Abstract
Purpose
4D flow MRI enables blood flow measurement in all major cerebral arteries with one scan. Clinical translational hurdles are time demanding post-processing and user-dependence induced variability during analysis. To overcome these shortcomings, a centerline processing scheme (CPS) for semi-automated segmentation and quantification is described, validated, and implemented in carotid siphons of healthy subjects.
Materials and Methods
A CPS for 4D flow data was developed to automatically separate cerebral artery trees. Flow parameters were quantified at planes along the centerline oriented perpendicular to the vessel path. At 3T, validation against 2D phase-contrast (PC) MR and 4D flow manual processing was performed on an intracranial flow phantom for constant flow, while pulsatile flow validation was performed in the internal carotid artery (ICA) of ten healthy volunteers. The CPS and 4D manual processing times were measured and compared. Flow and area measurements were also demonstrated along the length of the ICA siphon.
Results
Phantom measurements for area and flow were highly correlated between the CPS and 2D measurements (area: R = 0.95, flow: R = 0.94), while in-vivo waveforms were highly correlated (R = 0.93). Processing time was reduced by a factor of 4.6 compared with manual processing. Whole ICA measurements revealed significantly decreased area in the most distal segment of the carotid siphon (p = 0.0017), with flow unchanged (p = 0.84).
Conclusion
This study exhibits fast semi-automated analysis of intracranial 4D flow MRI. Internal consistency was shown through flow conservation along the tortuous ICA siphon, which is typically a difficult to assess.
Keywords: 4D flow MRI, vascular, segmentation, automated processing
Introduction
4D flow MRI is a 3-directional velocity encoding that is time-resolved to the cardiac cycle, enabling both volumetric angiographic and quantitative assessment of blood flow velocities (1). However, several translational obstacles for the clinical adaptation of 4D flow remain: 3D velocity acquisition through time results in large datasets and consequently long acquisition, reconstruction, and post-processing times.
To mitigate scan time, non-Cartesian trajectories and parallel acquisitions have been developed and implemented (2,3). The 4D flow MRI technique used here, Phase Contrast Vastly-undersampled Isotropic Projection Reconstruction (PC VIPR) (4,5), is an innovative, flow sensitive MRA sequence with a radial acquisition trajectory. Reduced scan time is achieved through k-space edge undersampling. Due to the high image contrast and sparse signal representation after background suppression, PC VIPR demonstrates benign undersampling artifacts that yield high temporal resolution capabilities, important in assessing blood velocity throughout the cardiac cycle. Most prominently, PC VIPR covers a large volume with high isotropic spatial resolution; in the cranial context whole brain vasculature coverage is achieved.
In the post-processing step for 4D flow MRI, accurate flow quantification and flow profile assessment generally requires manual segmentation and careful placements of double-oblique cutplanes orthogonal to the direction of the vessel path to localize measurements. Analysis time and reproducibility become concerns, especially when several measurement locations within the volume are assessed across a large patient population. In vessels with a tortuous path, such as in the siphon of the internal carotid artery (ICAsiphon), segmentations and measurements become increasingly user-dependent and time-consuming. Therefore, automated segmentation methods are preferred for robust and consistent measurements, but in the context of 4D flow MRI, have only been presented in large and relatively straight intrathoracic arteries (6,7).
This work aims to develop and implement a fast and internally consistent intracranial whole-vessel semi-automated segmentation and flow quantification centerline processing scheme (CPS) for 4D flow MRI.
Methods
Automated Post-processing
All processing was completed using an in-house graphical user interface developed in commercial software (Matlab, The Mathworks, MA, USA) and displayed in Figure 1. System hardware used for processing was a desktop computer, running Windows 7 64-bit (4 core Intel i7-4770 3.40 GHz processors, 32 GB RAM). After data loading, foreground vessels are separated from background noise using a time-maximum intensity projection (tMIP) image, which was formed by projecting the maximum complex difference signal of each voxel through all cardiac timeframes. Thus it is based on both phase and magnitude information. The complex difference approach provides high signal and good noise performance, allowing for background suppression and visualization of distal vessels (8).
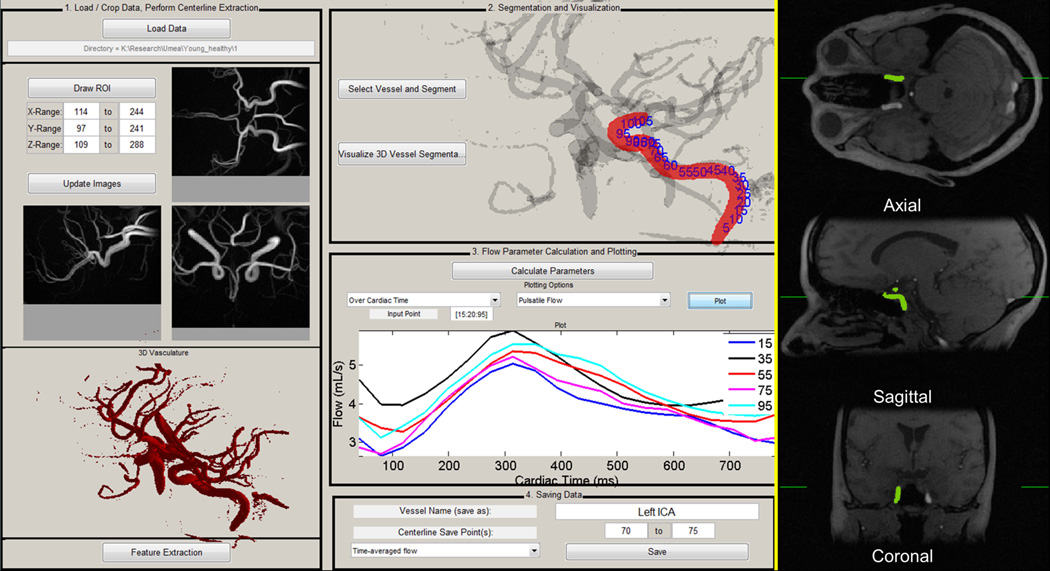
Figure 1.
4D flow centerline processing scheme (CPS) graphical user interface. After data loading (1), cropping, automatic thresholding, and visualization of 3D vasculature, vessel centerlines are extracted. The user selects the vessel for segmented via time-averaged magnitude images with time maximum intensity projection overlaid (right). The segmented 3D vessel with centerline point labels (2) is then visualized, and dynamic parameters can be calculated and plotted for all cardiac phases, shown for pulsatile flow here over 5 locations (3). Parameters can then be saved for specific centerline points (4).
Background signal is not completely nulled in the complex difference image, exhibiting a normal-like distribution. To threshold the tMIP and remove background signal during segmentation, a minimum variance unbiased estimator was used to fit a normal distribution to all voxels within the FOV, producing mean and standard deviation of the fit (µ, σ). After convergence, the tMIP threshold was automatically set as µ + 4σ. This process ensures that background noise will not be present in the whole volume vessel segmentation (Fig. 1 – left), making this step completely automatic and user-independent.
Skeletonization on the binary volume was performed according to a thinning procedure suitable for elongated objects such as blood vessels (9), resulting in a one-voxel wide vessel centerline representation. Vessel branch endpoints and junction points were automatically identified and labeled within the vascular tree, producing a unique branch identification for each vessel. Interactive localization via axial, coronal, and sagittal orthogonal magnitude images from 4D flow MRI (Fig. 1 - right) allowed for one-click selection of the vessel of interest, which was automatically segmented using its unique centerline identifier.
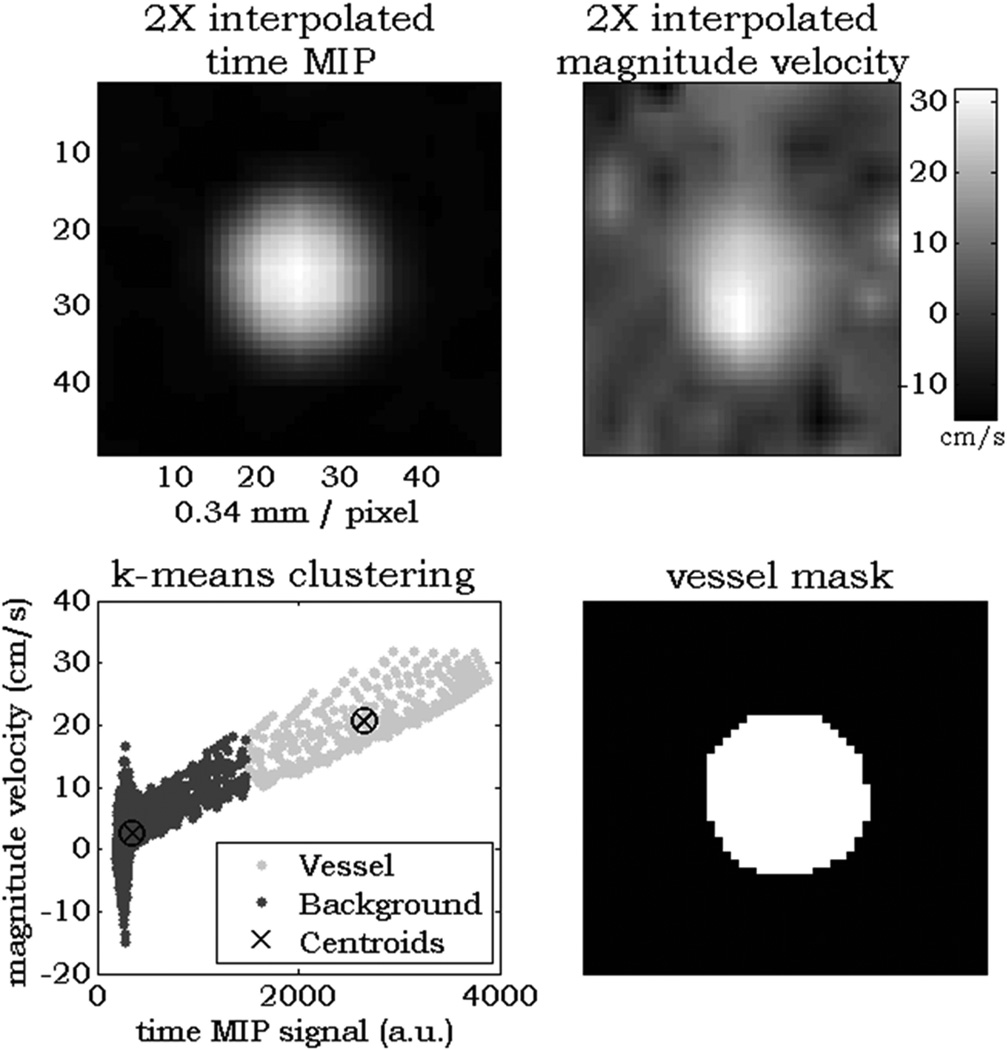
At each centerline voxel along the length of the vessel, two points before and after the currently analyzed centerline location were used to calculate and extract orthogonal cutplanes with respect to the propagation of the vessel. Within each cutplane, 2× linear interpolation was performed. Regions of interest (ROIs) were automatically contoured based on k-means clustering (Figure 2). Input features for clustering were tMIP and the sum-of-squares of the mean velocity across all timeframes within the extracted cutplane. These features were chosen under the assumption that any cross section will contain two regions: 1. low-signal background and 2. vessels. The clustering algorithm minimizes the sum of point-to-centroid distances to determine clusters; this approach has previously been utilized in whole volume automatic particle trace seeding for cardiac 4D flow MRI data visualization (10). Anatomical and blood flow parameters were then quantified from the ROI and acquired velocity data.
Figure 2.
Example cutplane k-means analysis in a healthy ICA. In each cutplane along the length of a centerline, 2× interpolated time MIP signal and sum-squared mean velocity (top row) are used as input features for the k-means clustering algorithm (bottom left). After the two clusters are separated, region growing from the center pixel ensures exclusion of extra vessels within the cutplane to produce a measurement region of interest (bottom right).
In contrast, 4D flow MRI manual processing includes data loading and conversion, visualization, manual placements of cutplanes, and manually drawn ROIs.
Phantom validation
For validation of the CPS, scans were performed using a clinical 3T system (Discovery MR750, GE Healthcare, Waukesha, WI, USA) on an intracranial flow phantom (Model H+N-R-A-010, Shelley Medical Imaging Technologies, London, ON, Canada) containing water doped with a single dose of gadolinium contrast equivalent to the amount used for a 70 kg (at 0.2 mmol/kg) subject; this was used to mimic clinical contrast. Six levels of constant flow input values (0.85, 0.95, 1.05, 1.15, 1.25, and 1.35 L/min) were assigned according to normal in-vivo ranges of total intra- plus extra-cerebral blood flow (11). Constant flow was chosen to compare area and flow measurements.
PC VIPR scans were performed with the following parameters: FOV = 22 × 22 × 22 cm3, acquired spatial resolution − 0.69 mm isotropic, velocity encoding (optimized to maximum signal for highest flow in the middle cerebral artery, MCA) = 70 cm/s, α = 15°, TR/TE = 11.4/3.9 ms, number of projections = 1500. Phase offsets from eddy currents were automatically corrected during reconstruction. For each scan, flow was calculated as cross sectional vessel area multiplied by mean blood velocity. 2D PC scans based on clinical protocols were used for comparison. These were performed in the basilar artery, left and right internal carotid artery, and right MCA of the flow phantom: FOV − 8.0 × 8.0 cm, 0.31 × 0.31 mm in-plane resolution, 3 mm slice thickness, velocity encoding = 50 cm/s for BA/ICA; 70 cm/s for MCA, α = 15°, TR/TE = 7.5/4.6 ms, 2 averages. No eddy current compensation was performed for 2D scans because static parts of the phantom displayed negligible velocity bias (< 1 cm/s). Time for processing each case (4D CPS and manual) was recorded.
In-vivo validation
For the assessment of the CPS in a clinical intracranial territory with pulsatility, ten healthy (7M, age 37 ± 9 years) subjects were imaged after informed consent and IRB approval. PC VIPR scans were performed on a 3T magnet of identical build. Scan parameters: 32 channel head coil, FOV = 22 × 22 × 22 cm3, (0.69 mm)3 isotropic resolution, scan time = 9.5 min, Venc = 110 cm/s, 20 reconstructed cardiac timeframes. Data were reconstructed from an acquisition matrix of (300)3 to (320)3 using zero padded interpolation during reconstruction. For comparison a single 2D PC scan was performed, covering the left and right ICA at the level of the carotid C3–C4 segment. Scan parameters: 0.35 × 0.35 mm in-plane resolution, 3 mm slice thickness, velocity encoding = 80 cm/s, α = 15°, TR/TE = 7.5/4.6 ms, 6 views per segment, 32 reconstructed cardiac timeframes. This data has been used in a previous study to compare pulsatile flow in PC VIPR versus 2D PC (12). Here we aim not to re-validate pulsatility measures between 2D PC and 4D flow MR, but to demonstrate the use of the CPS to provide accurate semi-automated results in pulsatile flow. CPS waveforms were interpolated to 32 timeframes, allowing one-to-one comparisons of flow throughout the cardiac cycle. For evaluation, waveforms from both 2D PC and the CPS were temporally aligned via cross correlation and averaged across all volunteers. An additional measure of pulsatility index (PI), calculated as:
was used to further compare waveforms.
Demonstration along vessel
To demonstrate the use of the CPS along the length of an entire vessel, whole ICAsiphon measurements of area and flow using the CPS were generated from the above in-vivo data.
Statistical Analysis
All processing of data was completed by a researcher (E.S.) with 4+ years of experience in MR flow image processing. In the validation analyses, 4 consecutive centerline points were averaged to closely match slice thickness in 2D PC scans (giving a roughly equivalent slice thickness of 4 × .69 mm = 2.75 mm). Both phantom and in-vivo validation results were compared using least squares regression and Bland-Altman analysis (13). In the whole vessel demonstration, left and right ICAsiphon area and flow were partitioned and averaged based on segment (C1 – C7) according to Bouthillier et al (14). Statistical comparisons between each ICAsiphon segment were tested using one-way ANOVA (15).
Results
Phantom validation
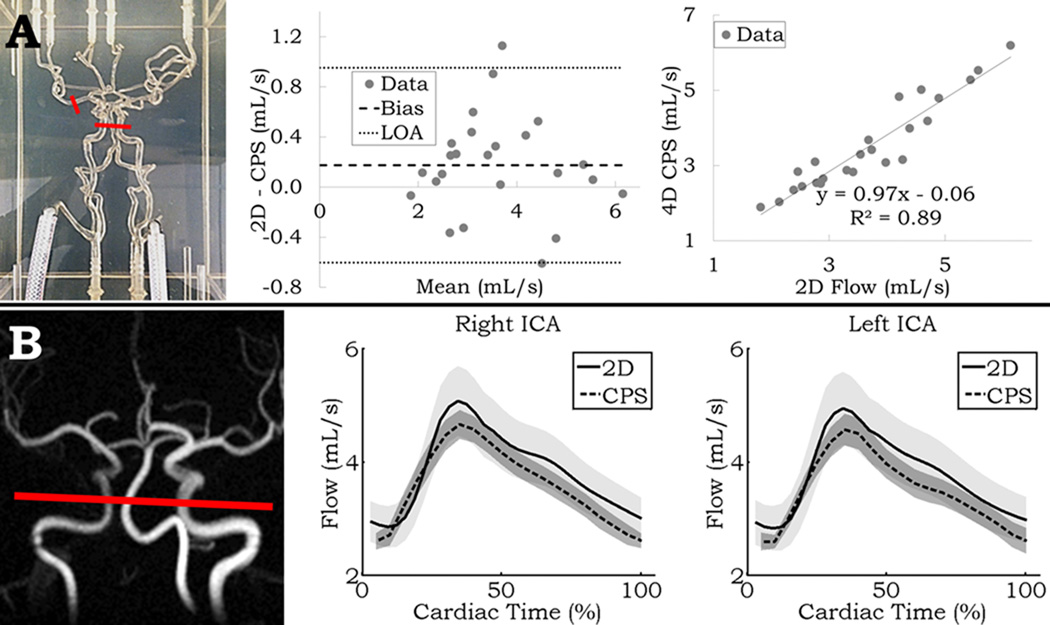
Linear regression and Bland-Altman analysis between 2D PC and the 4D CPS is shown in Figure 3A. Results comparing both area and flow calculated from the 2D PC scan, and both manual and the CPS 4D flow output are shown in Table 1 – top. Very high correlation from regression analysis was seen between different methods, and Bland-Altman results showed roughly 20% limits of agreement. Compared with the 2D PC output, the 4D flow MRI CPS outperformed the 4D manual processing (R = 0.94 vs. R = 0.88). Both forms of 4D processing overestimated the vessel area, yet were still highly correlated (4D manual bias = 2.61 mm2; R = 0.93, CPS bias = 4.77 mm2; R = 0.95). Average processing time per vessel across all flow settings using the CPS, including data loading, thinning and centerline extraction, vessel selection and segmentation, and quantification was 56.0 ± 3.6 s. This was less than the time required achieving a single measurement per vessel by processing the data manually (261.4 ± 23.1 s).
Figure 3.
A: left – Front view photo of the neurovascular phantom; red segments delineate measurement locations. middle/right – Linear regression and Bland-Altman analysis between the measurements from the 4D centerline processing scheme (CPS) and 2D PC MR. B: left – Limited MIP of PC VIPR in-vivo with 2D PC and CPS measurement location in red. middle/right – 2D PC and CPS flow waveforms averaged across all volunteers for the right and left ICA for 32 timeframes. Shading represents the standard deviation of the mean at each timeframe.
Table 1.
Top: Linear regression and Bland-Altman analysis results for 2D PC versus 4D (manual and the centerline processing scheme - CPS) comparison of flow (left) and area (right) from N = 24 measurements. Bottom: Linear regression and Bland-Altman analysis performed on in-vivo pulsatile data across ten healthy volunteers for pulsatility index in right and left ICA (N = 20, left) and flow waveform comparison in right and left ICA (frame by frame, N = 64, right). LOA - limit of agreement.
| Phantom Validation | |||||||
|---|---|---|---|---|---|---|---|
| Flow comparison | Area comparison | ||||||
| 2D vs. CPS | 2D vs. 4D manual |
CPS vs. 4D manual |
2D vs. CPS | 2D vs. 4D manual |
CPS vs. 4D manual |
||
| Slope | 0.97 | 0.94 | 0.89 | Slope | 1.42 | 1.54 | 0.98 |
| R | 0.94 | 0.88 | 0.85 | R | 0.95 | 0.93 | 0.86 |
| Bias (mL/s) [upper LOA, lower LOA] | 0.18 [0.95, −.60] | 0.56 [1.74, −0.61] | −0.39 [0.91, −1.69] | Bias (mm2) [upper LOA, lower LOA] | −4.77 [2.42, −11.96] | −2.61 [6.72,11.93] | 2.16 [7.91, −3.59] |
| In-vivo Validation | |||
|---|---|---|---|
| Pulsatility Index comparison | Flow waveform comparison | ||
| Slope | 0.85 | Slope | 0.93 |
| R | 0.48 | R | 0.93 |
| Bias [upper LOA, lower LOA] | −0.97 [0.26, −0.46] | Bias (mL/s) [upper LOA, lower LOA] | 0.26 [0.80, −0.27] |
In-vivo validation
Similar flow waveforms are observed when comparing between 2D PC and the 4D CPS in both the right and left ICA in-vivo (Figure 3B), with overall lower values from the 4D flow scans. Results comparing these flow waveforms, both throughout the waveforms and with PI are shown in Table 1 – bottom. A moderate agreement (R = 0.48) for PI was observed, and similar to the phantom results, a strong linear correlation (R = 0.93) was observed for the waveform comparison. The CPS slightly overestimated PI (bias = 0.097), while flow was underestimated (bias = 0.26 or ~ 7% lower than 2D PC).
Demonstration along vessel
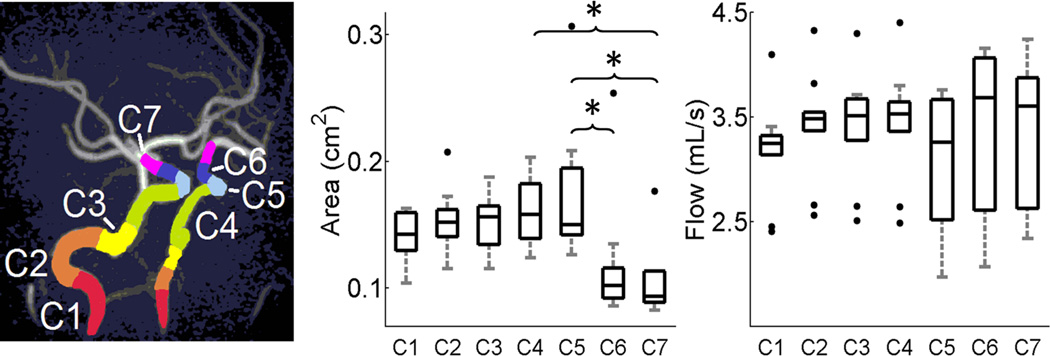
Figure 4 – middle displays boxplot results for left and right summed and segmented ICAsiphon averaged area across all volunteers. Figure 4 – right shows the same analysis performed on flow. In the C7 segment, area was significantly lower than in the C4–C5 segments, while area in the C6 segment was significantly lower than the C5 segment (p = 0.0017). Flow conservation along the length of the ICAsiphon was accurately represented by the CPS as no statistical differences were observed between segments (p = 0.84).
Figure 4.
Left: Time maximum intensity projection image of arterial vasculature with color overlaid segments. Middle and right: Boxplots of averaged area and flow measurements across all ICA segments shows decreased cross-sectional area in more superior segments while flow is conserved. Upper and lower box boundaries represent the 1st and 3rd quartile of the data, respectively. * - p < 0.05
Discussion
This paper presents a fast post-processing CPS for the assessment of blood flow related parameters derived from 4D flow MRI. Its value lies in the ability to perform user-independent segmentation and quantification in tortuous arterial segments which would be otherwise very difficult to study with other flow sensitive techniques such as transcranial Doppler ultrasound. Results through both Bland-Altman analysis and linear regression in both a phantom with constant flow and in-vivo pulsatile flow revealed the validity of using the CPS.
The CPS allows for faster analysis than can currently be performed for 4D flow manual post-processing in the brain. Direct comparison of processing times does not elucidate that as the number of locations of interest for interrogation within the neurovasculature increases, time per measurement will decrease. This is because the bulk of processing time is devoted to data loading, which is dependent on the number of reconstructed timeframes (for in-vivo dataset of 20 timeframes and hardware used here, roughly 2 minutes). By comparison, skeletonization and centerline extraction were much swifter (on the order of tens of seconds). After these steps are performed the user was free to select any number of vessels of interest for quantification quickly and efficiently.
Among automated segmentation techniques that have been published elsewhere, the active surface model method by van Pelt (6), while fast, accurate, and topologically stable, is affected by limited spatial resolution and can therefore not segment smaller arteries which are the target of cerebral 4D flow MRI scans. Although pulsatility likely changes luminal diameters in the ICA, such differences in the common carotid artery are on the order of 1 mm in healthy young volunteers (16). Thus active surface modelling would not be needed for these small luminal changes intracranially throughout the cardiac cycle. Additionally, the initial isosurface used in van Pelt (6) was generated with a varying manually-defined threshold on their version of the tMIP. These manual interactions potentially introduce user-dependence and are not part of our automated segmentation scheme.
A centerline approach, in which manual seed points are used for centerline calculation, has been presented before and is currently used for pathological aortic flow assessment (7). While this approach is efficient and integrated into vendor software, it has not been tested in small vascular structures, such as intracranial vessels shown here. Some user interaction of seed point placement for aortic centerline extraction is required, which may introduce user-dependent variability. The centerline extraction in our method was performed automatically on all vasculature so that the only user interaction is the selection of the vessel branch of interest. Vessel segmentation was performed through simple one-click selection on 2D orthogonal magnitude data. Blood flow over the cardiac cycle can be visualized at specific centerline points in the 3D volume. Because segmentation and flow quantification was automated, results are user-independent and completely repeatable.
Flow estimations were generated at each centerline point along a given arterial branch, which can then be averaged to produce a better estimate of the flow in the branch. As shown with the grouping of different segments of the ICAsiphon, several consecutive flow measurements can be combined and averaged to give a less noisy representation of blood flow. These combinations of points could be chosen arbitrarily so that particular locations or parameters of interest for the user may be further retrospectively investigated.
Among the ways to measure flow in-vivo, the choice of using 2D PC MR as the gold standard is supported by its high performance in larger vessels such as the ICA (17). There was particularly high correlation between 2D PC and the CPS for area, total flow, and across pulsatile waveforms. While good correlation was still observed between 2D PC and 4D manual output, it was lower than that from the CPS. Potentially confounding and user-dependent factors, including manual placement of orthogonal cutplanes and manual drawing of ROIs, likely contributed to this larger disagreement but are removed in the CPS. Moreover, while the manual processing measures flow through a one-voxel thick cutplane, the CPS results were averaged over consecutive cutplanes to match the slice thickness used in 2D PC measurements. This likely factored into higher correlation seen from the automated results, and illuminates another benefit of this technique.
The higher values and lower correlation in PI from CPS compared to 2D PC data likely stem from a number well-known factors when comparing 4D flow with 2D PC, including: lower temporal resolution, longer scan times resulting in temporal smoothing of beat to beat variations in waveforms, and lower spatial resolution causing partial volume effects at vessel lumens. Despite these effects, 4D flow and 2D PC have been shown to have the same measurement variability (12), pointing to 4D flow results having equal measurement consistency as 2D PC.
Whole ICAsiphon results from the CPS show that, on average, cross-sectional area of the ICAsiphon decreases significantly in the distal C7 segment, while blood flow is unchanged. Before bifurcation into the MCA and ACA, the ICA has at most two drainage pathways: the ophthalmic artery in the C5 segment and the posterior communicating artery in the C6 segment (in about 20% of adults). However, due to the small cross-sectional area and observed velocity in these vessels (18), flow was not expected to change significantly along the length of the ICA. This is substantiated by a previous study in the ophthalmic artery of healthy individuals assessing average blood flow to be 0.17 mL/s (19), far below total ICA flow observed in our study. The conservation of mass (i.e. flow) along the length of the ICAsiphon shown here is yet another validation of the internal consistency of the presented CPS.
This study has some limitations. First, the CPS relies heavily on good image quality within the tMIP for automated processing in: the global threshold, the vessel segmentation, and the quantification within a cross section using k-means clustering. While this was not an issue in the data for this study, in patients with atypically high or low flow with respect to the Venc, such as flow jets or in slow aneurysm flow, signal dropout in the reconstructed tMIP may cause missed areas of segmentation. This issue would still be present in the 4D manual processing, however, and can only be ameliorated through some phase unwrapping of the data or a dual-Venc approach in cases where a large flow range is expected (20). Second, this method was introduced and presented only in intracranial arteries for demonstration. It could in theory work in other complicated extracranial vasculature. Examples may include liver and abdominal vessels, or the draining cerebrospinal veins. Further studies are warranted to investigate these territories of interest.
In conclusion, this study exhibits the integration of a number of image processing methods into a fast and internally consistent 4D flow MRI intracranial segmentation and quantification CPS. By viewing vasculature as a tree of unique vessel branches, segmentation of any individual vessel (or concatenated vessels to follow blood along a certain route) provides comprehensive knowledge of flow along the entire vessel. The flexible functionality of the CPS may be of use in other vascular territories or in assessment of blood flow in neurovascular diseases such as stroke or vascular dementia.
Acknowledgments
Grant support:
NMSS fund RC1003-A-1
NIH grant R01HL072260 and GE Healthcare
References
- 1.Markl M, Frydrychowicz A, Kozerke S, Hope M, Wieben O. 4D flow MRI. Journal of magnetic resonance imaging : JMRI. 2012;36(5):1015–1036. doi: 10.1002/jmri.23632. [DOI] [PubMed] [Google Scholar]
- 2.Carlsson M, Toger J, Kanski M, et al. Quantification and visualization of cardiovascular 4D velocity mapping accelerated with parallel imaging or k-t BLAST: head to head comparison and validation at 1.5 T and 3 T. Journal of cardiovascular magnetic resonance : official journal of the Society for Cardiovascular Magnetic Resonance. 2011;13:55. doi: 10.1186/1532-429X-13-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigfridsson A, Petersson S, Carlhall CJ, Ebbers T. Four-dimensional flow MRI using spiral acquisition. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2012;68(4):1065–1073. doi: 10.1002/mrm.23297. [DOI] [PubMed] [Google Scholar]
- 4.Gu T, Korosec FR, Block WF, et al. PC VIPR: a high-speed 3D phase-contrast method for flow quantification and high-resolution angiography. AJNR American journal of neuroradiology. 2005;26(4):743–749. [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson KM, Lum DP, Turski PA, Block WF, Mistretta CA, Wieben O. Improved 3D phase contrast MRI with off-resonance corrected dual echo VIPR. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2008;60(6):1329–1336. doi: 10.1002/mrm.21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Pelt R, Nguyen H, ter Haar Romeny B, Vilanova A. Automated segmentation of blood-flow regions in large thoracic arteries using 3D-cine PC-MRI measurements. International journal of computer assisted radiology and surgery. 2012;7(2):217–224. doi: 10.1007/s11548-011-0642-9. [DOI] [PubMed] [Google Scholar]
- 7.Semaan EM, Carr M, Gulsun M, et al. Evaluation of an optimized post-processing tool for 4D flow MRI data analysis in healthy volunteers and patients with aortic stenosis, aortic insufficiency, and aortic aneurysm. 22nd Annual Meeting of International Society for Magnetic Resonance in Medicine; Milan, IT. 2014. p. 3948. [Google Scholar]
- 8.Anderson AG, Johnson KM, Bock J, Markl M, Wieben O. Comparison of Image Reconstructioin Algorithms for the Depiction of Vessel Anatomy in PC VIPR Datasets. 16th Annual Meeting of International Society for Magnetic Resonance in Medicine; Toronto, ON, CA. 2008. p. 934. [Google Scholar]
- 9.Palágyi KSE, Balogh E, Kuba A, Halmai Cs, Erdôhelyi B, Hausegger K. A sequential 3D thinning algorithm and its medical applications. Proc 17th Int Conf Information Processing in Medical Imaging, IPMI 2001; Davis, CA, USA. 2001. [Google Scholar]
- 10.Stalder AF, Gulsun MA, Greiser A, Jolly M-P. Fully automatic visualization of 4D Flow data. Internal Society of Magnetic Resonance in Medicine. Salt Lake City, UT, USA: 2013. [Google Scholar]
- 11.Uematsu S, Yang A, Preziosi TJ, Kouba R, Toung TJ. Measurement of carotid blood flow in man and its clinical application. Stroke a journal of cerebral circulation. 1983;14(2):256–266. doi: 10.1161/01.str.14.2.256. [DOI] [PubMed] [Google Scholar]
- 12.Wåhlin A, Ambarki K, Birgander R, et al. Measuring pulsatile flow in cerebral arteries using 4D phase-contrast MR imaging. AJNR Am J Neuroradiol. 2013;34(9):1740–1745. doi: 10.3174/ajnr.A3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bland JM, Altman DG. Agreement between methods of measurement with multiple observations per individual. J Biopharm Stat. 2007;17(4):571–582. doi: 10.1080/10543400701329422. [DOI] [PubMed] [Google Scholar]
- 14.Bouthillier A, van Loveren HR, Keller JT. Segments of the internal carotid artery: a new classification. Neurosurgery. 1996;38(3):425–432. doi: 10.1097/00006123-199603000-00001. discussion 432–423. [DOI] [PubMed] [Google Scholar]
- 15.Rosner B. Fundamentals of Biostatistics. 7 ed. Boston, MA: Brooks/Cole; 2010. pp. 562–567. [Google Scholar]
- 16.Studinger P, Lénárd Z, Kováts Z, Kocsis L, Kollai M. Static and dynamic changes in carotid artery diameter in humans during and after strenuous exercise. J Physiol. 2003;550(Pt 2):575–583. doi: 10.1113/jphysiol.2003.040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wåhlin A, Ambarki K, Hauksson J, Birgander R, Malm J, Eklund A. Phase contrast MRI quantification of pulsatile volumes of brain arteries, veins, and cerebrospinal fluids compartments: repeatability and physiological interactions. J Magn Reson Imaging. 2012;35(5):1055–1062. doi: 10.1002/jmri.23527. [DOI] [PubMed] [Google Scholar]
- 18.Klotzsch C, Popescu O, Berlit P. Assessment of the posterior communicating artery by transcranial color-coded duplex sonography. Stroke a journal of cerebral circulation. 1996;27(3):486–489. doi: 10.1161/01.str.27.3.486. [DOI] [PubMed] [Google Scholar]
- 19.Ambarki K, Hallberg P, Johannesson G, et al. Blood flow of ophthalmic artery in healthy individuals determined by phase-contrast magnetic resonance imaging. Investigative ophthalmology & visual science. 2013;54(4):2738–2745. doi: 10.1167/iovs.13-11737. [DOI] [PubMed] [Google Scholar]
- 20.Nett EJ, Johnson KM, Frydrychowicz A, et al. Four-dimensional phase contrast MRI with accelerated dual velocity encoding. J Magn Reson Imaging. 2012;35(6):1462–1471. doi: 10.1002/jmri.23588. [DOI] [PMC free article] [PubMed] [Google Scholar]