Abstract
Objective
MicroRNAs (miRNAs) are small non-coding RNAs that post-transcriptionally regulate gene expression and serve as potential mediators and markers of disease. Recently, plasma miR-24-3p and miR-125a-5p concentrations were shown to be elevated in rheumatoid arthritis (RA) and useful for RA diagnosis. We assessed the utility of seven candidate plasma miRNAs, selected for biological relevance, for RA diagnosis and use as markers of disease activity and subclinical atherosclerosis in RA.
Methods
The cross-sectional study included 168 patients with RA and 91 control subjects, of similar age, race and sex. Plasma concentrations of miR-15a-5p, miR-24-3p, miR-26a-5p, miR-125a-5p, miR-146a-5p, miR-155-5p, and miR-223-3p were measured by quantitative PCR. Utility of plasma miRNA concentrations for RA diagnosis was assessed by area under the receiver operating characteristic curve (AUROC). Association between plasma miRNA concentrations and RA disease activity and coronary artery calcium score were assessed by Spearman correlations.
Results
Plasma concentrations of miR-15a-5p, 24-3p, miR-26a-5p, miR-125a-5p, miR-146a-5p, miR-155-5p, and miR-223-3p were significantly increased in patients with RA. miR-24-3p had the highest AUROC for diagnosis of RA (AUROC=0.725), including among rheumatoid factor negative patients (AUROC=0.772). Among all patients with RA, the combination of miR-24-3p, miR-26a-5p and miR-125a-5p improved the model modestly (AUROC=0.747). miR-155-5p was weakly inversely associated with swollen joint count (P=0.024), but no other miRNAs were associated with disease activity or coronary artery calcium score.
Conclusion
The combination of miR-24-3p, miR-26a-5p and miR-125a-5p had strongest diagnostic accuracy for RA. Candidate miRNAs had little or no association with RA disease activity or subclinical atherosclerosis.
Keywords: rheumatoid arthritis, microRNA, atherosclerosis, disease activity, diagnosis, cardiovascular risk
MicroRNAs (miRNAs) are small (approximately 22 nucleotides in length) non-coding RNAs that bind to 3′ untranslated regions of messenger RNA (mRNA) and cause destabilization of transcription machinery or transcript degradation (1-3). Each miRNA has the capacity to bind to many different mRNA transcripts, thus potentially affecting multiple distinct pathways. miRNAs are differentially expressed within many cell types and are stable within extracellular fluids, such as plasma, where they are protected within exosomes (4),microvesicles (5), lipoproteins (6), and Argonaute2 protein complexes (7). The association of miRNAs with lipid and protein carriers in plasma confers extraordinary stability (8, 9). Their stability in plasma and known biological function within cells make them highly relevant to disease mechanisms and ideal candidates for diagnostic tests, disease biomarkers, and therapeutic targets.
Circulating miRNAs have great potential as biomarkers in patients with rheumatoid arthritis (RA) to aid in diagnosis and disease activity monitoring, as well as prediction of cardiovascular risk, which is the main cause of early death in patients with RA (10, 11). Currently, there is little information regarding plasma miRNAs in RA. More is known about cellular miRNA alterations and how these affect important cellular functions in RA. Examples include the role of miR-15a-5p in cellular apoptosis (12, 13), miR-24-3p in maintenance of autoreactive B cells (14), miR-125a-5p and miR-223-3p in macrophage cytokine production (15), and miR-26a-5p, miR-146a-5p and miR-155-5p in IL-17 producing T cells (16). Alterations of several of these miRNAs, such as miR-146a-5p and miR-155-5p, have been suggested to also play a role in the development of cardiovascular disease in RA (17).
Recently, miR-24-3p, and miR125a-5p were reported to have diagnostic potential for RA (18). We hypothesized that these two miRNAs, along with miR-15a-5p, miR-26a-5p, miR-146a-5p, miR-155-5p, and miR-223-3p, which as described above have biologically relevant roles in RA, may be useful biomarkers for RA diagnosis and disease activity. Since many of the miRNAs with a role in the pathogenesis of RA (19) overlap with those implicated in the development of atherosclerosis (20, 21), we also hypothesized that these miRNAs may also serve as biomarkers of subclinical atherosclerosis in patients with RA.
METHODS
Study population
This was a cross-sectional study that included 168 patients with RA and 91 control subjects. This cohort is part of a group of patients extensively characterized for cardiovascular risk (22). Recruitment and study procedures have previously been described (22). All subjects were older than 18 years of age and patients with RA fulfilled American College of Rheumatology 1987 classification criteria for RA (23). RA and control groups were frequency-matched for age, race and sex and control subjects did not have RA or other inflammatory disease. The study was approved by the Vanderbilt Institutional Review Board and all subjects gave written informed consent.
Clinical and laboratory data
Clinical information and laboratory measurements were obtained as previously described (22). Disease activity of RA was determined by the 28 joint count disease activity score (DAS28) using erythrocyte sedimentation rate (ESR) as the marker of inflammation (24). Body mass index (BMI) was calculated and expressed as kg/m2. Framingham risk score for 10-year percent risk of a cardiovascular event was determined as previously described (25, 26).
Fasting cholesterol panel, ESR and high-sensitivity C-reactive protein (CRP) were measured by the Vanderbilt University Medical Center Clinical Laboratory or by ELISA (Millipore, St. Charles, MO, USA). The degree of insulin resistance was calculated by the homeostatic model assessment of insulin resistance (HOMA) (fasting glucose [mmol/L] × fasting insulin [μU/ml]/22.5]) (27). Coronary artery calcium score was measured by electron beam computed tomography with an Imatron C-150 scanner (GE/Imatron, South San Francisco, CA, USA) as previously described (22) and quantified in Agatston units (28).
RNA isolation, cDNA synthesis, and quantitative PCR
Plasma (collected in EDTA-containing tubes) from each subject was stored at −80°C. RNA was extracted from 250 μl of plasma using a total RNA purification kit, in 96-well format (Norgen, Ontario, Canada). All plasma RNA samples were extracted at the same time. RNA was collected in a total volume of 100 μl in sequential 50 μl elutions. A volume of 5 μl of RNA from each subject was polyadenylated and used for first strand complementary DNA (cDNA) synthesis (Quanta Biosciences, Gaithersburg, MD , USA). The cDNA was diluted 1:20 in TE buffer and stored at −20°C.
All miRNA assays (miR-15a-5p, 24-3p, miR-26a-5p, miR-125-5p, miR-146a-5p, miR-155-5p, and miR-223-3p) used 5 μl diluted cDNA in 20 μl final reaction volume (Quanta Biosciences, Gaithersburg, MD , USA) in 96 well format on Bio-Rad CFX96 instruments. Cycling conditions consisted of a 3 minute incubation at 95°C; 50 cycles of amplification at 95°C for 10 seconds then 60°C for 30 seconds; 95°C for 1 minute; 55°C for 1 minute, followed by an 80 step melt curve in 0.5°C increments from 55°C to 95°C. Technical replicates were performed in triplicate. Proportional numbers of RA and control cDNA samples, a no cDNA template negative control, and a four-point, 10-fold dilution standard of whole blood cDNA were included on each of 10 PCR plates for each miRNA assay. Quality control results for the miRNA assays are shown in Supplemental Table 1.
Quantitative PCR (qPCR) miRNA data were converted to molar concentrations using 1 zeptomole to 1 attomole serial dilution of qPCR miRNA mimics. Single-stranded DNA mimics were synthesized for miR-15a-5p, miR-24-3p, and miR-223-3p, each of which included the universal PCR primer sequence, a two base dA spacer, and the reverse complement of the miRNA assay primer sequence (IDT, Coral City, IA, USA). Quality control results for the mimic standard dilutions are shown in Supplemental Table 2, with nearly identical results for each. A composite curve including the three mimic dilution standards was used to convert qPCR data to molar amounts for each miRNA assay. All assays and analyses were done using applicable minimum information for publication of qPCR experiment (MIQE) guidelines (29).
Statistical methods
Descriptive statistics were calculated as mean ± standard deviation for continuous variables, and frequency and proportions for categorical variables. Wilcoxon’s rank sum tests were used to compare continuous variables and Pearson’s chi-square test to compare categorical variables.
To determine the utility of plasma miRNAs for diagnosis of RA, receiver operating characteristic (ROC) curves were plotted and area under the curve (AUROC) was calculated for each miRNA. For combinations of miRNAs, logistic regression was used with the dependent variable as disease status (RA versus control), and the independent variable as the miRNA of interest. The predicted probability for RA diagnosis determined from the logistic regression was plotted in a ROC curve and AUROC was calculated. Spearman correlation coefficients for continuous variables and Wilcoxon’s rank sum tests for categorical variables were used to determine the association between plasma miRNA concentrations and variables of interest pertaining to RA disease activity and cardiovascular risk.
Statistical analyses were performed using IBM SPSS Statistics v22. Two-sided P values less than or equal to 0.05 were considered statistically significant.
RESULTS
Clinical characteristics
As previously described (22, 30), clinical characteristics of RA patients and control subjects are presented in Table 1. Patients and controls were of similar age, race and sex. Patients with RA had moderate disease activity with a mean DAS28 score of 3.79, and higher CRP than controls P<0.001 (Table 1). The majority of patients with RA were rheumatoid factor positive (69%), and taking methotrexate (71%) and prednisone (54%) (Table 1). Patients with RA had greater waist circumference, and lower LDL-C than control subjects (Table 1).
Table 1.
Patient characteristics
| RA (N=168) | Control (N=91) | P value | |
|---|---|---|---|
| Demographics | |||
| Age, years | 54 ± 12 | 53 ± 11 | 0.30 |
| Race, no. (%) Caucasian | 149 (89) | 77 (85) | 0.48 |
| Sex, no. (%) female | 116 (69) | 57 (63) | 0.30 |
| RA related | |||
| DAS28, units | 3.79 ± 1.61 | - | - |
| CRP, mg/L | 8.9 ± 13.1 | 2 ± 4.2 | <0.001 |
| ESR, mm/hr | 22 ± 20 | - | - |
| Disease duration, years | 10 ± 11 | - | |
| RF, no. (%) positive (N=161) | 116 (69) | - | - |
| Methotrexate use, no. (%) | 120 (71) | - | - |
| Anti-TNFα use, no. (%) | 35 (21) | - | - |
| Prednisone use, no. (%) | 91 (54) | - | - |
| CV related | |||
| Systolic BP, mmHg | 133 ± 20 | 129 ± 17 | 0.08 |
| Diastolic BP, mmHg | 75 ± 11 | 73 ± 9 | 0.12 |
| Waist circumference, cm | 95.6 ± 17.3 | 89.9 ± 14.4 | 0.01 |
| Total-C, mg/dl | 184 ± 40 | 193 ± 35 | 0.12 |
| HDL-C, mg/dl | 47 ± 14 | 47 ± 13 | 0.67 |
| LDL-C, mg/dl | 113 ± 34 | 122 ± 31 | 0.03 |
| Triglycerides, mg/dl | 145 ± 179 | 119 ± 62 | 0.26 |
Presented as mean ± standard deviation or number (percent). DAS28=disease activity score based on 28 joints, CRP=high sensitivity C-reactive protein, ESR=erythrocyte sedimentation rate, RF= rheumatoid factor (available in 161 RA patients), anti-TNFα= anti-tumor necrosis factor alpha, BP=blood pressure, C= cholesterol.
Diagnostic capacity of candidate miRNAs for rheumatoid arthritis
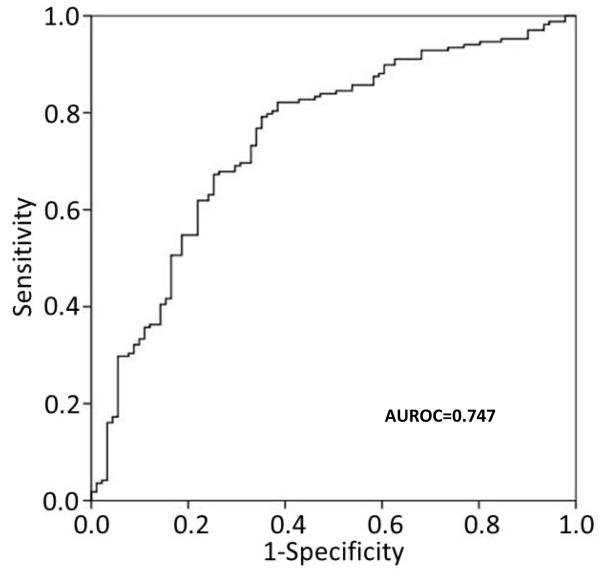
Patients with RA had higher plasma concentrations of miR-15a-5p (2.7 fold, P<0.001), miR-24-3p (3.5 fold, P<0.001), miR-26a-5p (1.8 fold, P<0.001), miR-125a-5p (1.2 fold, P=0.003), miR-146a-5p (2.5 fold, P<0.001), miR-155-5p (2.7 fold, P<0.001), and miR-223-3p (2.5 fold, P<0.001) compared to control subjects (Table 2). The AUROC for RA diagnosis using individual miRNAs ranged from 0.610 to 0.725, with miR-24-3p having the highest AUROC (Table 2). The combination of miR-24-3p, miR-26a-5p and miR-125a-5p had the highest AUROC for RA diagnosis (AUROC=0.747, 95% CI: 0.683-0.810) (Figure 1). The combination of miR-24-3p and miR-125a-5p, as previously reported (18), had a similar AUROC for RA diagnosis (AUROC=0.732, 95% CI: 0.669-0.796).
Table 2.
Candidate plasma miRNA concentrations in RA vs control plasma and area under the receiver operating characteristic curve for RA diagnosis
| Candidate miRNA |
miRNA picomoles/ml plasma (Mean ± SD) |
Fold difference |
P value | AUROC | |
|---|---|---|---|---|---|
| RA | Control | ||||
| miR-15a-5p | 11.85 ± 14.42 | 4.35± 7.89 | 2.7 | <0.001 | 0.716 (0.651-0.780) |
| miR-24-3p | 44.88± 82.51 | 12.97 ± 35.99 | 3.5 | <0.001 | 0.725 (0.661-0.788) |
| miR-26a-5p | 4.90 ± 5.64 | 2.67 ± 4.97 | 1.8 | <0.001 | 0.690 (0.621-0.759) |
| miR-125a-5p | 2.21 ± 1.62 | 1.77 ± 1.50 | 1.2 | 0.003 | 0.610 (0.535-0.685) |
| miR-146a-5p | 23.21 ± 41.59 | 9.38 ± 30.62 | 2.5 | <0.001 | 0.666 (0.599-0.732) |
| miR-155-5p | 0.038 ± 0.051 | 0.014 ± 0.032 | 2.7 | <0.001 | 0.702 (0.635-0.769) |
| miR-223-3p | 195.26 ± 290.52 | 78.78 ± 173.91 | 2.5 | <0.001 | 0.666 (0.598-0.734) |
miRNA concentrations presented as mean ± standard deviation. AUROC= area under the receiver operating characteristic curve, presented as AUROC (95% confidence interval).
Figure. Predictive capacity of the combination of miR-24-3p, miR-26a-5p and miR-125a-5p for RA diagnosis.
Receiver operating characteristic curve was made using the combination of plasma concentrations of miR-24-3p, miR-26a-5p and miR-125a-5p. The area under the curve (AUROC) = 0.747 for diagnosis of RA with this combination of miRNAs.
Comparing patients with RA who were rheumatoid factor negative to control subjects, all miRNAs were significantly elevated (all P<0.003) except for miR-125a-5p (Supplementary Table 3). Moreover, miR-24-3p maintained the highest AUROC for RA diagnosis (AUROC=0.772, 95% CI: 0.688-0.857) (Supplementary Table 3, and Supplementary Figure), though the combination of miR-24-3p, miR-26a-5p, and miR-125a-5p was similar (AUROC=0.764, 95% CI: 0.680-0.848).
miRNA expression and relationship to disease-related factors and medications
We evaluated the utility of the seven candidate miRNAs for use as biomarkers of disease activity. Only miR-155-5p was weakly inversely correlated with swollen joint count in patients with RA (Rho=-0.174, P=0.02), and no other miRNAs were correlated with DAS28 score, tender joint count, global health score, ESR, or CRP (all P > 0.08) (Supplementary Table 4). Moreover, medication use including methotrexate, leflunomide, hydroxychloroquine, anti-TNFα agents, corticosteroids, and non-steroidal anti-inflammatories were not associated with altered concentrations of any of the seven miRNAs evaluated (all P>0.1).
miRNA expression and relationship to subclinical atherosclerosis and CV risk factors
We next determined if these seven miRNAs were associated with cardiovascular risk factors in patients with RA. None of the seven miRNAs were associated with coronary artery calcium score in RA (all P>0.1) (Supplementary Table 5). miR-26a-5p was weakly associated with Framingham risk score (Rho=0.150, P=0.05). miR-125a-5p was weakly associated with systolic blood pressure (rho=0.198, P=0.01), but none with the diagnosis of hypertension (all P>0.1). miR-15a-5p and miR24-3p were modestly associated with waist circumference (Rho=0.185, P=0.02 for miR-15a-5p; and Rho=0.208, P=0.01 for miR-24-3p) (Supplementary Table 5). None of the candidate miRNAs were associated with traditional lipid profile (total cholesterol, LDL-C, HDL-C, or triglycerides), smoking status, degree of insulin resistance (HOMA) or diabetes in patients with RA (all P>0.05).
DISCUSSION
We found that miR-15a-5p, miR-24-3p, miR-26a-5p, miR-125a-5p, miR-146a-5p, miR-155-5p, and miR-223-3p concentrations were elevated in the plasma of patients with RA. The combination of miR-24-3p, miR-26a-5p and miR-125a-5p performed best to differentiate between patients with RA and control subjects, though the performance of the other miRNAs did not differ meaningfully from these. Among rheumatoid factor negative patients, miR-24-3p was a strong marker of RA. Among the miRNAs tested, only miR-155-5p was weakly inversely associated with RA disease activity (swollen joint count). Moreover, there was little to no association between the plasma concentrations of the candidate miRNAs and subclinical atherosclerosis or cardiovascular risk factors.
Our findings are consistent with a recent study by Murata et al in which concentrations of miR-24-3p and miR-125a-5p had the best diagnostic utility for RA, and miR-26a-5p was also helpful (18). However, we found that miR-24-3p rather than miR-125a-5p or a combination of miRNAs was a better diagnostic tool in seronegative RA, which differs slightly from the previous study which found miR-125a-5p or the combination of miR-24-3p and miR-125a-5p was more accurate than miR-24-3p alone in seronegative (anti-citrullinated peptide antibody) RA (18). Comparing the current study with Murata et al (18), the fold increase comparing RA to control subjects in miR-24-3p was 3.5 in the current study and 3.75 in the prior study; the fold change in miR-26a-5p was 1.8 in the current study and 2.85 in the prior study; and the fold change in miR-125a-5p was 1.2 in the current study and 2.7 in the prior study. In the same study, plasma concentrations of miR-125a-5p did not correlate with indices of inflammation or disease activity, consistent with our findings. In contrast to our results, miR-24-3p was positively correlated with some indices of disease activity including CRP, global health, and DAS28, but not ESR or tender or swollen joint count (18). The previous study (18) selected initial candidate miRNAs from profiling results of a very small number of subjects (3 patients with RA and 3 control subjects); thus, important miRNAs may not have been identified and studied. Therefore, we examined miRNAs that were most strongly associated with RA in the previous study and also selected additional candidate miRNAs that have been identified as having biological relevance to RA.
Two smaller studies compared selected circulating miRNAs in patients with RA and control subjects. One included 30 patients with RA, 30 patients with osteoarthritis, and 30 healthy control subjects in a study measuring plasma miRNAs (8), and the second included 34 patients with treatment-naïve early RA, 28 patients with late RA, and 16 healthy control subjects in a study measuring serum miRNAs (31). Both studies found that miR-146a-5p, miR-155-5p, and miR-223-3p levels were not different in patients with established RA compared to control subjects; however, both of these studies compared RA and control subjects without age or sex matching (e.g. approximately 15-18 year mean age difference between groups in both studies), and these factors may alter circulating miRNAs (32, 33). Also, there have been problems with the reproducibility of findings with miR-146a-5p, miR-155-5p and miR-223-3p in other studies. For example, in another inflammatory autoimmune disease, systemic lupus erythematosus, findings with plasma miR-146a-5p and miR-223-3p were not replicated in exploratory and validation cohorts (34). Moreover, the relatively low abundance of plasma miR-155-5p may make it less reproducible across studies.
A recently published study evaluated miRNAs which might predict response to therapy in RA. miRNA microarrays were performed on serum of 10 patients with RA before and six months after starting disease modifying antirheumatic drug (DMARD) or anti-TNFα therapy. A subset of differentially expressed miRNAs with relevance to inflammation, immune response, musculoskeletal system or connective tissue were chosen to validate in a larger cohort of 85 patients with RA on paired serum samples. In this study miR-146a-5p and miR-223-3p, among others were significantly increased after DMARD/anti-TNFα therapy, and miR-223-3p (along with miR-23) were predictors of response (35).
The plasma miRNAs which were most strongly associated with RA in this study (miR-24-3p, miR-26a-5p, and miR-125a-5p) appear to be reproducible (18) and relevant to processes underlying RA. miR-24-3p mediates plasma cell survival and is increased with plasma cell exposure to IL-6 (14). Thus, miR-24-3p may contribute to RA by maintaining autoreactive plasma cells. miR-26a-5p, along with other miRNAs like miR-155-5p and miR-146a-5p, is increased in IL-17 producing T cells (16), which may contribute to the high plasma levels of these miRNAs in patients with RA. miR-125a-5p may have a role in modulating the inflammatory response within macrophages. For example, in mouse bone marrow derived macrophages, activation of toll-like receptors 2 and 4 increased miR-125a-5p expression, and miR-125a-5p promoted an anti-inflammatory cell phenotype (36). Conversely, miR-125a-5p amplified the inflammatory response by repressing a negative regulator of NF-kB signaling in human THP-1 cells (37).
The other plasma miRNAs examined, which were elevated among patients with RA, also have important biologic function in RA. One of the most widely studied is miR-146a-5p, which is increased in many cells types in RA such as synovial fibroblasts, PBMCs, CD4+ T cells and Th17 cells, and acts as a negative regulator of NF-kB activation (reviewed(38)). Another is miR-155-5p, which is a major regulator of B-cell development and function, T-cell dependent antibody responses, and T cell functions (reviewed (39)). Also, miR-223-3p modifies inflammation through several pathways, by targeting NLRP3 decreasing IL-1b production in monocytes, and suppressing T cell IL-10 production (reviewed (39, 40)).
In addition to their association with RA and inflammation, many of the miRNAs we examined have a relationship to cardiovascular disease. Two key examples are miR-223-3p and miR-146a-5p. miR-223-3p decreases cholesterol biosynthesis and enhances cholesterol efflux (41). Moreover, delivery of miR-223-3p to endothelial cells decreased adhesion molecule expression (42), a key factor in the development of atherosclerosis. Concentrations of miR-146a-5p were increased in the plasma and peripheral blood mononuclear cells of patients with acute coronary syndrome (43), and miR-146a-5p affects endothelial function by regulation of nitric oxide production (44). Nevertheless, these plasma miRNAs were not associated with coronary artery calcium score.
The study had some limitations. For example, we used a candidate miRNA approach to evaluate key miRNAs thought to be important in the development of RA. A drawback of this method is that the miRNA field is rapidly expanding; therefore, potentially important miRNAs were likely not included. Also, we did not study patients with other inflammatory autoimmune diseases, such as systemic lupus erythematosus or psoriatic arthritis, to determine if the miRNAs examined here may differentiate between RA and other inflammatory autoimmune diseases. Future studies to evaluate this will be interesting, particularly given our findings in seronegative RA, a condition in which alternative diagnoses are often considered and an additional diagnostic tool would be very useful.
In summary, miR-15a-5p, miR-24-3p, miR-26a-5p, miR-125a-5p, miR-146a-5p, miR-155-5p, and miR-223-3p are all elevated in the plasma of patients with RA. miR-24-3p, miR-26a-5p, and miR-125a-5p had the strongest diagnostic accuracy for RA, and miR-24-3p had the strongest diagnostic accuracy for seronegative RA. Among the candidate miRNAs evaluated miR-155-5p was inversely associated with swollen joint count. None of the candidate miRNAs were associated with coronary artery atherosclerosis in RA.
Supplementary Material
Acknowledgments
Funding: Arthritis Foundation Clinical to Research Transition Award, ACR Rheumatology Research Foundation, NIH Grants: P60 AR056116, P01HL116263, KL2TR000446 and CTSA award UL1TR000445 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Study conception and design: Ormseth, Solus, Vickers, and Stein. Acquisition of data: Ormseth, Solus, Oeser, Raggi. Analysis and interpretation of data: Ormseth, Solus, Stein.
Disclosures: None
Contributor Information
Michelle J Ormseth, Department of Medicine and Division of Rheumatology, Vanderbilt University Medical Center, Nashville, TN, USA.
Joseph F Solus, Department of Medicine and Division of Clinical Pharmacology, Vanderbilt University Medical Center, Nashville, TN, USA.
Kasey C Vickers, Department of Medicine and Division of Cardiovascular Medicine, Vanderbilt University Medical Center, Nashville, TN, USA.
Annette M Oeser, Department of Medicine and Division of Clinical Pharmacology, Vanderbilt University Medical Center, Nashville, TN, USA.
Paolo Raggi, Department of Medicine and Division of Cardiology, University of Alberta, Edmonton, Canada.
C. Michael Stein, Department of Medicine and Division of Clinical Pharmacology, Vanderbilt University Medical Center, Nashville, TN, USA.
REFERENCES
- 1.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 5.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murata K, Yoshitomi H, Tanida S, Ishikawa M, Nishitani K, Ito H, et al. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2010;12:R86. doi: 10.1186/ar3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myllykangas-Luosujarvi RA, Aho K, Isomaki HA. Mortality in rheumatoid arthritis. Semin Arthritis Rheum. 1995;25:193–202. doi: 10.1016/s0049-0172(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 11.Pincus T, Sokka T, Wolfe F. Premature mortality in patients with rheumatoid arthritis: evolving concepts. Arthritis Rheum. 2001;44:1234–6. doi: 10.1002/1529-0131(200106)44:6<1234::AID-ART213>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 12.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagata Y, Nakasa T, Mochizuki Y, Ishikawa M, Miyaki S, Shibuya H, et al. Induction of apoptosis in the synovium of mice with autoantibody-mediated arthritis by the intraarticular injection of double-stranded MicroRNA-15a. Arthritis Rheum. 2009;60:2677–83. doi: 10.1002/art.24762. [DOI] [PubMed] [Google Scholar]
- 14.Gabler J, Wittmann J, Porstner M, Renz H, Jack HM, Abram M, et al. Contribution of microRNA 24-3p and Erk1/2 to interleukin-6-mediated plasma cell survival. Eur J Immunol. 2013;43:3028–37. doi: 10.1002/eji.201243271. [DOI] [PubMed] [Google Scholar]
- 15.Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB, et al. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J Immunol. 2012;189:3795–9. doi: 10.4049/jimmunol.1200312. [DOI] [PubMed] [Google Scholar]
- 16.Niimoto T, Nakasa T, Ishikawa M, Okuhara A, Izumi B, Deie M, et al. MicroRNA-146a expresses in interleukin-17 producing T cells in rheumatoid arthritis patients. BMC Musculoskelet Disord. 2010;11:209. doi: 10.1186/1471-2474-11-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Pedrera C, Perez-Sanchez C, Ramos-Casals M, Santos-Gonzalez M, Rodriguez-Ariza A, Cuadrado MJ. Cardiovascular risk in systemic autoimmune diseases: epigenetic mechanisms of immune regulatory functions. Clin Dev Immunol. 2012;2012:974648. doi: 10.1155/2012/974648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murata K, Furu M, Yoshitomi H, Ishikawa M, Shibuya H, Hashimoto M, et al. Comprehensive microRNA analysis identifies miR-24 and miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS One. 2013;8:e69118. doi: 10.1371/journal.pone.0069118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duroux-Richard I, Jorgensen C, Apparailly F. What do microRNAs mean for rheumatoid arthritis? Arthritis Rheum. 2012;64:11–20. doi: 10.1002/art.30651. [DOI] [PubMed] [Google Scholar]
- 20.Condorelli G, Latronico MV, Cavarretta E. microRNAs in cardiovascular diseases: current knowledge and the road ahead. J Am Coll Cardiol. 2014;63:2177–87. doi: 10.1016/j.jacc.2014.01.050. [DOI] [PubMed] [Google Scholar]
- 21.Madrigal-Matute J, Rotllan N, Aranda JF, Fernandez-Hernando C. MicroRNAs and atherosclerosis. Curr Atheroscler Rep. 2013;15:322. doi: 10.1007/s11883-013-0322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung CP, Oeser A, Raggi P, Gebretsadik T, Shintani AK, Sokka T, et al. Increased coronary-artery atherosclerosis in rheumatoid arthritis: relationship to disease duration and cardiovascular risk factors. Arthritis Rheum. 2005;52:3045–53. doi: 10.1002/art.21288. [DOI] [PubMed] [Google Scholar]
- 23.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 24.Prevoo ML, van ’t Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995;38:44–8. doi: 10.1002/art.1780380107. [DOI] [PubMed] [Google Scholar]
- 25.Wilson PW, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–47. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 26.Chung CP, Oeser A, Avalos I, Gebretsadik T, Shintani A, Raggi P, et al. Utility of the Framingham risk score to predict the presence of coronary atherosclerosis in patients with rheumatoid arthritis. Arthritis Res Ther. 2006;8:R186. doi: 10.1186/ar2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–9. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 28.Agatston AS, Janowitz WR, Hildner FJ, Zusmer NR, Viamonte M, Jr., Detrano R. Quantification of coronary artery calcium using ultrafast computed tomography. J Am Coll Cardiol. 1990;15:827–32. doi: 10.1016/0735-1097(90)90282-t. [DOI] [PubMed] [Google Scholar]
- 29.Shipley GL. The MIQE Guidelines Uncloaked. PCR Troubleshooting and Optimization: The Essential Guide. 2011.
- 30.Chung CP, Oeser A, Solus JF, Avalos I, Gebretsadik T, Shintani A, et al. Prevalence of the metabolic syndrome is increased in rheumatoid arthritis and is associated with coronary atherosclerosis. Atherosclerosis. 2008;196:756–63. doi: 10.1016/j.atherosclerosis.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Filkova M, Aradi B, Senolt L, Ospelt C, Vettori S, Mann H, et al. Association of circulating miR-223 and miR-16 with disease activity in patients with early rheumatoid arthritis. Ann Rheum Dis. 2014;73:1898–904. doi: 10.1136/annrheumdis-2012-202815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon LM, Edelstein LC, Nagalla S, Woodley AB, Chen ES, Kong X, et al. Human platelet microRNA-mRNA networks associated with age and gender revealed by integrated plateletomics. Blood. 2014;123:e37–45. doi: 10.1182/blood-2013-12-544692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meder B, Backes C, Haas J, Leidinger P, Stahler C, Grossmann T, et al. Influence of the confounding factors age and sex on microRNA profiles from peripheral blood. Clin Chem. 2014;60:1200–8. doi: 10.1373/clinchem.2014.224238. [DOI] [PubMed] [Google Scholar]
- 34.Carlsen AL, Schetter AJ, Nielsen CT, Lood C, Knudsen S, Voss A, et al. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum. 2013;65:1324–34. doi: 10.1002/art.37890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castro-Villegas C, Perez-Sanchez C, Escudero A, Filipescu I, Verdu M, Ruiz-Limon P, et al. Circulating miRNAs as potential biomarkers of therapy effectiveness in rheumatoid arthritis patients treated with anti-TNFalpha. Arthritis Res Ther. 2015;17:49. doi: 10.1186/s13075-015-0555-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banerjee S, Cui H, Xie N, Tan Z, Yang S, Icyuz M, et al. miR-125a-5p regulates differential activation of macrophages and inflammation. J Biol Chem. 2013;288:35428–36. doi: 10.1074/jbc.M112.426866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME. Identifying functional microRNAs in macrophages with polarized phenotypes. J Biol Chem. 2012;287:21816–25. doi: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan EK, Ceribelli A, Satoh M. MicroRNA-146a in autoimmunity and innate immune responses. Annals of the rheumatic diseases. 2013;72(Suppl 2):ii90–5. doi: 10.1136/annrheumdis-2012-202203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ammari M, Jorgensen C, Apparailly F. Impact of microRNAs on the understanding and treatment of rheumatoid arthritis. Current opinion in rheumatology. 2013;25:225–33. doi: 10.1097/BOR.0b013e32835d8385. [DOI] [PubMed] [Google Scholar]
- 40.Fulci V, Scappucci G, Sebastiani GD, Giannitti C, Franceschini D, Meloni F, et al. miR-223 is overexpressed in T-lymphocytes of patients affected by rheumatoid arthritis. Human immunology. 2010;71:206–11. doi: 10.1016/j.humimm.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Vickers KC, Landstreet SR, Levin MG, Shoucri BM, Toth CL, Taylor RC, et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc Natl Acad Sci U S A. 2014;111:14518–23. doi: 10.1073/pnas.1215767111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabet F, Vickers KC, Cuesta Torres LF, Wiese CB, Shoucri BM, Lambert G, et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat Commun. 2014;5:3292. doi: 10.1038/ncomms4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo M, Mao X, Ji Q, Lang M, Li S, Peng Y, et al. miR-146a in PBMCs modulates Th1 function in patients with acute coronary syndrome. Immunol Cell Biol. 2010;88:555–64. doi: 10.1038/icb.2010.16. [DOI] [PubMed] [Google Scholar]
- 44.Sun X, Belkin N, Feinberg MW. Endothelial MicroRNAs and Atherosclerosis. Curr Atheroscler Rep. 2013;15:372. doi: 10.1007/s11883-013-0372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.