Abstract
Purpose
Paclitaxel is used worldwide in the treatment of breast, lung, ovarian and other cancers. Sensory peripheral neuropathy is an associated adverse effect that cannot be predicted, prevented or mitigated. To better understand the contribution of germline genetic variation to paclitaxel-induced peripheral neuropathy, we undertook an integrative approach that combines genome-wide association study (GWAS) data generated from HapMap lymphoblastoid cell lines (LCLs) and Asian patients.
Methods
GWAS was performed with paclitaxel-induced cytotoxicity generated in 363 LCLs and with paclitaxel-induced neuropathy from 145 Asian patients. A gene-based approach was used to identify overlapping genes and compare to a European clinical cohort of paclitaxel-induced neuropathy. Neurons derived from human induced pluripotent stem cells were used for functional validation of candidate genes.
Results
SNPs near AIPL1 were significantly associated with paclitaxel-induced cytotoxicity in Asian LCLs (P < 10−6). Decreased expression of AIPL1 resulted in decreased sensitivity of neurons to paclitaxel by inducing neurite morphological changes as measured by increased relative total outgrowth, number of processes and mean process length. Using a gene-based analysis, there were 32 genes that overlapped between Asian LCL cytotoxicity and Asian patient neuropathy (P < 0.05) including BCR. Upon BCR knockdown, there was an increase in neuronal sensitivity to paclitaxel as measured by neurite morphological characteristics.
Conclusion
We identified genetic variants associated with Asian paclitaxel-induced cytotoxicity and functionally validated the AIPL1 and BCR in a neuronal cell model. Furthermore, the integrative pharmacogenomics approach of LCL/patient GWAS may help prioritize target genes associated with chemotherapeutic-induced peripheral neuropathy.
Keywords: paclitaxel, neuropathy, Asians, GWAS, iPSCs
Introduction
Paclitaxel, a microtubule-stabilizing agent that binds tubulin and disrupts cell division, represents standard of care for common cancers, including breast, lung and ovarian cancer (1). Unfortunately, about 20-30% of patients treated with paclitaxel experience clinically relevant sensory peripheral neuropathy (2, 3). Furthermore, there is substantial inter-patient variability in time to symptom onset, time to peak symptoms, severity of peak symptoms and reversibility (4, 5). Patients experiencing moderate to severe chemotherapy induced peripheral neuropathy (CIPN) report a reduced quality of life, chronic discomfort and disruption of physical abilities for general life activities that can be temporary or permanent (6). CIPN causes a significant symptom burden because many patients that undergo adjuvant treatment are long-term survivors. Moreover, CIPN can lead to dose reduction of the chemotherapeutic agent or possible cessation of treatment (7). This may have an adverse impact on cancer treatment and disease outcomes, particularly in the adjuvant setting.
There are few effective methods in clinical practice for managing peripheral neuropathy. ASCO recommendations, recently published (7) and based on a systematic evaluation of 48 randomized controlled trials, concluded that there are no agents recommended for the prevention of CIPN and supported a moderate recommendation of duloxetine for pain associated with CIPN (8). Physical therapy, acupuncture, massage and medications such as steroids, anti-epileptics, anti-depressants and opioids for pain management are often attempted. Unfortunately, the extent to which these interventions improve symptoms is limited and some of these interventions carry side effects of their own (7). An improved understanding of the genetic variants and genes contributing to paclitaxel induced neuropathy will assist in the development of future neuroprotective agents and in the design of novel therapies with improved toxicity profiles. Furthermore, finding predictive genetic biomarkers could enable identification of patients who should be treated with neuroprotective agents (when they are available) and/or be treated with reduced paclitaxel doses or switched to non-neuropathic treatments.
In attempts to identify genes relevant to CIPN, several genome-wide association studies (GWAS) have been conducted with the identification of single-nucleotide polymorphisms (SNP) in FGD4 (9), EPHA5 and XKR4 (10) associated with the risk of paclitaxel-induced sensory peripheral neuropathy. Because it is challenging to replicate large, well-controlled clinical trials to evaluate pharmacogenomics markers of toxicity (11, 12), we developed an integrative approach combining clinical studies and cell-based models enabling variant identification and followed with functional validation of the findings to help elucidate underlying mechanisms and pathways (13). This method has found utility for paclitaxel-induced peripheral neuropathy in individuals of European descent and capecitabine-induced hand-foot syndrome (14, 15).
Although paclitaxel is used globally, large clinical GWAS studies of paclitaxel-induced sensory peripheral neuropathy have been performed primarily in European patients (9, 10, 14, 16). However, pharmacoethnicity have been recognized as an important factor in chemotherapeutic response and toxicity (17). In this report, we compare genetic variants identified in a GWAS of sensitivity of LCLs derived from individuals of Asian, European and African ancestry to variants identified in an Asian patient GWAS of paclitaxel neuropathy. In addition to analyzing SNPs, we identified significant genes through expression quantitative trait loci (eQTL) data (18) and functionally validate a target gene using human iPSC-derived neurons.
Materials and Methods
Cytotoxicity assays
HapMap LCLs derived from Japanese individuals residing in Tokyo, Japan (HAPMAPPT02/05, JPT, n = 57) and a set derived from Han Chinese residing in Beijing, China (HAPMAPPT02/05, CHB, n = 59), purchased from the Coriell Cell Repositories (Camden, NJ, USA), were treated with increasing concentrations of paclitaxel (Sigma-Aldrich, St. Louis, MO, USA). Cytotoxicity was determined using an AlamarBlue (Invitrogen, nologies Inc.; Carlsbad, CA, USA) cellular growth inhibition assay as described (19). Mean area under the paclitaxel concentration-cell survival percentage curve (AUC) was determined by at least 6 replicates from 2 independent experiments and used as the phenotype in the GWAS. AUC values for each cell line were log2-transformed before statistical analysis to form an approximately normal distribution. Paclitaxel cytotoxicity data of CEU (n = 77), YRI (n = 87) and ASW (n = 83) are previously published (19). For each population, a linear regression was calculated to determine the correlation between the AUC for each replicate; the correlation for CEU, YRI, ASW, CHB and JPT was 0.83, 0.86, 0.81, 0.83 and 0.82, respectively.
Authentication of Cell Lines
The PAAR Cell Core routinely checks the identity of LCLs by genotyping 47 informative SNPs included in the Sequenom iPLEX Sample ID Plus Panel (San Diego, CA, USA). Using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) DNA was extracted from each sample. Genotyping was performed following the iPLEX Pro application guide and the iPLEX Pro reaction products were dispensed onto a 384-sample SpectroCHIP and run on a Sequenom MassARRAY system at The University of Chicago DNA Sequencing and Genotyping Facility. Genotypes passed quality control (formed clear genotype clusters) for 46/47 SNPs and all 46 genotypes matched the known HapMap (http://hapmap.ncbi.nlm.nih.gov/) genotypes for all samples. Twenty-four of 116 LCLs were randomly chosen from this study and all passed quality control.
LCL genome-wide association studies
A GWAS of paclitaxel-induced cytotoxicity was conducted by combining the JPT and CHB (Asian population: ASN). Phased genotype data of the 186 ASN cell lines sequenced in the 1000 Genomes Project phase 1 version 3 (20) were downloaded from the BEAGLE website (http://faculty.washington.edu/browning/beagle/beagle.html). An additional 15 ASN samples genotyped in HapMap r28 were imputed to 1000 Genomes using BEAGLE version 3.3.2 (21). Phased genotype data were also downloaded for the 85 CEU and 88 YRI sequenced in the 1000 Genomes Project and used to impute the additional 93 CEU and 90 YRI samples genotyped in HapMap r28 with BEAGLE, which considers the relatedness of the trios in the imputation. Within each population, approximately 4.6 million SNPs [imputation R2 > 0.8, population minor allele frequency (MAF) > 0.05, and in Hardy–Weinberg equilibrium (P > 1 × 10−6)] were tested for association with paclitaxel cytotoxicity using the linear mixed model Wald test, which accounts for the genetic relatedness of individuals, implemented in GEMMA version 0.94 (22).
Patient samples and GWAS
Germline DNA of 183 cancer patients treated with monotherapy paclitaxel stored in Biobank Japan (University of Tokyo) were genotyped using Illumina OmniExpress BeadChip that contained 733,202 SNPs (23). Of them, 24 patients experienced either grade 2 or higher paclitaxel-induced sensory peripheral neuropathy (PIPN), 121 patients revealed no neuropathy and the remaining 38 individuals had grade 1 neuropathy. For GWAS, we used 24 patients who developed ≥ grade 2 PIPN (cases), and compared them with 121 patients who did not show neuropathy (controls). The patient characteristics are described in Supplemental Table S1. The grade of toxicity was classified in accordance with the US National Cancer Institute’s Common Toxicity Criteria version 2.0. All participants provided written informed consent. This project was approved by the Institutional Review Board in the Institute of Medical Science, the University of Tokyo, and RIKEN Center for Genomic Medicine. Sample quality control was carried out by methods including identity-by-state to evaluate cryptic relatedness for each sample, and population stratification by the use of principal component (PC) analysis to exclude genetically heterogeneous samples from further analysis. Then, our standard SNP quality control was carried out by excluding SNPs deviating from the Hardy–Weinberg equilibrium (P ≤ 10−6), non-polymorphic SNPs, SNPs with a call rate of < 0.99, and those on the X chromosome. We performed a genome-wide case/control allelic association study using Fisher’s exact test.
LCL/Patient comparison
We used the SCAN database to determine which of the top SNPs significantly associated with paclitaxel-induced cytotoxicity in ASN (P < 10−6) were located in or near (within 2 kb) gene transcripts (24, 25). We also used the eQTL Browser (http://eqtl.uchicago.edu/Home.html) and GTEx Portal (http://www.gtexportal.org/home/) to detect whether SNPs genotyped in 1000 Genomes were eQTLs in LCLs and other tissues. The previous genomic studies using LCLs showed that Japanese and Han Chinese have similar genetic structure, but cluster separately from European and African-American individuals in principal components analysis (26). We used Sherlock, the computational algorithm that matches patterns of GWAS results with independent eQTL data to detect significant genes associated with the cytotoxicity and neuropathy phenotypes (18). We used the public HapMap CEU eQTL dataset (27) contained in Sherlock for gene association testing. Among the genes associated with each phenotype (P < 0.05) through Sherlock (18), the LCL/patient overlap genes were prioritized for functional analysis. Results were also compared with the published clinical GWAS of 855 genetic Europeans with breast cancer from CALGB40101, in which a dose-to-event analysis was conducted with an event defined as paclitaxel-induced sensory peripheral neuropathy ≥ grade 2 (9).
Functional studies in iPSC derived neurons
Human iPSC-derived cortical neurons, termed iCell Neurons (Cellular Dynamics International, Madison, WI, USA), were used for functional validation of the candidate genes. These cells have been previously used to model chemotherapy-induced neuropathy (28, 29).
RNA-Seq
RNA isolation from iPSC derived cortical neurons was performed after removal of media and addition of 50µL Qiazol per well plate for 5 minutes at room temperature to lyse cells. After vigorously pipetting each well several times, samples were collected and stored for further processing at -70°C. RNA was purified using the Ambion RNA protocol (15596026.PPS), only substituting Qiazol as the lysing reagent after consulting with the company. The final pellets were resuspended in 50 µl RNAase-free water dissolved at 55°C for 10 minutes, aliquoted and stored at -70°C. Nucleic acids quantification was performed using the Qubit dye for RNA kits (Molecular Probes, Life Technologies), as per manufacturer's specifications. 1 µg RNA from each time-point was submitted to the University of Chicago Genomics Core. RNA quality was checked on the Agilent Bio-analyzer 2100. RNA-SEQ libraries were generated in the core using Illumina RS-122-2101 TruSeq® Stranded mRNA LT Libraries and the final libraries checked again on the Agilent bio-analyzer 2100 which was followed by sequencing on the Illumina HiSeq2500. The raw sequencing reads were processed and analyzed using the RNA-Seq pipeline in the Galaxy Project (30). Briefly, the raw reads were mapped to the human genome reference (hg19) using TopHat2 (version 2.06) (31), which aligns RNA-Seq reads to the human genomes using the ultra high-throughput short read aligner Bowtie2 (32). Cufflinks (version 0.0.7) (33) was then used to quantify and quantile normalize the expression levels of the assembled transcripts.
siRNA knockdown studies
iCell Neurons (1.33×104) were seeded with 3.3μg/mL laminin (Sigma-Aldrich) in iCell Neuron complete maintenance media onto poly-D-lysine coated 96-well Greiner Bio-One plates (Monroe, NC, USA) according to manufactures recommendations. Dharmacon Accell technology (GE Dharmacon, Lafayette, CO, USA) was applied at 4 hours post plating using 1 µM each human siAIPL1, siPTPMT1, siBCR or siDDX54 SMARTpool or the non-targeting control (NTC) and then 24 hours later cells were either collected for real time reverse transcription polymerase chain reaction (RT-PCR) or an exchange of transfection media with paclitaxel at a dose range of 0.001-10 µM or 0.17% DMSO vehicle control. After 24 or 48 hours, the drug was removed and cells were stained with 1 µg/mL Hoechst 33342 (Sigma-Aldrich) and 2 µg/mL Calcein AM in dPBS (Molecular Probes, Life Technologies Inc.; Carlsbad, CA, USA). Image analysis was performed on the ImageXpress Micro imaging system (Molecular Devices, LLC, Sunnyvale, CA, USA) at the University of Chicago Cellular Screening Center. Individual cell measurements of neurite outgrowth including mean/median/maximum process length, total neurite outgrowth (sum of the length of all processes), number of processes, number of branches, cell body area, mean outgrowth intensity and straightness were analyzed using the MetaXpress software Neurite Outgrowth Application Module. Each experiment was replicated three times and the difference in the relative neurite outgrowth for each siRNA treatment was compared to NTC and tested for significance by two-way ANOVA analysis.
Cells for RT-PCR from 2 wells for each experiment were collected at 24 and 48 hours post-transfection using Cells to CT (Life Technologies) as recommended by manufacturer. Two independent reverse transcription reactions were performed and the efficiency of knockdown for AIPL1 (Hs04185077_m1), PTPMT1 (Hs00378514_m1), BCR (Hs01036532_m1) or DDX54 (Hs_00225320_m1) (Life Technologies) for triplicate wells was determined using Taqman fast chemistry on the Viia7 PCR machine (LifeTechnologies). A comparative delta delta CT analysis was used to determine the percent knockdown of each gene by comparing the CT difference relative to the beta-2-microglobulin control (NM_004048.2) and normalizing to NTC.
Results
Paclitaxel-induced cytotoxicity in LCLs
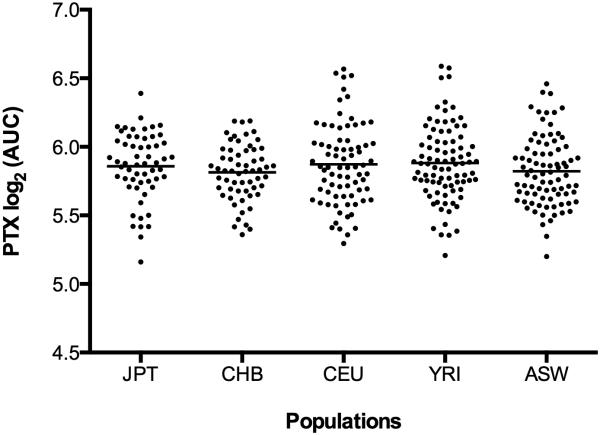
HapMap LCLs were treated with increasing concentrations of paclitaxel and cellular growth inhibition was measured. The log2-transformed area under the survival curve (AUC) was used as the paclitaxel-induced cytotoxicity phenotype. To evaluate ethnic differences in mean log2 AUC of paclitaxel, we compared results from LCLs derived from the Asian populations (JPT, n = 57; CHB, n = 59) to our previous data of three other HapMap LCL populations (CEU, n = 77; YRI, n = 87; ASW, n = 83) (19). The paclitaxel-induced log2-transformed cytotoxicity phenotype showed normal distributions in each population. Figure 1 illustrates the paclitaxel-induced cytotoxicity among 5 populations with no statistical differences among the populations observed.
Figure 1. Cellular sensitivity to paclitaxel in 363 HapMap LCLs.
LCLs from different world populations (JPT: n = 57, CHB: n = 59, CEU: n = 77, YRI: n = 87, ASW: n = 83) were phenotyped for the cellular growth inhibitory effects of paclitaxel. The cytotoxicity phenotype is log2-transformed mean area under the paclitaxel concentration-cell survival percentage curve (AUC). There were no statistically significant differences among the 5 populations.
Genome-wide association study in Asian LCLs
Figure 2 illustrates the Manhattan plot of paclitaxel-induced cytotoxicity in LCLs derived from the Asian populations (JPT, CHB). There were 248 SNPs associated with paclitaxel cytotoxicity in the ASN population at a suggestive significance of P < 10−5; however among them only 1 SNP (rs56763443) and 2 SNPs (rs8049617 and rs7598550) were significant in the CEU and YRI GWAS results at P < 0.05, respectively. Three of the most significant 6 SNPs in the ASN GWAS of paclitaxel-induced cytotoxicity were located near (within 2 kb) gene AIPL1 (aryl-hydrocarbon-interacting protein-like 1). The three AIPL1 SNPs are in perfect LD (r2=1, D’=1 in ASN) and thus represent one signal. We evaluated whether any of these 6 SNPs were expression quantitative trait loci eQTLs with r2 ≥ 0.8 in two distinct databases, University of Chicago database (http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/) and GTEx Portal (http://www.gtexportal.org); however they were not eQTLs in any tissues listed (generic genome browser version 1.68). We also downloaded the expression data collected from ASN LCLs in Stranger et al. (34). We found no relationship between AIPL1 expression and paclitaxel cytotoxicity (n = 88, β = -0.012, P = 0.66) or AIPL1 expression and rs3892315 genotype (n = 90, β = -0.009, P = 0.52). Table 1 illustrates these 3 SNPs trending toward significance in LCLs derived from individuals of Northern/Western Europe (CEU; P < 0.065-0.078) but there was no significance in African LCLs (YRI; p = 0.305-0.355). Thus, the genetic variation near AIPL1 may contribute to paclitaxel-induced cytotoxicity particularly in Asians. Supplemental Table S2 lists 28 SNPs in or near AIPL1 and their association with sensory neuropathy in Asian patients; one SNP (rs7214517) has a p-value of 0.028. The 3 LCL SNPs (rs56763443, rs8049617 and rs7598550) were not genotyped in the Asian study.
Figure 2. Asian LCL GWAS results.
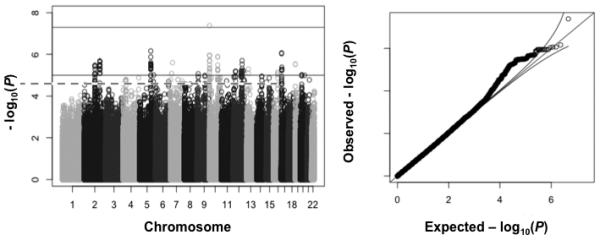
Paclitaxel-induced cytotoxicity in Asian LCLs. Top line in Manhattan plot is at the genome-wide significance threshold of P = 5×10−8 and below line is at the suggestive significance threshold of P = 10−5. Q-Q plots show some signal above the expected p-values (black lines represent the expected 95% confidence intervals) in the LCL GWAS.
Table 1.
SNPs significantly associated with paclitaxel-induced cytotoxicity in Asian LCLs (P < 10−6) and their level of significance in CEU LCLs and YRI LCLs.
| SNP | ASN | CEU | YRI | Host Gene |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Beta | P value | MAF | Beta |
P
value |
MAF | Beta |
P
value |
MAF | ||
|
| ||||||||||
| rs11595500 | −0.212 | 4.13E-08 | 0.167 | −0.036 | 0.463 | 0.346 | 0.171 | 0.149 | 0.058 | NA |
| rs264124 | 0.146 | 6.95E-07 | 0.343 | 0.020 | 0.694 | 0.463 | - | - | - | NA |
| rs3892315 | 0.207 | 8.72E-07 | 0.134 | 0.151 | 0.065 | 0.113 | −0.047 | 0.353 | 0.321 | AIPL1 |
| rs11651916 | 0.207 | 8.75E-07 | 0.134 | 0.144 | 0.078 | 0.113 | −0.050 | 0.307 | 0.317 | AIPL1 |
| rs3892316 | 0.206 | 9.23E-07 | 0.137 | 0.151 | 0.066 | 0.113 | −0.047 | 0.355 | 0.321 | AIPL1 |
Functional validation of AIPL1 using human iPSC-derived neurons
The effects of AIPL1 expression on paclitaxel-induced neurotoxicity was assessed in neurons derived from induced pluripotent stem cells (iPSC). To evaluate the effects of low AIPL1, expression on paclitaxel-induced neurotoxicity, we modulated the gene in neurons through transient siRNA transfection and verified with quantitative polymerase chain reaction (qPCR), a reduction of expression in AIPL1 to 62% and 14% relative to control at 24 and 48 hours post transfection relative to non-targeting control (Figure 3A). The modulation of expression of AIPL1 resulted in increased resistance of neurons to paclitaxel at 48 hours using two-way ANOVA analysis, as measured by significant increase in relative total outgrowth (Fig. 3B; P = 0.017), mean process length (Fig. 3C; P = 0.008) and relative number of processes (Fig. 3D; P = 0.029). Median process length (P = 0.005), max process length (P = 0.011), phenotypes related to mean process length and mean outgrowth intensity, a phenotype related to thickening of the neurite outgrowths (P= 0.0065) were also significantly different (data not shown). Figure 3E illustrates morphological changes for NTC and siAIPL1 treated cells for neurite outgrowth following 48 hours paclitaxel treatment of iPSC-derived neurons by high content imaging.
Figure 3. Decreased AIPL1 expression reduces sensitivity of human iPSC-derived neurons to paclitaxel.
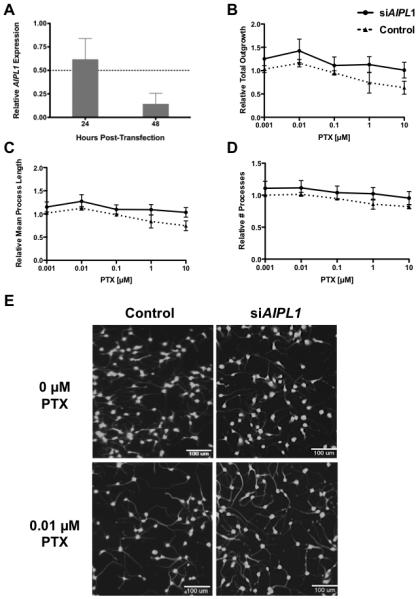
A) Relative AIPL1 expression 24 to 48 h post-transfection normalized to NTC. Neurite outgrowth phenotypes measured following 48 h paclitaxel exposure (0.001-10 µM) were tested for differences between siAIPL1 and NTC by two-way ANOVA including B) relative total neurite outgrowth (P = 0.017), C) relative mean process length (P = 0.008), and D) relative number of processes (P = 0.029). Error bars represent the standard error of the mean from three experiments of >2000 cells per dose. E) Representative high content images comparing human iPSC-derived neurons upon siRNA knockdown of AIPL1 and non-targeting control (NTC) 48 h post-treatment with 0.01 M paclitaxel. Images taken at 10× magnification, scale bar = 100 µm.
GWAS in patient samples of paclitaxel induced neuropathy
We performed GWAS of paclitaxel-induced neuropathy in 183 Asian cancer patients (Figure 4A). We identified 4 SNPs above a suggestive threshold of p<10−5 and no genome wide significant SNPs at p = 5×10−8. Gene based tests are commonly used following traditional SNP based analysis because they reduce multiple testing and provide genes as opposed to SNPs. Therefore, we evaluated LCL Asian GWAS (Figure 2), Asian cancer patient data (Figure 4A) and GWAS data from CALGB40101, a study of 855 Europeans evaluated for paclitaxel-induced neuropathy (9) using Sherlock, a program that enables gene-based sorting of targets by scoring alignment of genetic signatures of eQTLs and the phenotype (18). Comparison of results from Asian LCL cytotoxicity and Asian patient neuropathy at P < 0.05, had 32 overlap genes (Fig. 4B, Supplemental Table S3); comparison of Asian LCL cytotoxicity and European patient neuropathy at P < 0.05 had 37 overlap genes (Fig. 4B, Supplemental Table S4) and comparison of Asian and European patient GWAS cohorts at P < 0.05 had 10 overlap genes (Fig. 4B, Supplemental Table S5). Interestingly, PTPMT1 (protein tyrosine phosphatase, mitochondrial 1) overlapped all 3 analysis: Asian LCL cytotoxicity, Asian patient neuropathy, and European patient neuropathy at P < 0.05. Although we modulated gene expression of PTPMT1 in iCell neurons, resulting in 77 and 41% gene remaining after 24 and 48 hours, no significant difference was found in any of the neurite outgrowth phenotypes when compared to NTC (data not shown), indicating that some mechanism distinct from levels of gene expression may explain the role of PTPMT1 in paclitaxel-induced peripheral neuropathy. Alternatively, the effect of gene knockdown on other neuronal phenotypes such as electrical activity would not be captured in this system that is focused on morphological changes.
Figure 4. Asian patient GWAS results and LCL/patient overlap genes.
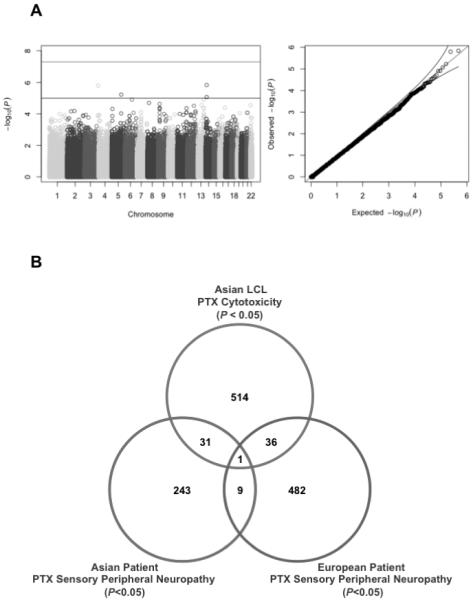
A) Paclitaxel-induced sensory peripheral neuropathy in Asian patients. Top line in Manhattan plot is at the genome-wide significance threshold of P = 5×10−8 and below line is at the suggestive significance threshold of P = 10−5. Q-Q plots show no signal above the expected p-values (black lines represent the expected 95% confidence intervals) in the patient GWAS. B) LCL/patient overlap genes associated with paclitaxel-induced phenotypes (P < 0.05, number: genes). Thirty-two genes were overlapped between Asian LCL cytotoxicity and Asian patient neuropathy (P < 0.05). PTPMT1 surpassed the threshold for association in all 3 analyses.
Functional validation of BCR and DDX54 using human iPSC-derived neurons
To prioritize the 32 Asian specific genes (Supplemental Table S3), we evaluated both their gene level expression in iPSC derived neurons (using RNA-Seq) and published literature using Pubmed search terms: gene name and CIPN or neuropathy or neurons done 2/2015 (Supplemental Figure 6). Breakpoint cluster region (Bcr) protein, highly expressed in iPSC derived neurons, is enriched in neurons and involved in neural activities and therefore was chosen for further studies (35-37). We found that a change in expression to 62% and 67% at 24 and 48 hours compared to NTC (Fig 5A), resulting in a decrease in the relative total outgrowth (P = 0.0097), number of processes (P = 0.0298), max process length (P = 0.0016) and number of branches (P = 0.0003) (Figs. 5B-D) after treatment with paclitaxel in three independent experiments. Ddx54 protein has been shown to play a critical role in central nervous system myelination, presumably in myelin sheath formation after the differentiation of oligodendrocytes (38). DDX54 (DEAD-box RNA helicase) was modulated to 66% and 61% at 24 and 48 hours, but resulted in no significant changes in any neurite phenotype measured including relative total outgrowth, number of processes, max process length and number of branches.
Figure 5. Decreased BCR expression reduces sensitivity of human iPSC-derived neurons to paclitaxel.
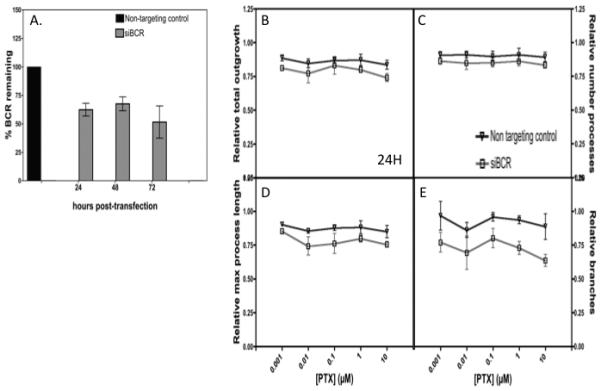
A) Relative BCR expression 24h post-transfection normalized to NTC. Neurite outgrowth phenotypes measured following 24 h paclitaxel exposure (0.001-10 µM) were tested for differences between siBCR and NTC by two-way ANOVA including relative B) total neurite outgrowth (P = 0.0097), C) number of processes (P = 0.0298) D) max process length (P = 0.0016), and E) number of branches (P = 0.0003). Error bars represent the standard error of the mean from three experiments of >2000 cells per dose.
Discussion
A comparison of paclitaxel-induced cytotoxicity in LCLs derived from patients of Asian, European and African ancestry demonstrates no difference in cellular sensitivity to paclitaxel. Traditional GWAS reveals only a small fraction (3 out of 248) SNPs suggestively significant in Asian LCLs for paclitaxel induced cytotoxicity are also significant in either CEU or YRI LCLs with the most significant genetic variant associations unique within populations. The top intragenic SNPs in the ASN GWAS of paclitaxel-induced cytotoxicity were located near AIPL1 gene. Upon knockdown of AIPL1 in human induced pluripotent stem cell derived neurons, decreased sensitivity to paclitaxel as measured by neurite outgrowth phenotypes. Using a gene based approach, a gene identified as Asian specific (overlapped in Asian LCLs and Asian clinical cohort of neuropathy) BCR (breakpoint cluster region), was validated using iPSC-derived neurons with knockdown corresponding to increased sensitivity to paclitaxel induced damage to neurons. The same approach identified PTPMT1 associated with paclitaxel-induced cytotoxicity in Asian LCLs, as well as Asian and European paclitaxel induced peripheral neuropathy clinical cohorts. By integrating gene-based analyses from clinical studies of paclitaxel-induced neuropathy in Asian and European patients with LCL results, population specific and across population target genes were identified.
The HapMap LCLs are advantageous for evaluating pharmacogenomics of anticancer agents because these samples have publicly available genotypes (39, 40) and sequencing data (20) for genome wide association studies. The samples also have gene expression (41), miRNA (42), modified cytosine (43) and protein data (44) that allow for the elucidation of potential function of the genetic variants associated with pharmacologic phenotypes. Our observation of a lack of population difference in paclitaxel-induced cytotoxicity is in contrast to population differences observed for platinating agents (45), and antimetabolites (46). In part, these population differences in drug sensitivity may be explained by population differences in gene expression (41). The role of gene expression in population differences in chemotherapeutic cytotoxicity is supported by the observation that pharmacologic SNPs for several different chemotherapeutic agents (etoposide, daunorubicin, cisplatin, carboplatin, cytarabine, capecitabine) are enriched in eQTLs or SNPs associated with gene expression (25). In contrast, pharmacologic SNPs associated with paclitaxel are not enriched in eQTLs; however enrichment in protein quantitative trait loci has been observed (47). These differences may be due to differences in mechanism of action of paclitaxel, a microtubule stabilizer compared to the other cytotoxic agents that target DNA.
In our Asian patient dataset, it appears that peripheral neuropathy is more common in females than males (males: 1 case/33 controls; females: 23 cases/88 controls); however, this gender difference is likely confounded by differences in cumulative dose, treatment schedule or age of the patients. Evidence suggests that age, race, and comorbid conditions, such as diabetes, and cumulative dose of drug administered may be associated with an increased risk of developing neuropathy (48).
In efforts to decrease multiple tests and identify genes instead of SNPs, we chose to utilize a gene-based approach to detect clinically significant genes with multiple trans eQTLs with moderate associations, which cannot be identified from the GWAS alone (18). We employed Sherlock to compare genes significant in two patient studies of paclitaxel induced neuropathy (855 European and 183 Japanese) and the LCL data we generated in the Asian populations. According to the gene-based test, 10 genes including PTPMT1 were found to overlap between Asians and Europeans along with hundreds of candidate gene associations that were unique to each population. At this threshold, PTPMT1 was revealed as a common target of paclitaxel-induced sensory peripheral neuropathy among populations. PTPMT1 is anchored in the inner mitochondrial membrane and is required for mitochondrial integrity and oxidative respiration (49). Because its ablation increased apoptosis in cancer cell lines and decreased differentiation and proliferation, but not survival in embryonic stem cells, its function may be cell-type specific in the regulation of cell growth and survival (50). Thus PTPMT1 in peripheral nerve mitochondria may play a critical role in paclitaxel-induced neurotoxicity across populations and is worth further studies. Our data indicates expression knockdown PTPMT1 in iPSC derived neurons did not alter sensitivity to paclitaxel as measured by morphological changes in neuronal outgrowths; however it is possible that the gene is important in electrical activity or other measures of neuronal damage.
Through LCL GWAS of paclitaxel-induced cytotoxicity, SNPs in AIPL1, were implicated as the most significant intragenic SNPs in the Asian LCLs. In evaluating SNPs within or close to AIPL1, rs7214517 was associated with Asian patient peripheral neuropathy (p = 0.028). AIPL1 expressed in rod and cone photoreceptors has a critical role in cell viability, and gene defects in AIPL1 cause heterogeneous degenerations of the human retina, such as retinitis pigmentosa (51). Interestingly, this disease is related to NARP syndrome (neuropathy, ataxia and retinitis pigmentosa) which is an inherited condition chiefly affecting nervous systems due to mitochondrial DNA dysfunction in creating adenosine triphosphate (52). Its worldwide prevalence is unknown and it remains unclear how this disruption in mitochondrial energy production leads to muscle weakness, vision loss and the other specific features of NARP syndrome. Kirschman et al. also found that transgenic expression of AIPL1 restored rod morphology and the rod-derived electroretinogram response in Aipl−/- mice, implicating a neuron component for this gene (53). Mitochondrial dysfunction has been implicated as a mechanism for neurotoxicity of paclitaxel (54) and our results demonstrated that decreased expression of AIPL1 significantly decreased sensitivity of neurons to paclitaxel, hence, there could be a cell-type specific function of AIPL1 in a mitochondrial component in peripheral neurons and AIPL1 could be a potential therapeutic target of paclitaxel-induced sensory peripheral neuropathy especially in Asians.
We identified a set of 32 Asian specific genes that overlapped in the Asian LCL and Asian patient datasets. One of those genes, BCR, was functionally validated in the iPSC derived neuronal model with increased sensitivity of neurons to paclitaxel upon knockdown of the gene. Bcr has been shown in rat PC12 cells to cooperate with Fes to activate Rho family GTPases as part of a novel pathway regulating neurite extension (55). Furthermore, Huntington’s-associated protein, 1A interacts with Bcr on microtubules to regulate neuronal differentiation (56). Increasingly, Bcr is emerging as an important regulator of excitatory synapse receptor expression and development. Our data indicates that BCR contributes to the protection of neurons from paclitaxel induced damage as measured by several neuronal phenotypes including total outgrowth of neurites.
Although CIPN is a common, potentially severe and dose-limiting adverse effect of many commonly used cancer therapeutics including paclitaxel, there are no effective genetic markers or treatment for this side effect (4, 7, 8). Therefore, to identify genetic variants and genes associated with CIPN, several large clinical GWAS have been conducted and revealed a few promising associations (9, 10, 14). However, there are some considerable issues in clinical GWAS of CIPN including: 1) lack of replication studies; 2) pharmacogenomics underlying ethnic specificity and; 3) lack of appropriate models to functionally validate important GWAS results. Therefore, combining cell-based data with multiple clinical genome wide association studies, as done in this study, is a way to identify the most promising genes. Furthermore, we utilized a novel technology, human iPSC-derived neurons for gene functional validation. The elucidation of variants specific to the Asian population may be helpful in developing personalized cancer treatments.
Supplementary Material
Statement of Translational Relevance.
Chemotherapy-induced peripheral neuropathy (CIPN) causes a significant symptom burden because many patients that undergo adjuvant treatment are long-term survivors. Moreover, CIPN can lead to dose reduction of the chemotherapeutic agent or possible cessation of treatment. This may have an adverse impact on cancer treatment and disease outcomes. In this study, we have used a genome wide approach in a clinical and preclinical setting to identify genetic variants associated with peripheral neuropathy in Asians. The findings can be used in a clinical setting to dose reduce patients at risk or alter therapy to avoid this devastating side effect. In addition, an improved understanding of the pathophysiology of paclitaxel-induced neuropathy in and across populations will assist in the development of future neuroprotective agents and in the design of novel therapies with improved toxicity profiles.
Acknowledgements
The authors thank Dr. R. Stephanie Huang, Bonnie LaCroix and Jamie Meyers of the Pharmacogenomics of Anticancer Agents Research LCL core for maintaining, distributing and authenticating LCLs. The authors also appreciate the imaging guidance provided by Sam Bettis, Siquan Chen and Elise Fletcher of the University of Chicago Cellular Screening Center.
Financial Support: This work is supported by the NIH/NIGMS Pharmacogenomics of Anticancer Agents Research Grant U01GM61393 (M.E.D.), NIH/NCI National Research Service Award F32CA165823 (H.E.W.), University of Chicago Breast Cancer SPORE Grant NCI P50 CA125183 (M.E.D.), NIH/NCI R01 CA136765 (M.E.D.), and NIH/NIGMS Pharmacogenomics of Membrane Transporters Grant U01GM61390. The Biobank Japan Project funded by the Japanese Ministry of Education, Culture, Sports, Science and Technology. This work is part of the NIH Pharmacogenomics Research Network-RIKEN Center for Genomic Medicine Global Alliance.
Footnotes
Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Crown J, O'Leary M. The taxanes: an update. Lancet. 2000;355:1176–8. doi: 10.1016/S0140-6736(00)02074-2. [DOI] [PubMed] [Google Scholar]
- 2.Jones SE, Erban J, Overmoyer B, Budd GT, Hutchins L, Lower E, et al. Randomized phase III study of docetaxel compared with paclitaxel in metastatic breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:5542–51. doi: 10.1200/JCO.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 3.Winer EP, Berry DA, Woolf S, Duggan D, Kornblith A, Harris LN, et al. Failure of higher-dose paclitaxel to improve outcome in patients with metastatic breast cancer: cancer and leukemia group B trial 9342. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004;22:2061–8. doi: 10.1200/JCO.2004.08.048. [DOI] [PubMed] [Google Scholar]
- 4.Cavaletti G, Alberti P, Marmiroli P. Chemotherapy-induced peripheral neurotoxicity in the era of pharmacogenomics. The Lancet Oncology. 2011;12:1151–61. doi: 10.1016/S1470-2045(11)70131-0. [DOI] [PubMed] [Google Scholar]
- 5.Loprinzi CL, Reeves BN, Dakhil SR, Sloan JA, Wolf SL, Burger KN, et al. Natural history of paclitaxel-associated acute pain syndrome: prospective cohort study NCCTG N08C1. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:1472–8. doi: 10.1200/JCO.2010.33.0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pachman DR, Barton DL, Watson JC, Loprinzi CL. Chemotherapy-induced peripheral neuropathy: prevention and treatment. Clinical pharmacology and therapeutics. 2011;90:377–87. doi: 10.1038/clpt.2011.115. [DOI] [PubMed] [Google Scholar]
- 7.Hershman DL, Lacchetti C, Dworkin RH, Lavoie Smith EM, Bleeker J, Cavaletti G, et al. Prevention and management of chemotherapy-induced peripheral neuropathy in survivors of adult cancers: American Society of Clinical Oncology clinical practice guideline. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:1941–67. doi: 10.1200/JCO.2013.54.0914. [DOI] [PubMed] [Google Scholar]
- 8.Smith EM, Pang H, Cirrincione C, Fleishman S, Paskett ED, Ahles T, et al. Effect of duloxetine on pain, function, and quality of life among patients with chemotherapy-induced painful peripheral neuropathy: a randomized clinical trial. Jama. 2013;309:1359–67. doi: 10.1001/jama.2013.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldwin RM, Owzar K, Zembutsu H, Chhibber A, Kubo M, Jiang C, et al. A genome-wide association study identifies novel loci for paclitaxel-induced sensory peripheral neuropathy in CALGB 40101. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:5099–109. doi: 10.1158/1078-0432.CCR-12-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leandro-Garcia LJ, Inglada-Perez L, Pita G, Hjerpe E, Leskela S, Jara C, et al. Genome-wide association study identifies ephrin type A receptors implicated in paclitaxel induced peripheral sensory neuropathy. Journal of medical genetics. 2013;50:599–605. doi: 10.1136/jmedgenet-2012-101466. [DOI] [PubMed] [Google Scholar]
- 11.Wheeler HE, Maitland ML, Dolan ME, Cox NJ, Ratain MJ. Cancer pharmacogenomics: strategies and challenges. Nature reviews Genetics. 2013;14:23–34. doi: 10.1038/nrg3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Low SK, Takahashi A, Mushiroda T, Kubo M. Genome-wide association study: a useful tool to identify common genetic variants associated with drug toxicity and efficacy in cancer pharmacogenomics. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:2541–52. doi: 10.1158/1078-0432.CCR-13-2755. [DOI] [PubMed] [Google Scholar]
- 13.Cox NJ, Gamazon ER, Wheeler HE, Dolan ME. Clinical translation of cell-based pharmacogenomic discovery. Clinical pharmacology and therapeutics. 2012;92:425–7. doi: 10.1038/clpt.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheeler HE, Gamazon ER, Wing C, Njiaju UO, Njoku C, Baldwin RM, et al. Integration of cell line and clinical trial genome-wide analyses supports a polygenic architecture of Paclitaxel-induced sensory peripheral neuropathy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:491–9. doi: 10.1158/1078-0432.CCR-12-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wheeler HE, Gonzalez-Neira A, Pita G, de la Torre-Montero JC, Alonso R, Lopez-Fernandez LA, et al. Identification of genetic variants associated with capecitabine-induced hand-foot syndrome through integration of patient and cell line genomic analyses. Pharmacogenetics and genomics. 2014;24:231–7. doi: 10.1097/FPC.0000000000000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider BPLL, Miller K, Flockhart DA, Radovich M, Hancock BA, et al. Genetic associations with taxane-induced neuropathy by a genome-wide association study in E5103. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(Suppl):1000. [Google Scholar]
- 17.O'Donnell PH, Dolan ME. Cancer pharmacoethnicity: ethnic differences in susceptibility to the effects of chemotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:4806–14. doi: 10.1158/1078-0432.CCR-09-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He X, Fuller CK, Song Y, Meng Q, Zhang B, Yang X, et al. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. American journal of human genetics. 2013;92:667–80. doi: 10.1016/j.ajhg.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Njiaju UO, Gamazon ER, Gorsic LK, Delaney SM, Wheeler HE, Im HK, et al. Whole-genome studies identify solute carrier transporters in cellular susceptibility to paclitaxel. Pharmacogenetics and genomics. 2012;22:498–507. doi: 10.1097/FPC.0b013e328352f436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. American journal of human genetics. 2007;81:1084–97. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nature genetics. 2012;44:821–4. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Low SK, Chung S, Takahashi A, Zembutsu H, Mushiroda T, Kubo M, et al. Genome-wide association study of chemotherapeutic agent-induced severe neutropenia/leucopenia for patients in Biobank Japan. Cancer science. 2013;104:1074–82. doi: 10.1111/cas.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO, Nicolae DL, et al. SCAN: SNP. and copy number annotation. Bioinformatics. 2010;26:259–62. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gamazon ER, Huang RS, Cox NJ, Dolan ME. Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9287–92. doi: 10.1073/pnas.1001827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamaguchi-Kabata Y, Nakazono K, Takahashi A, Saito S, Hosono N, Kubo M, et al. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. American journal of human genetics. 2008;83:445–56. doi: 10.1016/j.ajhg.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duan S, Huang RS, Zhang W, Bleibel WK, Roe CA, Clark TA, et al. Genetic architecture of transcript-level variation in humans. American journal of human genetics. 2008;82:1101–13. doi: 10.1016/j.ajhg.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wheeler HE, Wing C, Delaney SM, Komatsu M, Dolan ME. Modeling chemotherapeutic neurotoxicity with human induced pluripotent stem cell-derived neuronal cells. PloS one. 2015;10:e0118020. doi: 10.1371/journal.pone.0118020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diouf B, Crews KR, Lew G, Pei D, Cheng C, Bao J, et al. Association of an inherited genetic variant with vincristine-related peripheral neuropathy in children with acute lymphoblastic leukemia. Jama. 2015;313:815–23. doi: 10.1001/jama.2015.0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goecks J, Nekrutenko A, Taylor J, Galaxy T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, et al. Population genomics of human gene expression. Nature genetics. 2007;39:1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oh D, Han S, Seo J, Lee JR, Choi J, Groffen J, et al. Regulation of synaptic Rac1 activity, long-term potentiation maintenance, and learning and memory by BCR and ABR Rac GTPase-activating proteins. J Neurosci. 2010;30:14134–44. doi: 10.1523/JNEUROSCI.1711-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaartinen V, Nagy A, Gonzalez-Gomez I, Groffen J, Heisterkamp N. Vestibular dysgenesis in mice lacking Abr and Bcr Cdc42/RacGAPs. Dev Dyn. 2002;223:517–25. doi: 10.1002/dvdy.10071. [DOI] [PubMed] [Google Scholar]
- 37.Park AR, Oh D, Lim SH, Choi J, Moon J, Yu DY, et al. Regulation of dendritic arborization by BCR Rac1 GTPase-activating protein, a substrate of PTPRT. J Cell Sci. 2012;125:4518–31. doi: 10.1242/jcs.105502. [DOI] [PubMed] [Google Scholar]
- 38.Zhan R, Yamamoto M, Ueki T, Yoshioka N, Tanaka K, Morisaki H, et al. A DEAD-box RNA helicase Ddx54 protein in oligodendrocytes is indispensable for myelination in the central nervous system. J Neurosci Res. 2013;91:335–48. doi: 10.1002/jnr.23162. [DOI] [PubMed] [Google Scholar]
- 39.The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 40.Pastinen T, Ge B, Gurd S, Gaudin T, Dore C, Lemire M, et al. Mapping common regulatory variants to human haplotypes. Human molecular genetics. 2005;14:3963–71. doi: 10.1093/hmg/ddi420. [DOI] [PubMed] [Google Scholar]
- 41.Zhang W, Duan S, Kistner EO, Bleibel WK, Huang RS, Clark TA, et al. Evaluation of genetic variation contributing to differences in gene expression between populations. American journal of human genetics. 2008;82:631–40. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang RS, Gamazon ER, Ziliak D, Wen Y, Im HK, Zhang W, et al. Population differences in microRNA expression and biological implications. RNA biology. 2011;8:692–701. doi: 10.4161/rna.8.4.16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moen EL, Zhang X, Mu W, Delaney SM, Wing C, McQuade J, et al. Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics. 2013;194:987–96. doi: 10.1534/genetics.113.151381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hause RJ, Stark AL, Antao NN, Gorsic LK, Chung SH, Brown CD, et al. Identification and validation of genetic variants that influence transcription factor and cell signaling protein levels. American journal of human genetics. 2014;95:194–208. doi: 10.1016/j.ajhg.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang RS, Kistner EO, Bleibel WK, Shukla SJ, Dolan ME. Effect of population and gender on chemotherapeutic agent-induced cytotoxicity. Molecular cancer therapeutics. 2007;6:31–6. doi: 10.1158/1535-7163.MCT-06-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartford CM, Duan S, Delaney SM, Mi S, Kistner EO, Lamba JK, et al. Population-specific genetic variants important in susceptibility to cytarabine arabinoside cytotoxicity. Blood. 2009;113:2145–53. doi: 10.1182/blood-2008-05-154302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stark AL, Hause RJ, Jr., Gorsic LK, Antao NN, Wong SS, Chung SH, et al. Protein quantitative trait loci identify novel candidates modulating cellular response to chemotherapy. PLoS genetics. 2014;10:e1004192. doi: 10.1371/journal.pgen.1004192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rivera E, Cianfrocca M. Overview of neuropathy associated with taxanes for the treatment of metastatic breast cancer. Cancer Chemother Pharmacol. 2015;75:659–70. doi: 10.1007/s00280-014-2607-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pulido R, Stoker AW, Hendriks WJ. PTPs emerge as PIPs: protein tyrosine phosphatases with lipid-phosphatase activities in human disease. Human molecular genetics. 2013;22:R66–76. doi: 10.1093/hmg/ddt347. [DOI] [PubMed] [Google Scholar]
- 50.Niemi NM, Lanning NJ, Westrate LM, MacKeigan JP. Downregulation of the mitochondrial phosphatase PTPMT1 is sufficient to promote cancer cell death. PloS one. 2013;8:e53803. doi: 10.1371/journal.pone.0053803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan MH, Mackay DS, Cowing J, Tran HV, Smith AJ, Wright GA, et al. Leber congenital amaurosis associated with AIPL1: challenges in ascribing disease causation, clinical findings, and implications for gene therapy. PloS one. 2012;7:e32330. doi: 10.1371/journal.pone.0032330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duno M, Wibrand F, Baggesen K, Rosenberg T, Kjaer N, Frederiksen AL. A novel mitochondrial mutation m.8989G>C associated with neuropathy, ataxia, retinitis pigmentosa - the NARP syndrome. Gene. 2013;515:372–5. doi: 10.1016/j.gene.2012.12.066. [DOI] [PubMed] [Google Scholar]
- 53.Kirschman LT, Kolandaivelu S, Frederick JM, Dang L, Goldberg AF, Baehr W, et al. The Leber congenital amaurosis protein, AIPL1, is needed for the viability and functioning of cone photoreceptor cells. Human molecular genetics. 2010;19:1076–87. doi: 10.1093/hmg/ddp571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LaPointe NE, Morfini G, Brady ST, Feinstein SC, Wilson L, Jordan MA. Effects of eribulin, vincristine, paclitaxel and ixabepilone on fast axonal transport and kinesin-1 driven microtubule gliding: implications for chemotherapy-induced peripheral neuropathy. Neurotoxicology. 2013;37:231–9. doi: 10.1016/j.neuro.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laurent CE, Smithgall TE. The c-Fes tyrosine kinase cooperates with the breakpoint cluster region protein (Bcr) to induce neurite extension in a Rac- and Cdc42-dependent manner. Exp Cell Res. 2004;299:188–98. doi: 10.1016/j.yexcr.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 56.Huang PT, Chen CH, Hsu IU, Salim SA, Kao SH, Cheng CW, et al. Huntingtin-associated protein 1 interacts with breakpoint cluster region protein to regulate neuronal differentiation. PloS one. 2015;10:e0116372. doi: 10.1371/journal.pone.0116372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.