kidd JM, Newman TL, Tuzun E, et al. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet 2007;3:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry GH, Dominy NJ, Claw KG, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet 2007;39:1256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang TL, Chen XD, Guo Y, et al. Genome-wide copy-number-variation study identified a susceptibility gene, UGT2B17, for osteoporosis. Am J Hum Genet 2008;83:663–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarroll SA, Bradner JE, Turpeinen H, et al. Donor-recipient mismatch for common gene deletion polymorphisms in graft-versus-host disease. Nat Genet 2009;41:1341–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu W, Ramirez J, Gamazon ER, et al. Genetic factors affecting gene transcription and catalytic activity of UDP-glucuronosyltransferases in human liver. Hum Mol Genet 2014;23:5558–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinds DA, Kloek AP, Jen M, et al. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat Genet 2006;38:82–5. [DOI] [PubMed] [Google Scholar]
- 38.Gamazon ER, Nicolae DL, Cox NJ. A study of CNVs as trait-associated polymorphisms and as expression quantitative trait loci. PLoS Genet 2011;7:e1001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Craddock N, Hurles ME, Cardin N, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 2010;464:713–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh SB, Davis AS, Taylor GA, et al. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 2006;313:1438–41. [DOI] [PubMed] [Google Scholar]
- 41.McCarroll SA, Huett A, Kuballa P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet 2008;40:1107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet 2007;39:830–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horton R, Gibson R, Coggill P, et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics 2008;60:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Handsaker RE, Van Doren V, Berman JR, et al. Large multiallelic copy number variations in humans. Nat Genet 2015;47:296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cantsilieris S, Western PS, Baird PN, et al. Technical considerations for genotyping multi-allelic copy number variation (CNV), in regions of segmental duplication. BMC Genomics 2014;15:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sudmant PH, Kitzman JO, Antonacci F, et al. Diversity of human copy number variation and multicopy genes. Science 2010;330:641–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet 2006;7:85–97. [DOI] [PubMed] [Google Scholar]
- 48.Merla G, Howald C, Henrichsen CN, et al. Submicroscopic deletion in patients with Williams-Beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am J Hum Genet 2006;79:332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 2005;76:8–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kleinjan DJ, van Heyningen V. Position effect in human genetic disease. Hum Mol Genet 1998;7:1611–18. [DOI] [PubMed] [Google Scholar]
- 51.Jacquemont S, Reymond A, Zufferey F, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature 2011;478:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gheldof N, Witwicki RM, Migliavacca E, et al. Structural variation-associated expression changes are paralleled by chromatin architecture modifications. PLoS One 2013;8:e79973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hogart A, Leung KN, Wang NJ, et al. Chromosome 15q11-13 duplication syndrome brain reveals epigenetic alterations in gene expression not predicted from copy number. J Med Genet 2009;46:86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gamazon ER, Cox NJ, Davis LK. Structural architecture of SNP effects on complex traits. Am J Hum Genet 2014;95:477–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MacArthur DG, Balasubramanian S, Frankish A, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science 2012;335:823–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol 2010;20:R858–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014;32:347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]