Abstract
Triple-negative breast cancer (TNBC) patients have the highest risk of recurrence and metastasis. Because they cannot be treated with targeted therapies, and many do not respond to chemotherapy, they represent a clinically underserved group. TNBC is characterized by reduced expression of metastasis suppressors such as Raf Kinase Inhibitory Protein (RKIP), which inhibits tumor invasiveness. Mechanisms by which metastasis suppressors alter tumor cells are well characterized; however, their ability to regulate the tumor microenvironment, and the importance of such regulation to metastasis suppression is incompletely understood.
Here we use species-specific RNA sequencing to show that RKIP expression in tumors markedly reduces the number and metastatic potential of infiltrating TAMs. TAMs isolated from non-metastatic RKIP+ tumors, relative to metastatic RKIP− tumors, exhibit a reduced ability to drive tumor cell invasion and decreased secretion of pro-metastatic factors including PRGN and shed TNFR2. RKIP regulates TAM recruitment by blocking HMGA2, resulting in reduced expression of numerous macrophage chemotactic factors, including CCL5. CCL5 overexpression in RKIP+ tumors restores recruitment of pro-metastatic TAMs and intravasation, while treatment with the CCL5 receptor antagonist Maraviroc reduces TAM infiltration. These results highlight the importance of RKIP as a regulator of TAM recruitment through chemokines such as CCL5. The clinical significance of these interactions is underscored by our demonstration that a signature comprised of RKIP signaling and pro-metastatic TAM factors strikingly separates TNBC patients based on survival outcome. Collectively, our findings identify TAMs as a previously unsuspected mechanism by which the metastasis suppressor RKIP regulates tumor invasiveness, and further suggest that TNBC patients with decreased RKIP activity and increased TAM infiltration may respond to macrophage-based therapeutics.
Introduction
Of the approximately 40,000 women diagnosed with breast cancer each year, 15–20% will have triple-negative breast cancer (TNBC). The most aggressive subset of breast cancer, TNBCs lack expression of the estrogen, progesterone and HER2/neu receptors. While clinical outcomes have improved for many patients with breast cancer, TNBC patients have higher rates of metastasis, more aggressive tumors, higher disease burden, and early recurrence (1). Additionally, this disease disproportionately affects African-American women, with rates approximately three times higher in African-American women (2, 3). Moreover, only 30% of TNBC patients are responsive to platinum based chemotherapy (4). Therefore, there is interest in novel approaches for treating TNBC patients, including targeting of the tumor stroma (5).
One possible strategy is to mimic the action of physiological tumor metastasis suppressors such as Raf Kinase Inhibitory Protein (RKIP). RKIP, a member of the evolutionarily conserved phosphatidylethanolamine family, has been implicated as a metastasis suppressor for prostate, breast and other solid tumors (6–8). RKIP inhibits key signaling pathways including Raf/MAP kinase, GRK2-regulated ß-adrenergic receptor, and NFkB activation (6). Previously, RKIP was shown to suppress the expression of many pro-metastatic genes in TNBC cells by inhibiting transcriptional regulators such as HMGA2 (8–10). Because previous studies have focused on the effects of metastasis suppressors in tumor cells, their role in regulating the tumor microenvironment is unknown.
Multiple lines of evidence have shown that the microenvironment regulates both tumor progression and metastasis. In particular, macrophages have been shown to play a dual role in tumor growth, either driving tumor rejection or tumor progression depending on the type of macrophage activation (11). Classical activation of macrophages by IFNγ, lipopolysaccharide (LPS), or tumor necrosis factor- α (TNF α) leads to polarization of M1 macrophages that secrete inflammatory cytokines important in the body’s anti-tumor response. M2 macrophages, activated by factors such as IL4, play an essential role in wound healing. Secretion of factors from tumor-associated macrophages (TAMs), thought to be M2, leads to tumor growth, progression, and metastasis (12–14) as well drug resistance (15). However, recent evidence suggests that this division of macrophages into two discrete subtypes incompletely describes the range of macrophage phenotypes present in the tumor microenvironment (16). Importantly, studies of breast cancer patients show that CD163+ macrophage recruitment positively correlates with TNBC while negatively correlating with ER+ and luminal tumors (17). Therefore, recruitment of alternatively activated TAMs could play a significant role in the outcome of TNBC patients and explain their poor prognosis.
TAMs are recruited to mammary tumors through induction of a variety of cytokines and chemokines, where they play essential roles in driving metastasis. For example, TAMs recruited by CSF-1 express higher levels of VEGF-A, with increased angiogenesis in the polyoma middle T genetically engineered mouse model for breast cancer (18). Similarly, CCL2 was required for TAM infiltration in primary breast tumors as well as TAM-enabled metastatic colonization of lungs (19). Antagonists of the CCL5 receptor (CCR5) inhibited TAM recruitment in a syngeneic mouse model (20). Finally, recent work comparing breast tumors before and after EMT has shown that GM-CSF is able to recruit TAMs to the primary tumor (21, 22). Although factors enabling recruitment of pro- metastatic TAMs to mammary tumors have been identified, the regulation of these pathways by metastasis suppressors and the specific phenotypes of these TAMs are poorly understood.
Here we combine species-specific RNA sequencing, protein secretion profiling, functional assays, and gene knockdown studies in xenograft and syngeneic breast cancer models to characterize the effects of the metastasis suppressor RKIP on TAMs and to identify the molecular mechanisms that mediate these effects. Our findings demonstrate that RKIP blocks a subset of TAMs that secrete pro-metastatic factors and are enriched in human TNBC patients. These results suggest that one mechanism by which metastasis suppressors alter tumor invasiveness is by regulating TAMs.
Materials and Methods
See Supplementary Methods for additional description of methodology.
Cell Culture
MDA-MB-436 and 4T1.2 were obtained from ATCC. BM1 cells were obtained from Andy Minn. Numerous vials were frozen upon original receipt of the cells, and all work was done within 15 passages of the initially received lines. BM1, MDA-MD-436, and 4T1.2 cell lines were cultured in DMEM media supplemented with 10% fetal bovine serum, 50 U/ml penicillin, and 50 µg/ml streptomycin. Cells were transduced with lentiviral vectors for shRNA knockdown or overexpression from GE/Dharmacon. Cells were selected for 14 days using 3 µg/ml of puromycin or 10 µg/ml of blasticidin after lentiviral transduction before use.
Invasion Assays
2×105 BM1 cells were plated in 24-well BD trans-well inserts coated with growth factor depleted matrigel as previously described (8). After 24 hours inserts were transferred to a new well and stained with 4 ng/ul of Calcein AM for one hour. Stained cells were then dissociated using gentle shaking for one hour at 37 C and 150 RMP in dissociation buffer from Trevigen. Fluorescence was measured using a Victor X3 fluorescent plate reader with excitation at 465 nm and emission at 535 nm.
Tumor Associated Macrophage Isolations
Tumors were grown to approximately 0.2 g before being harvested. Tumors were dissociated using the Miltenyi Biotech human tumor dissociation kit using C-tubes. Cells were filtered through a 70 um mesh filter. Mononuclear cells were isolated using Ficoll-Paque PREMIUM (GE Healthcare) gradient centrifugation at 420 RPM for 40 minutes. Macrophages were then obtained using CD11b positive selection beads from Milteny Biotech. Flow cytometry with CD11b, F4/80, CD45, CD11c, CD205, and CCR5 was performed to determine the purity and heterogeneity of isolated TAMs.
Conditioned Media
For THP-1 conditioned media, 5×106 THP-1 cells were plated in a T-75 flask with 5 mL of 10% serum containing DMEM. Media was collected after 24 hours and cells and cell debris were removed by centrifugation.
For tumor derived macrophages, 5×105 TAMs were plated in one well of a 6-well plate. After 30 minutes, cells were washed with PBS to ensure only viable macrophages attached to the plate remained. Cells were incubated for 24 hours to obtain conditioned media in serum free DMEM. Cells and cell debris were removed by centrifugation.
Statistical Analysis
For all experiments, bar graphs represent the mean (±SEM) and * 0.05<p<0.01 ** 0.01≤p<0.001 *** 0.001≤p. Unless otherwise stated, statistical differences between means was determined using a Student’s T-test.
Results
Non-metastatic RKIP+ tumors contain fewer macrophages
To compare metastatic and non-metastatic tumors that were isogenic, we used highly metastatic BM1 derivatives of the MDA-MB-231 human TNBC cell line stably expressing either the metastasis suppressor Raf Kinase Inhibitory Protein (RKIP+) or a vector control (control) (Fig S1) (8, 9). Tumor cells were injected orthotopically into athymic nude mice, and RNA from the tumors was then isolated and sequenced. To overcome the challenge of distinguishing between tumor-specific and stroma-specific gene expression, we used next- generation RNA sequencing (RNAseq) in this xenograft mouse model to separate sequencing reads based on their species of origin (Fig S2).
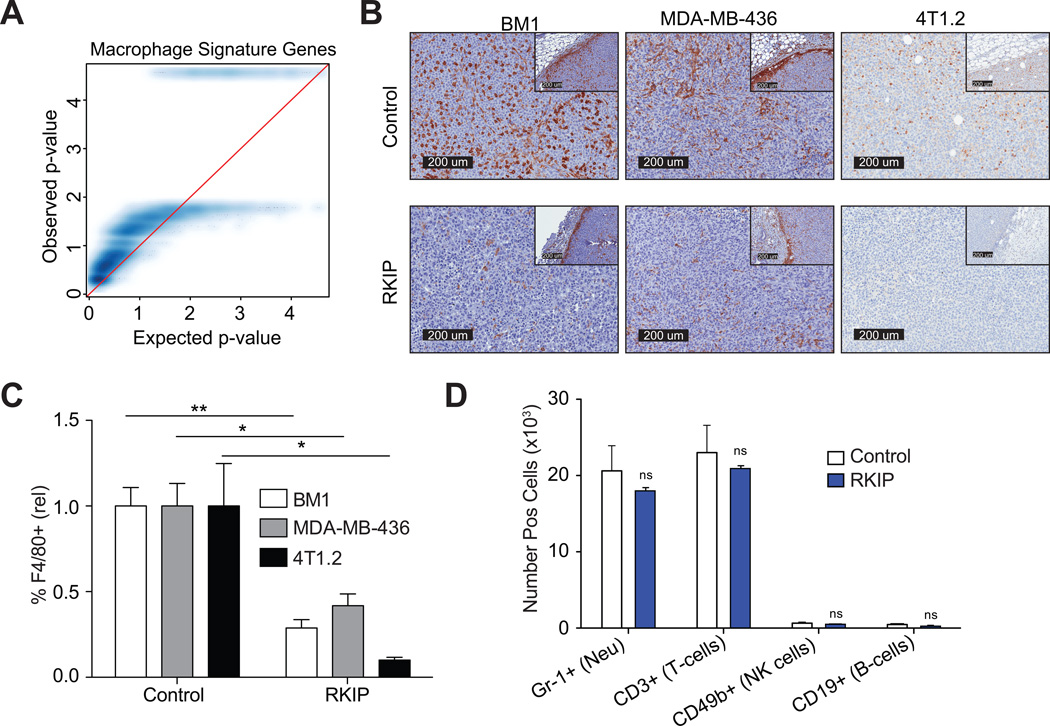
We observed dramatic changes in stromal gene expression profiles between RKIP+ and control tumors even though the tumors did not significantly differ in size (Fig. S3). Analysis of gene expression changes between RKIP+ and control tumor cells using GOseq revealed that the most significant difference was the immune response and, specifically, macrophage chemotaxis (Q-value = 2.2×10−4, Fig. S4). Using mouse gene sets characteristic of common immune cell types (23), we observed a clear depletion of gene expression associated with macrophages in the RKIP+ tumor microenvironment (Fig. 1A), and this was the most robust change observed (Fig. S5). Immunohistochemical staining confirmed a marked reduction in the number of tumor-associated macrophages (TAMs) at the primary tumor site in RKIP+ tumors relative to controls, both in xenograft (BM1, MDA-MB-436) and syngeneic (4T1.2) tumor models (Fig. 1B–C; S6). Moreover, the effect of RKIP on TAMs was quite specific, as we did not observe significant differences in the number of B cells (CD19+), T cells (CD3+), NK cells (CD49b+), or neutrophils (Gr-1+) when analyzed by flow cytometry (Fig. 1D).
Figure 1. Non-metastatic RKIP+ tumors contain fewer macrophages.
A) Quantile-quantile (qq)-plot showing ranked −log10 transformed p-values among macrophage-specific genes (y-axis) relative to a similar bootstrapped distribution in blue (x-axis). Distortion of the density above x = y (red line) indicates that the measured p-values are systematically lower than expected by chance. B) Representative images of relative macrophage presence in xenograft (BM1 and MDA-MB-436) and syngeneic (4T1.2) tumor models with and without RKIP expression were sectioned and immunostained for F4/80. C) Relative macrophage inflitration was quantified as the proportion of inner tumor mass positively stained with F4/80 via immunohistochemistry. D) Number of positively staining cells was determined by flow cytometry.
RKIP suppresses recruitment of a distinct TAM population that potentiates tumor cell invasion
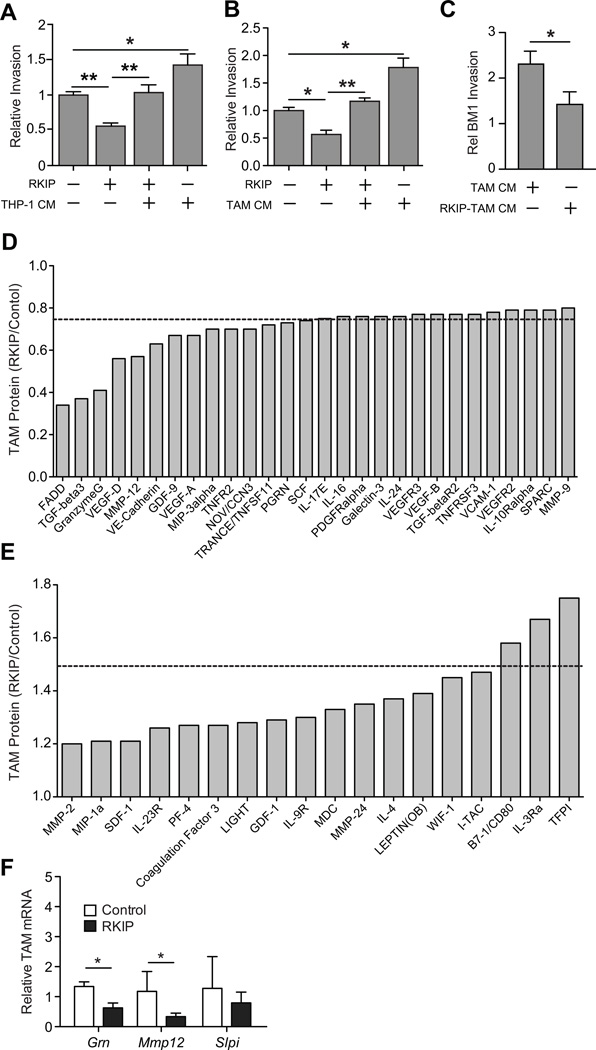
Since RKIP regulates the number of macrophages in tumors (Fig. 1A–C) and TAMs are known to play a significant role in tumor biology (11), we hypothesized that changes in TAMs may in part explain suppression of intravasation by RKIP. In support of this hypothesis, treating RKIP+ BM1 tumor cells with conditioned media (CM) from a human monocytic cell line (THP1) restored tumor cell invasion relative to levels observed in control BM1 tumor cells (Fig. 2A). Similar results were observed with the CM of TAMs purified from control BM1 tumors (Fig. 2B). These findings demonstrate that TAMs from metastatic tumors can overcome blockade of tumor cell invasion by RKIP.
Figure 2. RKIP suppresses recruitment of a distinct TAM population that potentiates tumor cell invasion.
A–C) BM1 or BM1+RKIP tumor cells were pretreated with conditioned media collected from various types of macrophages for 24h; A) THP1 human monocytic cell line (n=8 per group), B) TAMs isolated from BM1 tumors (n=5 per group), C) TAMs isolated from BM1 or BM1+RKIP tumors (n=6 per group). Relative invasion is against BM1 grown in a media control. P-values were calculated using an unpaired T-test with Welch’s correction. D–E) TAM conditioned media from four independent tumors was analyzed for protein levels using RayBiotech L308 Mouse Cytokine Arrays. Protein abundance for RKIP derived TAMs were normalized to control tumor TAMs. D) Proteins with 0.8 or lower relative abundance, E) Proteins with >1.2 relative abundance. F) Relative mRNA was measured from three independent TAM samples per group. Relative mRNA was calculated as compared to control TAMs, with Gapdh as the reference gene.
It is well established that, depending on environmental conditions, TAMs can adopt phenotypes with pro-tumor (“M2-like”) or anti-tumor (“M1-like”) properties (13). We therefore explored the possibility that, in addition to reducing the number of macrophages in tumors, RKIP might also alter their functional properties to suppress metastasis. To test this hypothesis, we purified TAMs from BM1 tumors (metastatic) and RKIP+ BM1 tumors (non-metastatic), which were uniformly CD45+, CD11b+, F4/80+, and CD205− (Fig. S7), and we assessed their functional phenotype using two interrelated approaches.
First, we determined the effect of TAM conditioned media on tumor cell invasion in vitro. Pretreating BM1 tumor cells with the conditioned media (CM) of TAMs isolated from control BM1 tumors, like THP1 cells, potentiated invasion (Fig. 2B). In sharp contrast, factors secreted by TAMs from RKIP+ BM1 tumors had no significant effect on tumor cell invasiveness (Fig. 2C). These results indicate that the TAMs from metastatic and non-metastatic (RKIP+) tumors have distinct phenotypes.
Second, we quantified the relative abundance of 400 proteins including inflammatory and tumorigenic factors (e.g. cytokines, growth factors) in the TAM conditioned media using the RayBiotech L308 mouse cytokine array. TAMs purified from RKIP+ BM1 tumors relative to control BM1 tumors were distinguished by reduced abundance of a number of pro-metastatic factors (11) including TGF-β3, VEGF-D, MMP-12, GDF-9, VEGF, sTNFR2, and PGRN (Fig. 2D). We also observed induction of secreted factors in the CM of RKIP+ BM1 tumors including CD80 and TFPI, two potential anti-tumor proteins (24–26). We confirmed differential regulation of Mmp12 and Grn (progranulin) transcripts in TAMs isolated from BM1 versus RKIP+ BM1 tumors by qRT- PCR (Fig. 2E).
Taken together, the direct functional evidence and protein expression analysis suggest that RKIP suppresses recruitment of a TAM population that secretes a set of pro-invasive and pro-metastatic proteins.
Overexpression of CCL5 restores TAM recruitment in RKIP+ tumors
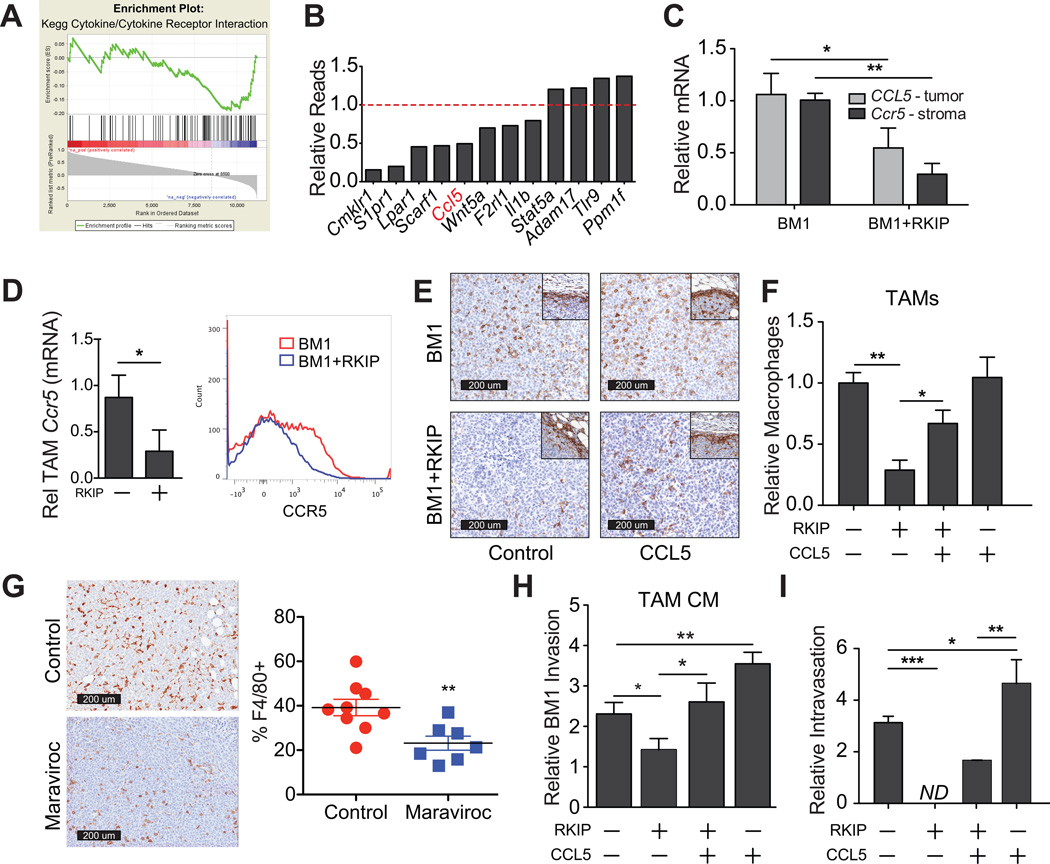
To determine the mechanism by which RKIP regulates TAM number and function, we examined our RNAseq data comparing BM1 and RKIP+ BM1 tumors. RKIP+ tumors suppressed numerous genes involved in cytokine/cytokine receptor interactions, particularly in relation to external stimulus and macrophage chemotaxis (Figs. 3A; S4) (FDR = 3.9×10−2). The foremost of the chemokine factors was CCL5 (Fig. 3B) (20, 27). We therefore analyzed CCL5 expression by species-specific qRT-PCR (Fig. 3C) and by ELISA (Fig. S8), and confirmed its down- regulation in RKIP+ tumors. Similar decreases in CCL5 transcripts were observed following RKIP expression in human MDA-MB-436 and mouse 4T1.2 tumor cell lines (Fig. S9). Thus, RKIP suppresses CCL5 expression in multiple human and murine tumor cell lines.
Figure 3. Overexpression of CCL5 restores TAMs and overcomes metastasis suppression in RKIP+ tumors.
A) GSEA identifies enrichment of genes involved in cytokine-cytokine receptor interactions (black lines) among genes with anticorrelated expression levels in tumors and surrounding stroma. B) Tumor genes differentially expressed in RKIP tumors relative to control (p<0.05) are shown from the external stimulus (GO) category from our RNAseq data. C) qRT-PCR was performed on mRNA purified from xenograft tumors (BM1 & BM1+RKIP). Species-specific primers were used to detect relative mRNA of CCL5 (Hs) and Ccr5 (Mm) in the tumor and stroma, respectively. Relative mRNA was normalized to GAPDH (Hs) or Rpl4 (Mm). D) Relative mRNA was calculated relative to BM1 TAMs with Gapdh as the reference gene. Flow cytometry of BM1 (red) and BM1+RKIP (blue) isolated TAMs are also shown for CCR5. E) Representative images of macrophage presence in BM1 tumors with and without RKIP and CCL5 expression. F) Relative macrophages in BM1 and BM1+RKIP tumors with or without exogenous CCL5 expression in tumor cells. Infiltration was quantified as the proportion of total tumor area positively stained with F4/80 (n=3 per group). G) Effect of Maraviroc on BM1 tumor macrophage numbers was assessed by immunostaining for F4/80. Data are displayed as the %F4/80+ cells in the core of the tumor. H) BM1 cells were pretreated with TAM conditioned media (BM1, BM1+RKIP, BM1+RKIP+CCL5, or BM1+CCL5 TAMs) for 24 hours prior to invasion assays, using TAMs from four independent tumors each. P-values were obtained using an unpaired T-test with Welch’s correction, n=6. I) Relative intravasation of tumor cells into blood 4 weeks following injection was estimated by quantifying the ratio of human GAPDH (tumor) to mouse Gapdh by qRT-PCR (n=4 per group).
We used three approaches to investigate whether suppression of CCL5 by RKIP plays an important role in regulating macrophage accumulation into tumors in vivo. First, since CCL5 recruits macrophages via interaction with its receptor CCR5 (20, 28), we measured Ccr5 expression in the stroma. We found that Ccr5 levels were significantly reduced in RKIP+ tumor stroma by RNAseq (Fig. 3C). In addition, when comparing the same number of TAMs, we observed significant decreases in the mean Ccr5 expression by qRT-PCR (Fig. 3D). Using flow cytometry, we determined that the decrease in mean Ccr5 was due to a reduction in the number of CCR5+ TAMs in RKIP tumors (Fig. 3D).
Second, we transfected CCL5 into BM1 cells stably expressing RKIP or control vector (Figs. S8, S10) and observed rescue of TAM infiltration in tumors expressing RKIP and CCL5 (RKIP+CCL5) compared to those just expressing RKIP alone (RKIP+) (Fig 3E,F). There was a corresponding increase in the number of CCR5+ macrophages in CCL5-rescued tumors (Fig. S7), consistent with CCL5 recruitment of TAMs.
Third, we determined if a reduction in CCL5 signaling alone was necessary for TAM infiltration and tumor growth. We treated mice with the orally bioavailable CCR5 inhibitor Maraviroc twice daily by oral gavage and found that antagonizing the CCL5 receptor significantly lowered the number of TAMs recruited into BM1 control tumors as well as decreased tumor growth (Figs. 3G, S11). To determine if the number of TAMs infiltrating into the tumor was simply due to a difference in tumor size, we performed a Pearson correlation between the tumor weight and the percent of F4/80+ cells in the tumor. We found that, whether we examined the total population of tumors or the control and Maraviroc-treated tumors individually, there was no correlation between the size of the tumor and the percent of F4/80+ cells in the tumor (Fig. S11). Together, these findings suggest that modulation of CCL5 expression is one important mechanism by which RKIP controls tumor macrophage recruitment.
Overexpression of CCL5 restores a prometastatic TAM phenotype and overcomes metastasis suppression in RKIP+ tumors
To determine if CCL5 overexpression in RKIP+ BM1 tumors could also restore a TAM phenotype that promotes tumor invasion, we first conducted functional assays. Whereas TAMs isolated from BM1 RKIP+ tumors had no effect on BM1 invasion; TAMs isolated from RKIP+ BM1 tumors overexpressing CCL5 induced tumor cell invasion with similar efficiency as TAMs isolated from metastatic BM1 tumors (Fig. 3H). Since invasion enables tumor cell entry into vessels, we investigated whether overexpression of CCL5 in RKIP+ BM1 tumor cells could overcome the inhibitory effect of RKIP on intravasation. Consistent with RKIP’s ability to suppress metastasis, RKIP expression in BM1 tumor cells potently inhibited intravasation into blood vessels (Fig. 3I) (9). Importantly, elevating CCL5 expression in RKIP+ BM1 cells produced a partial but significant recovery of tumor cell invasion (Fig. S12) and intravasation into blood vessels (Fig. 3I). CCL5 overexpression also potentiated both invasion (Fig. S12) and intravasation in control metastatic tumor cells (Fig. 3I).
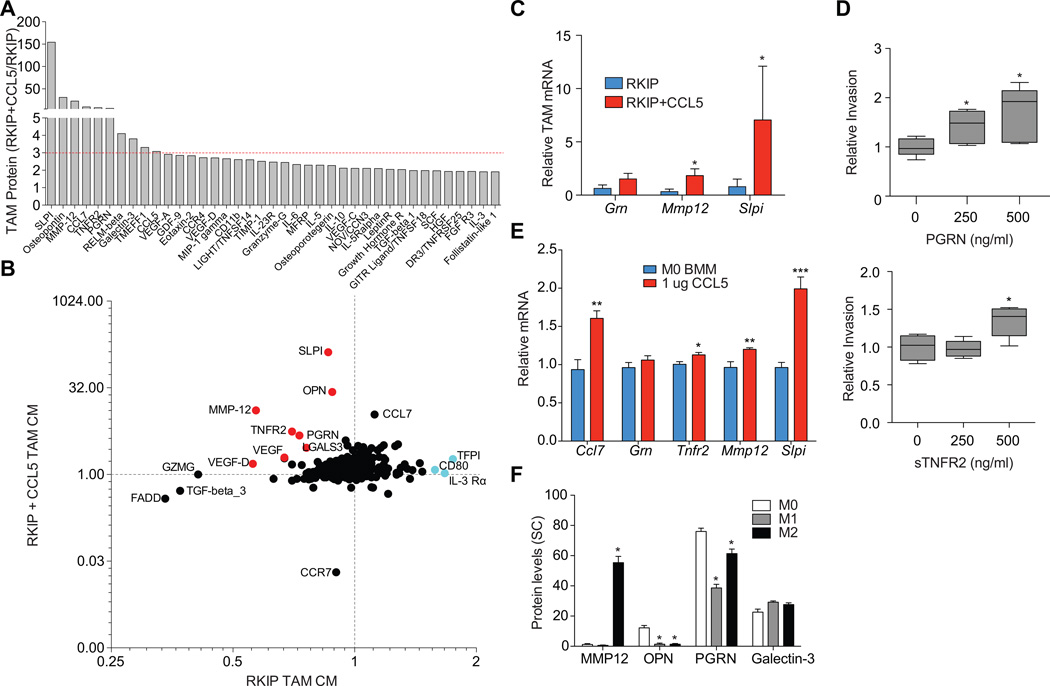
We then determined whether CCL5 overexpression in BM1+ RKIP tumors could enable recruitment of TAMs that secrete pro-metastatic factors. Analysis of proteins in the CM of isolated TAMs by cytokine arrays revealed robust induction of a number of factors that were suppressed in TAMs recruited to non-metastatic tumors (Fig. 4A, S13). For example, RKIP suppressed VEGF-A, VEGF-D, OPN, LGALS3, SLPI, MMP-12, sTNFR2, and PGRN expression by TAMs, and extracellular levels of these proteins were restored or even elevated in RKIP+CCL5 tumors relative to control tumors (Fig. 4B). We confirmed the induction of Mmp12, Slpi, and Grn in TAMs isolated from RKIP+CCL5 tumors by qRT-PCR (Fig. 4C).
Figure 4. RKIP blocked TAM phenotype rescued by CCL5.
A) TAM conditioned media from four independent tumors were analyzed for protein levels using RayBiotech L308 Mouse Cytokine Arrays. RKIP+CCL5 derived TAMs were normalized to RKIP derived TAMs with a cutoff set at greater than 3-fold expression. B) Relative protein is shown for each protein from the cytokine array; x-axis: RKIP TAMs relative to control TAMs, y-axis RKIP+CCL5 TAMs relative to RKIP TAMs. Both axes are shown on a log 2 scale. C) Relative mRNA was measured from three independent TAM samples per group. Relative mRNA was calculated as compared to control TAMs, with Gapdh as the reference gene. D) Relative invasion for BM1 cells pretreated with various concentrations of PGRN or sTNFR2 for 24 hours, results are plotted relative to an untreated control. Statistical significance was determined using a Student’s T-test, n=5 per group. E) Relative mRNA levels for bone marrow derived macrophages (BMMs) treated with or without 1 µg/ml of recombinant CCL5 for 24h. Gapdh is used as a reference gene. F) Protein levels (spectral counts) in the conditioned media collected from BMMs (M0), LPS -induced BMMs (M1), and IL-4-induced BMMs (M2) were quantified by mass spectrometry.
The group of TAM proteins suppressed by RKIP and induced by CCL5 included a number of potentially pro-metastatic factors. To confirm this possibility, we investigated whether PRGN or sTNFR2 were sufficient to drive invasion of TNBC cells in vitro. Treating BM1 cells with 500ng/mL recombinant sTNFR2 or PGRN significantly induced tumor cell invasion (Fig. 4D). These results provide empirical evidence that pro-metastatic factors counter-regulated by RKIP and CCL5 directly promote the invasiveness of human breast cancer cells.
The finding that CCL5 restored the pro-metastatic function of TAMs was surprising given that it is generally believed that CCL5 acts as a chemotactic agent to recruit macrophages to tissues (20). This observation led us to hypothesize that CCL5 might have direct actions on TAMs to promote the expression of pro-metastatic factors. Treating bone marrow-derived macrophages with CCL5 in vitro significantly induced the expression of several pro-metastatic factors including Ccl7, Tnfr2, Mmp12, and Slpi (Fig 4E). Thus, overexpressing CCL5 might overcome metastasis suppression in non-metastatic (RKIP+) tumors both by recruiting TAMs and directly programming them to overexpress pro-metastatic factors..
The difference between TAMs from metastatic (BM1 or BM1 RKIP+CCL5) and non-metastatic (RKIP+) tumors could reflect a switch from an M2 to an M1 phenotype. To examine this possibility, we analyzed proteins secreted by bone marrow-derived macrophages (M0), M1 macrophages (activated by LPS/IFNγ), and M2 macrophages (activated by IL4) using mass spectrometry. When we compared them to factors secreted by CCL5-recruited TAMs, MMP12 was significantly increased in M2 compared to M0 and M1 macrophages; however, GRN and LGALS3 were broadly expressed, and OPN was selectively expressed in M0 macrophages (Fig. 4F). These results suggest that the markers expressed in the CCL5-recruited TAMs (29) are not indicative of a classic M1 or M2 phenotype.
Collectively, these findings demonstrate that CCL5 overexpression can promote macrophage infiltration, macrophage function, and intravasation on a non-metastatic (RKIP+) background, suggesting that downregulation of CCL5 by RKIP, and the concomitant reduction in TAMs, may be an important mechanism by which RKIP suppresses metastasis.
Suppression of metastasis and TAMs by RKIP is coordinated though HMGA2 signaling
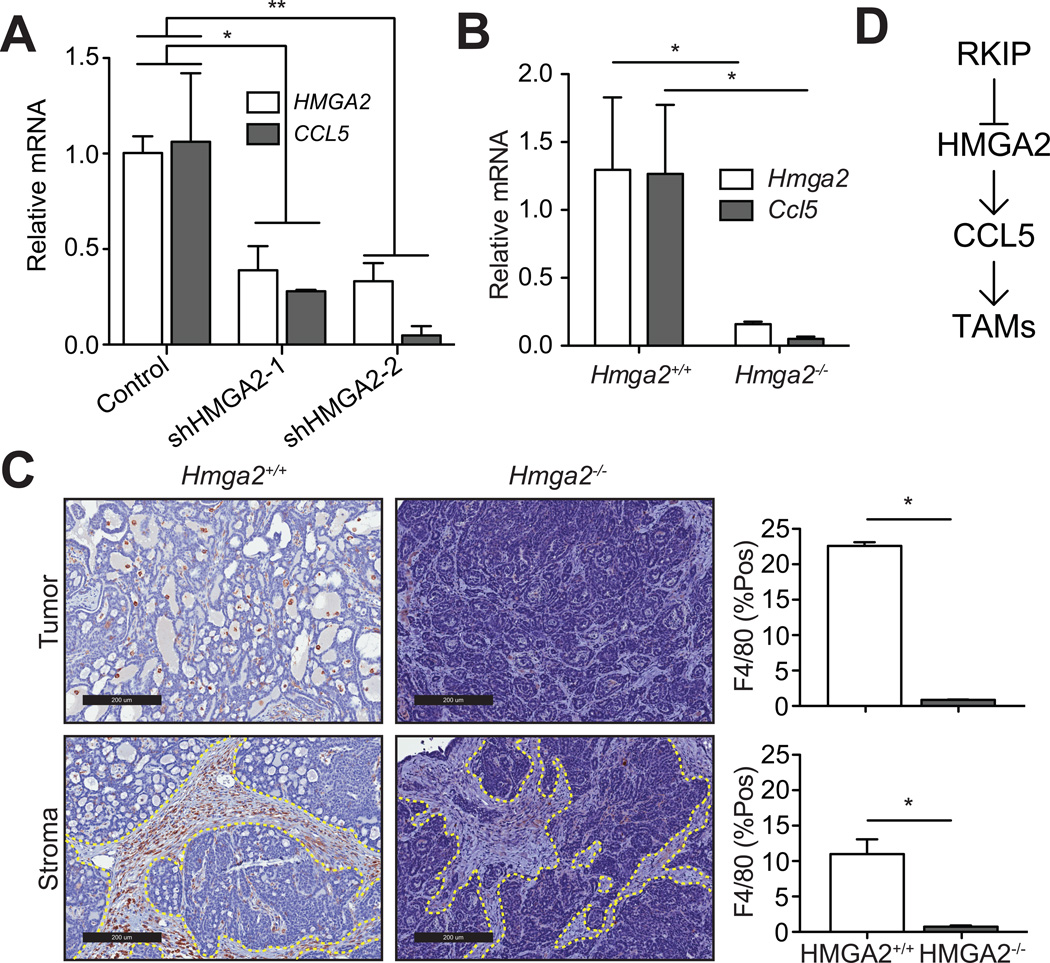
Our previous work showed that RKIP suppresses breast cancer metastasis in part by inhibiting the architectural transcription factor High-mobility group AT-hook 2 (HMGA2) (8, 9). We therefore determined whether RKIP suppresses macrophage recruitment via a similar mechanism. HMGA2 depletion in BM1 cells led to a significant decrease in CCL5 expression in vitro (Fig 5A). To test whether HMGA2 regulates macrophage accumulation in vivo, we crossed Hmga2−/− mice with the invasive MMTV-Wnt1 genetically engineered mouse (GEM; (29)). Similar to the RKIP− phenotype, Hmga2−/− GEM mice (relative to Hmga2+/+) had decreased Ccl5 expression in the mammary tumors (Fig. 5B) and a marked reduction in the number of macrophages present both in the tumor tissue as well as in the surrounding stroma (Fig 5C). Together with previous findings (8, 9), these results suggest that RKIP suppression of tumor cell CCL5 expression, macrophage recruitment and metastasis is coordinated through HMGA2 signaling (Fig 5D).
Figure 5. Suppression of metastasis and TAMs by RKIP is coordinated though HMGA2 signaling.
A) BM1 cells were transduced with two separate shRNAs targeting HMGA2. Relative expression of HMGA2 and CCL5 was quantified by qRT-PCR and normalized to GAPDH. Results (n=3 per group) are relative to BM1 cells transduced with control shRNA. B) qRT-PCR analysis of gene expression in tumors isolated from wild type and Hmga2−/− mice (n=4). C) Representative images of macrophage infiltration in wild type and Hmga2−/− mice as determined by F4/80+ staining. Stromal regions are delimited by the dashed lines. Macrophage infiltration was quantified as the %F4/80+ area in the tumor and stromal regions. D) A schematic showing the regulation of CCL5 by RKIP through HMGA2.
An RKIP-macrophage gene signature predicts metastasis-free survival
Our data suggest that an RKIP-HMGA2-CCL5 pathway regulates recruitment of a TAM population that promotes tumor metastasis in mice. To begin to validate this pathway in humans, we analyzed gene expression in human tumors obtained from TNBC (n=319) and non-TNBC (n=1631) patients. When we examined gene expression across 4 independent data sets from breast cancer patients, we found that RKIP was suppressed and HMGA2, CCL5 and CCR5 were induced in TNBC tumors relative to non-TNBC tumors (Fig. S14). Thus, an RKIP-HMGA2-CCL5 pathway can be used to classify metastatic versus non-metastatic tumors in both mouse models and human patients.
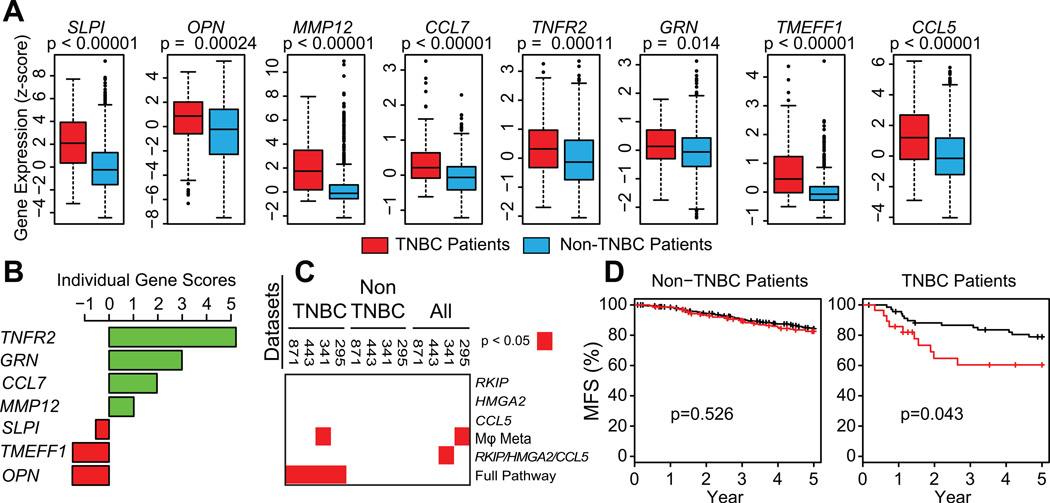
Since TAMs secrete regulators of metastasis in mice (Fig. 4), we examined the gene expression of these TAM-secreted proteins in human breast cancer patients and found that SLPI, OPN, MMP12, CCL7, TNFR2, GRN, TMEFF1, and CCL5 were all significantly increased in TNBC compared to non-TNBC patients (Fig 6A). Our results show that CCL5 recruits TAMs that secrete these factors. Therefore, we performed gene set analysis (GSA) as previously described (9) to identify which of these factors were consistently co-expressed with CCL5 in human TNBC tumors and found a strong correlation between the gene expression levels of CCL5 with TNFR2, GRN, and CCL7 in TNBC patients in all 4 datasets (Fig. 6B; S15). These results raise the possibility that the signaling pathway from RKIP to the three factors secreted by TAMs (TNFR2, GRN, and CCL7) defines a set of linked events that are prognostic for patient outcome.
Figure 6. An RKIP-HMGA2-CCL5-macrophage gene signature predicts metastasis-free survival.
A) Gene expression estimates from a set of 871 breast cancer patients, stratified into either TNBC or non-TNBC patients. B) Individual scores for each gene in comparison to CCL5 expression are plotted for 12 separate genes. C) Heatmap identifying data sets (top) where breast cancer metastasis free survival is significantly stratified by classifier (right). D) Kaplan-Meier plots are shown for a set of all 871 breast cancer patients.
To determine the clinical value of these genes, we developed a signature utilizing the expression levels of tumor genes regulating TAM recruitment (RKIP, HMGA2, CCL5) in combination with stromal TAM-secreted genes (a TAM metagene derived from TNFR2, GRN, and CCL7). When we examined all patients in the data sets or those categorized as non-TNBC using molecular phenotypes as classifiers (30), no significant relationship to clinical outcome was observed (Fig. 6C; S16). However, when we limited analyses to TNBC patients, a gene signature based upon the combination of RKIPlow, HMGA2high, CCL5high, and TAM-metagenehigh expression was significantly prognostic for poor metastasis-free survival (MFS) (Fig 6D). When considered alone, both the tumor-based gene signature (RKIP/HMGA2/CCL5) and the TAM genes (TNFR2, GRN, CCL7) were poor prognostic indicators for breast cancer outcome. Only the gene signature based on the combined tumor and TAM regulatory modules was significant across four independent sets of TNBC patients (Fig 6C). These results highlight the importance of tumor-stromal crosstalk in the metastatic progression of TNBCs.
Discussion
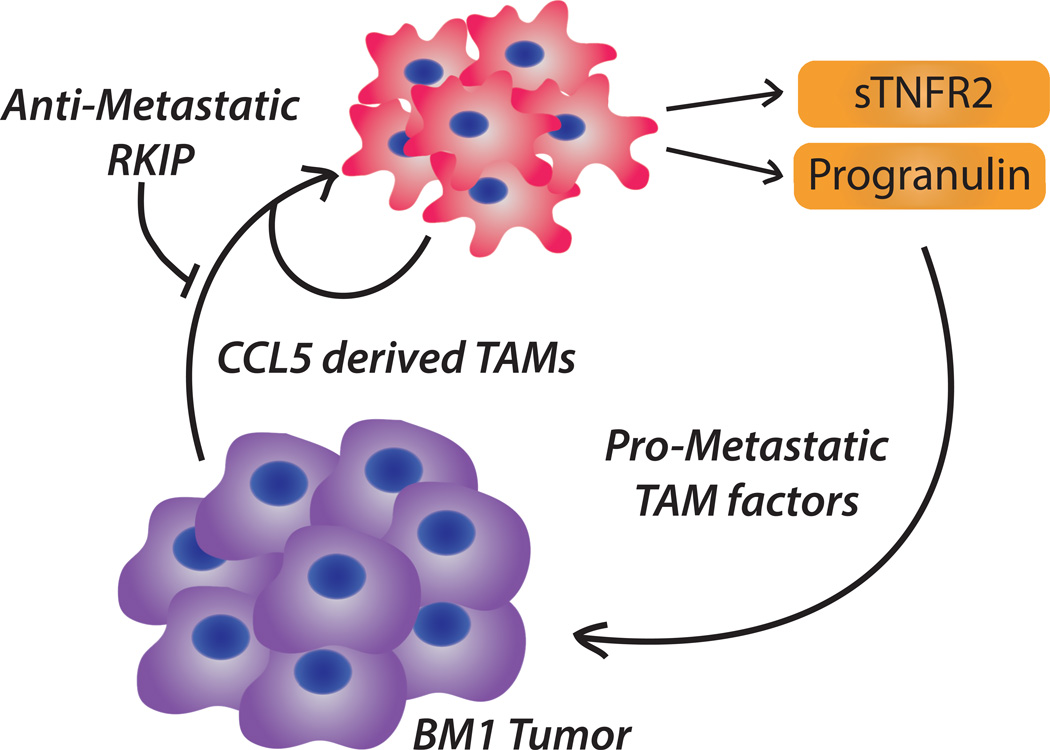
In this study, we identified a novel mechanism whereby RKIP regulates tumor invasiveness by inhibiting infiltration of a subset of TAMs that secrete pro-metastatic factors. We showed that TAMs recruited to metastatic RKIP− tumors, relative to non-metastatic RKIP+ tumors, had reduced ability to drive tumor invasion and decreased secretion of numerous pro-metastatic factors. We demonstrated that RKIP inhibits TAM recruitment by reducing CCL5 expression. CCL5 overexpression was sufficient to rescue recruitment of pro-metastatic TAMs and tumor cell intravasation on a non-metastatic RKIP+ background, and CCL5 inhibition reduced TAM infiltration. Interestingly, a gene signature based on the RKIP regulatory pathway combined with pro-metastatic TAM factors regulated by RKIP was prognostic for metastasis-free survival of TNBC patients. Thus, suppression of RKIP, through direct effects in tumor cells and indirect on TAM recruitment in the microenvironment, may partially explain the aggressive tumors observed in TNBC patients (Fig. 7).
Figure 7. TNBC-TAM crosstalk.
A schematic displaying the circular interplay between TNBC cells and TAMs.
Our demonstration that RKIP expression in tumors markedly attenuates infiltration of pro-metastatic TAMs suggests that metastasis suppressors play a more extensive role in regulating the tumor microenvironment than previously realized. TAMs are known to promote metastatic progression through secretion of growth factors, MMPs, and suppression of the immune system (11). Our protein array analysis of secreted factors suppressed in TAMs from RKIP+ tumors and restored in TAMs from RKIP+CCL5 tumors revealed similar categories including angiogenesis, extracellular matrix organization, growth factor activity, immune system development and regulation of locomotion (Tables S1,2) (31). Thus, one important mechanism by which RKIP suppresses metastasis is by reducing the number and pro-metastatic phenotype of TAMs.
On a molecular level, RKIP regulates TAM recruitment, in part, by attenuating CCL5 expression. Although the CCL5-CCR5 axis has been implicated in breast cancer metastasis, the role of macrophages in this process and the molecular and cellular mechanisms of action have been controversial (20, 27, 28). Our study shows that paracrine CCL5 signaling recruits TAMs and perhaps directly programs their pro-metastatic function. This process is further promoted by an autocrine loop leading to CCL5 expression in the TAMs themselves. Expression of CCL5 and the presence of CCL5-recruited TAMs are insufficient by themselves to predict outcome, consistent with the fact that CCL5 cannot completely rescue the TAM phenotype. However, the combination of the RKIP tumor-signaling pathway with the CCL5-TAMs enables generation of a prognostic gene signature for TNBC patients. This result highlights the crosstalk between tumor cells and TAMs (Fig. 7) and suggests that taking into account both tumor and stromal factors may be effective for prognosis and therapeutic efficacy in TNBC patients.
Consistent with this hypothesis, previous work has shown that shed TNFR2 (sTNFR2) protein is higher in the plasma of pancreatic, endometrial, and breast cancer patients (32–34) and is associated with an increased risk of cancer (34). Progranulin (PGRN) expression blocks TNFR2-mediated inflammation and has been shown to drive migration, invasion and VEGF expression in breast cancer (35, 36). PGRN is highly expressed in a number of tumors including breast, and has also been targeted using biologics in hepatocellular carcinoma (37, 38). Moreover, we provide direct evidence that PGRN and sTNFR2 can promote the invasiveness of human TNBC tumor cells in vitro. Because of the strong evidence of pro-invasive action and the clinical relevance of these factors, CCL5, PGRN, and sTNFR2 are all potential targets for anti-TNBC drug treatment.
While our studies revealed a role for RKIP in macrophage recruitment to tumors in both xenograft and syngeneic mouse models, other metastasis suppressors might have unique roles in regulating additional cell types in the stroma. Furthermore, we identified factors secreted by pro-metastatic TAMs in this study using immune-compromised nude mice that lack mature T-cells. It is possible that, in TNBC patients, RKIP might also play a role in regulating T- cells through factors such as CD80 (upregulated in RKIP+ BM1-derived TAMs) and is a potential therapeutic tool in the treatment of breast cancer patients (25).
Understanding how the tumor cells and TAMs interact could lead to novel strategies for blocking TAM recruitment to TNBCs and tumor metastatic progression. In the case of CCL5 - recruited TAMs, there is an array of pro-metastatic genes that support the tumor. Elegant work on TAMs in breast cancer by Pollard and others has implicated CCL2 and GM-CSF in TAM recruitment in breast cancer. However, the models used to study CCL2 are largely based on the luminal/HER2+ MMTV-PyVT GEM model (19) and are likely to display a unique set of molecular interactions. In the present study, no inhibition of CCL2 or GM- CSF expression by RKIP was observed by RNAseq analysis. Instead, we show that expression of a metastasis suppressor in tumor cells regulates recruitment of a CCL5-responsive TAM population secreting pro-metastatic factors. Future studies will be necessary to determine which inhibitors of these pathways will be most effective therapeutically either alone or in combination in triple- negative breast cancer.
Supplementary Material
Acknowledgements
This work was supported by grant GM087630 (M.R.R.). We thank Kelly Schoenfelt for technical assistance.
Footnotes
None of the authors have conflicts of interest to declare.
References
- 1.Howlader N, Noone A, Krapcho M, Garshell J, Miller D, Altekruse S, et al. Cancer Statistics Review, 1975–2011-SEER Satistics. SEER Cancer Statistics Review. 2011 Available from: http://seer.cancer.gov/csr/1975_2011/
- 2.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 3.Anders CK, Carey LA. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin Breast Cancer. 2009;9(Suppl 2):S73–S81. doi: 10.3816/CBC.2009.s.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 5.Tchou J, Conejo-Garcia J. Targeting the tumor stroma as a novel treatment strategy for breast cancer: shifting from the neoplastic cell-centric to a stroma-centric paradigm. Advances in pharmacology. 2012;65:45–61. doi: 10.1016/B978-0-12-397927-8.00003-8. [DOI] [PubMed] [Google Scholar]
- 6.Zeng L, Imamoto A, Rosner MR. Raf kinase inhibitory protein (RKIP): a physiological regulator and future therapeutic target. Expert Opin Ther Targets. 2008;12:1275–1287. doi: 10.1517/14728222.12.10.1275. [DOI] [PubMed] [Google Scholar]
- 7.Fu Z, Smith PC, Zhang L, Rubin MA, Dunn RL, Yao Z, et al. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J Natl Cancer Inst. 2003;95:878–889. doi: 10.1093/jnci/95.12.878. [DOI] [PubMed] [Google Scholar]
- 8.Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, Hammond SM, et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347–358. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yun J, Frankenberger CA, Kuo WL, Boelens MC, Eves EM, Cheng N, et al. Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 2011;30:4500–4514. doi: 10.1038/emboj.2011.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeung KC, Rose DW, Dhillon AS, Yaros D, Gustafsson M, Chatterjee D, et al. Raf kinase inhibitor protein interacts with NF-kappaB-inducing kinase and TAK1 and inhibits NF-kappaB activation. Mol Cell Biol. 2001;21:7207–7217. doi: 10.1128/MCB.21.21.7207-7217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agarwal S, Gertler FB, Balsamo M, Condeelis JS, Camp RL, Xue X, et al. Quantitative assessment of invasive mena isoforms (Menacalc) as an independent prognostic marker in breast cancer. Breast Cancer Res. 2012;14:R124. doi: 10.1186/bcr3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green CE, Liu T, Montel V, Hsiao G, Lester RD, Subramaniam S, et al. Chemoattractant signaling between tumor cells and macrophages regulates cancer cell migration, metastasis and neovascularization. PLoS One. 2009;4:e6713. doi: 10.1371/journal.pone.0006713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Palma M, Lewis CE. Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell. 2013;23:277–286. doi: 10.1016/j.ccr.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Meissner F, Scheltema RA, Mollenkopf HJ, Mann M. Direct proteomic quantification of the secretome of activated immune cells. Science. 2013;340:475–478. doi: 10.1126/science.1232578. [DOI] [PubMed] [Google Scholar]
- 17.Medrek C, Ponten F, Jirstrom K, Leandersson K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer. 2012;12:306. doi: 10.1186/1471-2407-12-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin EY, Li JF, Gnatovskiy L, Deng Y, Zhu L, Grzesik DA, et al. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res. 2006;66:11238–11246. doi: 10.1158/0008-5472.CAN-06-1278. [DOI] [PubMed] [Google Scholar]
- 19.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson SC, Scott KA, Wilson JL, Thompson RG, Proudfoot AE, Balkwill FR. A chemokine receptor antagonist inhibits experimental breast tumor growth. Cancer Res. 2003;63:8360–8365. [PubMed] [Google Scholar]
- 21.Lu H, Clauser KR, Tam WL, Frose J, Ye X, Eaton EN, et al. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat Cell Biol. 2014;16:1105–1117. doi: 10.1038/ncb3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su S, Liu Q, Chen J, Chen J, Chen F, He C, et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell. 2014;25:605–620. doi: 10.1016/j.ccr.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 23.Hijikata A, Kitamura H, Kimura Y, Yokoyama R, Aiba Y, Bao Y, et al. Construction of an open-access database that integrates cross-reference information from the transcriptome and proteome of immune cells. Bioinformatics. 2007;23:2934–2941. doi: 10.1093/bioinformatics/btm430. [DOI] [PubMed] [Google Scholar]
- 24.Peach RJ, Bajorath J, Naemura J, Leytze G, Greene J, Aruffo A, et al. Both extracellular immunoglobin-like domains of CD80 contain residues critical for binding T cell surface receptors CTLA-4 and CD28. J Biol Chem. 1995;270:21181–21187. doi: 10.1074/jbc.270.36.21181. [DOI] [PubMed] [Google Scholar]
- 25.Dols A, Smith JW, 2nd, Meijer SL, Fox BA, Hu HM, Walker E, et al. Vaccination of women with metastatic breast cancer, using a costimulatory gene (CD80)-modified, HLA-A2-matched, allogeneic, breast cancer cell line: clinical and immunological results. Human gene therapy. 2003;14:1117–1123. doi: 10.1089/104303403322124828. [DOI] [PubMed] [Google Scholar]
- 26.Stavik B, Skretting G, Aasheim HC, Tinholt M, Zernichow L, Sletten M, et al. Downregulation of TFPI in breast cancer cells induces tyrosine phosphorylation signaling and increases metastatic growth by stimulating cell motility. BMC Cancer. 2011;11:357. doi: 10.1186/1471-2407-11-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 28.Velasco-Velazquez M, Jiao X, De La Fuente M, Pestell TG, Ertel A, Lisanti MP, et al. CCR5 antagonist blocks metastasis of basal breast cancer cells. Cancer Res. 2012;72:3839–3850. doi: 10.1158/0008-5472.CAN-11-3917. [DOI] [PubMed] [Google Scholar]
- 29.Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell. 1988;55:619–625. doi: 10.1016/0092-8674(88)90220-6. [DOI] [PubMed] [Google Scholar]
- 30.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Grote VA, Kaaks R, Nieters A, Tjonneland A, Halkjaer J, Overvad K, et al. Inflammation marker and risk of pancreatic cancer: a nested case-control study within the EPIC cohort. Br J Cancer. 2012;106:1866–1874. doi: 10.1038/bjc.2012.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dossus L, Becker S, Rinaldi S, Lukanova A, Tjonneland A, Olsen A, et al. Tumor necrosis factor (TNF)-alpha, soluble TNF receptors and endometrial cancer risk: the EPIC study. Int J Cancer. 2011;129:2032–2037. doi: 10.1002/ijc.25840. [DOI] [PubMed] [Google Scholar]
- 34.Gross AL, Newschaffer CJ, Hoffman-Bolton J, Rifai N, Visvanathan K. Adipocytokines, inflammation, and breast cancer risk in postmenopausal women: a prospective study. Cancer Epidemiol Biomarkers Prev. 2013;22:1319–1324. doi: 10.1158/1055-9965.EPI-12-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Z, Ismail A, Kriazhev L, Sadvakassova G, Bateman A. Progranulin (PC-cell-derived growth factor/acrogranin) regulates invasion and cell survival. Cancer Res. 2002;62:5590–5596. [PubMed] [Google Scholar]
- 36.Tangkeangsirisin W, Serrero G. PC cell-derived growth factor (PCDGF/GP88, progranulin) stimulates migration, invasiveness and VEGF expression in breast cancer cells. Carcinogenesis. 2004;25:1587–1592. doi: 10.1093/carcin/bgh171. [DOI] [PubMed] [Google Scholar]
- 37.He Z, Bateman A. Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. Journal of molecular medicine. 2003;81:600–612. doi: 10.1007/s00109-003-0474-3. [DOI] [PubMed] [Google Scholar]
- 38.Ho JC, Ip YC, Cheung ST, Lee YT, Chan KF, Wong SY, et al. Granulin-epithelin precursor as a therapeutic target for hepatocellular carcinoma. Hepatology. 2008;47:1524–1532. doi: 10.1002/hep.22191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.